Abstract
Biological membranes are vital, active contributors to cell function. In addition to specific interactions of individual lipid molecules and lateral organization produced by membrane domains, the bulk physicochemical properties of biological membranes broadly regulate protein structure and function. Therefore, these properties must be homeostatically maintained within a narrow range that is compatible with cellular physiology. Although such adaptiveness has been known for decades, recent observations have dramatically expanded its scope by showing the breadth of membrane properties that must be maintained, and revealing the remarkable diversity of biological membranes, both within and between cell types. Cells have developed a broad palette of sense-and-respond machineries to mediate physicochemical membrane homeostasis, and the molecular mechanisms of these are being discovered through combinations of cell biology, biophysical approaches, and computational modeling.
Introduction: Cell membranes are active, responsive biomaterials
Membranes are central to cellular architecture and deeply integrated into their physiology. Much more than simple passive solvents for protein-mediated activity, membranes contribute to functionality at all relevant scales: individual membrane lipids act as substrates for enzymes and signaling molecules [1], lipid assemblies regulate protein recruitment and interactions [2], and bulk membrane properties determine protein structure and function [3].
Despite their central role in cell function, our understanding of membrane physiology continues to lag behind most other aspects of cell biology. Part of the reason is that lipids are not amenable to the powerful tools developed for dissection of genes and proteins. Further, lipids are rarely lone wolves - membranes are inherently collectives, comprised of tens of thousands of individual molecules representing hundreds of different species [4]. Finally, membranes are highly plastic and adaptive, able to maintain their bioactivity in spite of external perturbations. In this brief review, we highlight recent advances in defining molecular machineries responsible for sensing membrane properties and mediating homeostatic responses.
The many faces of membrane homeostasis
The classical understanding of membrane homeostasis was informed by a series of studies in the 1970s, which revealed that cells adapt their membrane compositions to changes in ambient temperature to maintain membrane physical properties [5,6]. For example, E. coli shifted to cooler growth temperatures substitute loosely-packing unsaturated lipids for tightly-packing saturated ones, in order to maintain membrane viscosity [5]. Such “homeoviscous adaptation” to temperature variations has since been observed broadly, from prokaryotes to ectothermic animals [7,8] (Figure 1A). For a small subset of these responses, the molecular mechanisms have been elucidated. For example, the DesK/DesR system from B. subtilis is proposed to sense membrane thickness as a proxy for membrane viscosity, and feed-back onto lipid desaturation as a response to perturbations [9,10].
Figure 1. Physicochemical membrane homeostasis.
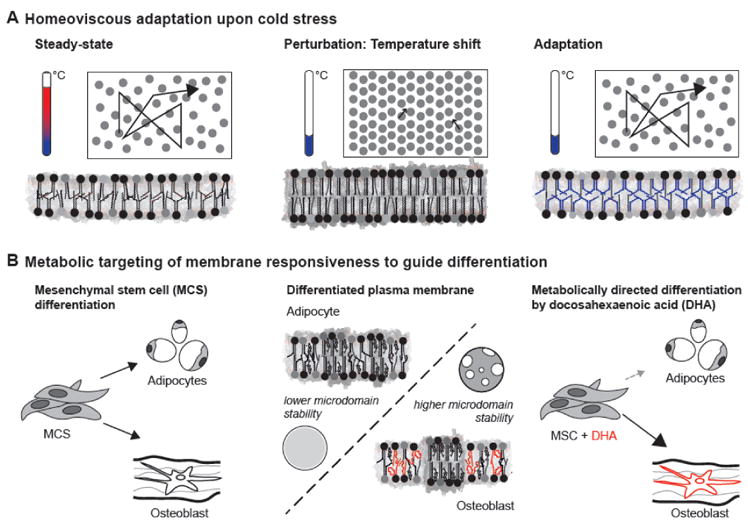
(A) Classical experiments by Michael Sinensky [5] established that when the membrane of cells (left) becomes less fluid upon a drop of temperature (middle), cells adjust their lipid composition to reestablish membrane fluidity (right). (B) Physicochemical membrane properties can direct differentiation processes [22]. Mesenchymal stem cells can differentiate into fat cells or osteoblasts (left). The PMs of these cells show lineage-specific lipidomic remodeling affecting membrane phase behavior (middle), with osteoblast PMs characterized by a higher polyunsaturated lipid content and higher tendency to phase segregate. Mimicking the compositional and biophysical remodeling of the PM by supplementation with polyunsaturated fatty acid (DHA) directs osteoblast differentiation (right).
The scope of membrane homeostasis has been dramatically expanded by findings over the last two decades (Figure 1B). For example, in contrast to prokaryotes, eukaryotic cells have an abundance of intracellular membranes, each with distinct lipid compositions and physical properties [11,12]. The best characterized variations occur in the secretory pathway, where the endoplasmic reticulum (ER), Golgi apparatus, and plasma membrane (PM) present a gradient of increasing cholesterol and sphingolipid concentrations, which lead to increasing membrane stiffness and thickness. These physical properties are believed to underlie the distinct functionalities of organelles and may be harnessed for the sorting of membrane proteins and lipids between them [13–16].
