Abstract
Pioneer transcription factors have the unique and important role of unmasking chromatin domains during development to allow the implementation of new cellular programs. Compared with those of other transcription factors, this activity implies that pioneer factors can recognize their target DNA sequences in so-called compacted or “closed” heterochromatin and can trigger remodeling of the adjoining chromatin landscape to provide accessibility to nonpioneer transcription factors. Recent studies identified several steps of pioneer action, namely rapid but weak initial binding to heterochromatin and stabilization of binding followed by chromatin opening and loss of cytosine-phosphate-guanine (CpG) methylation that provides epigenetic memory. Whereas CpG demethylation depends on replication, chromatin opening does not. In this Minireview, we highlight the unique properties of this transcription factor class and the challenges of understanding their mechanism of action.
Keywords: chromatin remodeling, epigenetics, cell differentiation, DNA demethylation, gene transcription, development
Introduction
In the late 1970s when chromatin structure was beginning to be probed with tools such as DNase (hyper)-sensitivity, the concept of pioneer factors emerged. These were factors that would have the capacity to bind specific DNA sequences within compacted heterochromatin and initiate the opening of this chromatin. This opening would be required for implementation of major developmental fate decisions. At the same time, Drosophila geneticists developed the notion of selector genes for early developmental regulators that in some way specify the outcome of future cell fates through their action on broad embryonic domains (1). In this context, the notion of pioneer factors offered a possible mechanism to achieve the purpose of selector genes, but these ideas remained more in the domain of evening conversations than experimental reality. For clarity, it should be mentioned that in more recent years, the term “selector” has been used by some to identify factors that have the opposite effect in the differentiation scheme compared with the original definition, namely factors that trigger the ultimate step in cell-fate decisions (2).
The idea of pioneer action was revived in the late 1990s when the transcription factor (TF)2 FoxA was shown to have the unique ability to bind its target sequence within nucleosomal DNA (3). This unique ability contrasted with many other TFs that will only bind efficiently naked or more readily accessible DNA as observed within active regulatory sequences. However, pioneer factors do not have completely unrestricted access to heterochromatin sites but do exhibit cell-specific actions (4). In parallel, the old binary view of chromatin as either hetero- or euchromatin changed dramatically as the enormous diversity of histone modifications became known, eventually leading to the concept of a histone code (5) that defines a continuum of chromatin flavors associated with regulatory and structural functions. The complexity of this code and the limited tools available to characterize chromatin limit our present ability to define the permissive or restrictive chromatin states that are targeted by pioneer factors. Despite this limitation, the basic features that define pioneer factors (Fig. 1) are as follows: 1) the ability to bind specific DNA sequences within “closed” or unmarked chromatin where genomic DNA is not readily accessible; 2) the ability to initiate chromatin remodeling leading to DNA accessibility; 3) consequently to allow binding of other transcription factors; and 4) finally to establish stable changes in chromatin structure associated with DNA accessibility and epigenetic stability. Collectively, these features imply that the “act of pioneering” may be a one-shot affair, i.e. once enacted, its effect on chromatin remains stable. Mechanisms for maintenance of chromatin state at pioneered sites may also exist. This Minireview will discuss the unique aspects of pioneer action and attempt to separate these from the transcriptional actions of the same factors because pioneers do act as transcriptional regulators like other TFs and often at the pioneered as well as other target sites. The list of TFs that share at least some features of pioneers is provided in Table 1.
Figure 1.

Salient properties of pioneer factors.
Table 1.
List of transcription factors that share at least some features of pioneers
For each feature listed at the top of the table are references within the table that provide supporting evidence.
| Factor | Binding to heterochromatin | Chromatin activation | Epigenetic memory: DNA demethylation | Cell fate reprogramming | Nucleosome binding | Mitotic bookmarking |
|---|---|---|---|---|---|---|
| Ascl1/Mash1 | 102 | 102 | 102, 103 | |||
| C/EBPα | 43 | 104 | ||||
| Ebf1 | 47, 48 | 47, 48 | 48 | |||
| Esrrb | 105 | |||||
| Foxa | 3 | 3, 4, 28, 31, 32 | 4, 69, 85 | 38, 39 | 28 | 77 |
| Gata | 59 | 59 | 38, 39 | 29 | 76 | |
| GR/AR | 18 | 18 | ||||
| Klf4 | 21, 22 | 21, 22 | 106, 107 | 53 | ||
| Neurod1 | 50, 70 | |||||
| Nrf1 | 70 | 70 | Inhibitory (70) | |||
| Oct4 | 21, 22 | 21, 22 | 106, 107 | 53 | ||
| p53 | 100, 101 | 100, 101 | ||||
| Pax7 | 44, 45 | 44, 45, 61 | 45 | 44 | ||
| PU.I | 41, 42 | 41, 42 | 104 | |||
| Sox2 | 21, 22 | 21, 22 | 106, 107 | 53 | 78, 79 |
Because the measure of chromatin features such as DNA accessibility and chromatin marks are not just absent versus present but are also present on a continuous scale, the expectation of pioneer function must be more clearly defined. Indeed, DNA accessibility (whether measured by DNase sensitivity (6), formaldehyde-assisted isolation of regulatory elements (FAIRE (7)), or the ATAC procedure (8)) or histone marks, such as histone H3K4me1 that marks active enhancer sequences (9), exhibit greater values as the activity of enhancers or the number of bound TFs increases (10). Increments in these marks may reflect quantitative changes in enhancer activity rather that the switch from “closed” naive chromatin to a state of accessibility. The label “pioneer” should thus be reserved for factors and actions shown to elicit chromatin opening from a state of complete absence of accessibility marks to the presence of such marks. On the genome scale, it is thus very important to separate the targets of pioneer action from those where the same pioneer factors only exert classical transcriptional activity at already accessible regulatory sequences; this requires an assessment of chromatin status before and after pioneer factor action in an experimental system dependent on cells that have never been exposed (in their developmental history) to the pioneer if epigenetic memory is indeed a pioneer property. Failing that, a pioneer activity may be inferred, but formal demonstration requires the before and after comparison.
Pioneers set the stage: Assisted-loading and settler factors
Pioneers appear to share the property of interacting with other TFs as do most TFs. Although very important from the biological perspective, this property is not a defining feature of pioneers. For example, the pioneer FoxA interacts with nuclear receptors such as glucocorticoid (GR/Nr3c1), estrogen (ER/Nr3a1), or androgen (AR/Nr3c4) receptors, and this allows recruitment of these nuclear receptors at subsets of enhancers (11–14) that establish hormone-responsive gene regulatory networks (15). In this context, FoxA pioneers the opening of subsets of enhancers targeted by the hormone-responsive receptors. This subsequent binding of nuclear receptors has been labeled as “assisted loading” (16), and the factors that require the open chromatin state were labeled as “settler factors” (17). The binding of a settler factor may be essential in the biological context, but it does not constitute the core pioneer activity that is restricted to initiation of chromatin opening. However, for this specific example, it appears that the interaction between FoxA and nuclear receptor may be reciprocal as nuclear receptors can also recruit FoxA to specific subsets of enhancers (18).
