Abstract
Misfolded proteins of the endoplasmic reticulum (ER) are discarded by a conserved process, called ER-associated protein degradation (ERAD). ERAD substrates are retro-translocated into the cytosol, polyubiquitinated, extracted from the ER membrane, and ultimately degraded by the proteasome. Recent in vitro experiments with purified components have given insight into the mechanism of ERAD. ERAD substrates with misfolded luminal or intra-membrane domains are moved across the ER membrane through a channel formed by the multi-spanning ubiquitin ligase Hrd1. Following polyubiquitination, substrates are extracted from the membrane by the Cdc48/p97 ATPase complex and transferred to the proteasome. We discuss the molecular mechanism of these processes and point out remaining open questions.
Introduction
Misfolded ER proteins are removed by ERAD, an evolutionarily conserved process in which substrates are retro-translocated into the cytosol, polyubiquitinated, and degraded by the proteasome (for review, see ref. [1]). Work in S. cerevisiae resulted in the simple concept that misfolded ER proteins are degraded by three different ERAD pathways (ERAD-L, -M, and -C), depending on whether their misfolded domain is localized in the ER lumen, within the membrane, or on the cytosolic side of the ER membrane [2–4]. These pathways involve distinct ubiquitin ligases. ERAD-L substrates use the RING-finger ligase Hrd1 in complex with three other membrane proteins (Hrd3, Usa1, and Der1) and a luminal protein (Yos9) [2,5,6]. ERAD-M substrates also use Hrd1 and Hrd3, but not Der1 [2], and only in some cases Usa1 [7]. ERAD-C substrates require Doa10, another RING-finger ligase [8]. However, it should be noted that Doa10 also recognizes some substrates by features within their membrane-spanning region (e.g. ref. [9]). A fourth pathway is responsible for the degradation of misfolded inner nuclear membrane proteins and utilizes a ubiquitin ligase consisting of three proteins (Asi1,2,3) [10,11]. Again, the Asi complex seems to recognize substrate features within the membrane. It remains unclear how proteins are targeted to the different pathways, as ERAD-M, -C and Asi all handle trans-membrane protein substrates. All ERAD pathways converge on the cytosolic side of the ER membrane, where they require other components of the ubiquitination machinery and an ATPase complex consisting of the AAA+ ATPase Cdc48 (called p97 or VCP in mammals) and a heterodimeric cofactor, consisting of Ufd1 and Npl4 [12–17]. In this review, we focus on the Hrd1-dependent ERAD-L and –M pathways. Although much of the mechanistic work has been done with S. cerevisiae components, the conclusions are likely applicable to all eukaryotes.
Overview of ERAD-L and –M
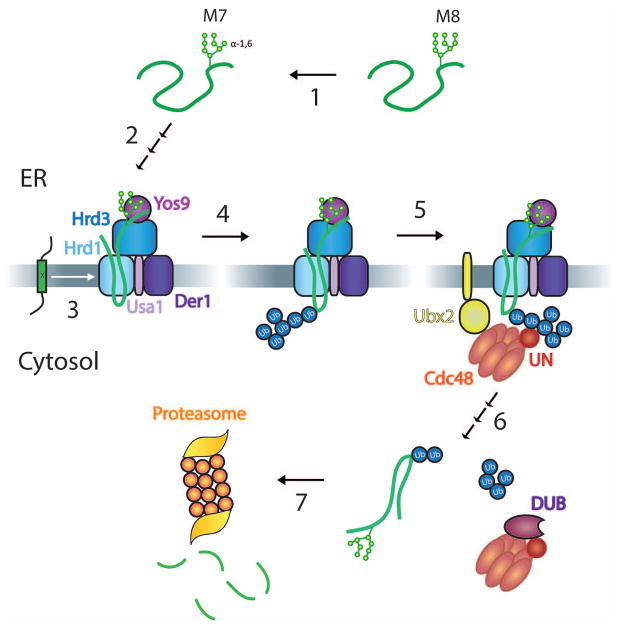
ERAD-L begins with the recognition of a misfolded protein domain in the ER lumen, which is best understood for misfolded glycoproteins (for review, see [18]). When these proteins linger too long in the ER, the N-linked glycan is trimmed to generate a terminal α-1,6-linked mannose residue (Figure 1, step 1). This sugar is recognized by the lectin Yos9p (step 2) [19,20], which in turn is bound to the luminal domain of Hrd3p [19,20]. In addition, Hrd3 probably recognizes a misfolded substrate segment around the glycan-attachment site, thus providing a second binding site for the substrate. Hrd3 also recognizes non-glycosylated ERAD-L substrates, but whether Yos9 is also involved in this case is unclear. How a substrate is transferred to downstream components is unknown, but ultimately the polypeptide inserts into the membrane channel formed by the Hrd1 ligase (step 2; see below). Membrane insertion also requires the multi-spanning membrane protein Der1 [21,22]. ERAD-M substrates probably enter the Hrd1 channel sideways from the lipid phase (step 3). In both pathways, substrates are then polyubiquitinated on the cytoplasmic side of the ER membrane by Hrd1p [5,6], which recruits ubiquitin-conjugating enzymes, with Ubc7 being the best studied (step 4). Ubc7 cooperates with Cue1, which serves as membrane anchor and activator.