Subcellular organelles have characteristic lipid compositions and membrane properties that are reflective of their physiological purpose, and therefore these must be homeostatically maintained. The most widely characterized example is in the ER, which is equipped with sensory machineries to maintain its membrane fluidity [17,18], and to balance production of proteins and lipids for membrane biogenesis [19,20]. These examples also highlight an emerging theme: the physicochemical properties of biological membranes are not determined solely by lipids, but are also reflective of membrane protein abundance and composition. This is most clearly demonstrated by observations that lipids isolated from biological membranes sometimes form non-lamellar phases, whereas they form bilayers in their native, protein-rich context [8]. However, the lipidome of cellular organelles can be remodeled much faster than the proteome, and it is therefore unsurprising that lipid compositions are responsive to perturbations such as cellular differentiation [21,22], metabolic challenge [23], and acute stress [24]. For example, it has been shown that yeast membrane compositions are highly plastic, with lipidome-wide changes induced by variations in common growth conditions [23]. Lipidomic flexibility has also been demonstrated in more complex organisms. For example, synaptic membranes isolated from mammalian neurons undergo dramatic compositional remodeling during early post-natal development [21]. Despite these lipid changes, the fluidity and packing of the neural membranes remain unchanged, suggesting that physical homeostasis is enforced on top of the compositional changes [21]. This inference is supported by observations of mammalian cells which are supplemeneted in culture with polyunsaturated fatty acids: these exogenous fatty acids incorporate into membrane lipids and produce significant changes to the lipidome without affecting membrane lipid packing in the membrane [25]. This flexibility of the lipidome may give the impression that membrane properties are not vitally important for physiology – after all, conservation is the hallmark of evolutionary value. However, this would be missing the forest for the trees: we propose that rather than conserving membrane compositions, cells seek to maintain membrane properties by tuning the multifactorial lipidome.
The problem of physicochemical membrane homeostasis in multicellular organisms is compounded by the variety of physiological demands imposed by distinct cell types and the presence of communicating organelles therein. In many cases, these demands require specific membrane compositions and physical properties. An extreme example are the photoreceptor membranes of the mammalian visual systems [26], and more generally neuronal PMs [27], which maintain remarkably high levels of omega-3 polyunsaturated fatty acids (up to 50%), which are present at minimal levels in most non-neuronal cell types. However, these extreme examples should not overshadow subtler though still physiologically important variations. For example, it was recently shown that in vitro differentiation of human mesenchymal stem cells results in robust remodeling of both PM and whole cell lipidomes, with two different differentiated cell types (adipocytes and osteoblasts) acquiring membrane lipidomes and biophysical properties distinct from both the precursor stem cells and each other [22]. Most importantly, this compositional and biophysical remodeling appears to be akin to a differentiation checkpoint, as inducing an osteoblast-like membrane phenotype promoted osteoblast differentiation [22].
Sense-and-response systems for membrane homeostasis
Membrane adaptiveness requires mechanisms to sense and control bulk membrane features. Given the complexity of cellular lipidomes, dedicated sensory machineries for each possible lipid species seem unlikely. These may not constitute efficient mechanisms for membrane homeostasis, as the above examples suggest that collective membrane properties, rather than exact lipid compositions, are required for functional membranes. Examples of membrane properties potentially under homeostatic regulation include viscosity [5], surface charge density [28], lipid packing in the headgroup region [29,30] and the membrane core [17], phase behavior and domain formation [31], the lateral pressure profile [32], thickness or compressibility [20,33], stiffness / bending rigidity [34], and intrinsic curvature stress [30] (Table 1). A complicating factor is that most, if not all, of these properties are interdependent - membranes composed of tightly packing lipids tend to be more ordered, more viscous, thicker, and have fewer hydrophobic defects.
Table 1.