Pluripotency factors
The reprogramming of diverse cells such as fibroblasts into induced pluripotent stem cells(iPS) revealed the unique ability of a group of factors to reverse the differentiation process (19) toward a pluripotent state. These pluripotency factors (OSK for Oct4, Sox2, and Klf4) initiate the remodeling (opening) of both enhancers and transcription start sites (TSS). This was revealed by deposition of H3K4me1/2 at targeted enhancers and of H3K4me2/3 at TSS (20). The initial binding of these factors occurs widely at unmarked (“closed”) chromatin to initiate their remodeling; they thus act as pioneer factors (21). The initial binding of OSK factors is followed by a lengthy period (weeks) of iPS cell selection that leads to the remodeling of large chromatin domains of the epigenome from a somatic to a pluripotent state. Interestingly, it was also reported that some of the iPS cell-reprogrammed enhancers require the expression and binding of more than one of the OSK factors. This suggests that pioneers may also require a cooperative action to remodel the epigenome (22). This process opens new sites for OSK binding together with sites for the accessory factor c-Myc (21). There are broad domains where OSK factors cannot bind early in reprogramming but only in the iPS cell state. These domains have high levels of H3K9me3, and this may thus constitute a barrier that contributes to refractoriness to OSK binding. Indeed, knockdown of the histone methyltransferases SUV39H1/H2 that are responsible for H3K9me3 deposition allows binding at previously inaccessible sites (20, 21, 23). The pluripotency factors Oct4 and Sox2 have critical roles in normal development to activate the zygote genome (24). In Drosophila, the factor Zelda has a similar role for induction of the zygote genome (25), and this is achieved through a pioneer mechanism of chromatin opening (26).
Lineage-specifying pioneer factors
The first indication that FoxA factors have pioneer activity came from showing that liver-specific FoxA-binding sites are occupied in the endoderm before liver specification (27). FoxA was then shown to bind nucleosomal DNA (28) and to open compacted chromatin (29). Genome-wide studies then showed its chromatin-remodeling activity (4, 30, 31) as well as the associated nucleosome depletion (32). In Caenorhabditis elegans, the FoxA-related factor PHA-4 is also critical for foregut development, and this is achieved through pioneer action (33). Interestingly, the pioneer action of PHA-4 is mostly exerted over promoter regions, and this leads to recruitment of RNA polymerase II (34). This recruitment initially leads to a poised state where RNA polymerase is paused on the promoter early on, and transcription only occurs later in foregut development.
GATA4 is also present at liver-specific enhancers in early endoderm, but its binding appeared supported by FoxA and did not show as strong an ability to bind nucleosomal DNA (29). GATA4 together with GATA6 are required for early liver development (35–37). Hence, GATA factors appear to have pioneer properties, although it may not be as effective as FoxA. Nonetheless, GATA4, like FoxA, can induce trans-differentiation into hepatocytes (38, 39).
Specification of the lymphoid, in particular macrophage, lineages depends on the factors PU.1 and C/EBPα. PU.1 is critical for development of these lineages (40). It initiates chromatin remodeling and is associated with deposition of active enhancer marks (41). Indeed, PU.1 increases chromatin accessibility and promotes nucleosome depletion (42). C/EBPα can also trigger trans-differentiation into that lineage and its binding to macrophage enhancers during that process is associated with deposition of the active enhancer marks H3K4me1 and H3K27ac. Thus, C/EBPα and PU.1 independently act as pioneer for the other during macrophage differentiation (43).
The pituitary intermediate lobe is specified to a unique developmental fate by the pioneer factor Pax7 (44). This is achieved through binding and chromatin remodeling of a subset of de novo active melanotrope-specific enhancers. The opening of these enhancers allows for recruitment of the differentiation determination factor Tpit that achieves terminal differentiation of this lineage. Pax7 pioneering results in appearance of DNA accessibility together with deposition of active enhancer histone marks (45).
Establishment of the B cell lineage requires the transcription factor EBF1 (46), and this was associated with chromatin remodeling and increased enhancer H3K4me2 (47). Some EBF1 pioneer actions were shown to depend on an EBF1 C-terminal domain that is required to trigger DNA accessibility and deposition of active chromatin marks at a specific subset of enhancers. Both C-terminally dependent and independent pioneer sites were enriched for the same EBF1 motif suggesting that the EBF1 DNA-binding site is not the defining factor between dependence and independence on the C-terminal domain. This supports a model where different pioneer interacting proteins may define functionally distinct subsets of pioneered enhancers (48).
Two neurogenic basic helix-loop-helix transcription factors shown to reprogram fibroblast to the neuronal fate appear to have pioneer activity. Indeed, Ascl1 is the driver of neuronal differentiation in association with Brn2 and Myt11, and its recruitment was associated with increased DNA accessibility (FAIRE) and with increased active chromatin marks H3K4me1 and H3K27ac together with decreases in the repressive mark H3K9me3 (49). The neurogenic factor NeuroD1 was also shown to induce similar chromatin changes at enhancers and promoters during neuronal reprogramming (50).
Pioneer interactions with DNA and chromatin
The pioneer factor activity was inferred from in vitro and in vivo footprinting experiments that showed FoxA and GATA site co-occupancy prior to hepatic specification (51). Flanking TF sites were only occupied once cells are specified toward liver identity, suggesting that pioneer factors have the unique ability to bind “closed” or naive chromatin (21, 44). Despite many genome-wide studies, the nature of this naive or closed chromatin remains vague, and the ability of pioneers to bind specific chromatin states is still defined by the negative, i.e. the absence of recognizable chromatin marks, and in some cases the presence of methylated cytosines in DNA. Indeed, as discussed below, some pioneers can bind methylated target DNA, whereas others appear to be methylation-sensitive.
Pioneer factors tend to have higher residency time or chromatin mobility than other TFs (18, 52) suggesting that stable chromatin–pioneer interactions may be critical for pioneer function. These stable chromatin–pioneer interactions may be explained by direct nucleosome binding, as shown for FoxA and the OSK pluripotency factors (29, 53). For FoxA, nucleosomal interaction may partly rely on a FoxA domain that resembles a linker histone H1 structure (31, 54). For the OSK factors, their ability to target partial consensus motifs may allow their DNA-binding domains to interact directly with nucleosomes (53).
In one instance, target DNA motif preference may play a role in binding stability; indeed, the pioneer Pax7 preferentially recognizes a composite motif composed of binding sites for its two DNA-binding domains, the homeodomain and paired domains, leading to greater binding stability and possibly allowing for pioneer action (44).
Epigenetic remodeling by pioneer factors
Pioneer factors provide competency for gene expression, but their binding to closed chromatin is not in itself sufficient. Indeed, chromatin remodeling is required to allow nonpioneer TF binding and transcriptional activation at newly competent regulatory sequences, primarily enhancers. The remodeling or activation of regulatory sequences from a naive or “closed” chromatin state appears to be a stepwise process (Fig. 2). None of the pioneers characterized so far have unrestricted access to the genome in heterochromatin; this was shown for FoxA, Pax7, and the pluripotency factors. This aspect is discussed below.
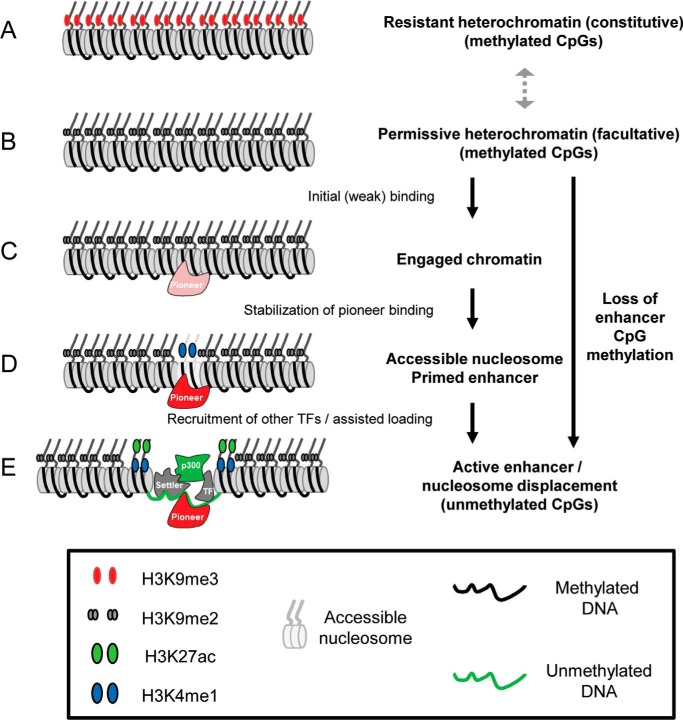
Figure 2.