Figure 1. Overview of ERAD-L and –M.
The scheme shows different steps in ERAD-L. Step 1: The N-glycan chain of a misfolded luminal glycoprotein is trimmed from eight to seven mannoses (M8 and M7, respectively) by glycosidases. Step 2: The generated terminal α1,6-linked mannose residue binds to Yos9, and the misfolded segment around the glycan attachment site binds to Hrd3. The substrate inserts into the Hrd1 channel with the help of Der1, which associates with Hrd1 through Usa1. Step 3: ERAD-M substrates are misfolded in their membrane-spanning segments (indicated by an “x”) and enter Hrd1 sideways. Step 4: Both ERAD-L and ERAD-M substrates are polyubiquitinated by Hrd1. Step 5: The Cdc48 ATPase is recruited to the ER membrane by binding of the Ufd1/Npl4 (UN) cofactor to the ubiquitin chain and by Cdc48 binding to Ubx2. Step 6: Cdc48 uses ATP hydrolysis to pull the polypeptide substrate out of the membrane, the complex of Cdc48 ATPase and substrate leaves the membrane, and a DUB trims the ubiquitin chain, allowing release of the substrate from Cdc48. Step 7: The substrate is degraded by the proteasome.
In the next step, the cytosolic Cdc48 ATPase complex is recruited to the membrane by an interaction of the polyubiquitin chain with the cofactor Ufd1/Npl4 (step 5). Recruitment of Cdc48 is also facilitated by an interaction with the membrane protein Ubx2 [23,24]. The Cdc48 ATPase then uses the energy of ATP hydrolysis to progressively pull the substrate out of the membrane [25,26]. Once the substrate is fully removed from the ER, Cdc48 probably dissociates from the Ubx2 anchor, diffusing away from the membrane with its associated substrate (step 6). The substrate is then released from Cdc48 in a process that requires trimming of the polyubiquitin chain by a deubiquitinase (DUB) (step 6). Finally, the substrate is passed on to the proteasome for degradation (step 7). Below, we discuss several of these steps in more detail.
Hrd1 forms a retro-translocation channel
The identity of the retro-translocation channel was controversial for many years. Based on mutations, pull-down experiments, and interaction with the proteasome, it was postulated that the Sec61 channel, which normally allows polypeptides to move from the cytosol into the ER lumen, can function in reverse (for review, see [27]). However, Sec61 mutations may indirectly affect the biosynthesis of ERAD components, and ERAD substrates interact only weakly with Sec61 [28]. Furthermore, the Sec61 channel contains a plug domain that is normally displaced by a signal sequence when a secretory protein inserts from the cytosolic side [29,30]. It is difficult to envision how the channel would open from the luminal side of the ER membrane in ERAD. Derlin-1, a mammalian homolog of Der1, was also proposed as a channel, based on the fact that it is a multi-spanning membrane protein that has interaction partners on both sides of the ER membrane [31,32]. However, Hrd1 has clearly emerged as the top candidate for forming a retro-translocation channel.
Initial evidence for Hrd1 forming a channel came from the observation that its overexpression in yeast makes the other components of the Hrd1 complex dispensable for the degradation of ERAD-L and –M substrates [28,33,34]; all downstream components, such as the ubiquitination machinery and the Cdc48 ATPase complex were still required. The existence of a Hrd1 channel was also supported by photo-crosslinking experiments that showed substrate in close proximity of Hrd1 during retro-translocation [28].
In vitro experiments with purified proteins later showed that Hrd1 binds and polyubiquitinates soluble ERAD-L substrates [26]. Retro-translocation was recapitulated with proteoliposomes that contained both Hrd1 and a single-spanning substrate protein with a large misfolded domain [35]. When the ubiquitination machinery was added, a segment that was initially inside the lumen of the vesicles was polyubiquitinated by Hrd1, suggesting that it moved across the membrane to the outside of the liposomes. Hrd1 also underwent efficient auto-ubiquitination in the same reaction. Auto-ubiquitination of Hrd1 was postulated to be the trigger for retro-translocation of the substrate, because there was no lysine of the substrate accessible to Hrd1 at the beginning of the reaction, but ubiquitination per se was required. Indeed, several studies showed that ubiquitination is still required when all lysines in a substrate are removed [35–38]. The crucial modification seems to occur in the RING finger of Hrd1, as mutation of lysines in this domain prevented retro-translocation in vitro and the degradation of ERAD-L substrates in vivo [35].