Bulk membrane properties, their determinants, and their (potential) sensors
| Bulk membrane property | Major determinant | Potential sensing mechanisms / Exemplary sensor | Sensor class |
|---|---|---|---|
| Surface charge density | Concentration of charged lipids (e.g. PA, PS, and phosphoinositides) | Sensing of electrostatics by amphipathic helix with basic residues / CCT [38], Pah1 [39] | Class I |
| Lateral pressure at membrane surface related to lipid packing | Lipid headgroups and acyl chains, Lipid shape | Gradual folding of an AH in packing defects / CCT [38], Squalene monooxygenase [55] | Class I |
| Lateral pressure in membrane core related to lipid packing | Lipid headgroups and acyl chains, Sterol content, Lipid shape | Conformational changes (e.g. helix:helix rotations) sensitive to the lateral pressure profile / Mga2 [17,18] | Class II |
| Lipid packing / Membrane order | Lipid saturation, Sterol content | Mga2 [17,18], CCT [38], Squalene monooxygenase [55] | Class I/II |
| Membrane thickness | Hydrophobic thickness of proteins, Length of lipids, Sterol content | Hydrophilic residues drive conformational changes when thickness increases / DesK [9,10] | Class III |
| Membrane compressibility | Lipid headgroup interactions, Lipid packing, Sterol content | Local membrane compression stabilizes oligomeric state when compressibility is reduced / Ire1 [29] | Class III |
| Membrane fluidity/viscosity | Lipid packing | Sensing the frequency of collisions of membrane constituents | Class I/II/III |
| Intrinsic curvature (of each leaflet) | Lipid shape | Sensing at surfaces and across the bilayer is required | Class I/III |
| Membrane phase behavior | in TGN/PM: Sphingolipid and sterol content, Lipid saturation | Challenging the line tension between coexisting lipid nanodomains A large protein might sample changes in its local environment |
Class II/III |
| Stiffness / bending rigidity | Lipid headgroups and acyl chains, Lipid shape, Electrostatics | Motor proteins could challenge membrane rigidity upon membrane bending and tubulation | Class I/III |
A vast number of proteins and cellular processes are dependent on bulk membrane properties. However, most of them do not control homeostatic responses. We consider a membrane-sensitive protein as a sensor only if it is linked to an effector module capable of mounting a homeostatic response [35,36]. According to this stringent definition, the molecular mechanisms of only a few membrane property sensors have so far been elucidated [37]. Based on these few examples, it appears that membrane homeostatic mechanisms in the early secretory pathway (i.e. ER) often control the activity of lipid biosynthetic enzymes per se or via transcriptional programs that rewire lipid and protein metabolic networks [18–20,38,39]. In contrast, sense-and-respond systems in the late secretory pathway (e.g. PM) are more likely to control lipid remodeling processes and fast, non-vesicular lipid transport at membrane contact sites [40–43]. We propose that membrane property sensors can be categorized into three distinct classes, based on their fundamental molecular mechanisms. Class I senses at membrane surfaces, class II senses properties within the membrane, class III senses membrane properties across the lipid bilayer (Figure 2) (Table 1).
Figure 2. Fundamental mechanisms of membrane property sensors.
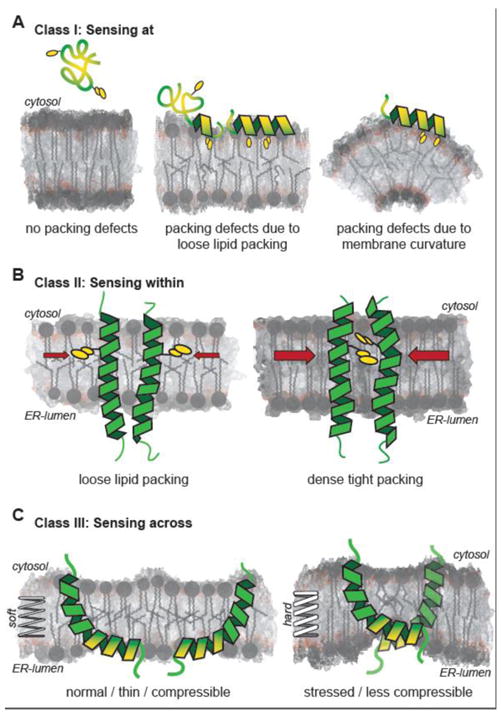
(A) Sensing at membrane surfaces: Proteins can sense the properties of the membrane surface via lipid packing defects that promote folding and inserting amphipathic helices into the interfacial region of a membrane. Lipid packing defects are indicated as voids between schematically illustrated lipid molecules and increased packing densities are denoted by darker shading of the bilayer. Oxygen atoms are shaded red. Exemplary hydrophobic residues inserting into lipid packing defects are indicated. (B) Sensing within the membrane core. A lipid packing sensor from yeast senses packing of the membrane core to control membrane fluidity [17]. The mechanism involves a selection of distinct rotational conformations driven by membrane environment. (C) Sensing across the lipid bilayer. Membrane compressibility, an important property affecting membrane protein sorting along the secretory pathway, can be sensed only across the lipid bilayer. The transducer or the unfolded protein response, Ire1, senses membrane rigidity and thickness by locally deforming the bilayer, leading to membrane-mediated oligomerization in a stressed ER membrane [20].
Class I - Sensing at membrane surfaces
The identity of organellar membranes is established by the presence of small G-proteins (e.g. Rabs) and rare signaling lipids (e.g. phosphoinositides), in combination with bulk membrane properties. For example, the early secretory pathway is characterized by abundant hydrophobic ‘voids’ in the water membrane interface that result from poor lipid packing. In contrast, the late secretory pathway is defined by high surface charge [28,44,45] resulting from the active enrichment of anionic lipids (primarily phosphatidylserine) in the cytosolic leaflet of these membranes [46,47]. Thus, membrane homeostasis necessitates mechanisms that can sense the presence of these features at membrane surfaces (Figure 2A). A notable example for sensing hydrophobic voids (inversely related to headgroup packing) is the folding of amphipathic helices (AH) into membranes [48]. The amphiphatic lipid packing sensor (ALPS) motifs found in proteins of the early secretory pathway feature AHs composed of small polar residues on the hydrophilic side and large aromatic residues penetrating into interfacial voids. These features impart strong sensitivity to lipid packing defects and membrane curvature, but low sensitivity to surface charge [48,49]. In contrast, AHs of late secretory pathway sensors typically have basic residues in their AHs, to interact with the high anionic surface charge density unique to those membranes [48,49].