Current scheme of pioneer action. The permissive chromatin state for pioneer action appears to be facultative heterochromatin. Following initial weak binding of the pioneer, target site chromatin (mostly characterized at enhancers) undergoes a first transition where a central nucleosome becomes more accessible, and this may (or not) overlap with a state of Primed enhancer characterized by a weak H3K4me1 signal. Complete activation of enhancers is characterized by nucleosome depletion, bimodal distribution of H3K4me1 and H3K27ac, together with recruitment of the general coactivator p300 and other transcription factors. Whereas the ability to bind methylated DNA target sites is not a unique feature of all pioneers, for most pioneers the current evidence correlates pioneer-dependent chromatin remodeling with loss of CpG methylation at the newly accessible DNA/enhancers.
The first step in pioneer action is the initial binding (Fig. 2C) to permissive heterochromatin (Fig. 2B), and it appears to be rapid (e.g. less than 30 min for Pax7 (45)). This is followed by a phase of binding stabilization (within 24 h for Pax7) that may or may not be paralleled by nucleosomal changes that increase accessibility (31) and to the appearance of low levels of the H3K4me1 mark in the center of target enhancers (Fig. 2D). These “accessible” or “primed” enhancers can undergo the final step of enhancer activation that involves the binding of other nonpioneer TFs, nucleosome depletion, and deposition of the active enhancer mark H3K27ac that is associated with the histone acetylase activity of the general coactivator p300 (Fig. 2E).
As most TFs, pioneers interact with chromatin remodeling proteins that are found within large complexes. These complexes have been associated with the process of transcription itself and/or its activation/initiation; the same complexes or different ones may be critical for the initial act of pioneering as well as for continued transcriptional action of pioneers. The challenge is thus to find experimental systems to separate these two actions. For example, the BRG1 ATPase of the SWI/SNF complex co-occupies many sites together with Oct4 in ES cells (55), and knockdown of BRG1 affects ES cell pluripotency (56). Oct4 is required for maintenance of open chromatin at enhancers in ES cells, and its inactivation leads to loss of accessibility at these enhancers (57). Oct4 pioneer function is thus dependent on the chromatin remodeler Brg1. Similarly, the INO80 remodeling complex co-occupies many sites in common with pluripotency factors, and its knockdown decreases chromatin accessibility at those sites (58), suggesting that the complex may increase accessibility following recruitment by the pluripotency factors. Similarly, GATA3 was shown to require BRG1 for cell reprogramming through pioneer action (59).
The Trithorax (Drosophila) complex (COMPASS in yeast and MLL in mammals) is involved in activation (opening) of chromatin structure (60). Pax7 was suggested to recruit the MLL1/2 complex through interaction with its component protein WDR5 (61), and FoxA1 directs H3K4me1 deposition through recruitment of MLL3 at enhancers (62). Indeed, this complex has H3K4me1 methylation activity and thus may lead to enhancer activation. It may also be implicated in pioneering as its component protein Ash2l is recruited to sites of Pax7 pioneering (45).
For transcriptional activation, chromatin accessibility is increased at both promoters and enhancers by recruitment of the variant histones H2A.Z and H3.3 that form unstable nucleosomes (63). FoxA factors (32, 64, 65) and CLOCK:BMAL1 (66) promote recruitment of H2A.Z. This likely contributes to nucleosome instability and loss, but it is not clear that this is critical for pioneer action per se. Indeed, FoxA-dependent nucleosome instability is not correlated with H2A.Z deposition, and in this particular case, increased nucleosome accessibility may result from displacement of the linker histone H1 (31). FoxA factors have the unique property of containing a H1 mimic region that binds nucleosomes (28).
Barriers to pioneer binding and action
Although pioneers have the unique ability to bind their target sequence within nucleosomes in contrast to many TF that cannot, this does not mean that pioneers can bind all their target sequences in the genome. Indeed, pioneers show different binding repertoires in different cell types. For example, Sox2 binds different target subsets in mouse cortex and spinal cord (67), indicating that there are additional constraints on pioneer binding. Furthermore, the pluripotency factors OSK have a large subset of targets that only become accessible in the late phase of reprogramming toward iPS (21). The OSK-binding sites within these latter binding regions initially have higher levels of the repressive histone mark H3K9me3, and knockdown of the histone-modifying enzymes SUV39H1/H2, and SETDB1 to a lesser extent was sufficient to allow early binding of Oct4 and Sox2 to these sites in fibroblasts. Thus, the mark H3K9me3 associated with constitutive heterochromatin can constitute a barrier to OSK and possibly other pioneer binding. In addition, maintenance of heterochromatin by the histone chaperone CAF-1 is important for stable somatic cell identity as its knockdown accelerates cellular re-programming by pioneer factors (68) Other pioneers such as Pax7,3 FoxA, and GATA (69) also exhibit lineage-specific binding repertoires. It remains to be seen whether all pioneers are subject to the same barriers or whether some may have unique limitations, and hence different permissive chromatin environments.
Whereas DNA binding by some pioneers like Pax7 is insensitive to CpG methylation within their DNA-binding site (45), DNA methylation may be an impediment to binding of TFs that have some properties of pioneers. Indeed, the factor Nrf1, predicted on theoretical bases to have pioneer action (17), will trigger chromatin access (DNase sensitivity) only if its DNA-binding site is unmethylated (70). The Nrf1 DNA-binding site is very GC-rich and contains two CpG motifs; its DNA interaction may thus be more sensitive to methylation. Another factor with methylation-insensitive DNA binding may thus be required to prime target enhancers through DNA demethylation to allow Nrf1 binding and action. There may thus be a hierarchy of pioneers with differing potencies; “true” pioneers may be considered to be those with methylation-insensitive DNA binding and an ability to induce DNA demethylation, but the biological context may provide an argument to consider factors such as Nrf1 as pioneer. For example, global DNA de-methylation occurs at two critical stages of mammalian development, in the pre-implantation embryo and during primordial germ cell proliferation and migration (71, 72). DNA methylation-sensitive pioneers may thus act as classical TFs in most cells but transiently behave as pioneers during development. Such limitation on pioneer action could explain specific roles played by pioneers in distinct cell types. The detailed assessment of the pioneer mode of action is thus critical to understand their role in lineage specification.
Many pioneers exhibit extensive binding site subsets of low affinity that are resistant to remodeling (45, 53). Some of these sites appear to have degenerate DNA-binding site sequences and were proposed to represent a mechanism for scanning targets. Notwithstanding this possibility, this mechanism does not provide an explanation for selection of specific pioneering sites.