These results led to a model in which auto-ubiquitination of Hrd1 opens the channel for ERAD-L substrates [35]. A polypeptide substrate would then be able to slide back and forth through the channel, but attachment of a polyubiquitin chain could prevent its back-sliding into the ER lumen. Interestingly, the lysine mutations in the RING finger domain do not have a strong effect on ERAD-M substrates (R. Baldridge, personal communication). Thus, ERAD-M substrates may not require auto-ubiquitination of Hrd1, perhaps because a segment of the polypeptide is already in the cytosol.
Structure of the Hrd1 channel
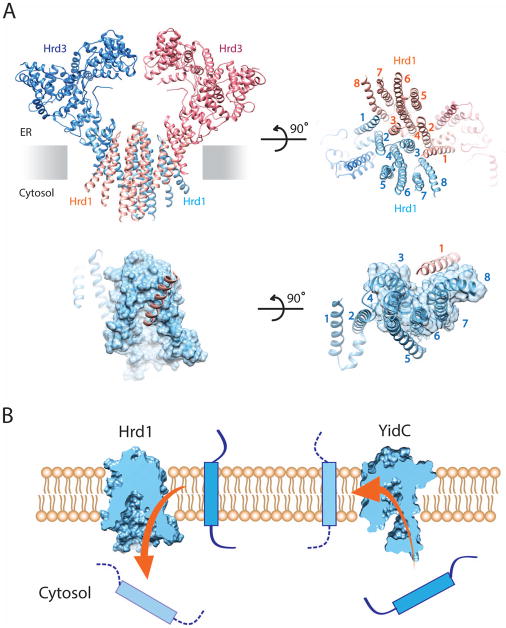
An electron cryo-microscopy (cryo-EM) structure was recently determined for Hrd1 bound to the luminal domain of Hrd3, a complex that is sufficient for ERAD-M [39]. The complex contains two symmetry-related molecules of Hrd1 and Hrd3 (Figure 2A). The two Hrd3 molecules form an arch on the luminal side. Hrd1 contains eight trans-membrane (TM) segments (Figure 2A), rather than six, as previously assumed [40]. Five of these TM segments form a funnel that extends from the cytosol almost to the luminal side of the membrane (Figure 2A). Trans-membrane segment 1 (TM1) of each Hrd1 molecule laterally seals the funnel of the neighboring Hrd1 molecule on the cytosolic side (Figure 2A; lower panels). The aqueous cavity of Hrd1 is reminiscent of that in the Sec61 channel and its prokaryotic homolog, the SecY channel, which allow protein transport in the opposite direction [29,30]. Deep cytosolic invaginations are also seen in the bacterial YidC protein and its homologs in plants and mitochondria (Figure 2B) [41,42]. These proteins allow hydrophobic TM segments to move from the cytosol into the lipid bilayer, whereas Hrd1 facilitates the reverse process during ERAD-M (Figure 2B). Given that the known protein-conducting conduits have large aqueous cavities, it seems that the thinning of the membrane might be a general principle to reduce the energy barrier for polypeptide movement in or out of a membrane.
Figure 2. Structure of a Hrd1/Hrd3 complex.
(A) Model of Hrd1 bound to the luminal domain of Hrd3, based on cryo-EM single-particle analysis [38]. The upper left panel shows a cartoon of the Hrd1/Hrd3 dimer, with the Hrd1 molecules in light blue and salmon, and the Hrd3 molecules in dark blue and red. The upper right panel shows a view from the cytosol. The lower left panel shows a space-filling model of the funnel of one Hrd1 molecule together with TM1 of the other. The lower right panel shows a view from the cytosol. (B) The left panel shows a cut through a space-filling model of Hrd1. Hrd1/Hrd3 allows an ERAD-M substrate to move into the cytosol (arrow). The right panel shows a cut through a space-filling model of the bacterial YidC protein, which allows TM segments to move from the cytosol into the membrane (arrow).
Open questions about the function of Hrd1 and its partners
Although Hrd1 has emerged as the best channel candidate for ERAD-L and –M, its function is still poorly understood. One issue is whether the same Hrd1 complex is involved in both ERAD pathways. In fact, it seems possible that a complex comprising Hrd1, Hrd3, Usa1, Der1, and Yos9 would function in ERAD-L, whereas a distinct Hrd1/Hrd3 sub-complex would mediate ERAD-M. Whether Hrd1 functions as a dimer or monomer in these pathways is also unclear. In the current Hrd1/Hrd3 structure, two Hrd1 channels are paired [39], but Hrd1 oligomerization in vivo actually requires Usa1 [7,28]. Perhaps, Hrd1 auto-ubiquitination and substrate binding cause the dissociation of the Hrd1 dimer into monomers [28].