A prominent example of a membrane sensor in the early secretory pathway that uses an ALPS motif for homeostasis is CCTα, a rate-limiting enzyme for production of phosphatidylcholine (PC) from phosphatidylethanolamine (PE) [38]. Owing to this activity, CCTα is a central modulator of intrinsic curvature of the ER membrane and the monolayer around lipid droplets [50,51]. A perturbed ratio of PC to PE causes cellular stress and severe morphologic changes of intracellular organelles [52]. CCTα uses an ALPS motif to sense the interfacial membrane voids that inherently occur in membranes with an overabundance of PE due to its small hydrophilic headgroup [38,51]. Membrane recruitment of CCTα via the ALPS motif induces PE-to-PC conversion for the alleviation of membrane stress. In addition to CCTα, a number of other proteins use AHs to sense surface properties in order to control membrane biogenesis [53], the activity of lipases on lipid droplets [54], and the turnover of lipid metabolic enzymes [55] (Table 1). We propose that the detailed structural and physicochemical properties of these AHs determine the relative sensitivity of the protein to electrostatics, lipid packing defects, or both.
Class II - Sensing within the membrane
A separate class of proteins senses the membrane properties within the hydrophobic core of the bilayer. Lipid saturation is a key determinant of membrane phase behavior and bulk viscosity. As an integral part of the homeoviscous response, lipid acyl chain saturation is actively modulated by dedicated machineries in bacteria, cyanobacteria, and fungi [7]. In yeast, the sole fatty acid desaturase gene OLE1 is controlled by two transcription factors Mga2 and Spt23 responsible for sensing the lipid packing density in the core of the ER membrane [18]. Their sensory mechanism relies on highly dynamic homodimers of the transmembrane helices (TMHs) and a bulky tryptophan sensor residue [17] (Figure 2B). The TMHs rotate against each other and the population of distinct rotational states is determined by the interplay of the sensor residue with the lipid environment. When the lipid packing density is high, the Trp is forced to ‘hide’ into the dimer interface; when unsaturated lipid content is high and the membrane is less packed, the Trp swings out into the lipid core [17]. This novel mechanism is certainly not the only way to sense membrane packing - ALPS-based sensors can also report on the lipid density (Table 1). However, these may also be sensitive to membrane curvature, whereas the TMH-based Mga2 is a more direct reporter for lipid packing in the hydrophobic core.
The example of Mga2 highlights an important theme that may also be valid for other membrane homeostasis sensors. By sensing a bulk property (here, lipid packing in hydrophobic core), Mga2 uses the ER membrane as platform for signal integration [7]. Because fatty acid desaturation is dependent on molecular oxygen, ER lipid unsaturation and the resulting lipid packing can be sensed by Mga2 to regulate the hypoxic response. Importantly, while the availability of molecular oxygen can change within seconds, membrane lipid composition integrates over much longer time scales enabling extraction of temporal information about environment conditions.
Class III - Sensing across the bilayer – challenging elastic membrane properties
Diverse perturbations of the ER membrane lipid composition, including the accumulation of saturated membrane lipids [24,56], a perturbed ratio of PC to PE [52], and aberrant sterol levels [57] activate the so-called unfolded protein response (UPR) [58]. The UPR represents a large-scale transcriptional program (more than 5% of all genes) that controls cellular secretory capacity and protein homeostasis. The UPR broadly lowers the rate of protein translation, but selectively upregulates machinery involved in lipid biosynthesis, ER protein folding, and secretion [58]. It was recently discovered that the most conserved transducer of the UPR, Ire1, is sensitive to membrane perturbations [59], and does so by locally compressing the ER membrane [20] (Figure 2C). This capability is mediated by an unusual architecture of the transmembrane region: the single TMH of Ire1 is particularly short, but is extended by a juxta-membrane AH that inserts into hydrophobic core of the membrane, thereby disordering the local environment and causing a thinning of the bilayer [20] (Table 1). Deformation of the bilayer comes at a significant energetic cost in the stressed ER membrane, and this cost can be minimized by coalescing the regions of deformation, which in turn drives Ire1 oligomerization.
The energetic cost of elastic membrane deformations can be substantial and dependent strongly on the lipid composition – higher in aberrantly stiff bilayers, lower in the normally soft ER membrane [20]. Thus, the oligomeric state and resulting activity of Ire1 is sensitive to membrane material properties, irrespective of the presence of unfolded proteins. This insight explains how apparently unrelated perturbations of lipid metabolism result in an identical response, namely UPR activation [20]. It is apparent that the UPR, which was originally identified as a stress response to unfolded proteins accumulating the lumen of the ER, integrates two types of information: the presence of unfolded proteins in the ER lumen and membrane compressibility, both of which drive clustering and activation of Ire1. This dual capacity to sense unfolded proteins and aberrant lipid states allows Ire1 balance the relative rate of protein and lipid biosynthesis to conduct the orchestra of membrane biogenesis.
These examples highlight the relevance of bulk membrane properties in the regulation of lipid and protein metabolism, hypoxia, and the UPR. The lack of stereo-specificity of these sensory systems is compensated by their inherent versatility. Yeast cells, for example, can adapt their UPR and lipid metabolism to the properties of fatty acids they cannot even produce [17].
