Abstract
PHR (PAM/Highwire/RPM-1) proteins are conserved RING E3 ubiquitin ligases that function in developmental processes, such as axon termination and synapse formation, as well as axon degeneration. At present, our understanding of how PHR proteins form ubiquitin ligase complexes remains incomplete. Although genetic studies indicate NMNAT2 is an important mediator of PHR protein function in axon degeneration, it remains unknown how PHR proteins inhibit NMNAT2. Here, we decipher the biochemical basis for how the human PHR protein PAM, also called MYCBP2, forms a noncanonical Skp/Cullin/F-box (SCF) complex that contains the F-box protein FBXO45 and SKP1 but lacks CUL1. We show FBXO45 does not simply function in substrate recognition but is important for assembly of the PAM/FBXO45/SKP1 complex. Interestingly, we demonstrate a novel role for SKP1 as an auxiliary component of the target recognition module that enhances binding of FBXO45 to NMNAT2. Finally, we provide biochemical evidence that PAM polyubiquitinates NMNAT2 and regulates NMNAT2 protein stability and degradation by the proteasome.
Keywords: E3 ubiquitin ligase, neuron, axon, proteasome, fluorescence resonance energy transfer (FRET), Caenorhabditis elegans (C. elegans), neurodegeneration, FBXO45, MYCBP2, NMNAT, PAM, RPM-1, PHR protein, ubiquitin ligase, FSN-1, F-box, axon degeneration, ubiquitination
Introduction
Post-translational modification of proteins by ubiquitination is a prominent mechanism for regulating protein trafficking, localization, stability, and activation. Ubiquitination plays an important role in cell cycle control (1, 2) and oncogenesis (3, 4). In the nervous system, ubiquitination impacts both neuronal development and function (5, 6) and is implicated in neurodegenerative disease (7). Proteins can be monoubiquitinated by the addition of a single ubiquitin molecule or polyubiquitinated with multiple ubiquitin species. Polyubiquitination occurs from successive reactions by a series of ubiquitin ligases called E1, E2, and E3. One outcome of polyubiquitination is degradation by the 26S proteasome.
Many E3 ubiquitin ligases form multisubunit complexes, some of which contain a Cullin scaffolding protein, a Skp adaptor protein, and an F-box protein that mediates target recognition. One example of these canonical Skp/Cullin/F-box (SCF)2 E3 ligase complexes is formed by Rbx1 with Cul1, Skp1, and the F-box Skp2 (8–10). Cul1 binds to Rbx1 on its C terminus and to the F-box protein, which recognizes substrates, on the N terminus. SCF complexes allow targets to be brought in close proximity to the RING domain of a given E3 ligase, such as Rbx1. Other examples of important canonical SCF complexes include Rbx1/Cul1/Skp1/β-TrCP, which degrades β-catenin (1, 11, 12), Rbx1/Cul1/Skp1/Cdc4, which regulates the cell cycle (10, 13, 14), and Rbx1/Cul1/Skp1/Fbw7, which is a prominent tumor suppressor (4, 15, 16).
The Pam/Highwire/RPM-1 (PHR) proteins, which include human PAM/MYCBP2, mouse Phr1, Drosophila Highwire, and Caenorhabditis elegans RPM-1, have been suggested to form atypical SCF complexes that lack a Cullin (17). PHR proteins are RING domain E3 ligases that play conserved functions in the developing nervous system where they regulate axon termination, axon guidance, and synapse formation (18–26). In mature neurons, PHR proteins take on an important role in axon degeneration following injury (27, 28). Outside of the nervous system, PHR proteins are implicated in oncogenesis (29–34). Despite growing recognition of the functional importance of PHR proteins, relatively little is known about how components of atypical PHR ubiquitin ligase complexes assemble.
Previous work using model organisms and cell-based biochemistry identified a conserved F-box protein, called FBXO45 in mammals or FSN-1 in C. elegans, that binds PHR ubiquitin ligases (35–37). At present, we have the best understanding of how C. elegans RPM-1 forms a complex with FSN-1. Biochemistry with 293 cells and in vivo biochemistry from the worm nervous system indicate that RPM-1 contains multiple FSN-1–binding domains that interact with FSN-1 (38, 39). It remains unknown whether a similar biochemical mechanism underpins assembly of ubiquitin ligase complexes formed by other PHR proteins, including human PAM. Furthermore, we do not know whether PHR proteins bind directly or indirectly to F-box proteins.
Proteomics and genetics in flies, as well as biochemistry in 293 cells, have suggested PHR proteins can form noncanonical SCF complexes, which contain SKP1 but lack a Cullin (37, 40). Although these studies provided evidence that individual components can interact, biochemistry showing that FBXO45 and SKP1, but not CUL1, assemble into a complex with PAM remains notably absent. We also know nothing about the biochemical function of SKP1 in PHR ubiquitin ligase complexes. Is SKP1 an adaptor that mediates FBXO45 binding to PAM? Does SKP1 influence FBXO45 binding to substrates?
Studies with mouse Phr1 and fly Highwire indicate NMNAT2 mediates PHR protein effects on axon degeneration (17, 27, 28). NMNAT2 is part of the nicotinamide mononucleotide adenylyltransferase protein family that catalyzes formation of NAD+ (41–43). NAD is an essential coenzyme for several biological process, such as cell death, energy metabolism, and calcium mobilization. NMNAT2 is highly expressed in the brain, and along with axon protection following injury (44–49), it is implicated in neurodegenerative disease (50–53). Although functional genetics indicate PHR proteins can inhibit NMNAT2, the biochemical mechanism by which this occurs remains untested. For example, we lack evidence that PHR proteins interact biochemically with NMNAT2. Furthermore, it is unknown whether PHR proteins polyubiquitinate NMNAT2 and if so whether ubiquitination results in NMNAT2 degradation by the proteasome.
In this study, we comprehensively evaluate the biochemistry that underpins assembly of the PAM ubiquitin ligase complex. We demonstrate that PAM assembles into a noncanonical SCF complex that contains FBXO45 and SKP1 but not CUL1. Importantly, our results indicate the biochemical mechanism behind this is a direct interaction between FBXO45 and PAM, which facilitates recruitment of SKP1 into the ligase complex. This precludes the need for a Cullin scaffold.
Interestingly, we uncover a novel role for SKP1 in this atypical SCF complex. SKP1 acts as part of the target recognition module with FBXO45 rather than acting as an adaptor to mediate formation of the PAM/FBXO45/SKP1 complex. Thus, SKP1 plays a very different role in the noncanonical PAM/FBXO45/SKP1 complex compared with traditional SCF complexes formed by the Rbx1 E3 ligase.
Increased insight into the biochemistry underpinning assembly of the PAM/FBXO45/SKP1 ubiquitin ligase complex prompted us to dissect the biochemical relationship between PAM and NMNAT2. Our results indicate NMNAT2 binds to the PAM/FBXO45/SKP1 complex via FBXO45. PAM polyubiquitinates NMNAT2 thereby regulating NMNAT2 protein stability and degradation by the proteasome. Importantly, experiments with PAM overexpression and endogenous PAM knockout cell lines indicate that PAM E3 ubiquitin ligase activity regulates NMNAT2 protein turnover. Our findings have interesting implications, given the importance of PHR proteins and NMNAT2 in axon degeneration, and growing links between NMNAT2 and neurodegenerative disease.
Results
FBXO45 binds directly to PAM via a conserved mechanism
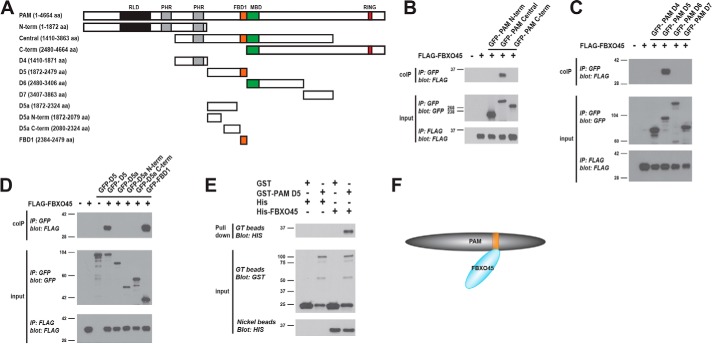
Previous biochemistry using HEK 293 cells and in vivo biochemistry from transgenic animals showed that C. elegans RPM-1 binds the F-box protein FSN-1 via three domains in RPM-1 called FBD1, FBD2, and FBD3 (38, 39). We tested whether human PAM/MYCBP2 relies upon a similar conserved mechanism to bind the human F-box protein FBXO45. To do so, we began by generating three fragments of PAM, and we tested their binding to FBXO45 in HEK 293 cells (Fig. 1A). To minimize the possibility that we might miss FBXO45 interaction sites, we generated PAM fragments that overlapped with one another. As shown in Fig. 1B, only the central fragment of PAM (GFP–PAM Central) coprecipitated with FLAG-tagged FBXO45 (FLAG–FBXO45). To further map this interaction, we transfected 293 cells with FLAG–FBXO45 and four smaller domains within the central PAM fragment, termed D4 to D7 (Fig. 1A). We only observed an interaction between PAM D5 and FBXO45 (Fig. 1C).
Figure 1.