Other unresolved questions concern the path of the substrate across the membrane and the role of the other components of the Hrd1 complex. Der1 is an inactive relative of rhomboid proteases [43] and recruits substrates from the ER lumen. Perhaps, it replaces the second Hrd1 molecule and sits at the lateral gate of a monomeric Hrd1 channel, allowing luminal substrates to enter sideways, rather than directly from the luminal aqueous phase. Because Usa1 physically links Der1 and Hrd1, it may position Der1 for substrate transfer [7,22,28]. The exact function of Yos9 and Hrd3 in substrate recognition also needs to be defined. Hrd3 probably not only binds substrate, but also inhibits the activity of Hrd1, as in its absence, Hrd1 undergoes excessive auto-ubiquitination and degradation [33,44]. How Hrd3 would regulate Hrd1 activity is unclear, particularly because the RING finger domains of the Hrd1 molecules are invisible in the density map of the Hrd1/Hrd3 complex (Figure 2A). In one model, substrate binding to Hrd3 would relieve the inhibition of Hrd1, allowing auto-ubiquitination and opening of the channel. Finally, it remains unclear how auto-ubiquitination of Hrd1 would be reversed to reset the channel for the next round of ERAD-L. The unidentified DUB must be distinct from the relatively well characterized enzyme Otu1, a DUB involved in the trimming of polyubiquitinated substrate on the Cdc48 ATPase complex (see below).
The answer to several of these questions will likely come from cryo-EM structures of the Hrd1 complex and from experiments that test the binding and translocation of defined substrates. It will be a major challenge to generate translocation intermediates, which have been instrumental in elucidating the mechanism of “forward translocation” through the Sec61/SecY channel [45].
Hrd1 is probably a member of a larger class of channel-forming ubiquitin ligases in the ER, which includes Doa10 and the Asi complex. In mammals, Hrd1 has a close homolog (gp78), and both proteins show sequence similarity in their putative channel-forming TMs 3–6 to the ligases RNF145 and RNF139 (alternately called TRC8) [39]. Future experiments need to test whether they all form protein-conducting channels that allow proteins to move from the ER into the cytosol.
Mechanism of the Cdc48 ATPase complex
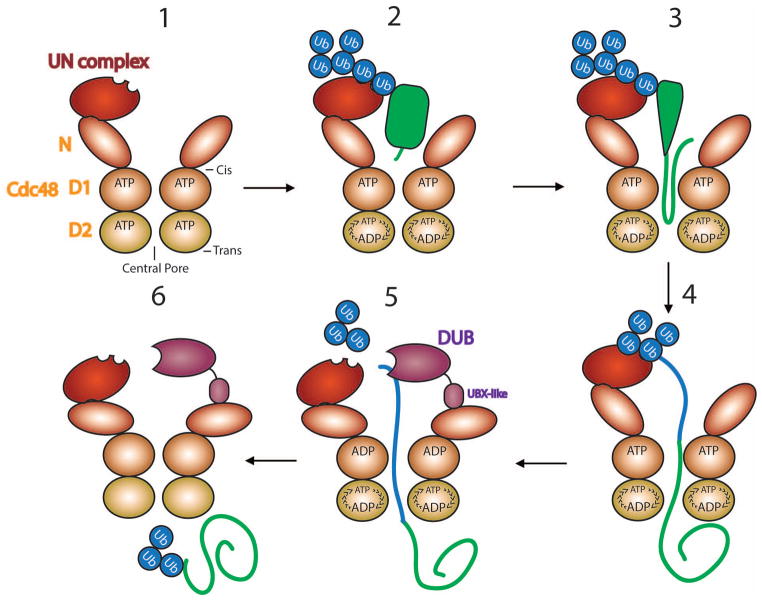
The Cdc48 ATPase consists of an N-terminal domain (N domain) and two ATPase domains (D1 and D2) [46]. Six Cdc48 monomers form a double-ring structure, with a “cis” and a “trans” side and a central pore (Figure 3). The N domains are coplanar with the D1 ring when the D1 ATPases are in the ADP-bound state (“down-conformation”), and in an “up-conformation” when in the ATP-bound state (Figure 3; stage 1) [47]. The hexamer associates on its “cis side” with one copy of the Ufd1/Npl4 (UN) cofactor complex [48].
Figure 3. Substrate processing by the Cdc48 ATPase.