Conclusions and perspectives
A variety of physicochemical membrane properties affect cellular physiology, both individually and jointly. Homeostasis of these crucial features necessitates a variety of membrane sensors and response modules. To date, only a few of these have been described in detail. As membrane properties are often interdependent, it is inherently challenging to establish the exact property being sensed. Does a homeostatic program that responds to changes in lipid acyl chain saturation sense lipid packing, lateral pressure, membrane compressibility, or a combination of these? Similarly, it is difficult to control these properties individually, as changes in lipid composition necessarily have multifactorial effects. Moreover, it is important to distinguish between the sensor and downstream effector functions. For example, Ole1 activity is a key cog in the machinery for membrane homeostasis, but only as an effector. Also, not every membrane-sensitive protein is a membrane sensor. Some proteins may rely on the properties of a given membrane for their activity without necessarily regulating the property that is being sensed.
We propose that all organellar membranes harbor sense-and-control machineries to maintain membrane properties. As a product of co-evolution, the sensor modules are likely to reflect characteristic properties of the membrane being interrogated. This is illustrated by the distinct physicochemical features of AHs binding to membranes of the early and the late secretory pathway, respectively [49]. A similar trend may be expected regarding the effector modules. The effectors of the early secretory pathway affect key lipid metabolic reactions or control transcriptional responses to homeostatically feed-back on membrane properties. In contrast, correcting membrane homeostasis in the signaling-active organelles of the late secretory pathway is particularly time-critical. Here, the effectors more likely reside at membrane contact sites acting as lipid transfer proteins, as exemplified by E-SYT [42], ORP5/8 [46] and Osh4 [40] involved in the homeostasis of PS and cholesterol in the, the TGN and the ER, respectively.
As is clear from the above discussion, we have just begun to scratch the surface of membrane homeostatic machineries. Accumulating evidence reveals that these mechanisms are likely to be diverse and much more numerous than previously appreciated. To uncover mechanisms of membrane sensing, it is necessary to combine classical molecular cell biology with computational simulations and in vitro experiments on isolated sensors in defined membrane environments. Categorizing a putative sensor molecule into one of the three sensor classes delineated above may narrow the experimental space.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft to R.E. and S.B. (SFB807 Transport and Communication across Biological Membranes). I.L. is supported by NIH/National Institute of General Medical Sciences Grant No. R01GM114282, R01GM100078, R01GM124072. This work was supported by the Volkswagen Foundation [grant no. 93089] to R.E., and [grant no. 93091] to I.L..
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Moravcevic K, Oxley CL, Lemmon MA. Conditional Peripheral Membrane Proteins: Facing up to Limited Specificity. Structure. 2012;20:15–27. doi: 10.1016/j.str.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sezgin E, Levental I, Mayor S, Eggeling C. The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat Rev Mol Cell Biol. 2017;18:361–374. doi: 10.1038/nrm.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marsh D. Protein modulation of lipids, and vice-versa, in membranes. Biochim Biophys Acta. 2008;1778:1545–75. doi: 10.1016/j.bbamem.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 4.Shevchenko A, Simons K. Lipidomics: coming to grips with lipid diversity. Nat Rev Mol Cell Biol. 2010;11:593–598. doi: 10.1038/nrm2934. [DOI] [PubMed] [Google Scholar]
- 5.Sinensky M. Homeoviscous adaptation--a homeostatic process that regulates the viscosity of membrane lipids in Escherichia coli. Proc Natl Acad Sci U S A. 1974;71:522–525. doi: 10.1073/pnas.71.2.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sinensky M. Adaptive alteration in phospholipid composition of plasma membranes from a somatic cell mutant defective in the regulation of cholesterol biosynthesis. J Cell Biol. 1980;85:166–9. doi: 10.1083/jcb.85.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ernst R, Ejsing CS, Antonny B. Homeoviscous Adaptation and the Regulation of Membrane Lipids. J Mol Biol. 2016;428:4776–4791. doi: 10.1016/j.jmb.2016.08.013. [DOI] [PubMed] [Google Scholar]
- 8.Hazel JR. Thermal Adaptation in Biological-Membranes - Is Homeoviscous Adaptation the Explanation. Annu Rev Physiol. 1995;57:19–42. doi: 10.1146/annurev.ph.57.030195.000315. [DOI] [PubMed] [Google Scholar]
