Abstract
Background
Haemophilia A (HA) and Haemophilia B (HB) are X-linked blood disorders that are caused by various mutations in the factor VIII (F8) and factor IX (F9) genes respectively. Identification of mutations is essential as some of the mutations are associated with the development of inhibitors. This study is the first comprehensive study of the F8 mutational profile in Malaysia.
Materials and methods
We analysed 100 unrelated HA and 15 unrelated HB patients for genetic alterations in the F8 and F9 genes by using the long-range PCR, DNA sequencing, and the multiplex-ligation-dependent probe amplification assays. The prediction software was used to confirm the effects of these mutations on factor VIII and IX proteins.
Results
44 (53%) of the severe HA patients were positive for F8 intron 22 inversion, and three (3.6%) were positive for intron one inversion. There were 22 novel mutations in F8, including missense (8), frameshift (9), splice site (3), large deletion (1) and nonsense (1) mutations. In HB patients, four novel mutations were identified including the splice site (1), small deletion (1), large deletion (1) and missense (1) mutation.
Discussion
The mutational spectrum of F8 in Malaysian patients is heterogeneous, with a slightly higher frequency of intron 22 inversion in these severe HA patients when compared to other Asian populations. Identification of these mutational profiles in F8 and F9 genes among Malaysian patients will provide a useful reference for the early detection and diagnosis of HA and HB in the Malaysian population.
Keywords: Factor VIII, Factor IX, Genetic mutation, Haemophilia A, Haemophilia B
Introduction
Haemophilia is an inherited X-linked blood disorder which causes prolonged bleeding time after injuries or trauma.1 Haemophilia A (HA) and Haemophilia B (HB) are due to the deficiency of coagulation factor VIII (gene, F8) and factor IX (gene, F9) respectively.2 Upon activation, factor VIII and factor IX form an active complex (tenase complex) which activates factor X and the following factors in the coagulation pathway.1 Thus, a deficiency or dysfunction of any of these factors can impair clot formation and consequently causes bleeding diathesis.
In HA cases, the most recurrent genetic mutations are the inversion of intron 22 (IVS22), which accounts for about 45% of severe patients,3 and intron 1 (IVS1) which accounts for 2–5% of severe patients.4 As for HB patients, the most common mutations identified in the F9 gene are the missense mutations (74%).5 Apart from these mutations, there is a wide range of different genetic alterations spread throughout F8 and F9 genes, including single nucleotide substitutions, small and large deletions.5–7 Until now, about 1968 unique variants of F8 are listed in the factor VIII database,6,7 and 1094 unique variants of F9 in the factor IX database.5
The current standard of treatment of HA and HB is primary prophylaxis, with regular infusion of factor VIII or factor IX respectively to prevent joint bleeding and damage.8 However, the development of inhibitors in these patients is a severe complication of this infusion therapy.9 Such inhibitory response happens in 25–30% of HA patients10,11 and 1–4% of HB patients,12,13 and these incidences may be higher depending on the ethnicity. 14,15 A systematic review showed that factor VIII inhibitory response was strongly associated with large deletions and nonsense mutations compared to IVS22 mutation,16 therefore suggesting a strong genetic predisposition, hence the importance of identifying such mutations before commencing the infusion treatment. In Malaysia, despite the prevalence of HA and HB are around 5.9/100000 males and 1.0/100000 males respectively,17 there have been only small studies on the mutational status of F8 (only in exon 14)18 and in F919,20 genes. Therefore, this study aimed to investigate comprehensively the mutational spectrums of F8 and F9 genes in a representative cohort of Malaysian patients corresponding to their disease severity as well as the inhibitory response.
Materials and Methods
Sample collection
This study was approved by the Universiti Kebangsaan Malaysia Ethics Committee and the ethics committee of the Ministry of Health of Malaysia. Written informed consent taken from all patients with confirmed non-familial HA (n=100) and HB (n=15) who were being followed-up at the National Blood Centre, Kuala Lumpur. Detailed clinical history along with pedigree data were taken, and the disease severity classification was as the following: 1) mild HA (FVIII/FIX:C:>5–40%), 2) moderate HA (FVIII/FIX:C:1–5%), and 3) severe HA (FVIII/FIX:C:<1%).21 Venous blood (10mL) was collected in EDTA Vacutainer collection tubes (BD, New Jersey, USA) and proceeded to DNA extraction using the salting-out extraction method.22 DNA quality and concentration were determined by using NanoDrop Spectrophotometry and gel electrophoresis according to the manufacturer’s instruction.