Stability of pioneer-induced chromatin remodeling
During development, pioneers stably reprogram the chromatin landscape leading to a stable cell identity. As such, they would implement a memory for long-term maintenance of cell identity. During mitosis, chromatin is disassembled and reconstituted after replication. There are mechanisms to reconstitute the daughter cell chromatin landscape as in the mother cell (73). It was proposed that pioneers, and possibly other TFs, bookmark the chromatin during mitosis to allow re-establishment of active regulatory networks. Indeed, although many TFs were shown to be excluded from mitotic chromosomes (74, 75), some pioneers appear to remain bound to mitotic chromosomes; for example, GATA1 binding is maintained in mitotic chromosome at a tissue-specific subset of 5% of its chromatin targets (76). Surprisingly, this study also identified mitosis-specific binding of GATA1 at sites that do not contain the consensus GATA motif. Both specific and nonspecific binding sites on mitotic chromosomes were also observed for FoxA1 (77) where specific FoxA1 binding occurs at 15% of its interphase targets. Recently, Sox2 and Oct4 were also shown to remain bound during mitosis (78, 79). In this last study, the authors also show, using live imaging techniques, that cross-linking with formaldehyde leads to eviction of most TFs from mitotic chromosomes. They proposed a model where most TFs remain bound during mitosis to maintain the original program despite only showing this for the well-characterized pioneer Sox2.
The most stable epigenetic mark associated with inactive heterochromatin is DNA methylation (80). Indeed, promoters and CpG-rich promoter regions (CpG islands) that are transcriptionally active are largely demethylated, and this is required for activity. Similarly, active enhancer sequences are hypomethylated, and the patterns of enhancer hypomethylation are associated with cell-specific gene–expression programs (81). Following replication, hemi-methylated CpG dinucleotides are recognized and methylated by the Dnmt1–Uhrf1 complex (82, 83). Maintenance of DNA methylation patterns by this mechanism thus ensures stability of lineage-specific gene-expression programs. As inactive (closed) regions of chromatin that are targeted by pioneers have high DNA methylation, it is expected that pioneers should bind their target sequence independently of DNA methylation, and this is indeed the case for FoxA and Pax7, although there may be exceptions as for Nrf1 discussed above (70).
Whereas direct DNA binding by FoxA and Pax7 is not impaired by CpG methylation of their binding site, their action leads to local demethylation of flanking enhancer sequences beyond the DNA-binding site (45, 69). This demethylation is associated with epigenetic memory and maintenance of an open/accessible chromatin environment (45).
A few pioneer factors were investigated for their impact on DNA methylation. FoxA1 can induce DNA demethylation (4) thus demonstrating its impact on the DNA methylation landscape. Active DNA demethylation can be achieved by the Tet enzymes (84), but for FoxA-dependent demethylation activity, it was rather suggested to require recruitment of a FoxA1 DNA repair complex (85). Also, EBF1 and Pax7 pioneer actions lead to loss of DNA methylation (45, 48). The mechanism of pioneer-induced DNA demethylation remains uncertain as the known DNA demethylation Tet pathway could not be implicated in either FoxA or EBF1 action (48, 85). It is noteworthy that FoxA-dependent chromatin remodeling can occur independently of replication, whereas DNA demethylation is impaired by blockade of replication (69). These data clearly separate two steps in pioneer action, and these are consistent with the time frames of action defined in an inducible system for Pax7 (45).
Pioneer factors in cancer
In view of their chromatin remodeling activities, pioneer TFs have the potential for significant epigenetic alterations as seen in cancer. Indeed, FOX family genes are involved in several cancers (86). Overexpression of Foxa1 is associated with a poor prognosis in prostate cancer (87), although it is generally a good prognosis of breast cancer (88). Point mutations of FOXA1 were also found in some prostate cancers, and this was associated with decreased androgen signaling and increased tumor growth (89).
In ER+ breast cancer cells, ER binding requires FOXA1 at many binding sites showing the role of FOXA1 in driving hormone response of these tumors (90). Similarly, AR binding is also influenced by FOXA1; indeed, some AR-binding sites are lost in cells depleted of FOXA1; however, many sites are also gained suggesting a more complex relationship of FOXA1 with AR than with ER (87, 91).
Also, FOXM1 is amplified in some breast cancers (92), in non-Hodgkin's lymphomas (93), or in malignant peripheral nerve sheath tumors (94). FOXM1 is activated through post-translational phosphorylation by ERK, and FOXM1 activation is associated with a poor prognosis for many human cancers such as lung, medulloblastoma, breast, gastric, and pancreatic cancers.
Chromosomal translocations leading to fusion of the N-terminal DNA-binding domain of PAX3 or PAX7 with the C-terminal transactivation domain of FOXO1 (FKHR) were found in rhabdomyosarcomas. These fusion proteins act as much more potent activators than the native PAX3 or PAX7 (95). PAX3–FOXO1 was shown to lead to activation of genes involved in cancer development and to inappropriate expression of developmental TFs (96). PAX7 and FOXO1 both have pioneer activity (44, 97). As such, these fusion proteins may also function as pioneers. Furthermore, FOXO3 or FOXO4 are trans-located to MLL gene in acute lymphoblastic leukemia leading to increased cell proliferation (98). FOXO proteins function as tumor suppressors (99), and their loss of activity due translocation or deletion may also lead to increased tumorigenesis.
Finally, two studies showed that the tumor suppressor p53 (TP53) can engage inaccessible chromatin. In one study, p53 binding led to deposition of H4K16ac together with H3K27ac at non-TSS sites. However, neither gain of chromatin accessibility nor deposition of H3K4me1 accompanied these changes, thus possibly defining a unique chromatin environment specific to p53 (100). A recent study showed that after DNA damage, a subset of p53-binding sites are associated with de novo accessibility assessed by ATACseq possibly highlighting a canonical pioneer action of p53 (101).
Perspective
As exemplified in this Minireview, the critical aspects of pioneer action are still the least understood. First and foremost, the molecular basis for pioneer access to their target DNA sequences in closed chromatin remains obscure. There may be more than one underlying mechanism as the mechanism proposed for FoxA interaction with nucleosomal DNA, namely its putative linker H1 mimicry binding interactions, does not seem to apply to other pioneers. The question remains whether all pioneers use the same molecular strategies to elicit chromatin remodeling. They may also may differ in their ability to access to various “flavors” of heterochromatin.
The initial binding and action of pioneers to closed chromatin regions and the initiation of chromatin remodeling are the critical features that distinguish pioneers from other TFs. Is there something unique about pioneer action on chromatin at this initiating event, or is the recruitment of chromatin remodeling complexes at that initiating event the same as those that occur during activation of enhancer function in transcription? This latter possibility would imply that the only unique aspect of pioneer action is the ability to recognize target sites in “closed” chromatin. Alternatively, this ability may be operating in conjunction with recruitment of a unique set of chromatin remodelers involved in initiating chromatin opening but not necessarily involved in maintenance of this accessible state. To answer these difficult questions requires the availability of experimental systems where the specific steps of pioneer action can be followed and investigated. Is there something unique about the maintenance of chromatin accessibility at pioneer sites, or does this simply result from recruitment of enhancer machinery (combination of TFs, chromatin remodelers, and chromatin modifiers) leading ultimately to changes in the most stable epigenetic mark, demethylation of DNA cytosines?
Addressing these questions is paramount to understand pioneer action and to use this knowledge in the context of cell fate reprogramming. Understanding the nature of the cell fate reprogramming that may occur during tumorigenic processes will be critical for therapeutic development of cell therapies.
This is the third article in the Thematic Minireview series “Chromatin and Transcription.” The authors declare that they have no conflicts of interest with the contents of this article.