- 9.Saita E, Albanesi D, De Mendoza D. Sensing membrane thickness: Lessons learned from cold stress. Biochim Biophys Acta - Mol Cell Biol Lipids. 2016;1861:837–846. doi: 10.1016/j.bbalip.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Cybulski LE, Martín M, Mansilla MC, Fernández A, De Mendoza D. Membrane thickness cue for cold sensing in a bacterium. Curr Biol. 2010;20:1539–1544. doi: 10.1016/j.cub.2010.06.074. [DOI] [PubMed] [Google Scholar]
- 11.Holthuis JCM, Menon AK. Lipid landscapes and pipelines in membrane homeostasis. Nature. 2014;510:48–57. doi: 10.1038/nature13474. [DOI] [PubMed] [Google Scholar]
- 12.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diaz-Rohrer BB, Levental KR, Simons K, Levental I. Membrane raft association is a determinant of plasma membrane localization. Proc Natl Acad Sci. 2014;111:8500–8505. doi: 10.1073/pnas.1404582111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diaz-Rohrer B, Levental KR, Levental I. Rafting through traffic: Membrane domains in cellular logistics. Biochim Biophys Acta. 2014;1838:3003–3013. doi: 10.1016/j.bbamem.2014.07.029. [DOI] [PubMed] [Google Scholar]
- 15.Sharpe HJ, Stevens TJ, Munro S. A Comprehensive Comparison of Transmembrane Domains Reveals Organelle-Specific Properties. Cell. 2010;142:158–169. doi: 10.1016/j.cell.2010.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorent JH, Diaz-Rohrer B, Lin X, Spring K, Gorfe AA, Levental KR, Levental I. Structural determinants and functional consequences of protein affinity for membrane rafts. Nat Commun. 2017;8:1219. doi: 10.1038/s41467-017-01328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Covino R, Ballweg S, Stordeur C, Michaelis JB, Puth K, Wernig F, Bahrami A, Ernst AM, Hummer G, Ernst R. A Eukaryotic Sensor for Membrane Lipid Saturation. Mol Cell. 2016;63:49–59. doi: 10.1016/j.molcel.2016.05.015. of outstanding interest – Covino et al. – By combining of genetic experiments, biochemical reconstitutions, and molecular dynamics simulations, this study identified the first eukaryotic lipid packing sensor in the core of the ER membrane that controls fatty acid desaturation and ultimately membrane fluidity. [DOI] [PubMed] [Google Scholar]
- 18.Ballweg S, Ernst R. Control of membrane fluidity: The OLE pathway in focus. Biol Chem. 2017;398:215–228. doi: 10.1515/hsz-2016-0277. [DOI] [PubMed] [Google Scholar]
- 19.Loewen CJR. Phospholipid Metabolism Regulated by a Transcription Factor Sensing Phosphatidic Acid. Science. 2004;304:1644–1647. doi: 10.1126/science.1096083. [DOI] [PubMed] [Google Scholar]
- 20.Halbleib K, Pesek K, Covino R, Hofbauer HF, Wunnicke D, Hänelt I, Hummer G, Ernst R. Activation of the Unfolded Protein Response by Lipid Bilayer Stress. Mol Cell. 2017;67:673–684e8. doi: 10.1016/j.molcel.2017.06.012. of outstanding interest – Halbleib et al. – This study establishes the molecular basis for the activation of the unfolded protein response by lipid bilayer stress. Using a combination of experiment and molecular dynamics simulations the interdependency of the proteostasis network and physicochemical properties of the ER membrane are established. [DOI] [PubMed] [Google Scholar]
- 21.Tulodziecka K, Diaz-Rohrer BB, Farley MM, Chan RB, Di Paolo G, Levental KR, Waxham MN, Levental I. Remodeling of the postsynaptic plasma membrane during neural development. Mol Biol Cell. 2016;27:3480–3489. doi: 10.1091/mbc.E16-06-0420. of interest – Tulodziecka et al. – A study on the lipidomic and biophysical changes that occur in mammalian synaptic membranes during early development shows that dramatic lipidomic remodeling during the first weeks of life. Remarkably, despite wholesale lipid changes, membrane fluidity remains unchanged. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levental KR, Surma MA, Skinkle AD, Lorent JH, Zhou Y, Klose C, Chang JT, Hancock JF, Levental I. ω-3 polyunsaturated fatty acids direct differentiation of the membrane phenotype in mesenchymal stem cells to potentiate osteogenesis. Sci Adv. 2017;3:eaao1193. doi: 10.1126/sciadv.aao1193. of outstanding interest – Levental et al. – Demonstration of the diversity of mammalian lipidomes showing that even closely related cell types can have significant, functionally relevant membrane differences. Differentiation of mesenchymal stem cells into adipocytes versus osteoblasts produces unique lipidomes and membrane physical properties, which can be used to affect lineage specification. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klose C, Surma MA, Gerl MJ, Meyenhofer F, Shevchenko A, Simons K. Flexibility of a eukaryotic lipidome - insights from yeast lipidomics. PLoS One. 2012;7:e35063. doi: 10.1371/journal.pone.0035063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Surma MA, Klose C, Peng D, Shales M, Mrejen C, Stefanko A, Braberg H, Gordon DE, Vorkel D, Ejsing CS, et al. A lipid E-MAP identifies Ubx2 as a critical regulator of lipid saturation and lipid bilayer stress. Mol Cell. 2013;51:519–530. doi: 10.1016/j.molcel.