The scheme shows different stages. Stage 1: The Cdc48 ATPase, containing an N domain and two ATPase domains (D1 and D2), forms a hexameric, double-ring structure that associates with one copy of the Ufd1/Npl4 (UN) cofactor. The central pore and the cis- and trans-sides are indicated. Stage 2: The ubiquitin (Ub) chain attached to a substrate (in green) binds to UN. The D1 ATPases are locked in the ATP-bound state with the N domains in the up-conformation, while the activity of the D2 ATPases is stimulated. Stage 3: The substrate is pulled through the central pore, causing polypeptide unfolding. Stage 4: The substrate is moved entirely to the trans-side of the ATPase ring. Ubiquitin is also unfolded and follows the substrate (blue line). Stage 5: ATP hydrolysis in D1 causes movement of the N domains into the down-conformation, allowing a DUB to trim the ubiquitin chain. The DUB Otu1 binds through its UBX-like domain to the N domain. Stage 6: Ubiquitin molecules emerging at the trans-side presumably refold, allowing the substrate to be recognized by shuttling factors and the proteasome.
Much of our knowledge about the mechanism of the Cdc48 ATPase in ERAD comes from the use of simplified in vitro systems, in which artificial, polyubiquitinated substrates were used [25,49]. These and other experiments resulted in a model for substrate processing by Cdc48 (Figure 3). First, the UN cofactor binds the polyubiquitin chain attached to the substrate (Figure 4; stage 2). This interaction decreases ATP hydrolysis of the D1 domains, thus favoring their ATP-bound state with the N domains in the up-conformation. At the same time, ATP hydrolysis by D2 is stimulated, resulting in the translocation and unfolding of fluorescent model substrates (stages 2–4) [25,50]. The polypeptide is pulled through the central pore of the double-ring ATPase, as demonstrated by photocrosslinking to internal amino acid positions in the rings [25]. Furthermore, when Cdc48 was fused to the protease domain of FtsH to generate an artificial proteasome, the substrate was degraded [25], indicating that the polypeptide completely exited Cdc48 at its trans-side and entered the associated protease.
Substrate release from the Cdc48 complex requires a DUB, as complete translocation is otherwise counteracted by the association of the polyubiquitin chain with the UN cofactor at the cis side. In vitro experiments show that Otu1, a DUB which binds to the N domain via its Ubx-like domain, trims the polyubiquitin chain [25]. However, there must be other DUBs that can replace Otu1, as the deletion of OTU1 in S. cerevisiae has no effect on ERAD [26]. Surprisingly, only a minority of the substrate molecules released from Cdc48 in vitro lose all ubiquitins; most retain an oligoubiquitin chain with up to ten ubiquitin molecules, which is also pulled through the central pore (stages 4–6). These ubiquitin molecules are probably sequentially unfolded and refold when they emerge on the trans-side. These results also imply that at least two polypeptide strands can be accommodated inside the pore, indicating that the pore is wider during translocation than seen in current structures of the resting ATPase. In addition, the carbohydrate chain of glycosylated ERAD substrates must also be accommodated inside the pore, because the N-glycanase Png1 binds to the C-terminus of Cdc48 and probably has only access to the chain after its translocation through the ATPase rings [51].
How substrates are transferred from Cdc48 to the proteasome is also still unclear. Many of the substrate molecules released from Cdc48 in vitro contain four or more ubiquitin molecules [25], and could therefore be processed by the 26S proteasome [52]. They are probably transferred from Cdc48 to the proteasome indirectly through the shuttling factors Rad23 and Dsk2, which each have both ubiquitin- and proteasome-binding domains [53,54]. Archaeal Cdc48 homologs and certain mutants of eukaryotic p97 can cooperate with the 20S proteasome to degrade substrate, without involvement of the 19S regulatory subunit [55,56]. Future experiments need to address whether this pathway is used by wild type p97 for some polyubiquitinated substrates. The transfer of substrates to the proteasome probably involves other Cdc48 cofactors. For example, Ufd3 binds to the C-terminal tail of Cdc48 and prevents abnormal ubiquitin degradation [57,58], perhaps by interfering with the Cdc48-20S interaction. The Ufd2 cofactor extends the polyubiquitin chain on substrates [59] and might thus rescue substrates whose chains are too short for degradation by the 26S proteasome. Future experiments need to clarify how substrates are processed by these cofactors and transferred to the proteasome.