A. Mayran and J. Drouin, unpublished data.
- TF
- transcription factor
- TSS
- transcription start site
- iPS
- induced pluripotent stem
- FAIRE
- formaldehyde-assisted isolation of regulatory elements
- ER
- estrogen receptor
- AR
- androgen receptor
- ES
- embryonic stem cell
- CpG
- cytosine-phosphate-guanine
- C/EBPα
- CCAAT/enhancer-binding protein α.
References
- 1. Garcia-Bellido A. (1975) Genetic control of wing disc development in Drosophila. Ciba Found. Symp. 0, 161–182 [DOI] [PubMed] [Google Scholar]
- 2. Hobert O. (2008) Regulatory logic of neuronal diversity: terminal selector genes and selector motifs. Proc. Natl. Acad. Sci. U.S.A. 105, 20067–20071 10.1073/pnas.0806070105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cirillo L. A., and Zaret K. S. (1999) An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol. Cell 4, 961–969 10.1016/S1097-2765(00)80225-7 [DOI] [PubMed] [Google Scholar]
- 4. Sérandour A. A., Avner S., Percevault F., Demay F., Bizot M., Lucchetti-Miganeh C., Barloy-Hubler F., Brown M., Lupien M., Métivier R., Salbert G., and Eeckhoute J. (2011) Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res. 21, 555–565 10.1101/gr.111534.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gardner K. E., Allis C. D., and Strahl B. D. (2011) Operating on chromatin, a colorful language where context matters. J. Mol. Biol. 409, 36–46 10.1016/j.jmb.2011.01.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Song L., Zhang Z., Grasfeder L. L., Boyle A. P., Giresi P. G., Lee B. K., Sheffield N. C., Gräf S., Huss M., Keefe D., Liu Z., London D., McDaniell R. M., Shibata Y., Showers K. A., et al. (2011) Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 21, 1757–1767 10.1101/gr.121541.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Giresi P. G., and Lieb J. D. (2009) Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (formaldehyde assisted isolation of regulatory elements). Methods 48, 233–239 10.1016/j.ymeth.2009.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., and Greenleaf W. J. (2013) Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Heintzman N. D., Stuart R. K., Hon G., Fu Y., Ching C. W., Hawkins R. D., Barrera L. O., Van Calcar S., Qu C., Ching K. A., Wang W., Weng Z., Green R. D., Crawford G. E., and Ren B. (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311–318 10.1038/ng1966 [DOI] [PubMed] [Google Scholar]
- 10. Langlais D., Couture C., Balsalobre A., and Drouin J. (2012) The Stat3/GR interaction code: predictive value of direct/indirect DNA recruitment for transcription outcome. Mol. Cell 47, 38–49 10.1016/j.molcel.2012.04.021 [DOI] [PubMed] [Google Scholar]
- 11. Laganière J., Deblois G., Lefebvre C., Bataille A. R., Robert F., and Giguère V. (2005) From the cover: location analysis of estrogen receptor α target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc. Natl. Acad. Sci. U.S.A. 102, 11651–11656 10.1073/pnas.0505575102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lupien M., Eeckhoute J., Meyer C. A., Wang Q., Zhang Y., Li W., Carroll J. S., Liu X. S., and Brown M. (2008) FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132, 958–970 10.1016/j.cell.2008.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carroll J. S., Liu X. S., Brodsky A. S., Li W., Meyer C. A., Szary A. J., Eeckhoute J., Shao W., Hestermann E. V., Geistlinger T. R., Fox E. A., Silver P. A., and Brown M. (2005) Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122, 33–43 10.1016/j.cell.2005.05.008 [DOI] [PubMed] [Google Scholar]
- 14. Wang Q., Li W., Zhang Y., Yuan X., Xu K., Yu J., Chen Z., Beroukhim R., Wang H., Lupien M., Wu T., Regan M. M., Meyer C. A., Carroll J. S., Manrai A. K., et al. (2009) Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell 138, 245–256 10.1016/j.cell.2009.04.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hurtado A., Holmes K. A., Ross-Innes C. S., Schmidt D., and Carroll J. S. (2011) FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat. Genet. 43, 27–33 10.1038/ng.730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Voss T. C., Schiltz R. L., Sung M. H., Yen P. M., Stamatoyannopoulos J. A., Biddie S. C., Johnson T. A., Miranda T. B., John S., and Hager G. L. (2011) Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell 146, 544–554 10.1016/j.cell.2011.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sherwood R. I., Hashimoto T., O'Donnell C. W., Lewis S., Barkal A. A., van Hoff J. P., Karun V., Jaakkola T., and Gifford D. K. (2014) Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat. Biotechnol. 32, 171–178 10.1038/nbt.2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Swinstead E. E., Miranda T. B., Paakinaho V., Baek S., Goldstein I., Hawkins M., Karpova T. S., Ball D., Mazza D., Lavis L. D., Grimm J. B., Morisaki T., Grøntved L., Presman D. M., and Hager G. L. (2016) Steroid receptors reprogram FoxA1 occupancy through dynamic chromatin transitions. Cell 165, 593–605 10.1016/j.cell.2016.02.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Okita K., Ichisaka T., and Yamanaka S. (2007) Generation of germline-competent induced pluripotent stem cells. Nature 448, 313–317 10.1038/nature05934 [DOI] [PubMed] [Google Scholar]
- 20. Koche R. P., Smith Z. D., Adli M., Gu H., Ku M., Gnirke A., Bernstein B. E., and Meissner A. (2011) Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell 8, 96–105 10.1016/j.stem.2010.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Soufi A., Donahue G., and Zaret K. S. (2012) Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome. Cell 151, 994–1004 10.1016/j.cell.2012.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chronis C., Fiziev P., Papp B., Butz S., Bonora G., Sabri S., Ernst J., and Plath K. (2017) Cooperative binding of transcription factors orchestrates reprogramming. Cell 168, 442–459.e20 10.1016/j.cell.2016.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sridharan R., Tchieu J., Mason M. J., Yachechko R., Kuoy E., Horvath S., Zhou Q., and Plath K. (2009) Role of the murine reprogramming factors in the induction of pluripotency. Cell 136, 364–377 10.1016/j.cell.2009.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee M. T., Bonneau A. R., Takacs C. M., Bazzini A. A., DiVito K. R., Fleming E. S., and Giraldez A. J. (2013) Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature 503, 360–364 10.1038/nature12632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liang H. L., Nien C. Y., Liu H. Y., Metzstein M. M., Kirov N., and Rushlow C. (2008) The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 456, 400–403 10.1038/nature07388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schulz K. N., Bondra E. R., Moshe A., Villalta J. E., Lieb J. D., Kaplan T., McKay D. J., and Harrison M. M. (2015) Zelda is differentially required for chromatin accessibility, transcription factor binding, and gene expression in the early Drosophila embryo. Genome Res. 25, 1715–1726 10.1101/gr.192682.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gualdi R., Bossard P., Zheng M., Hamada Y., Coleman J. R., and Zaret K. S. (1996) Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev. 10, 1670–1682 10.1101/gad.10.13.1670 [DOI] [PubMed] [Google Scholar]