Identification of a domain in the E3 ligase PAM that binds directly to the F-box protein FBXO45. A, schematic of PAM with annotated protein domains and PAM constructs tested for binding to FBXO45: RCC-1 like domain (RLD, black); PHR family–specific domain (PHR, gray); FSN-1 binding domain 1 (FBD1, orange); MYC-binding domain (MBD, green); and RING-H2 ubiquitin ligase domain (RING, red). B, coIP of FLAG–FBXO45 with GFP–PAM fragments using transfected HEK 293 cells. FLAG–FBXO45 coprecipitates with GFP–PAM central fragment. C, coIP of FLAG–FBXO45 with GFP–PAM domains using 293 cells. FLAG–FBXO45 coprecipitates with GFP–PAM D5. D, Further mapping of subdomains in GFP–PAM D5 that bind FLAG–FBXO45. FBD1 domain of PAM is sufficient for binding FBXO45. E, GST-pulldown experiment with recombinant proteins expressed in E. coli. His6-FBXO45 is pulled down by GST–PAM D5 but not GST. F, summary showing FBXO45 interacts directly with PAM via the FBD1 domain (orange). B–E, representative is shown for results from three or more independent experiments.
Interestingly, D5 contains the conserved region of PAM called FBD1, which was previously shown to be sufficient for binding to C. elegans FSN-1 (Fig. 1A) (38). This prompted us to further determine whether the FBD1 of PAM was sufficient for binding to FBXO45. Coimmunoprecipitation (coIP) experiments with several smaller constructs derived from PAM D5 showed only GFP–FBD1 coprecipitated with FLAG–FBXO45 (Fig. 1D). Thus, FBD1 of PAM is capable of binding FBXO45.
Next, we wanted to determine whether the interaction between PAM and FBXO45 was direct. We expressed GST–PAM D5, GST, His6-FBXO45, or His6 in Escherichia coli. Lysates from E. coli expressing GST–PAM D5 or the negative control GST were incubated with glutathione (GT) beads. E. coli lysates expressing His–FBXO45 or His alone were incubated with GT beads bound with GST–PAM D5 or GST. Pulldown of His–FBXO45 was only observed when GST–PAM D5 was used as bait (Fig. 1E). As a control, lysates expressing His–FBXO45 or His alone were also precipitated with nickel beads and immunoblotted with anti-His antibody. This ensured equal amounts of His–FBXO45 were applied to GT beads bound with GST–PAM D5 or GST (Fig. 1E). Given that homologs of PAM and FBXO45 are not present in E. coli, these results show that PAM D5 is sufficient for direct binding to FBXO45 via the FBD1 domain (Fig. 1F).
RPM-1 and FSN-1 interact in subcellular axonal compartments in vivo
Our results here show that the FBD1 domain of PAM binds directly to FBXO45 (Fig. 1). Similarly, prior work showed the PAM ortholog RPM-1 utilizes FBD1 to bind the F-box protein FSN-1 (38). Because these findings suggest a conserved, direct interaction occurs between PHR proteins and F-box proteins, we wanted to explore where this interaction occurs in neurons in vivo. Therefore, we turned to in vivo two-photon FLIM–FRET (2pFLIM) using C. elegans RPM-1 and FSN-1.
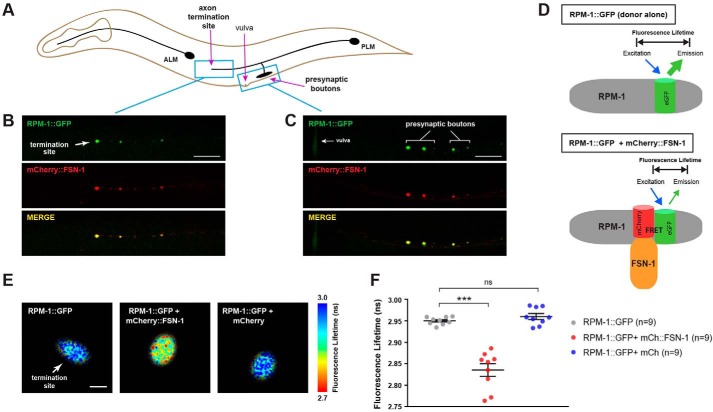
Prior to evaluating FLIM–FRET in worms, we initially validated reagents needed for these experiments. First, we generated transgenic worms that coexpress RPM-1::GFP and mCherry::FSN-1 specifically in mechanosensory neurons (Fig. 2A). Previous work showed that RPM-1 and FSN-1 function together to regulate axon termination and chemical synapse formation in these neurons (21, 54). Consistent with this, we observed robust colocalization of RPM-1::GFP with mCherry::FSN-1 at the axon termination site (Fig. 2B) and presynaptic boutons (Fig. 2C) using confocal microscopy.
Figure 2.
FLIM–FRET shows RPM-1 and FSN-1 interact in axonal compartments in vivo. A, schematic of C. elegans mechanosensory neurons. Blue boxes highlight axon termination site and presynaptic boutons of PLM neuron. Note vulva is an anatomical reference point. B and C, transgenic animals expressing RPM-1::GFP and mCherry::FSN-1 in PLM neurons show strong colocalization at axon termination site (B) and presynaptic boutons (C). Scale bars, 10 μm. D, schematic of FLIM–FRET in control animals expressing RPM-1::GFP (donor alone) where no FRET occurs, and animals coexpressing both RPM-1::GFP and mCherry::FSN-1 where FRET occurs shortening the RPM-1::GFP fluorescence lifetime. E, 2pFLIM with indicated genotypes. Fluorescence lifetime imaging of RPM-1::GFP is reduced at the axon termination site of PLM neurons when mCherry::FSN-1 is coexpressed. Scale bar, 0.5 μm. F, quantitation of 2pFLIM. Scatterplots show mean fluorescence lifetime of RPM-1::GFP at axon termination sites of indicated genotypes. RPM-1::GFP lifetime is significantly reduced when mCherry::FSN-1 is coexpressed compared with RPM-1::GFP alone. RPM-1::GFP lifetime is not reduced by coexpression of mCherry cell fill. Data are shown for nine animals (n = 9) from two independent experiments for all genotypes. Significance was determined using Student's t test, and error bars represent standard error of mean. ***, p ≤ 0.001; ns = not significant.
Having shown RPM-1 and FSN-1 colocalize in different subcellular compartments, we performed 2pFLIM using these reagents (Fig. 2D). Fluorescence lifetime imaging is a method of quantifying FRET, which provides many advantages, including enhanced sensitivity over ratiometric imaging or other intensity-based FRET measurements (55, 56). This approach allows us to test whether both proteins are in close molecular proximity. Importantly, FLIM only measures the fluorescence lifetime of the donor, which becomes shortened when the fluorophore undergoes FRET. 2pFLIM revealed that the fluorescence lifetime of GFP (donor) fused to RPM-1 was shorter when mCherry::FSN-1 was present at the axon termination site of mechanosensory neurons (Fig. 2E). In contrast, RPM-1::GFP fluorescence lifetime was not affected by expression of mCherry alone (Fig. 2E). Quantitation of 2pFLIM showed that RPM-1::GFP fluorescence lifetime was significantly reduced in the presence of mCherry::FSN-1 but not mCherry (Fig. 2F). These results show for the first time that RPM-1 physically interacts with FSN-1 at axon termination sites in vivo and is consistent with RPM-1 binding directly to FSN-1.
Conserved amino acids in FBD1 are sufficient for PAM binding to FBXO45
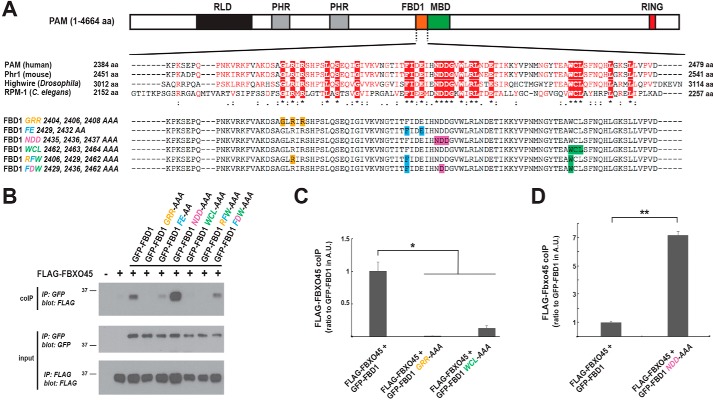
To further evaluate the mechanism by which the FBD1 domain of PAM mediates direct binding to FBXO45, we performed coIP with several FBD1 point mutants. Fig. 3A highlights several conserved motifs in FBD1 that we tested by alanine screening. Conserved amino acids in either individual motifs or residues across a combination of different motifs were mutated to alanine (Fig. 3A). GFP-tagged FBD1 mutants were cotransfected with FLAG–FBXO45 in 293 cells and tested for binding (Fig. 3B). GFP–FBD1 alanine mutants were expressed at similar levels, and FBXO45 coIP was reduced when three different motifs were mutated in FBD1, GRR (G2404A/R2406A/R2408A), FE (F2429A/E2432A), and WCL (W2462A/C2463A/L2464A) (Fig. 3B). Quantitation showed FBXO45 binding to FBD1 GRR and WCL alanine mutants was significantly reduced compared with WT FBD1 (Fig. 3C).
Figure 3.
Conserved motifs in the FBD1 domain of PAM are required for binding to FBXO45. A, schematic of PAM, and alignment of FBD1 sequences from PHR proteins across species generated using ESPript (66). Identical amino acids (aa) across species (red box underscored by asterisk), conserved amino acids of high similarity (red letters underscored by two dots), and conserved amino acids with weak similarity (red letters underscored by single dot). Shown below the alignment are FBD1 point mutants. Highlighted with single colors are residues in individual motifs that are mutated. B, coIP from 293 cells tests binding of FLAG–FBXO45 with different GFP–FBD1 point mutants. Decreased binding occurs with GFP–FBD1 GRR, FE, WCL, RFW, and FDW mutants. GFP–FBD1 NDD mutant shows increased binding to FLAG–FBXO45. C, quantitation of coIP (shown in arbitrary units (A.U.)) indicates FBD1 GRR and WCL point mutants have reduced binding to FBXO45. D, quantitation of coIP (A.U.) shows FBD1 NDD has increased binding to FBXO45. B, representative of at least three independent experiments is shown. C and D, quantitation was done with a minimum of three replicates. Significance was determined using Student's t test, and error bars represent standard error of mean. *, p < 0.05; **, p < 0.01; IP, immunoprecipitation.