Acknowledgments
We thank Pedro Carvalho and Nick Bodnar for critical reading of the manuscript. The work in the authors’ laboratory was supported by an NIH grant (GM 052586). X.W. is a recipient of a Jane Coffin Childs fellowship and T.A.R. is a Howard Hughes Medical Institute investigator.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Christianson JC, Ye Y. Cleaning up in the endoplasmic reticulum: ubiquitin in charge. Nature structural & molecular biology. 2014;21:325–335. doi: 10.1038/nsmb.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 3.Vashist S, Ng DTW. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. The Journal of cell biology. 2004;165:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huyer G, Piluek WF, Fansler Z, Kreft SG, Hochstrasser M, Brodsky JL, Michaelis S. Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. The Journal of biological chemistry. 2004;279:38369–38378. doi: 10.1074/jbc.M402468200. [DOI] [PubMed] [Google Scholar]
- 5.Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nature cell biology. 2001;3:24–9. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
- 6.Bordallo J, Plemper RK, Finger A, Wolf DH. Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Molecular biology of the cell. 1998;9:209–222. doi: 10.1091/mbc.9.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horn SC, Hanna J, Hirsch C, Volkwein C, Schutz A, Heinemann U, Sommer T, Jarosch E. Usa1 functions as a scaffold of the HRD-ubiquitin ligase. Molecular cell. 2009;36:782–793. doi: 10.1016/j.molcel.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 8.Swanson R, Locher M, Hochstrasser M. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 2001;15:2660–2674. doi: 10.1101/gad.933301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Habeck G, Ebner FA, Shimada-Kreft H, Kreft SG. The yeast ERAD-C ubiquitin ligase Doa10 recognizes an intramembrane degron. The Journal of cell biology. 2015;209:261–273. doi: 10.1083/jcb.201408088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foresti O, Rodriguez-Vaello V, Funaya C, Carvalho P. Quality control of inner nuclear membrane proteins by the Asi complex. Science. 2014;346:751–755. doi: 10.1126/science.1255638. [DOI] [PubMed] [Google Scholar]
- 11.Khmelinskii A, Blaszczak E, Pantazopoulou M, Fischer B, Omnus DJ, Le Dez G, Brossard A, Gunnarsson A, Barry JD, Meurer M, et al. Protein quality control at the inner nuclear membrane. Nature. 2014;516:410–413. doi: 10.1038/nature14096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bays NW, Wilhovsky SK, Goradia A, Hodgkiss-Harlow K, Hampton RY. HRD4/NPL4 Is Required for the Proteasomal Processing of Ubiquitinated ER Proteins. Molecular biology of the cell. 2001;12:4114–4128. doi: 10.1091/mbc.12.12.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braun S, Matuschewski K, Rape M, Thoms S, Jentsch S. Role of the ubiquitin-selective CDC48(UFD1/NPL4 )chaperone (segregase) in ERAD of OLE1 and other substrates. The EMBO journal. 2002;21:615–621. doi: 10.1093/emboj/21.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jarosch E, Taxis C, Volkwein C, Bordallo J, Finley D, Wolf DH, Sommer T. Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nature cell biology. 2002;4:134–139. doi: 10.1038/ncb746. [DOI] [PubMed] [Google Scholar]
- 15.Rabinovich E, Kerem A, Frohlich KU, Diamant N, Bar-Nun S. AAA-ATPase p97/Cdc48p, a Cytosolic Chaperone Required for Endoplasmic Reticulum-Associated Protein Degradation. Molecular and cellular biology. 2002;22:626–34. doi: 10.1128/MCB.22.2.626-634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- 17.Ye Y, Meyer HH, Rapoport TA. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. The Journal of cell biology. 2003;162:71–84. doi: 10.1083/jcb.200302169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie W, Ng DT. ERAD substrate recognition in budding yeast. Semin Cell Dev Biol. no date doi: 10.1016/j.semcdb.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 19.Denic V, Quan EM, Weissman JS. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- 20.Gauss R, Jarosch E, Sommer T, Hirsch C. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nature cell biology. 2006;8:849–854. doi: 10.1038/ncb1445. [DOI] [PubMed] [Google Scholar]
- 21.Knop M, Finger A, Braun T, Hellmuth K, Wolf DH. Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. The EMBO journal. 1996;15:753–63. [PMC free article] [PubMed] [Google Scholar]
- 22.Mehnert M, Sommer T, Jarosch E. Der1 promotes movement of misfolded proteins through the endoplasmic reticulum membrane. Nature cell biology. 2014;16:77–86. doi: 10.1038/ncb2882. [DOI] [PubMed] [Google Scholar]
- 23.Neuber O, Jarosch E, Volkwein C, Walter J, Sommer T. Ubx2 links the Cdc48 complex to ER-associated protein degradation. Nature cell biology. 2005;7:993–998. doi: 10.1038/ncb1298. [DOI] [PubMed] [Google Scholar]