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levental KR, Lorent JH, Lin X, Skinkle AD, Surma MA, Stockenbojer EA, Gorfe AA, Levental I. Polyunsaturated Lipids Regulate Membrane Domain Stability by Tuning Membrane Order. Biophys J. 2016;110:1800–1810. doi: 10.1016/j.bpj.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shindou H, Koso H, Sasaki J, Nakanishi H, Sagara H, Nakagawa KM, Takahashi Y, Hishikawa D, Iizuka-Hishikawa Y, Tokumasu F, et al. Docosahexaenoic acid preserves visual function by maintaining correct disc morphology in retinal photoreceptor cells. J Biol Chem. 2017;292:12054–12064. doi: 10.1074/jbc.M117.790568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin X, Lorent JH, Skinkle AD, Levental KR, Waxham MN, Gorfe AA, Levental I. Domain Stability in Biomimetic Membranes Driven by Lipid Polyunsaturation. J Phys Chem B. 2016;120:11930–11941. doi: 10.1021/acs.jpcb.6b06815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeung T, Gilbert GE, Shi J, Silvius J, Kapus A, Grinstein S. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 2008;319:210–3. doi: 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- 29.Saenz JP, Sezgin E, Schwille P, Simons K. Functional convergence of hopanoids and sterols in membrane ordering. Proc Natl Acad Sci. 2012;109:14236–14240. doi: 10.1073/pnas.1212141109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boumann HA. Depletion of Phosphatidylcholine in Yeast Induces Shortening and Increased Saturation of the Lipid Acyl Chains: Evidence for Regulation of Intrinsic Membrane Curvature in a Eukaryote. Mol Biol Cell. 2005;17:1006–1017. doi: 10.1091/mbc.E05-04-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burns M, Wisser K, Wu J, Levental I, Veatch SL. Miscibility Transition Temperature Scales with Growth Temperature in a Zebrafish Cell Line. Biophys J. 2017;113:1212–1222. doi: 10.1016/j.bpj.2017.04.052. of interest – Burns et al. – This paper suggests that separation of the plasma membrane into coexisting ordered and disordered phases is homeostatically regulated. Fish cells can be grown at different temperatures, and remodel their lipidomes, with the apparent purpose of maintaining phase separation temperature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marsh D. Lateral pressure profile, spontaneous curvature frustration, and the incorporation and conformation of proteins in membranes. Biophys J. 2007;93:3884–99. doi: 10.1529/biophysj.107.107938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cybulski LE, Ballering J, Moussatova A, Inda ME, Vazquez DB, Wassenaar TA, de Mendoza D, Tieleman DP, Killian JA. Activation of the bacterial thermosensor DesK involves a serine zipper dimerization motif that is modulated by bilayer thickness. Proc Natl Acad Sci. 2015;112:6353–6358. doi: 10.1073/pnas.1422446112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pinot M, Vanni S, Pagnotta S, Lacas-Gervais S, Payet L-a, Ferreira T, Gautier R, Goud B, Antonny B, Barelli H. Polyunsaturated phospholipids facilitate membrane deformation and fission by endocytic proteins. Science. 2014;345:693–697. doi: 10.1126/science.1255288. [DOI] [PubMed] [Google Scholar]
- 35.Stordeur C, Puth K, Sáenz JP, Ernst R. Crosstalk of lipid and protein homeostasis to maintain membrane function. Biol Chem. 2014;395:313–326. doi: 10.1515/hsz-2013-0235. [DOI] [PubMed] [Google Scholar]
- 36.Radanović T, Reinhard J, Ballweg S, Pesek K, Ernst R. An Emerging Group of Membrane Property Sensors Controls the Physical State of Organellar Membranes to Maintain Their Identity. Bioessays. 2018 doi: 10.1002/bies.201700250. [DOI] [PubMed] [Google Scholar]
- 37.Puth K, Hofbauer HF, Saénz JP, Ernst R. Homeostatic control of biological membranes by dedicated lipid and membrane packing sensors. Biol Chem. 2015;396:1043–1058. doi: 10.1515/hsz-2015-0130. [DOI] [PubMed] [Google Scholar]
- 38.Cornell RB. Membrane lipid compositional sensing by the inducible amphipathic helix of CCT. Biochim Biophys Acta - Mol Cell Biol Lipids. 2016;1861:847–861. doi: 10.1016/j.bbalip.2015.12.022. [DOI] [PubMed] [Google Scholar]
- 39.O’Hara L, Han GS, Sew PC, Grimsey N, Carman GM, Siniossoglou S. Control of phospholipid synthesis by phosphorylation of the yeast lipin Pah1p/Smp2p Mg2+-dependent phosphatidate phosphatase. J Biol Chem. 2006;281:34537–34548. doi: 10.1074/jbc.M606654200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mesmin B, Bigay J, Polidori J, Jamecna D, Lacas-Gervais S, Antonny B. Sterol transfer, PI4P consumption, and control of membrane lipid order by endogenous OSBP. EMBO J. 2017;36:3156–3174. doi: 10.15252/embj.201796687. of outstanding interest – Mesmin et al. – This study provides insights into the orchestration of sterol transfer between the ER to the trans-Golgi network (TGN). It shows sterol transport by OSBP, phosphatidylinositol 4-phosphate turnover, and the gradient of lipid order along the secretory pathway are inherently coupled in living cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mesmin B, Bigay J, Moser Von Filseck J, Lacas-Gervais S, Drin G, Antonny B. A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi Tether OSBP. Cell. 2013;155:830–843. doi: 10.1016/j.cell.2013.09.056. [DOI] [PubMed] [Google Scholar]