We also simultaneously mutated single residues from multiple motifs of FBD1 and examined FBXO45 coIP (Fig. 3B). Reduced coprecipitation was observed for FBD1 RFW (R2406A/F2429A/W2462A) and FBD1 FDW (F2429A/D2436A/W2462A) alanine mutants compared with WT FBD1 (Fig. 3B).
Interestingly, FBD1 NDD (N2435A/D2436A/D2437A) showed increased FBXO45 binding (Fig. 3B). Quantitation demonstrated FBD1 NDD binding to FBXO45 was significantly increased compared with WT FBD1 (Fig. 3D).
Collectively, these observations support two conclusions. 1) Several conserved motifs in the FBD1 domain of PAM are necessary for binding to FBXO45. 2) The NDD motif of FBD1 restricts binding to FBXO45.
FBXO45 recruits SKP1 into a complex with PAM
Previous biochemical and proteomic experiments using cultured cells showed SKP1 binds to FBXO45 (37, 57). Affinity purification proteomics with Highwire in flies also identified the SKP1 ortholog SkpA (40). These observations invoke several interesting questions. How is SKP1 recruited to PHR ubiquitin ligase complexes? Does SKP1 function as an adaptor that helps stabilize the interaction between FBXO45 and PAM? Can SKP1 influence target recognition?
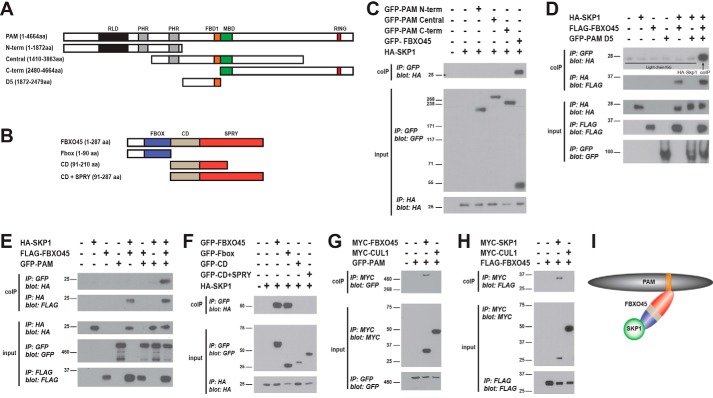
To answer these questions, we began by testing whether SKP1 binds PAM. HEK 293 cells were cotransfected with HA–SKP1 and GFP–PAM fragments (N-term, Central, and C-term, Fig. 4A). We did not observe coprecipitation of HA–SKP1 with any GFP–PAM fragments (Fig. 4C). Consistent with prior work (37, 57), HA–SKP1 did coIP with GFP–FBXO45 (Fig. 4C). These results indicate FBXO45 and SKP1 interact, and this is likely to be independent of PAM.
Figure 4.
FBXO45 facilitates assembly of a ubiquitin ligase complex containing SKP1 and PAM. A and B, schematics for different constructs tested for PAM (A) and FBXO45 (B). Annotated and highlighted in color are domains in PAM and FBXO45. C, coIP from transfected 293 cells showing HA–SKP1 binds GFP–FBXO45 but fails to bind GFP–PAM N-terminal (-term), GFP–PAM central, and GFP–PAM C-terminal constructs. D, coIP of HA–SKP1 with GFP–PAM D5 only occurs in the presence of FLAG–FBXO45 (upper coIP panel). HA–SKP1 coIPs with FLAG–FBXO45 in the presence or absence of GFP–PAM D5 (lower coIP panel). E, HA–SKP1 does not coIP with full-length GFP–PAM, but binding occurs in the presence of FLAG–FBXO45 (upper coIP panel). HA–SKP1 coIPs with FLAG–FBXO45 in the presence or absence of GFP–PAM (lower coIP panel). F, coIP showing HA–SKP1 binds the F-box domain of FBXO45. GFP–PAM (G) and FLAG–FBXO45 (H) fail to coIP with MYC–CUL1. I, summary showing interactions between SKP1, FBXO45, and PAM. C–H, shown are representatives of at least three independent experiments IP, immunoprecipitation.
Because SKP1 binds FBXO45 but not PAM, we tested whether FBXO45 can recruit SKP1 into a complex with PAM. To do so, we cotransfected different combinations of GFP–PAM D5, FLAG–FBXO45, and HA–SKP1, and we analyzed binding by coIP. Although GFP–PAM D5 and HA–SKP1 did not coprecipitate, we did observe binding of these two proteins when FLAG–FBXO45 was included in transfections (Fig. 4D). Similar results occurred with full-length PAM, as we only observed coprecipitation of HA–SKP1 with GFP–PAM when FLAG–FBXO45 was coexpressed (Fig. 4E).
Next, we mapped the interaction between FBXO45 and SKP1. FBXO45 is highly conserved across species (Fig. S1) and contains three domains: an F-box domain, a conserved domain (CD) of unknown function, and a SPRY domain (Fig. 4B and Fig. S1). We generated three FBXO45 constructs: the F-box domain alone, the CD domain alone, and a construct containing both CD and SPRY domains (Fig. 4B). The SPRY domain alone was not generated because it was problematic to construct. We cotransfected GFP-tagged FBXO45 domains or full-length GFP–FBXO45 with HA–SKP1 in 293 cells, and evaluated binding by coIP. We only observed an interaction between SKP1 and the F-box domain of FBXO45. This indicates the F-box domain is sufficient for FBXO45 to bind SKP1 (Fig. 4I). Previous work by Nakayama and colleagues (37) showed that the SPRY domain of FBXO45 mediates binding to PAM. Our results taken with this prior observation demonstrate that FBXO45 uses different biochemical mechanisms to bind SKP1 and PAM. Our results now provide an explanation for why FBXO45 is sufficient to recruit SKP1 into a complex with PAM (Fig. 4I). This provides new and more comprehensive insight into how human PAM forms a noncanonical SCF complex with SKP1 and FBXO45.
CUL1 is not part of the PAM/FBXO45/SKP1 complex
In traditional SCF complexes formed by the Rbx1 E3 ligase, the Cullin scaffolding protein binds directly to Rbx1 (8, 10). Skp1 then acts as an adaptor between the F-box protein and the Cullin. In contrast to traditional SCF complexes, we showed FBXO45 binds directly to PAM (Fig. 1), and SKP1 is recruited by FBXO45 into a complex with PAM (Fig. 4, C–F). These results are consistent with the lack of the Cullin CUL1 in this complex. To directly test this, we cotransfected GFP–PAM and MYC–CUL1 into 293 cells. We did not observe coIP of PAM with CUL1, but PAM did coIP with our positive control FBXO45, which was expressed at similar levels to CUL1 (Fig. 4G; Fig. S2). We then determined whether CUL1 and FBXO45 interact by coexpressing MYC–CUL1 and FLAG–FBXO45. We observed no coIP of FLAG–FBXO45 with MYC–CUL1 (Fig. 4H). CoIP of FLAG–FBXO45 with MYC–SKP1 was used as a positive control for binding (Fig. 4H). These results confirm that CUL1 fails to bind PAM or FBXO45 and does not serve as a scaffold for the atypical SCF complex formed by PAM, FBXO45, and SKP1 (Fig. 4I).
PAM polyubiquitination of NMNAT2 results in proteasome-mediated degradation
Genetic and cellular experiments in flies and rodents have shown PAM inhibits NMNAT2, but the biochemical mechanism of how this occurs remains untested. We hypothesized that PAM ubiquitinates NMNAT2, which results in degradation by the proteasome.
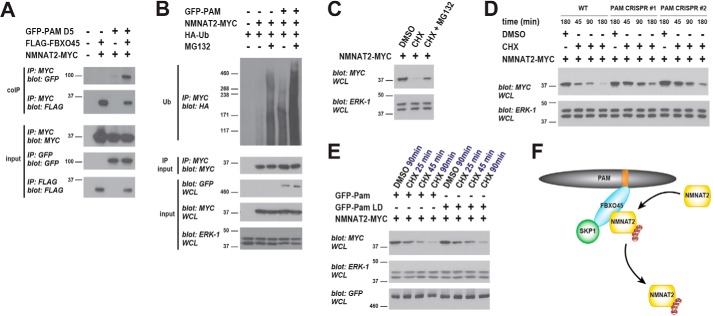
To test this hypothesis, we initially determined whether the PAM ubiquitin ligase complex binds NMNAT2. We cotransfected 293 cells with NMNAT2–MYC and GFP–PAM D5 in the presence or absence of FLAG–FBXO45. Consistent with a prior observation (28), we observed robust coIP between FLAG–FBXO45 and NMNAT2–MYC (Fig. 5A). When GFP–PAM D5 and NMNAT2–MYC were expressed together, we detected a low level of coIP, which is likely to be mediated by endogenous FBXO45 (Fig. 5A). Alternatively, PAM could interact at low levels with NMNAT2 in the absence of FBXO45. More importantly, binding of PAM D5 to NMNAT2 was much stronger when FLAG–FBXO45 was coexpressed (Fig. 5A). These results indicate FBXO45 recruits NMNAT2 into a protein complex with PAM (Fig. 5F).
Figure 5.