- 24.Schuberth C, Buchberger A. Membrane-bound Ubx2 recruits Cdc48 to ubiquitin ligases and their substrates to ensure efficient ER-associated protein degradation. Nature cell biology. 2005;7:999–1006. doi: 10.1038/ncb1299. [DOI] [PubMed] [Google Scholar]
- *25.Bodnar NO, Rapoport TA. Molecular mechanism of substrate processing by the Cdc48 ATPase complex. Cell. 2017;169:722–735e9. doi: 10.1016/j.cell.2017.04.020. This paper elucidates how the Cdc48 ATPase complex processes its substrates. Substrates are first bound through the attached polyubiquitin chain to the UN cofactor and then pulled through the central pore of the hexameric ATPase ring. The ubiquitin chain is trimmed by a DUB, and the remaining oligoubiquitin chain then follows the substrate through the pore. This paper and ref. 49 show that the ATPase exerts a pulling force that leads to substrate unfolding. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stein A, Ruggiano A, Carvalho P, Rapoport TA. Key steps in ERAD of luminal ER proteins reconstituted with purified components. Cell. 2014;158:1375–1388. doi: 10.1016/j.cell.2014.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Römisch K. A Case for Sec61 Channel Involvement in ERAD. Trends in Biochemical Sciences. 2017;42:171–179. doi: 10.1016/j.tibs.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 28.Carvalho P, Stanley AM, Rapoport TA. Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell. 2010;143:579–591. doi: 10.1016/j.cell.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van den Berg B, Clemons WMJ, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 30.Voorhees RM, Fernandez IS, Scheres SH, Hegde RS. Structure of the mammalian ribosome-Sec61 complex to 3. 4 A resolution. Cell. 2014;157:1632–1643. doi: 10.1016/j.cell.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lilley BN, Ploegh HL. A membrane protein required for dislocation of misfolded proteins from the ER. Nature. 2004;429:834–840. doi: 10.1038/nature02592. [DOI] [PubMed] [Google Scholar]
- 32.Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- 33.Gardner RG, Swarbrick GM, Bays NW, Cronin SR, Wilhovsky S, Seelig L, Kim C, Hampton RY. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. The Journal of cell biology. 2000;151:69–82. doi: 10.1083/jcb.151.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plemper RK, Bordallo J, Deak PM, Taxis C, Hitt R, Wolf DH. Genetic interactions of Hrd3p and Der3p/Hrd1p with Sec61p suggest a retro-translocation complex mediating protein transport for ER degradation. Journal of cell science. 1999;112:4123–34. doi: 10.1242/jcs.112.22.4123. [DOI] [PubMed] [Google Scholar]
- *35.Baldridge RD, Rapoport TA. Autoubiquitination of the Hrd1 Ligase Triggers Protein Retrotranslocation in ERAD. Cell. 2016;166:394–407. doi: 10.1016/j.cell.2016.05.048. This paper shows that membrane translocation of a polypeptide during ERAD-L can be recapitulated with proteoliposomes containing purified Hrd1 ubiquitin ligase. Hrd1 seems to form a ubiquitin-gated protein-conducting channel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hassink GC, Barel MT, Van Voorden SB, Kikkert M, Wiertz EJ. Ubiquitination of MHC class I heavy chains is essential for dislocation by human cytomegalovirus-encoded US2 but not US11. The Journal of biological chemistry. 2006;281:30063–30071. doi: 10.1074/jbc.M602248200. [DOI] [PubMed] [Google Scholar]
- 37.Wang X, Yu YY, Myers N, Hansen TH. Decoupling the role of ubiquitination for the dislocation versus degradation of major histocompatibility complex (MHC) class I proteins during endoplasmic reticulum-associated degradation (ERAD) The Journal of biological chemistry. 2013;288:23295–23306. doi: 10.1074/jbc.M113.482018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu H, Kopito RR. The role of multiubiquitination in dislocation and degradation of the alpha subunit of the T cell antigen receptor. The Journal of biological chemistry. 1999;274:36852–8. doi: 10.1074/jbc.274.52.36852. [DOI] [PubMed] [Google Scholar]
- *39.Schoebel S, Mi W, Stein A, Ovchinnikov S, Pavlovicz R, DiMaio F, Baker D, Chambers MG, Su H, Li D, et al. Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3. Nature. 2017;548:352–355. doi: 10.1038/nature23314. This paper reports a cryo-EM structure of a complex of the Hrd1 ligase and its luminal binding partner Hrd3. Hrd1 has a deep cytoplasmic cavity, reminiscent of other protein-conducting channels. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deak PM, Wolf DH. Membrane topology and function of Der3/Hrd1p as a ubiquitin-protein ligase (E3) involved in endoplasmic reticulum degradation. The Journal of biological chemistry. 2001;276:10663–10669. doi: 10.1074/jbc.M008608200. [DOI] [PubMed] [Google Scholar]
- 41.Kumazaki K, Chiba S, Takemoto M, Furukawa A, Nishiyama K-I, Sugano Y, Mori T, Dohmae N, Hirata K, Nakada-Nakura Y, et al. Structural basis of Sec-independent membrane protein insertion by YidC. Nature. 2014;509:516–520. doi: 10.1038/nature13167. [DOI] [PubMed] [Google Scholar]