- 28. Cirillo L. A., McPherson C. E., Bossard P., Stevens K., Cherian S., Shim E. Y., Clark K. L., Burley S. K., and Zaret K. S. (1998) Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J. 17, 244–254 10.1093/emboj/17.1.244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cirillo L. A., Lin F. R., Cuesta I., Friedman D., Jarnik M., and Zaret K. S. (2002) Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell 9, 279–289 10.1016/S1097-2765(02)00459-8 [DOI] [PubMed] [Google Scholar]
- 30. Li Z., Schug J., Tuteja G., White P., and Kaestner K. H. (2011) The nucleosome map of the mammalian liver. Nat. Struct. Mol. Biol. 18, 742–746 10.1038/nsmb.2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Iwafuchi-Doi M., Donahue G., Kakumanu A., Watts J. A., Mahony S., Pugh B. F., Lee D., Kaestner K. H., and Zaret K. S. (2016) The pioneer transcription factor FoxA maintains an accessible nucleosome configuration at enhancers for tissue-specific gene activation. Mol. Cell 62, 79–91 10.1016/j.molcel.2016.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li Z., Gadue P., Chen K., Jiao Y., Tuteja G., Schug J., Li W., and Kaestner K. H. (2012) Foxa2 and H2A.Z mediate nucleosome depletion during embryonic stem cell differentiation. Cell 151, 1608–1616 10.1016/j.cell.2012.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fakhouri T. H., Stevenson J., Chisholm A. D., and Mango S. E. (2010) Dynamic chromatin organization during foregut development mediated by the organ selector gene PHA-4/FoxA. PLoS Genet. 6, e1001060 10.1371/journal.pgen.1001060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hsu H. T., Chen H. M., Yang Z., Wang J., Lee N. K., Burger A., Zaret K., Liu T., Levine E., and Mango S. E. (2015) TRANSCRIPTION. Recruitment of RNA polymerase II by the pioneer transcription factor PHA-4. Science 348, 1372–1376 10.1126/science.aab1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Holtzinger A., and Evans T. (2005) Gata4 regulates the formation of multiple organs. Development 132, 4005–4014 10.1242/dev.01978 [DOI] [PubMed] [Google Scholar]
- 36. Zhao R., Watt A. J., Li J., Luebke-Wheeler J., Morrisey E. E., and Duncan S. A. (2005) GATA6 is essential for embryonic development of the liver but dispensable for early heart formation. Mol. Cell. Biol. 25, 2622–2631 10.1128/MCB.25.7.2622-2631.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Watt A. J., Zhao R., Li J., and Duncan S. A. (2007) Development of the mammalian liver and ventral pancreas is dependent on GATA4. BMC Dev. Biol. 7, 37 10.1186/1471-213X-7-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang P., He Z., Ji S., Sun H., Xiang D., Liu C., Hu Y., Wang X., and Hui L. (2011) Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature 475, 386–389 10.1038/nature10116 [DOI] [PubMed] [Google Scholar]
- 39. Sekiya S., and Suzuki A. (2011) Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature 475, 390–393 10.1038/nature10263 [DOI] [PubMed] [Google Scholar]
- 40. Carotta S., Wu L., and Nutt S. L. (2010) Surprising new roles for PU.1 in the adaptive immune response. Immunol. Rev. 238, 63–75 10.1111/j.1600-065X.2010.00955.x [DOI] [PubMed] [Google Scholar]
- 41. Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., and Glass C. K. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Barozzi I., Simonatto M., Bonifacio S., Yang L., Rohs R., Ghisletti S., and Natoli G. (2014) Coregulation of transcription factor binding and nucleosome occupancy through DNA features of mammalian enhancers. Mol. Cell 54, 844–857 10.1016/j.molcel.2014.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. van Oevelen C., Collombet S., Vicent G., Hoogenkamp M., Lepoivre C., Badeaux A., Bussmann L., Sardina J. L., Thieffry D., Beato M., Shi Y., Bonifer C., and Graf T. (2015) C/EBPα activates pre-existing and de novo macrophage enhancers during induced pre-B cell transdifferentiation and myelopoiesis. Stem Cell Rep. 5, 232–247 10.1016/j.stemcr.2015.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Budry L., Balsalobre A., Gauthier Y., Khetchoumian K., L'honoré A., Vallette S., Brue T., Figarella-Branger D., Meij B., and Drouin J. (2012) The selector gene Pax7 dictates alternate pituitary cell fates through its pioneer action on chromatin remodeling. Genes Dev. 26, 2299–2310 10.1101/gad.200436.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mayran A., Khetchoumian K., Hariri F., Pastinen T., Gauthier Y., Balsalobre A., and Drouin J. (2018) Pioneer factor Pax7 deploys a stable enhancer repertoire for specification of cell fate. Nat. Genet. 50, 259–269 10.1038/s41588-017-0035-2 [DOI] [PubMed] [Google Scholar]
- 46. Györy I., Boller S., Nechanitzky R., Mandel E., Pott S., Liu E., and Grosschedl R. (2012) Transcription factor Ebf1 regulates differentiation stage-specific signaling, proliferation, and survival of B cells. Genes Dev. 26, 668–682 10.1101/gad.187328.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Treiber T., Mandel E. M., Pott S., Györy I., Firner S., Liu E. T., and Grosschedl R. (2010) Early B cell factor 1 regulates B cell gene networks by activation, repression, and transcription-independent poising of chromatin. Immunity 32, 714–725 10.1016/j.immuni.2010.04.013 [DOI] [PubMed] [Google Scholar]
- 48. Boller S., Ramamoorthy S., Akbas D., Nechanitzky R., Burger L., Murr R., Schübeler D., and Grosschedl R. (2016) Pioneering activity of the C-terminal domain of EBF1 shapes the chromatin landscape for B cell programming. Immunity 44, 527–541 10.1016/j.immuni.2016.02.021 [DOI] [PubMed] [Google Scholar]
- 49. Wapinski O. L., Vierbuchen T., Qu K., Lee Q. Y., Chanda S., Fuentes D. R., Giresi P. G., Ng Y. H., Marro S., Neff N. F., Drechsel D., Martynoga B., Castro D. S., Webb A. E., Sudhof T. C., et al. (2013) Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell 155, 621–635 10.1016/j.cell.2013.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pataskar A., Jung J., Smialowski P., Noack F., Calegari F., Straub T., and Tiwari V. K. (2016) NeuroD1 reprograms chromatin and transcription factor landscapes to induce the neuronal program. EMBO J. 35, 24–45 10.15252/embj.201591206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bossard P., and Zaret K. S. (1998) GATA transcription factors as potentiators of gut endoderm differentiation. Development 125, 4909–4917 [DOI] [PubMed] [Google Scholar]
- 52. Sekiya T., Muthurajan U. M., Luger K., Tulin A. V., and Zaret K. S. (2009) Nucleosome-binding affinity as a primary determinant of the nuclear mobility of the pioneer transcription factor FoxA. Genes Dev. 23, 804–809 10.1101/gad.1775509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Soufi A., Garcia M. F., Jaroszewicz A., Osman N., Pellegrini M., and Zaret K. S. (2015) Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161, 555–568 10.1016/j.cell.2015.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Clark K. L., Halay E. D., Lai E., and Burley S. K. (1993) Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364, 412–420 10.1038/364412a0 [DOI] [PubMed] [Google Scholar]