NMNAT2 is polyubiquitinated by PAM and degraded by the proteasome. A, coIP from transfected 293 cells showing FLAG–FBXO45 increases binding of NMNAT2–MYC to GFP–PAM D5. B, PAM strongly stimulates polyubiquitination of NMNAT2 when the proteasome is inhibited with MG132. C, NMNAT2–MYC protein turnover visualized using the translation inhibitor cycloheximide (CHX). NMNAT2 turnover is reduced by MG132 inhibition of the proteasome. D, NMNAT2 turnover is reduced in two PAM knockout cell lines generated by CRISPR. E, NMNAT2 protein turnover is faster in the presence of WT PAM than catalytically inactive PAM LD. F, summary showing PAM/FBXO45/SKP1 complex polyubiquitinating NMNAT2. A–C and E, representative is shown of at least three independent experiments. D, shown is a representative of two independent experiments, in which PAM knockout cell lines were derived de novo each time using two different PAM gRNAs. IP, immunoprecipitation.
We then evaluated whether PAM can stimulate ubiquitination of NMNAT2. 293 cells were cotransfected with NMNAT2–MYC and HA-tagged ubiquitin (HA–Ub) with or without full-length GFP–PAM. As shown in Fig. 5B, PAM coexpression triggered mild increases in polyubiquitination of NMNAT2–MYC (size >171 kDa). When the proteasome was inhibited with MG132, PAM caused further large increases in polyubiquitination of NMNAT2 (Fig. 5B). We note in this experiment that it is likely PAM utilizes endogenous FBXO45 to recruit and ubiquitinate transfected NMNAT2–MYC. These results demonstrate that PAM can stimulate polyubiquitination of NMNAT2, and ubiquitination is enhanced when the 26S proteasome is inhibited.
Because PAM polyubiquitinates NMNAT2 in a proteasome-dependent manner, we evaluated how proteasome activity and PAM affect the stability of NMNAT2. Initially, we evaluated whether NMNAT2 is degraded by the proteasome. 293 cells were transfected with NMNAT2–MYC and treated for 3 h with DMSO, the protein synthesis inhibitor cycloheximide (CHX), or a combination of CHX and MG132. As shown in Fig. 5C, NMNAT2–MYC levels are reduced following cycloheximide treatment due to NMNAT2 protein turnover in the absence of new protein synthesis. When MG132 is used to inhibit the proteasome, NMNAT2 turnover was partially blocked. Thus, NMNAT2 protein turnover occurs, in part, via the proteasome.
Because PAM is expressed in 293 cells (37), we examined how NMNAT2 protein stability is affected by endogenous PAM. To do so, we generated two independent 293 cell lines in which endogenous PAM was knocked out using CRISPR/Cas9 gene editing and nonhomologous end joining (58). To validate the efficiency of CRISPR with two different PAM single guide RNAs (sgRNAs), we cotransfected individual plasmids that express a single PAM sgRNA and Cas9, as well as a plasmid expressing GFP–PAM. Following puromycin selection for CRISPR editing, which induces deletions via nonhomologous end joining, we observed a decrease in GFP–PAM levels (Fig. S3). This did not occur when a nonspecific sgRNA was used (Fig. S3). This showed that PAM sgRNAs successfully recognize the PAM sequence, allow CRISPR cleavage of the plasmid encoding GFP–PAM, and result in reduced GFP–PAM expression.
We used these validated PAM sgRNAs to generate two independent PAM knockout cell lines. NMNAT2–MYC was transfected into PAM knockout cells, and we tested the level of NMNAT2 protein turnover when new NMNAT2 synthesis was blocked by cycloheximide. We observed that NMNAT2 turnover was reduced in both PAM knockout lines compare with WT HEK 293 in which endogenous PAM is expressed normally (Fig. 5D). This result demonstrates that endogenous PAM regulates turnover of NMNAT2.
To further test whether PAM ubiquitin ligase activity affects NMNAT2 turnover, we coexpressed NMNAT2–MYC with WT PAM or a PAM ligase-dead (LD) point mutant, which lacks catalytic RING activity. Interestingly, NMNAT2 turnover was not as rapid in the presence of PAM LD compared with WT PAM (Fig. 5E). This is consistent with PAM ubiquitin ligase activity affecting NMNAT2 protein stability.
Taken as a whole, our results demonstrate that the PAM ubiquitin ligase complex can bind and polyubiquitinate NMNAT2 (Fig. 5F). Polyubiquitination of NMNAT2 by PAM results in NMNAT2 degradation by the proteasome and NMNAT2 turnover. Finally, our results provide the first evidence that endogenous human PAM regulates NMNAT2 protein stability in human cells.
SKP1 increases FBXO45 binding to NMNAT2
Our observation that FBXO45 recruits SKP1 into a complex with PAM (Fig. 4) indicates SKP1 does not play its traditional role as an adaptor protein in the noncanonical PAM/FBXO45/SKP1 complex. As a first step in deciphering the function of SKP1 in the PAM/FBXO45/SKP1 complex, we assessed whether SKP1 binds to NMNAT2, which we showed is a ubiquitination substrate for this ubiquitin ligase complex (Fig. 5).
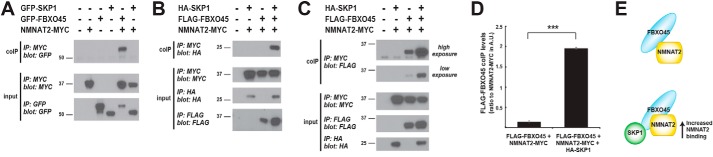
We coexpressed GFP–SKP1 with NMNAT2–MYC in 293 cells and examined binding by coIP. As shown in Fig. 6A, SKP1 did not coprecipitate with NMNAT2. However, we did observe robust coIP between GFP–FBXO45 and NMNAT2–MYC (Fig. 6A). These results demonstrate that FBXO45, but not SKP1, is sufficient to bind NMNAT2.
Figure 6.
SKP1 increases FBXO45 binding to NMNAT2. A, coIP from 293 cells shows GFP–FBXO45 binds NMNAT2–MYC, and GFP–SKP1 fails to bind NMNAT2–MYC. B, coIP showing HA–SKP1 will bind NMNAT2–MYC in the presence of FLAG–FBXO45. C, coexpression of HA–SKP1 increases binding of FLAG–FBXO45 to NMNAT2–MYC. D, quantitation, in arbitrary units (A.U.), of coIP between FLAG–FBXO45 and NMNAT2–MYC in the presence or absence of HA–SKP1. Significant increase in FLAG–FBXO45 coIP with NMNAT2–MYC occurs when HA–SKP1 is present. E, summary showing SKP1 increases NMNAT2 binding to FBXO45. B and C, shown are representatives of at least three independent experiments. D, quantitation was done with a minimum of three replicates. Significance was determined using Student's t test, and error bars represent standard error of mean. ***, p < 0.001; IP, immunoprecipitation.
There are a few possible explanations for our observation. 1) SKP1 does not play a role in target recognition. 2) SKP1 can modify the level of binding between FBXO45 and NMNAT2. 3) SKP1 is target-specific and might recognize other targets of PAM, such as DLK/MAP3K12 (59, 60). To differentiate between these possibilities, we examined protein interactions when FLAG–FBXO45, HA–SKP1, and NMNAT2–MYC were coexpressed. Interestingly, when FBXO45 was present, we detected coprecipitation of SKP1 with NMNAT2 (Fig. 6B). This demonstrates that FBXO45 can form a complex with SKP1 and NMNAT2.
Next, we tested whether SKP1 influences the interaction between FBXO45 and NMNAT2. As shown in Fig. 6C, FLAG–FBXO45 binding to NMNAT2–MYC is greatly increased when HA–SKP1 is present. Quantitation confirmed that FBXO45 binding to NMNAT2 is significantly increased severalfold when SKP1 is coexpressed compared with when it is absent (Fig. 6D).
These observations suggest that in the noncanonical PAM/FBXO45/SKP1 complex SKP1 plays a novel function as an auxiliary component of the substrate recognition module and facilitates FBXO45 binding to NMNAT2 (Fig. 6E). This contrasts with traditional SCF complexes where SKP1 functions as an adaptor between the Cullin and F-box protein. However, we also note that the level of FBXO45 was increased slightly with coexpression of SKP1 (Fig. 6C, 2nd panel from bottom). Thus, we cannot rule out that NMNAT2 binding to FBXO45 is increasing when SKP1 is coexpressed because SKP1 stabilizes FBXO45 expression.
Discussion
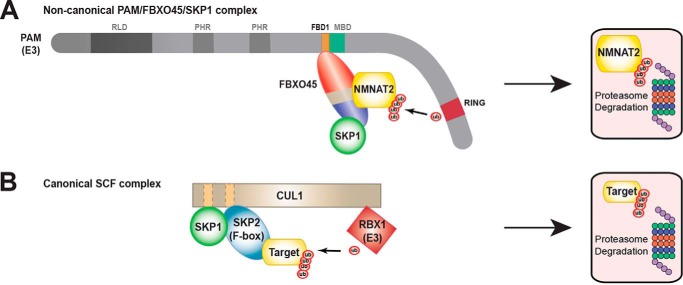
In this study, we show that the human PHR protein PAM assembles into a noncanonical SCF complex that contains FBXO45 and SKP1 but lacks CUL1 (Fig. 7). The SCF complex formed by PAM, FBXO45, and SKP1 displays further atypical characteristics, as SKP1 does not act as an adaptor protein in complex assembly but rather is an auxiliary subunit that enhances substrate binding to the F-box protein FBXO45. Furthermore, we provide the first biochemical evidence that PAM polyubiquitinates NMNAT2 thereby triggering NMNAT2 degradation by the proteasome.
Figure 7.
Comparison of noncanonical PAM/FBXO45/SKP1 complex with traditional SCF ubiquitin ligase complex. A, summary of biochemistry underpinning formation of the PAM/FBXO45/SKP1 complex, which ubiquitinates NMNAT2 and targets it for proteasomal degradation. The F-box protein FBXO45 binds directly to the FBD1 domain of PAM and recognizes NMNAT2 as a target for ubiquitination. SKP1 acts as an auxiliary component that increases FBXO45 binding to NMNAT2. B, diagram of traditional SCF complex formed by RBX1/CUL1/SKP1/SKP2 (adapted from Zheng et al. (8)).