- 42.Bian X, Saheki Y, De Camilli P. Ca 2+ releases E-Syt1 autoinhibition to couple ER-plasma membrane tethering with lipid transport. EMBO J. 2018;37:219–234. doi: 10.15252/embj.201797359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Drin G. Topological regulation of lipid balance in cells. Annu Rev Biochem. 2014;83:51–77. doi: 10.1146/annurev-biochem-060713-035307. [DOI] [PubMed] [Google Scholar]
- 44.Bigay J, Antonny B. Curvature, Lipid Packing, and Electrostatics of Membrane Organelles: Defining Cellular Territories in Determining Specificity. Dev Cell. 2012;23:886–895. doi: 10.1016/j.devcel.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 45.Fairn GD, Schieber NL, Ariotti N, Murphy S, Kuerschner L, Webb RI, Grinstein S, Parton RG. High-resolution mapping reveals topologically distinct cellular pools of phosphatidylserine. J Cell Biol. 2011;194:257–275. doi: 10.1083/jcb.201012028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chung J, Torta F, Masai K, Lucast L, Czapla H, Tanner LB, Narayanaswamy P, Wenk MR, Nakatsu F, De Camilli P. PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER-plasma membrane contacts. Science. 2015;349:428–32. doi: 10.1126/science.aab1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pomorski T, Menon AK. Lipid flippases and their biological functions. Cell Mol Life Sci. 2006;63:2908–2921. doi: 10.1007/s00018-006-6167-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drin G, Antonny B. Amphipathic helices and membrane curvature. FEBS Lett. 2010;584:1840–1847. doi: 10.1016/j.febslet.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 49.Antonny B. Mechanisms of membrane curvature sensing. Annu Rev Biochem. 2011;80:101–23. doi: 10.1146/annurev-biochem-052809-155121. [DOI] [PubMed] [Google Scholar]
- 50.Krahmer N, Guo Y, Wilfling F, Hilger M, Lingrell S, Heger K, Newman HW, Schmidt-Supprian M, Vance DE, Mann M, et al. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:Phosphocholine cytidylyltransferase. Cell Metab. 2011;14:504–515. doi: 10.1016/j.cmet.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prévost C, Sharp ME, Kory N, Lin Q, Voth GA, Farese RV, Walther TC. Mechanism and Determinants of Amphipathic Helix-Containing Protein Targeting to Lipid Droplets. Dev Cell. 2018;44:73–86e4. doi: 10.1016/j.devcel.2017.12.011. of interest – Prévost et al. – By combining genetic and biophysical approaches with molecular dynamics simulation this study establishes the molecular basis for targeting AH-containing proteins to the surface of lipid droplets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thibault G, Shui G, Kim W, McAlister GC, Ismail N, Gygi SP, Wenk MR, Ng DT. The Membrane Stress Response Buffers Lethal Effects of Lipid Disequilibrium by Reprogramming the Protein Homeostasis Network. Mol Cell. 2012;48:16–27. doi: 10.1016/j.molcel.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karanasios E, Han G-S, Xu Z, Carman GM, Siniossoglou S. A phosphorylation-regulated amphipathic helix controls the membrane translocation and function of the yeast phosphatidate phosphatase. Proc Natl Acad Sci U S A. 2010;107:17539–44. doi: 10.1073/pnas.1007974107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rowe ER, Mimmack ML, Barbosa AD, Haider A, Isaac I, Ouberai MM, Thiam AR, Patel S, Saudek V, Siniossoglou S, et al. Conserved Amphipathic Helices Mediate Lipid Droplet Targeting of Perilipins 1–3. J Biol Chem. 2016;291:6664–6678. doi: 10.1074/jbc.M115.691048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chua NK, Howe V, Jatana N, Thukral L, Brown AJ. A Conserved Degron Containing an Amphipathic Helix Regulates the Cholesterol-Mediated Turnover of Human Squalene Monooxygenase, a Rate-Limiting Enzyme in Cholesterol Synthesis. J Biol Chem. 2017;292:19959–19973. doi: 10.1074/jbc.M117.794230. of interest – Chua et al. – This study identifies a membrane-sensitive degron that controls the proteolytic turnover of a key enzyme in cholesterol biosynthesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deguil J, Pineau L, Rowland Snyder EC, Dupont S, Beney L, Gil A, Frapper G, Ferreira T. Modulation of Lipid-Induced ER Stress by Fatty Acid Shape. Traffic. 2011;12:349–362. doi: 10.1111/j.1600-0854.2010.01150.x. [DOI] [PubMed] [Google Scholar]
- 57.Pineau L, Colas J, Dupont S, Beney L, Fleurat-Lessard P, Berjeaud JM, Bergès T, Ferreira T. Lipid-induced ER stress: Synergistic effects of sterols and saturated fatty acids. Traffic. 2009;10:673–690. doi: 10.1111/j.1600-0854.2009.00903.x. [DOI] [PubMed] [Google Scholar]
- 58.Walter P, Ron D. The Unfolded Protein Response: From stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 59.Volmer R, Ron D. Lipid-dependent regulation of the unfolded protein response. Curr Opin Cell Biol. 2015;33:67–73. doi: 10.1016/j.ceb.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]