- 55. de Dieuleveult M., Yen K., Hmitou I., Depaux A., Boussouar F., Bou Dargham D., Jounier S., Humbertclaude H., Ribierre F., Baulard C., Farrell N. P., Park B., Keime C., Carrière L., Berlivet S., et al. (2016) Genome-wide nucleosome specificity and function of chromatin remodellers in ES cells. Nature 530, 113–116 10.1038/nature16505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kidder B. L., Palmer S., and Knott J. G. (2009) SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells 27, 317–328 10.1634/stemcells.2008-0710 [DOI] [PubMed] [Google Scholar]
- 57. King H. W., and Klose R. J. (2017) The pioneer factor OCT4 requires the chromatin remodeller BRG1 to support gene regulatory element function in mouse embryonic stem cells. eLife 2017;6:e22631 10.7554/eLife.22631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang L., Du Y., Ward J. M., Shimbo T., Lackford B., Zheng X., Miao Y. L., Zhou B., Han L., Fargo D. C., Jothi R., Williams C. J., Wade P. A., and Hu G. (2014) INO80 facilitates pluripotency gene activation in embryonic stem cell self-renewal, reprogramming, and blastocyst development. Cell Stem Cell 14, 575–591 10.1016/j.stem.2014.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Takaku M., Grimm S. A., Shimbo T., Perera L., Menafra R., Stunnenberg H. G., Archer T. K., Machida S., Kurumizaka H., and Wade P. A. (2016) GATA3-dependent cellular reprogramming requires activation-domain dependent recruitment of a chromatin remodeler. Genome Biol. 17, 36 10.1186/s13059-016-0897-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schuettengruber B., Martinez A. M., Iovino N., and Cavalli G. (2011) Trithorax group proteins: switching genes on and keeping them active. Nat. Rev. Mol. Cell Biol. 12, 799–814 10.1038/nrm3230 [DOI] [PubMed] [Google Scholar]
- 61. McKinnell I. W., Ishibashi J., Le Grand F., Punch V. G., Addicks G. C., Greenblatt J. F., Dilworth F. J., and Rudnicki M. A. (2008) Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nat. Cell Biol. 10, 77–84 10.1038/ncb1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jozwik K. M., Chernukhin I., Serandour A. A., Nagarajan S., and Carroll J. S. (2016) FOXA1 directs H3K4 monomethylation at enhancers via recruitment of the methyltransferase MLL3. Cell Rep. 17, 2715–2723 10.1016/j.celrep.2016.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jin C., Zang C., Wei G., Cui K., Peng W., Zhao K., and Felsenfeld G. (2009) H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat. Genet. 41, 941–945 10.1038/ng.409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Updike D. L., and Mango S. E. (2006) Temporal regulation of foregut development by HTZ-1/H2A.Z and PHA-4/FoxA. PLoS Genet. 2, e161 10.1371/journal.pgen.0020161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gévry N., Hardy S., Jacques P. E., Laflamme L., Svotelis A., Robert F., and Gaudreau L. (2009) Histone H2A.Z is essential for estrogen receptor signaling. Genes Dev. 23, 1522–1533 10.1101/gad.1787109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Menet J. S., Pescatore S., and Rosbash M. (2014) CLOCK:BMAL1 is a pioneer-like transcription factor. Genes Dev. 28, 8–13 10.1101/gad.228536.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hagey D. W., Zaouter C., Combeau G., Lendahl M. A., Andersson O., Huss M., and Muhr J. (2016) Distinct transcription factor complexes act on a permissive chromatin landscape to establish regionalized gene expression in CNS stem cells. Genome Res. 26, 908–917 10.1101/gr.203513.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cheloufi S., Elling U., Hopfgartner B., Jung Y. L., Murn J., Ninova M., Hubmann M., Badeaux A. I., Euong Ang C., Tenen D., Wesche D. J., Abazova N., Hogue M., Tasdemir N., Brumbaugh J., et al. (2015) The histone chaperone CAF-1 safeguards somatic cell identity. Nature 528, 218–224 10.1038/nature15749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Donaghey J., Thakurela S., Charlton J., Chen J. S., Smith Z. D., Gu H., Pop R., Clement K., Stamenova E. K., Karnik R., Kelley D. R., Gifford C. A., Cacchiarelli D., Rinn J. L., Gnirke A., et al. (2018) Genetic determinants and epigenetic effects of pioneer-factor occupancy. Nat. Genet. 50, 250–258 10.1038/s41588-017-0034-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Domcke S., Bardet A. F., Adrian Ginno P., Hartl D., Burger L., and Schübeler D. (2015) Competition between DNA methylation and transcription factors determines binding of NRF1. Nature 528, 575–579 10.1038/nature16462 [DOI] [PubMed] [Google Scholar]
- 71. Seisenberger S., Andrews S., Krueger F., Arand J., Walter J., Santos F., Popp C., Thienpont B., Dean W., and Reik W. (2012) The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell 48, 849–862 10.1016/j.molcel.2012.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Messerschmidt D. M., Knowles B. B., and Solter D. (2014) DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev. 28, 812–828 10.1101/gad.234294.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bernstein B. E., Meissner A., and Lander E. S. (2007) The mammalian epigenome. Cell 128, 669–681 10.1016/j.cell.2007.01.033 [DOI] [PubMed] [Google Scholar]
- 74. Gottesfeld J. M., and Forbes D. J. (1997) Mitotic repression of the transcriptional machinery. Trends Biochem. Sci. 22, 197–202 10.1016/S0968-0004(97)01045-1 [DOI] [PubMed] [Google Scholar]
- 75. Martínez-Balbás M. A., Dey A., Rabindran S. K., Ozato K., and Wu C. (1995) Displacement of sequence-specific transcription factors from mitotic chromatin. Cell 83, 29–38 10.1016/0092-8674(95)90231-7 [DOI] [PubMed] [Google Scholar]
- 76. Kadauke S., Udugama M. I., Pawlicki J. M., Achtman J. C., Jain D. P., Cheng Y., Hardison R. C., and Blobel G. A. (2012) Tissue-specific mitotic bookmarking by hematopoietic transcription factor GATA1. Cell 150, 725–737 10.1016/j.cell.2012.06.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Caravaca J. M., Donahue G., Becker J. S., He X., Vinson C., and Zaret K. S. (2013) Bookmarking by specific and nonspecific binding of FoxA1 pioneer factor to mitotic chromosomes. Genes Dev. 27, 251–260 10.1101/gad.206458.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Deluz C., Friman E. T., Strebinger D., Benke A., Raccaud M., Callegari A., Leleu M., Manley S., and Suter D. M. (2016) A role for mitotic bookmarking of SOX2 in pluripotency and differentiation. Genes Dev. 30, 2538–2550 10.1101/gad.289256.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Teves S. S., An L., Hansen A. S., Xie L., Darzacq X., and Tjian R. (2016) A dynamic mode of mitotic bookmarking by transcription factors. eLife 5, e22280 10.7554/eLife.22280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Almouzni G., and Cedar H. (2016) Maintenance of epigenetic information. Cold Spring Harb. Perspect. Biol. 8, a019372 10.1101/cshperspect.a019372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Varley K. E., Gertz J., Bowling K. M., Parker S. L., Reddy T. E., Pauli-Behn F., Cross M. K., Williams B. A., Stamatoyannopoulos J. A., Crawford G. E., Absher D. M., Wold B. J., and Myers R. M. (2013) Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 23, 555–567 10.1101/gr.147942.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hermann A., Goyal R., and Jeltsch A. (2004) The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J. Biol. Chem. 279, 48350–48359 10.1074/jbc.M403427200 [DOI] [PubMed] [Google Scholar]