Biochemistry underpinning assembly of the noncanonical PAM/FBXO45/SKP1 ubiquitin ligase complex
Results from previous studies suggested that SCF ubiquitin ligase complexes formed by PHR proteins, including human PAM and fly Highwire, do not require Cullin as a scaffold protein (37, 40).
Our results confirm that PAM forms an atypical SCF complex that contains the F-box protein FBXO45 and SKP1 but lacks CUL1 (Fig. 7A). We performed extensive biochemical analysis that now provides a biochemical mechanism for how this noncanonical SCF complex assembles. Our results show that the F-box protein FBXO45 binds directly to PAM, the RING E3 ubiquitin ligase (Fig. 1). This interaction is mediated by the FBD1 domain of PAM, which utilizes at least three conserved motifs, GRR, WCL, and FE, to bind FBXO45 (Fig. 3, B and C). Previous work showed these same conserved motifs mediate binding of the C. elegans PHR protein RPM-1 to the F-box protein FSN-1 (38). Thus, the FBD1 domain represents a conserved biochemical mechanism by which PHR proteins interact with F-box proteins. Interestingly, one motif, NDD, appears to limit binding to FBXO45 and could be important for releasing ubiquitination substrates (Fig. 3, B and D).
Because human PAM relies upon a conserved mechanism to bind directly to FBXO45, we explored where the interaction between orthologous proteins in C. elegans occurs in neurons in vivo. Our results with two photon FLIM–FRET indicate that RPM-1 and FSN-1 physically interact at the axon termination site of C. elegans mechanosensory neurons (Fig. 2). This observation expands upon prior work showing RPM-1 and FSN-1 form a ubiquitin ligase complex that regulates axon termination (21, 38, 39). Our findings now provide the first in vivo biochemical evidence that RPM-1/FSN-1 ubiquitin ligase activity is likely to occur locally at axon termination sites.
When taken together, previous work (37) and results from our study (Fig. 4, G and H) show convincingly that the CUL1 scaffolding protein is not present in the atypical SCF complex formed by PAM, FBXO45, and SKP1. Several of our results now provide a molecular explanation for why the CUL1 scaffold is absent. First, PAM binds directly to FBXO45 (Fig. 1), which removes the necessity of CUL1 to facilitate binding to the F-box protein. Second, FBXO45 binds SKP1 via its F-box domain and recruits SKP1 into a complex with PAM (Fig. 4, D–F). Thus, another role CUL1 normally plays in canonical SCF complexes, binding to SKP1, is facilitated by the F-box protein FBXO45. Finally, because FBD1 is located near the middle of the extremely large PAM protein, it is plausible PAM has sufficient distance between its catalytic RING domain and FBXO45 to ubiquitinate targets without a CUL1 scaffold (Fig. 7, compare A and B). Consistent with this model, PAM (4664 amino acids) is extremely large compared with CUL1 (776 amino acids), which forms a canonical SCF complex with the E3 ligase Rbx1 (108 amino acids) (8).
Interestingly, we found that SKP1 plays a novel, very different role in the noncanonical SCF complex formed by PAM compared with traditional SCF complexes containing Rbx1. In traditional SCF complexes, SKP1 functions as an adaptor protein to facilitate binding between the F-box protein and the CUL1 scaffold. In contrast, in the PAM ubiquitin ligase complex, SKP1 interacts only with the F-box protein FBXO45 and facilitates increased binding of FBXO45 to the ubiquitination substrate NMNAT2. This suggests that in the atypical PAM/FBXO45/SKP1 SCF complex, SKP1 is likely to function as an auxiliary protein in the target recognition module.
PAM/FBXO45/SKP1 complex polyubiquitinates NMNAT2, targeting it for degradation
Previous knockdown and loss of function genetic experiments in fly and rodent neurons have shown that PHR proteins, such as Drosophila Highwire and mouse Phr1, regulate axon degeneration by inhibiting Nmnat2 (27, 28, 61). These findings implied a model in which Nmnat2 ubiquitination by Phr1 results in Nmnat2 degradation.
To date, biochemistry supporting this model has remained notably absent (43). We address this gap in our knowledge via a range of biochemical findings with human PAM, FBXO45, and NMNAT2. 1) We show FBXO45 binds to NMNAT2, and this interaction is increased in the presence of SKP1 (Fig. 6). 2) FBXO45 recruits NMNAT2 into a complex with PAM (Fig. 5A). 3) NMNAT2 is turned over by proteasomal degradation (Fig. 5C). 4) PAM polyubiquitinates NMNAT2, and polyubiquitination is increased when the proteasome is inhibited (Fig. 5B). 5) CRISPR knockout of endogenous PAM decreases the turnover rate of NMNAT2 (Fig. 5D). 6) Finally, catalytically inactive PAM shows reduced ability to trigger turnover of NMNAT2 compared with WT PAM (Fig. 5E). Collectively, these results support the conclusion that PAM polyubiquitinates NMNAT2, which leads to proteasomal degradation and NMNAT2 turnover (Fig. 7A). Our findings now provide a biochemical explanation for how PHR proteins inhibit NMNAT2 during axon degeneration.
An intriguing recent study, published during revision of this manuscript, showed that PAM is an unusual RING E3 ligase that can ubiquitinate threonine and serine residues, as well as lysine (62). Although our results indicate that PAM polyubiquitination of NMNAT2 (which would occur on lysine residues) leads to proteasomal degradation, it is entirely plausible that PAM monoubiquitination of NMNAT2 on threonine or serine residues could act as a second molecular mechanism to restrain NMNAT2. If PAM indeed mediates both these types of ubiquitin modifications of NMNAT2, it is plausible PAM could regulate both local activity of NMNAT2 via monoubiquitination and NMNAT2 protein turnover via polyubiquitination. Importantly, prior work in worms looking at axon and synapse development showed that the PHR protein RPM-1 utilizes two molecular mechanisms to inhibit the DLK-1 MAP3K: recruitment of the PPM-2 phosphatase to dephosphorylate DLK-1, and polyubiquitination and degradation of DLK-1 (59, 63). It is an interesting possibility that PAM might also employ multiple molecular mechanisms, i.e. different types of ubiquitination, to regulate NMNAT2 degradation and activity during axon degeneration. If this occurs, PHR proteins could potentially exercise sophisticated temporal and spatial control over NMNAT2 in neurons following injury or during neurodegenerative disease.
Materials and methods
Molecular cloning
Constructs used in this study were cloned using PCR8 TOPO TA and Gateway recombination, unless stated otherwise. All constructs were confirmed to be free of mutations by sequencing or repaired by point mutagenesis (QuikChange II XL or QuikChange Lightning, Agilent Technologies) before use. The cytomegalovirus promoter was used for expression of all constructs. Here we list plasmids used in this study: GFP–PAM N-term (pBG-GY743), GFP–PAM Central (pBG-GY642), and GFP–PAM C-term (pBG-GY750); PAM D4 (pBG-GY643), D5 (pBG-GY644), D6 (pBG-645), and D7 (pBG-646); GFP–PAM D5a (pBG-GY664), GFP–PAM D5a N-term (pBG-GY665), GFP–PAM D5a C-term (pBG-GY666), and GFP–FBD1 (pBG-GY663); GST–PAM D5 (bacterial expression) (pBG-GY694); human GFP–PAM full-length (pBG-GY765); human HA–SKP1 (pBG-273; kind gift of Dr. Manfred Gessler (64)); GFP–SKP1 (pBG-740); MYC–SKP1 (pBG-GY871); human NMNAT2–MYC–His6 (pBG-289; kind gift of Dr. Aaron DiAntonio (28)); MYC–CUL1 (pBG-GY873; original human CUL1 sequence from Addgene (29518)).
Original sequence of mouse F-box45 was codon-optimized for expression in human cells by Genewiz (South Plainfield, NJ) and used to generate the following: FLAG–FBXO45 (pBG-GY655); GFP–FBXO45 (pBG-GY667); MYC-FBXO45 (pBG-GY870); GFP-F-box (pBG-GY914); GFP-CD (pBG-GY912); GFP-CD + SPRY (pBG-GY913); and His6–FBXO45 (bacterial expression) (pBG-GY696).
Point mutagenesis (QuikChange II XL or QuikChange Lightning, Agilent Technologies) was used to generate several constructs. PAM LD (pBG-GY849), which is mutated at the C4432A, C4434A, and C4437A residues of the RING domain and FBD1 point mutants G2404A, R2406, R2408A, F2429A, E2432A, N2435A, D2436A, D2437A, W2462A, C2463A, and L2464A.
Biochemistry
For all experiments, HEK293T cells were transfected using Lipofectamine 3000 (Invitrogen). Transfections were performed with plasmids of interest and pBluescript as needed to reach an equal amount of total DNA per transfection. Detailed descriptions of DNA amounts transfected for each experiment can be found in Table S1.