- 42.Dalbey RE, Kuhn A. Membrane Insertases Are Present in All Three Domains of Life. Structure. 2015;23:1559–1560. doi: 10.1016/j.str.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 43.Greenblatt EJ, Olzmann JA, Kopito RR. Derlin-1 is a rhomboid pseudoprotease required for the dislocation of mutant α-1 antitrypsin from the endoplasmic reticulum. Nature structural & molecular biology. 2011;18:1147–1152. doi: 10.1038/nsmb.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *44.Vashistha N, Neal SE, Singh A, Carroll SM, Hampton RY. Direct and essential function for Hrd3 in ER-associated degradation. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:5934–5939. doi: 10.1073/pnas.1603079113. This paper shows that Hrd3 regulates the enzymatic activity of Hrd1 and is an integral part of the retro-translocation machinery. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mothes W, Prehn S, Rapoport TA. Systematic probing of the environment of a translocating secretory protein during translocation through the ER membrane. The EMBO journal. 1994;13:3937–3982. doi: 10.1002/j.1460-2075.1994.tb06713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeLaBarre B, Brunger AT. Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nature structural biology. 2003;10:856–863. doi: 10.1038/nsb972. [DOI] [PubMed] [Google Scholar]
- 47.Banerjee S, Bartesaghi A, Merk A, Rao P, Bulfer SL, Yan Y, Green N, Mroczkowski B, Neitz RJ, Wipf P, et al. 2. 3 Å resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition. Science. 2016;351:871–875. doi: 10.1126/science.aad7974. This paper reports high-resolution cryo-EM structures of the p97 ATPase, which demonstrate that the N-terminal domains move up and down in response to the nucleotide state of the ATPase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bebeacua C, Förster A, McKeown C, Meyer HH, Zhang X, Freemont PS. Distinct conformations of the protein complex p97-Ufd1-Npl4 revealed by electron cryomicroscopy. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1098–1103. doi: 10.1073/pnas.1114341109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *49.Blyth E, Olson K, Chau V, Deshaies R. Ubiquitin and ATP_dependent unfoldase activity of p97/VCP-NPLOC4-UFD1L1 is enhanced by a mutation that causes mutlisystem proteinopathy. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:E4380–E4388. doi: 10.1073/pnas.1706205114. This paper, together with ref. 25, shows that the p97/Cdc48 ATPase exerts a pulling force on its substrates, thereby leading to their unfolding. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blythe EE, Olson KC, Chau V, Deshaies RJ. Ubiquitin- and ATP-dependent unfoldase activity of P97/VCP•NPLOC4•UFD1L is enhanced by a mutation that causes multisystem proteinopathy. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:E4380–E4388. doi: 10.1073/pnas.1706205114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li G, Zhao G, Zhou X, Schindelin H, Lennarz WJ. The AAA ATPase p97 links peptide N-glycanase to the endoplasmic reticulum-associated E3 ligase autocrine motility factor receptor. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8348–8353. doi: 10.1073/pnas.0602747103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. The EMBO journal. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Medicherla B, Kostova Z, Schaefer A, Wolf DH. A genomic screen identifies Dsk2p and Rad23p as essential components of ER-associated degradation. EMBO reports. 2004;5:692–697. doi: 10.1038/sj.embor.7400164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim I, Mi K, Rao H. Multiple interactions of rad23 suggest a mechanism for ubiquitylated substrate delivery important in proteolysis. Molecular biology of the cell. 2004;15:3357–3365. doi: 10.1091/mbc.E03-11-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barthelme D, Sauer RT. Identification of the Cdc48•20S proteasome as an ancient AAA+ proteolytic machine. Science. 2012;337:843–846. doi: 10.1126/science.1224352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barthelme D, Sauer RT. Bipartite determinants mediate an evolutionarily conserved interaction between Cdc48 and the 20S peptidase. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:3327–3332. doi: 10.1073/pnas.1300408110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Böhm S, Lamberti G, Fernández-Sáiz V, Stapf C, Buchberger A. Cellular functions of Ufd2 and Ufd3 in proteasomal protein degradation depend on Cdc48 binding. Molecular and cellular biology. 2011;31:1528–1539. doi: 10.1128/MCB.00962-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson ES, Ma PC, Ota IM, Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. The Journal of biological chemistry. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- 59.Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]