- 83. Bostick M., Kim J. K., Estève P. O., Clark A., Pradhan S., and Jacobsen S. E. (2007) UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317, 1760–1764 10.1126/science.1147939 [DOI] [PubMed] [Google Scholar]
- 84. Rasmussen K. D., and Helin K. (2016) Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 30, 733–750 10.1101/gad.276568.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zhang Y., Zhang D., Li Q., Liang J., Sun L., Yi X., Chen Z., Yan R., Xie G., Li W., Liu S., Xu B., Li L., Yang J., He L., and Shang Y. (2016) Nucleation of DNA repair factors by FOXA1 links DNA demethylation to transcriptional pioneering. Nat. Genet. 48, 1003–1013 10.1038/ng.3635 [DOI] [PubMed] [Google Scholar]
- 86. Katoh M., Igarashi M., Fukuda H., Nakagama H., and Katoh M. (2013) Cancer genetics and genomics of human FOX family genes. Cancer Lett. 328, 198–206 10.1016/j.canlet.2012.09.017 [DOI] [PubMed] [Google Scholar]
- 87. Sahu B., Laakso M., Ovaska K., Mirtti T., Lundin J., Rannikko A., Sankila A., Turunen J. P., Lundin M., Konsti J., Vesterinen T., Nordling S., Kallioniemi O., Hautaniemi S., and Jänne O. A. (2011) Dual role of FoxA1 in androgen receptor binding to chromatin, androgen signalling and prostate cancer. EMBO J. 30, 3962–3976 10.1038/emboj.2011.328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Thorat M. A., Marchio C., Morimiya A., Savage K., Nakshatri H., Reis-Filho J. S., and Badve S. (2008) Forkhead box A1 expression in breast cancer is associated with luminal subtype and good prognosis. J. Clin. Pathol. 61, 327–332 [DOI] [PubMed] [Google Scholar]
- 89. Grasso C. S., Wu Y. M., Robinson D. R., Cao X., Dhanasekaran S. M., Khan A. P., Quist M. J., Jing X., Lonigro R. J., Brenner J. C., Asangani I. A., Ateeq B., Chun S. Y., Siddiqui J., Sam L., et al. (2012) The mutational landscape of lethal castration-resistant prostate cancer. Nature 487, 239–243 10.1038/nature11125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Carroll J. S., Meyer C. A., Song J., Li W., Geistlinger T. R., Eeckhoute J., Brodsky A. S., Keeton E. K., Fertuck K. C., Hall G. F., Wang Q., Bekiranov S., Sementchenko V., Fox E. A., Silver P. A., et al. (2006) Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 38, 1289–1297 10.1038/ng1901 [DOI] [PubMed] [Google Scholar]
- 91. Pomerantz M. M., Li F., Takeda D. Y., Lenci R., Chonkar A., Chabot M., Cejas P., Vazquez F., Cook J., Shivdasani R. A., Bowden M., Lis R., Hahn W. C., Kantoff P. W., Brown M., et al. (2015) The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat. Genet. 47, 1346–1351 10.1038/ng.3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Curtis C., Shah S. P., Chin S. F., Turashvili G., Rueda O. M., Dunning M. J., Speed D., Lynch A. G., Samarajiwa S., Yuan Y., Gräf S., Ha G., Haffari G., Bashashati A., Russell R., et al. (2012) The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 10.1038/nature10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Green M. R., Aya-Bonilla C., Gandhi M. K., Lea R. A., Wellwood J., Wood P., Marlton P., and Griffiths L. R. (2011) Integrative genomic profiling reveals conserved genetic mechanisms for tumorigenesis in common entities of non-Hodgkin's lymphoma. Genes Chromosomes Cancer 50, 313–326 10.1002/gcc.20856 [DOI] [PubMed] [Google Scholar]
- 94. Yu J., Deshmukh H., Payton J. E., Dunham C., Scheithauer B. W., Tihan T., Prayson R. A., Guha A., Bridge J. A., Ferner R. E., Lindberg G. M., Gutmann R. J., Emnett R. J., Salavaggione L., Gutmann D. H., et al. (2011) Array-based comparative genomic hybridization identifies CDK4 and FOXM1 alterations as independent predictors of survival in malignant peripheral nerve sheath tumor. Clin. Cancer Res. 17, 1924–1934 10.1158/1078-0432.CCR-10-1551 [DOI] [PubMed] [Google Scholar]
- 95. Bennicelli J. L., Advani S., Schäfer B. W., and Barr F. G. (1999) PAX3 and PAX7 exhibit conserved cis-acting transcription repression domains and utilize a common gain of function mechanism in alveolar rhabdomyosarcoma. Oncogene 18, 4348–4356 10.1038/sj.onc.1202812 [DOI] [PubMed] [Google Scholar]
- 96. Cao L., Yu Y., Bilke S., Walker R. L., Mayeenuddin L. H., Azorsa D. O., Yang F., Pineda M., Helman L. J., and Meltzer P. S. (2010) Genome-wide identification of PAX3-FKHR binding sites in rhabdomyosarcoma reveals candidate target genes important for development and cancer. Cancer Res. 70, 6497–6508 10.1158/0008-5472.CAN-10-0582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Hatta M., and Cirillo L. A. (2007) Chromatin opening and stable perturbation of core histone:DNA contacts by FoxO1. J. Biol. Chem. 282, 35583–35593 10.1074/jbc.M704735200 [DOI] [PubMed] [Google Scholar]
- 98. So C. W., and Cleary M. L. (2003) Common mechanism for oncogenic activation of MLL by forkhead family proteins. Blood 101, 633–639 10.1182/blood-2002-06-1785 [DOI] [PubMed] [Google Scholar]
- 99. Dansen T. B., and Burgering B. M. (2008) Unravelling the tumor-suppressive functions of FOXO proteins. Trends Cell Biol. 18, 421–429 10.1016/j.tcb.2008.07.004 [DOI] [PubMed] [Google Scholar]
- 100. Sammons M. A., Zhu J., Drake A. M., and Berger S. L. (2015) TP53 engagement with the genome occurs in distinct local chromatin environments via pioneer factor activity. Genome Res. 25, 179–188 10.1101/gr.181883.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Younger S. T., and Rinn J. L. (2017) p53 regulates enhancer accessibility and activity in response to DNA damage. Nucleic Acids Res. 45, 9889–9900 10.1093/nar/gkx577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Wapinski O. L., Lee Q. Y., Chen A. C., Li R., Corces M. R., Ang C. E., Treutlein B., Xiang C., Baubet V., Suchy F. P., Sankar V., Sim S., Quake S. R., Dahmane N., Wernig M., and Chang H. Y. (2017) Rapid chromatin switch in the direct reprogramming of fibroblasts to neurons. Cell Rep. 20, 3236–3247 10.1016/j.celrep.2017.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Raposo A. A., Vasconcelos F. F., Drechsel D., Marie C., Johnston C., Dolle D., Bithell A., Gillotin S., van den Berg D. L., Ettwiller L., Flicek P., Crawford G. E., Parras C. M., Berninger B., Buckley N. J., et al. (2015) Ascl1 coordinately regulates gene expression and the chromatin landscape during neurogenesis. Cell Rep. pii: S2211–1247(15)00171–0 10.1016/j.celrep.2015.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Feng R., Desbordes S. C., Xie H., Tillo E. S., Pixley F., Stanley E. R., and Graf T. (2008) PU.1 and C/EBPα/β convert fibroblasts into macrophage-like cells. Proc. Natl. Acad. Sci. U.S.A. 105, 6057–6062 10.1073/pnas.0711961105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Festuccia N., Dubois A., Vandormael-Pournin S., Gallego Tejeda E., Mouren A., Bessonnard S., Mueller F., Proux C., Cohen-Tannoudji M., and Navarro P. (2016) Mitotic binding of Esrrb marks key regulatory regions of the pluripotency network. Nat. Cell Biol. 18, 1139–1148 10.1038/ncb3418 [DOI] [PubMed] [Google Scholar]
- 106. Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., Aoi T., Okita K., Mochiduki Y., Takizawa N., and Yamanaka S. (2008) Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 26, 101–106 10.1038/nbt1374 [DOI] [PubMed] [Google Scholar]
- 107. Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., and Yamanaka S. (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]