22–26 h after transfection, cells were lysed with 0.1% Nonidet P-40 buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 10% glycerol, 1 mm DTT, and 1× Pierce HALT protease inhibitor mixture or EDTA-free protease inhibitor tablet, Roche Applied Science). 0.25 to 1 mg of total protein lysate was used for coIPs. For coIP, lysates were incubated with one of the following primary antibodies for 30 min: 1.5 μl (1.6 μg) of M2 anti-FLAG (mouse monoclonal, Sigma); 1.5–3 μl (0.6 μg) of 3E6 anti-GFP (mouse monoclonal, MP Biomedicals); 1.5–3 μl (0.75 μg) of anti-HA (rabbit polyclonal, Invitrogen 715500); or 1.5 μl (4 μg) of 9E10 anti-MYC (mouse monoclonal, Sigma). Antibody complexes were immunoprecipitated by applying 10 μl of protein G-agarose (Roche Applied Science) for 4 h at 4 °C. Precipitates were boiled in Laemmli sample buffer containing β-mercaptoethanol (Bio-Rad) and run on 4–12% BisTris gels (Invitrogen). An exception to this was full-length PAM, which was run on a 3–8% NuPAGE Tris acetate gel. 20–25% of IPs were run for input blots and 40–75% of IPs were run for coIP blots. Gels were transferred to PVDF membranes in Tris acetate transfer buffer (1 h at 100 V or 16 h at 30 V for large molecular weight proteins) and immunoblotted. The following primary antibodies were diluted 1:1000 in 5% nonfat milk in TBST and applied to blots overnight at 4 °C: mixture of mouse monoclonal anti-GFP antibodies (Roche Applied Science); rabbit polyclonal anti-FLAG (Cell Signaling); mouse monoclonal 9B11 anti-MYC (Sigma); or mouse monoclonal 6E2 anti-HA antibody (Cell Signaling). Blots were visualized with HRP-conjugated anti-mouse (GE Healthcare or Veriblot, Abcam), anti-rabbit (ThermoFisher Scientific), or light chain reactive anti-mouse (Millipore) secondary antibodies. Secondaries were detected using enhanced chemiluminescent reagent (1:5 dilution of Supersignal West Femto (ThermoFisher Scientific) in TBS), and visualized with X-ray film.
Quantitation of immunoblots was done using Fiji software from NIH image (http://rsb.info.nih.gov/ij/). Quantitation was done from three or more replicates for a given condition run on the same gel. Band intensity (values in arbitrary units) of coprecipitating proteins was normalized to the intensity of bands corresponding to the target of the IP. Statistical significance was determined using a Student's t test.
Turnover of NMNAT2 by the proteasome
HEK 293T cells were transfected with 1 μg of NMNAT2. 20 h after transfection, cells were treated for 3 h with DMSO, CHX (10 μg/ml), or a mix of CHX (10 μg/ml) and MG132 (50 μm). Following drug treatment, cells were lysed with 1% Triton RIPA buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 5 mm N-ethylmaleimide, 1× phosphatase inhibitor mixture (Halt, Pierce) and 1× protease inhibitor mixture (Halt, Pierce)). For immunoblots, 30 μg of WCL were run on gels. ERK-1 was used as a nontransfected loading control. ERK-1 was detected by re-probing blots with K23 anti-ERK-1 antibody (rabbit polyclonal, Santa Cruz Biotechnology) after they were quenched with sodium azide (1 mm) in 5% nonfat milk in TBST.
Turnover of NMNAT2 by PAM
HEK 293T cells were transfected with 1 μg of NMNAT2–MYC and 7 μg of GFP–PAM WT or GFP–PAM LD. 20 h after transfection, cells were treated for the indicated length of time with DMSO or CHX (10 μg/ml). Following drug treatment, cells were lysed with 1% Triton RIPA buffer. For immunoblots, 30 μg of WCL was run on gels. ERK-1 was used as a nontransfected loading control and was used to re-probe blots quenched with sodium azide.
Establishment of PAM knockout cell lines
The two PAM sgRNA CRISPR plasmids were generated by GenScript and designed to target the PAM start site and generate deletions via nonhomologous end joining (58). 5 μg of PAM sgRNA pLentiCRISPR version 2 were transfected into HEK293T cells. 24 h after transfection, 2.5 μg/ml of puromycin was added to plates. Cells were cultured with puromycin for 6 days, and experiments were performed 3–4 days after drug selection.
Ubiquitination of NMNAT2
HEK 293T cells were transfected with 0.5 μg of HA–ubiquitin (pBG-325), 0.5–1.5 μg of NMNAT2–MYC or control MYC plasmid, and 1.5 μg of GFP–PAM or GFP control plasmid. 22–24 h after transfection, cells were treated for 1 h with DMSO or MG132 (50 μm) and lysed using 1% Triton RIPA buffer. To detect ubiquitination of NMNAT2, 0.5 mg of total protein lysate was used for anti-MYC IPs, which were run on a 3–8% Tris acetate gradient gel (Invitrogen) and then blotted for HA-ubiquitin.
Expression of GFP–PAM, NMNAT2–MYC, and HA–Ub was assessed by immunoblotting 12.5–25 μg of WCL. ERK-1 was used as a nontransfected loading control, and it was used to re-probe blots quenched with sodium azide.
Pulldown assays
Constructs were expressed in BL21-CodonPlus (DE3)-RILP cells (Agilent technology). 100 ml of LB media + antibiotic was inoculated from overnight cultures and grown to 0.6–0.8 an A600. Recombinant protein expression was induced with isopropyl 1-thio-β-d-galactopyranoside (0.05 mm) at 16 °C for 20–25 h. Bacterial lysates were prepared using two different buffers: 1) His binding buffer (300 mm NaCl, 20 mm sodium phosphate, pH 7.4, 40 mm imidazole, 10% glycerol, 1 mm DTT, and 1× protease inhibitor mixture (Pierce Halt)), or 2) GST binding buffer (50 mm Tris, pH 8.0, 150 mm NaCl, 10% glycerol, 1 mm DTT and 1× protease inhibitor mixture). Lysates were generated by sonication. GSH beads and nickel beads were washed (six times) with PBS or His-binding buffer, respectively. GST–PAM (1.5–3 mg) or GST (0.15–0.3 mg) lysates were incubated with the GSH beads for 2 h at 4 °C. Beads with bound protein were washed (three times) with binding buffer and incubated for 2 h at 4 °C with lysate expressing His6–FBXO45 (1.5 mg) or His6. Subsequently, beads were washed (six times) with binding buffer, boiled in sample buffer, and run on stain-free gel (Mini-PROTEAN® TGX Stain-FreeTM precast gels). Proteins were transferred to PVDF membranes using Tris acetate transfer buffer (1 h at 100 V). GST-tagged proteins were immunoblotted with Z-5 anti-GST polyclonal mouse antibody (Santa Cruz Biotechnology). His-tagged proteins were immunoblotted with an H8 anti-His monoclonal mouse antibody (ThermoFisher Scientific). Appropriate secondary antibodies were applied and detected using Supersignal West Pico ECL (ThermoFisher Scientific) and visualized with X-ray film.
C. elegans strains and confocal imaging
Integrated transgene bggIs34 (Pmec-3::RPM-1::GFP + Pmyo-2::mCherry) was shown previously in rescue experiments to be fully functional (21). bggEx131 and bggEx132 transgenic extrachromosomal arrays were generated by injecting Pmec-7::mCherry::FSN-1 (100 ng/μl) into bggIs34. To generate bggEx133 and bggEx134, Pmec-7::mCherry (25–100 ng/μl) was injected into bggIs34. For extrachromosomal arrays, Pttx-3::RFP (50 ng/μl) was used as a coinjection marker. All transgenic lines and arrays were generated on a glo-1 (zu391) background, which removes gut autofluorescence and is optimal for 2pFLIM.
For confocal microscopy, young adults were immobilized on 2% agarose pads in levamisole (5 μm). Animals were imaged on a Leica SP8 confocal microscope under ×25 objective and ×4 scan zoom factor. Leica Application Suite (LAS) software defined Z-stacks (0.8–1.0-μm intervals), which are shown as maximum intensity projections.
Two-photon FLIM
The interaction between RPM-1::GFP and mCherry::FSN-1 at axon termination sites of PLM mechanosensory neurons was measured using 2pFLIM. Briefly, mEGFP and mCherry were excited with a Ti:sapphire laser (Chameleon, Coherent) at a wavelength of 920 nm and a power of 1.4–1.7 milliwatts. Fluorescence lifetime images were obtained using a time-correlated single photon counting board (SPC-150; Becker and Hickl) controlled with custom software in Matlab (55). Fluorescence lifetime images were collected by 128 × 128 pixels acquired at 2 ms/line and averaged over 24 frames. The fluorescence lifetime of mEGFP was measured by curve fitting as described previously (65).
Unpaired Student's t tests (two-tailed, 95% confidence) compared RPM-1::mEGFP alone with RPM-1::mEGFP + mCherry::FSN-1 or with RPM-1::mEGFP + mCherry expressed in the mechanosensory neurons. p values (****, p ≤ 0.0001; ns, not significant). Number of independent measurements (n) was nine individual worms per genotype from two independent experiments.
Author contributions
M. D., O. C., S. T. B., R. Y., and B. G. conceptualization; M. D., O. C., P. R. E., and S. T. B. data curation; M. D., P. R. E., and S. T. B. formal analysis; M. D., P. R. E., I. M., and B. G. methodology; M. D., O. C., P. R. E., R. Y., and B. G. writing-original draft; M. D., O. C., P. R. E., R. Y., and B. G. writing-review and editing; R. Y. and B. G. supervision; B. G. funding acquisition; B. G. project administration.
Supplementary Material
Acknowledgments
We thank Dr. Manfred Gessler for HA–SKP1 plasmid, Dr. Aaron DiAntonio for NMNAT2–MYC-HIS6 plasmid, and Dr. Ning Zheng for CUL1 plasmid (Addgene 29518). We thank Drs. Samuel Young, Jr., and Kirill Martemyanov for helpful discussions. We also thank Dr. Dipak Patil for technical discussions.
This work was supported by National Institutes of Health Grant R01 NS072129 (to B. G.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S3 and Table S1.
- SCF
- Skp/Cullin/F-box
- IP
- immunoprecipitation
- coIP
- coimmunoprecipitation
- GST
- glutathione S-transferase
- PHR
- PAM/Highwire/RPM-1
- WCL
- whole-cell lysate
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- PVDF
- polyvinylidene difluoride
- CHX
- cycloheximide
- EGFP
- enhanced green fluorescent protein
- CD
- conserved domain
- sgRNA
- single guide RNA
- LD
- ligase-dead
- GT
- glutathione.
References
- 1. Nakayama K. I., and Nakayama K. (2006) Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer 6, 369–381 10.1038/nrc1881 [DOI] [PubMed] [Google Scholar]
- 2. Chang L., and Barford D. (2014) Insights into the anaphase-promoting complex: a molecular machine that regulates mitosis. Curr. Opin. Struct. Biol. 29, 1–9 10.1016/j.sbi.2014.08.003 [DOI] [PubMed] [Google Scholar]
- 3. Kirkin V., and Dikic I. (2011) Ubiquitin networks in cancer. Curr. Opin. Genet. Dev. 21, 21–28 10.1016/j.gde.2010.10.004 [DOI] [PubMed] [Google Scholar]
- 4. Davis R. J., Welcker M., and Clurman B. E. (2014) Tumor suppression by the Fbw7 ubiquitin ligase: mechanisms and opportunities. Cancer Cell 26, 455–464 10.1016/j.ccell.2014.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. DiAntonio A., and Hicke L. (2004) Ubiquitin-dependent regulation of the synapse. Annu. Rev. Neurosci. 27, 223–246 10.1146/annurev.neuro.27.070203.144317 [DOI] [PubMed] [Google Scholar]
- 6. Goo M. S., Scudder S. L., and Patrick G. N. (2015) Ubiquitin-dependent trafficking and turnover of ionotropic glutamate receptors. Front. Mol. Neurosci. 8, 60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tai H. C., and Schuman E. M. (2008) Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat. Rev. Neurosci. 9, 826–838 10.1038/nrn2499 [DOI] [PubMed] [Google Scholar]
- 8. Zheng N., Schulman B. A., Song L., Miller J. J., Jeffrey P. D., Wang P., Chu C., Koepp D. M., Elledge S. J., Pagano M., Conaway R. C., Conaway J. W., Harper J. W., and Pavletich N. P. (2002) Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416, 703–709 10.1038/416703a [DOI] [PubMed] [Google Scholar]
- 9. Reitsma J. M., Liu X., Reichermeier K. M., Moradian A., Sweredoski M. J., Hess S., and Deshaies R. J. (2017) Composition and regulation of the cellular repertoire of SCF ubiquitin ligases. Cell 171, 1326–1339.e14 10.1016/j.cell.2017.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deshaies R. J., and Joazeiro C. A. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 10.1146/annurev.biochem.78.101807.093809 [DOI] [PubMed] [Google Scholar]
- 11. Latres E., Chiaur D. S., and Pagano M. (1999) The human F box protein β-Trcp associates with the Cul1/Skp1 complex and regulates the stability of β-catenin. Oncogene 18, 849–854 10.1038/sj.onc.1202653 [DOI] [PubMed] [Google Scholar]
- 12. Wu G., Xu G., Schulman B. A., Jeffrey P. D., Harper J. W., and Pavletich N. P. (2003) Structure of a β-TrCP1-Skp1-β-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol. Cell 11, 1445–1456 10.1016/S1097-2765(03)00234-X [DOI] [PubMed] [Google Scholar]
- 13. Nash P., Tang X., Orlicky S., Chen Q., Gertler F. B., Mendenhall M. D., Sicheri F., Pawson T., and Tyers M. (2001) Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414, 514–521 10.1038/35107009 [DOI] [PubMed] [Google Scholar]
- 14. Petroski M. D., and Deshaies R. J. (2005) Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell 123, 1107–1120 10.1016/j.cell.2005.09.033 [DOI] [PubMed] [Google Scholar]
- 15. Welcker M., Orian A., Jin J., Grim J. A., Harper J. W., Eisenman R. N., and Clurman B. E. (2004) The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. U.S.A. 101, 9085–9090 10.1073/pnas.0402770101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Davis M. A., Larimore E. A., Fissel B. M., Swanger J., Taatjes D. J., and Clurman B. E. (2013) The SCF-Fbw7 ubiquitin ligase degrades MED13 and MED13L and regulates CDK8 module association with mediator. Genes Dev. 27, 151–156 10.1101/gad.207720.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grill B., Murphey R. K., and Borgen M. A. (2016) The PHR proteins: intracellular signaling hubs in neuronal development and axon degeneration. Neural Dev. 11, 8 10.1186/s13064-016-0063-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhen M., Huang X., Bamber B., and Jin Y. (2000) Regulation of presynaptic terminal organization by C. elegans RPM-1, a putative guanine nucleotide exchanger with a RING-H2 finger domain. Neuron 26, 331–343 10.1016/S0896-6273(00)81167-8 [DOI] [PubMed] [Google Scholar]
- 19. Schaefer A. M., Hadwiger G. D., and Nonet M. L. (2000) rpm-1, a conserved neuronal gene that regulates targeting and synaptogenesis in C. elegans. Neuron 26, 345–356 10.1016/S0896-6273(00)81168-X [DOI] [PubMed] [Google Scholar]
- 20. Wan H. I., DiAntonio A., Fetter R. D., Bergstrom K., Strauss R., and Goodman C. S. (2000) Highwire regulates synaptic growth in Drosophila. Neuron 26, 313–329 10.1016/S0896-6273(00)81166-6 [DOI] [PubMed] [Google Scholar]
- 21. Opperman K. J., and Grill B. (2014) RPM-1 is localized to distinct subcellular compartments and regulates axon length in GABAergic motor neurons. Neural Dev. 9, 10 10.1186/1749-8104-9-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. D'Souza J., Hendricks M., Le Guyader S., Subburaju S., Grunewald B., Scholich K., and Jesuthasan S. (2005) Formation of the retinotectal projection requires Esrom, an ortholog of PAM (protein associated with Myc). Development 132, 247–256 10.1242/dev.01578 [DOI] [PubMed] [Google Scholar]
- 23. Lewcock J. W., Genoud N., Lettieri K., and Pfaff S. L. (2007) The ubiquitin ligase Phr1 regulates axon outgrowth through modulation of microtubule dynamics. Neuron 56, 604–620 10.1016/j.neuron.2007.09.009 [DOI] [PubMed] [Google Scholar]
- 24. Bloom A. J., Miller B. R., Sanes J. R., and DiAntonio A. (2007) The requirement for Phr1 in CNS axon tract formation reveals the corticostriatal boundary as a choice point for cortical axons. Genes Dev. 21, 2593–2606 10.1101/gad.1592107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Borgen M., Rowland K., Boerner J., Lloyd B., Khan A., and Murphey R. (2017) Axon termination, pruning, and synaptogenesis in the giant fiber system of Drosophila melanogaster is promoted by highwire. Genetics 205, 1229–1245 10.1534/genetics.116.197343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Borgen M. A., Wang D., and Grill B. (2017) RPM-1 regulates axon termination by affecting growth cone collapse and microtubule stability. Development 144, 4658–4672 10.1242/dev.154187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xiong X., Hao Y., Sun K., Li J., Li X., Mishra B., Soppina P., Wu C., Hume R. I., and Collins C. A. (2012) The Highwire ubiquitin ligase promotes axonal degeneration by tuning levels of Nmnat protein. PLoS Biol. 10, e1001440 10.1371/journal.pbio.1001440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Babetto E., Beirowski B., Russler E. V., Milbrandt J., and DiAntonio A. (2013) The Phr1 ubiquitin ligase promotes injury-induced axon self-destruction. Cell Rep. 3, 1422–1429 10.1016/j.celrep.2013.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gao M. X., Liao E. H., Yu B., Wang Y., Zhen M., and Derry W. B. (2008) The SCF FSN-1 ubiquitin ligase controls germline apoptosis through CEP-1/p53 in C. elegans. Cell Death Differ. 15, 1054–1062 10.1038/cdd.2008.30 [DOI] [PubMed] [Google Scholar]
- 30. Xu M., Zhu C., Zhao X., Chen C., Zhang H., Yuan H., Deng R., Dou J., Wang Y., Huang J., Chen Q., Jiang B., and Yu J. (2015) Atypical ubiquitin E3 ligase complex Skp1-Pam-Fbxo45 controls the core epithelial-to-mesenchymal transition-inducing transcription factors. Oncotarget 6, 979–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Scaravilli M., Porkka K. P., Brofeldt A., Annala M., Tammela T. L., Jenster G. W., Nykter M., and Visakorpi T. (2015) MiR-1247–5p is overexpressed in castration resistant prostate cancer and targets MYCBP2. Prostate 75, 798–805 10.1002/pros.22961 [DOI] [PubMed] [Google Scholar]
- 32. Chen X., Sahasrabuddhe A. A., Szankasi P., Chung F., Basrur V., Rangnekar V. M., Pagano M., Lim M. S., and Elenitoba-Johnson K. S. (2014) Fbxo45-mediated degradation of the tumor-suppressor Par-4 regulates cancer cell survival. Cell Death Differ. 21, 1535–1545 10.1038/cdd.2014.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hebbar N., Burikhanov R., Shukla N., Qiu S., Zhao Y., Elenitoba-Johnson K. S. J., and Rangnekar V. M. (2017) A naturally generated decoy of the prostate apoptosis response-4 protein overcomes therapy resistance in tumors. Cancer Res. 77, 4039–4050 10.1158/0008-5472.CAN-16-1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tricoli J. V., Boardman L. A., Patidar R., Sindiri S., Jang J. S., Walsh W. D., McGregor P. M. 3rd., Camalier C. E., Mehaffey M. G., Furman W. L., Bahrami A., Williams P. M., Lih C. J., Conley B. A., and Khan J. (2018) A mutational comparison of adult and adolescent and young adult (AYA) colon cancer. Cancer 124, 1070–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liao E. H., Hung W., Abrams B., and Zhen M. (2004) An SCF-like ubiquitin ligase complex that controls presynaptic differentiation. Nature 430, 345–350 10.1038/nature02647 [DOI] [PubMed] [Google Scholar]
- 36. Wu C., Daniels R. W., and DiAntonio A. (2007) DFsn collaborates with highwire to down-regulate the Wallenda/DLK kinase and restrain synaptic terminal growth. Neural Dev. 2, 16 10.1186/1749-8104-2-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Saiga T., Fukuda T., Matsumoto M., Tada H., Okano H. J., Okano H., and Nakayama K. I. (2009) Fbxo45 forms a novel ubiquitin ligase complex and is required for neuronal development. Mol. Cell. Biol. 29, 3529–3543 10.1128/MCB.00364-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sharma J., Baker S. T., Turgeon S. M., Gurney A. M., Opperman K. J., and Grill B. (2014) Identification of a peptide inhibitor of the RPM-1.FSN-1 ubiquitin ligase complex. J. Biol. Chem. 289, 34654–34666 10.1074/jbc.M114.614065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baker S. T., and Grill B. (2017) Defining minimal binding regions in regulator of presynaptic morphology 1 (RPM-1) using Caenorhabditis elegans neurons reveals differential signaling complexes. J. Biol. Chem. 292, 2519–2530 10.1074/jbc.M116.748004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brace E. J., Wu C., Valakh V., and DiAntonio A. (2014) SkpA restrains synaptic terminal growth during development and promotes axonal degeneration following injury. J. Neurosci. 34, 8398–8410 10.1523/JNEUROSCI.4715-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Coleman M. P., and Freeman M. R. (2010) Wallerian degeneration, wld(s), and nmnat. Annu. Rev. Neurosci. 33, 245–267 10.1146/annurev-neuro-060909-153248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Conforti L., Gilley J., and Coleman M. P. (2014) Wallerian degeneration: an emerging axon death pathway linking injury and disease. Nat. Rev. Neurosci. 15, 394–409 10.1038/nrn3680 [DOI] [PubMed] [Google Scholar]
- 43. Brazill J. M., Li C., Zhu Y., and Zhai R. G. (2017) NMNAT: it's an NAD(+) synthase. It's a chaperone. It's a neuroprotector. Curr. Opin. Genet. Dev. 44, 156–162 10.1016/j.gde.2017.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mack T. G., Reiner M., Beirowski B., Mi W., Emanuelli M., Wagner D., Thomson D., Gillingwater T., Court F., Conforti L., Fernando F. S., Tarlton A., Andressen C., Addicks K., Magni G., et al. (2001) Wallerian degeneration of injured axons and synapses is delayed by a Ube4b/Nmnat chimeric gene. Nat. Neurosci. 4, 1199–1206 10.1038/nn770 [DOI] [PubMed] [Google Scholar]
- 45. Avery M. A., Sheehan A. E., Kerr K. S., Wang J., and Freeman M. R. (2009) WldS requires Nmnat1 enzymatic activity and N16-VCP interactions to suppress Wallerian degeneration. J. Cell Biol. 184, 501–513 10.1083/jcb.200808042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gilley J., and Coleman M. P. (2010) Endogenous Nmnat2 is an essential survival factor for maintenance of healthy axons. PLoS Biol. 8, e1000300 10.1371/journal.pbio.1000300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hicks A. N., Lorenzetti D., Gilley J., Lu B., Andersson K. E., Miligan C., Overbeek P. A., Oppenheim R., and Bishop C. E. (2012) Nicotinamide mononucleotide adenylyltransferase 2 (Nmnat2) regulates axon integrity in the mouse embryo. PLoS ONE 7, e47869 10.1371/journal.pone.0047869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kitay B. M., McCormack R., Wang Y., Tsoulfas P., and Zhai R. G. (2013) Mislocalization of neuronal mitochondria reveals regulation of Wallerian degeneration and NMNAT/WLD(S)-mediated axon protection independent of axonal mitochondria. Hum. Mol. Genet. 22, 1601–1614 10.1093/hmg/ddt009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Neukomm L. J., Burdett T. C., Seeds A. M., Hampel S., Coutinho-Budd J. C., Farley J. E., Wong J., Karadeniz Y. B., Osterloh J. M., Sheehan A. E., and Freeman M. R. (2017) Axon death pathways converge on axundead to promote functional and structural axon disassembly. Neuron 95, 78–91.e5 10.1016/j.neuron.2017.06.031 [DOI] [PubMed] [Google Scholar]
- 50. Ljungberg M. C., Ali Y. O., Zhu J., Wu C. S., Oka K., Zhai R. G., and Lu H. C. (2012) CREB-activity and nmnat2 transcription are down-regulated prior to neurodegeneration, while NMNAT2 over-expression is neuroprotective, in a mouse model of human tauopathy. Hum. Mol. Genet. 21, 251–267 10.1093/hmg/ddr492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ruan K., Zhu Y., Li C., Brazill J. M., and Zhai R. G. (2015) Alternative splicing of Drosophila Nmnat functions as a switch to enhance neuroprotection under stress. Nat. Commun. 6, 10057 10.1038/ncomms10057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Matarin M., Salih D. A., Yasvoina M., Cummings D. M., Guelfi S., Liu W., Nahaboo Solim M. A., Moens T. G., Paublete R. M., Ali S. S., Perona M., Desai R., Smith K. J., Latcham J., Fulleylove M., et al. (2015) A genome-wide gene-expression analysis and database in transgenic mice during development of amyloid or τ pathology. Cell Rep. 10, 633–644 10.1016/j.celrep.2014.12.041 [DOI] [PubMed] [Google Scholar]
- 53. Ali Y. O., Allen H. M., Yu L., Li-Kroeger D., Bakhshizadehmahmoudi D., Hatcher A., McCabe C., Xu J., Bjorklund N., Taglialatela G., Bennett D. A., De Jager P. L., Shulman J. M., Bellen H. J., and Lu H. C. (2016) NMNAT2:HSP90 complex mediates proteostasis in proteinopathies. PLoS Biol. 14, e1002472 10.1371/journal.pbio.1002472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Grill B., Bienvenut W. V., Brown H. M., Ackley B. D., Quadroni M., and Jin Y. (2007) C. elegans RPM-1 regulates axon termination and synaptogenesis through the Rab GEF GLO-4 and the Rab GTPase GLO-1. Neuron 55, 587–601 10.1016/j.neuron.2007.07.009 [DOI] [PubMed] [Google Scholar]
- 55. Yasuda R. (2006) Imaging spatiotemporal dynamics of neuronal signaling using fluorescence resonance energy transfer and fluorescence lifetime imaging microscopy. Curr. Opin. Neurobiol. 16, 551–561 10.1016/j.conb.2006.08.012 [DOI] [PubMed] [Google Scholar]
- 56. Yasuda R. (2012) Studying signal transduction in single dendritic spines. Cold Spring Harb. Perspect. Biol. 4, a005611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chung F. Z., Sahasrabuddhe A. A., Ma K., Chen X., Basrur V., Lim M. S., and Elenitoba-Johnson K. S. (2014) Fbxo45 inhibits calcium-sensitive proteolysis of N-cadherin and promotes neuronal differentiation. J. Biol. Chem. 289, 28448–28459 10.1074/jbc.M114.561241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., and Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nakata K., Abrams B., Grill B., Goncharov A., Huang X., Chisholm A. D., and Jin Y. (2005) Regulation of a DLK-1 and p38 MAP kinase pathway by the ubiquitin ligase RPM-1 is required for presynaptic development. Cell 120, 407–420 10.1016/j.cell.2004.12.017 [DOI] [PubMed] [Google Scholar]
- 60. Huntwork-Rodriguez S., Wang B., Watkins T., Ghosh A. S., Pozniak C. D., Bustos D., Newton K., Kirkpatrick D. S., and Lewcock J. W. (2013) JNK-mediated phosphorylation of DLK suppresses its ubiquitination to promote neuronal apoptosis. J. Cell Biol. 202, 747–763 10.1083/jcb.201303066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yamagishi Y., and Tessier-Lavigne M. (2016) An atypical SCF-like ubiquitin ligase complex promotes Wallerian degeneration through regulation of axonal Nmnat2. Cell Rep. 17, 774–782 10.1016/j.celrep.2016.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pao K. C., Wood N. T., Knebel A., Rafie K., Stanley M., Mabbitt P. D., Sundaramoorthy R., Hofmann K., van Aalten D. M. F., and Virdee S. (2018) Activity-based E3 ligase profiling uncovers an E3 ligase with esterification activity. Nature 556, 381–385 10.1038/s41586-018-0026-1 [DOI] [PubMed] [Google Scholar]
- 63. Baker S. T., Opperman K. J., Tulgren E. D., Turgeon S. M., Bienvenut W., and Grill B. (2014) RPM-1 uses both ubiquitin ligase and phosphatase-based mechanisms to regulate DLK-1 during neuronal development. PLoS Genet. 10, e1004297 10.1371/journal.pgen.1004297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Salat D., Winkler A., Urlaub H., and Gessler M. (2015) Hey bHLH proteins interact with a FBXO45 containing SCF ubiquitin ligase complex and induce its translocation into the nucleus. PLoS ONE 10, e0130288 10.1371/journal.pone.0130288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chang J. Y., Parra-Bueno P., Laviv T., Szatmari E. M., Lee S. R., and Yasuda R. (2017) CaMKII autophosphorylation is necessary for optimal integration of Ca2+ signals during LTP induction, but not maintenance. Neuron 94, 800–808.e4 10.1016/j.neuron.2017.04.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Robert X., and Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.