Detection of intron 22 inversion (IVS22) in F8
Detection of IVS22 in F8 performed by using the Long-Range PCR kit (QIAGEN, Hilden, USA) according to previously published methods23,24 with slight modifications. Primers P, Q, A & B (Life Technologies, Wien, Austria) were utilised to amplify the region of interests.23,24 Briefly, a total of 25 μl PCR reaction which contained 20ng genomic DNA, LD-PCR reaction master-mix (Qiagen LD-PCR kit, Hilden, USA), 7.5% DMSO, 10 mM of 7-deaza-dGTP and 10 pmol of primers P, Q, A & B in each single-tube PCR reaction. Conditions for the PCR reaction: initial denaturation at 95 °C for 2 min and 15 s; followed by 30 cycles of denaturation at 95 °C for 12 s, annealing at 65 °C for 30 s and elongation at 68 °C for 12 min for the first ten cycles. The remaining 20 cycles had 20 s addition to each cycle step. Confirmation of the PCR products was visualised using agarose gel electrophoresis.
Detection of intron 1 inversion (IVS1) in F8
Detection of IVS1 in F8 done by using the PCR Core kit (Roche Diagnostics, Indiana, USA) according to the previously published methods and primers.4 The PCR products were visualised using agarose gel electrophoresis.
Detections of other mutations in F8 and F9
F8 coding regions (26 exons) including the intron/exon boundaries and the promoter regions were amplified using 26 sets of previously published primers and methods.25 The entire F9 coding region (8 exons) including the intron/exon boundaries, the promoter region, and the polyadenylation site was amplified using eight sets of previously published primers and methods.26,27 The confirmation of the PCR products was performed using gel electrophoresis and sequenced using the BigDye Terminator v3.1 sequencing kit (Applied Biosystems, California, USA) on an ABI 3130xl Genetic Analyzer (Applied Biosystems, California, USA) according to the manufacturer’s instructions.
Detection of large deletions in F8 and F9
Samples that did not show any exon amplification (but flanking exon amplification) or did not show any mutations were suspected of having large deletions. Detection of large deleted regions in F8 and F9 was performed using the multiplex-ligation-independent probe amplification (MLPA) kits, namely the SALSA MLPA P178 F8 and SALSA MLPA P207-C1 F9 probe mix kits (MRC-Holland, Amsterdam, Netherlands), according to the manufacturer’s instructions. Amplified PCR products were separated by ABI 3130xl Genetic Analyzer (Applied Biosystems, California, USA) with LIZ-500 (Applied Biosystems, California, USA) as the size standard. The data was analysed using Coffalyser.Net (MRC-Holland, Amsterdam, Netherlands) according to the provided guidelines. Probe ratios <0.7 were considered as deletions and probe ratios >1.3 as duplications. Negative controls were the donor DNA samples from healthy males.
Molecular genetic analysis and nomenclature
F8 and F9 nucleotide numbering (c.) is designated according to coding bases from A (nucleotide+1) from the initiation codon for methionine (ATG) at position −171 (F8:ref. NM_000132.3) and (ATG) at position −29 (F9:ref. NM_000133.3) respectively. While the protein numbering (p.) follows the amino acid sequences that assign the first residue methionine as +1 in each factor VIII and IX sequences (FVIII: NP_000123.1 and FIX: NP_000124.1 respectively) according to the Human Genome Variation Society guidelines. 28 Sequence variants were aligned with the corresponding wild-type sequences using BLAST (NCBI) and compared to the HA and HB mutation databases (Factor 8,6,7 Factor 9,5 Human Gene Mutation Database and CDC Haemophilia A Mutation Project database29). Novel variants further analysed for their effects on the factor VIII and IX protein by using multiple software, including Sorting Intolerant From Tolerant (SIFT) and Polymorphism Phenotyping (PolyPhen2),30,31 PROVEAN (Protein Variation Effect Analyzer)32 and Mutation Taster2.33 A SIFT score ranges from deleterious (< or equal to 0.05) to tolerated SNP (> 0.05). For PolyPhen2, the score ranges from 0.0 (tolerated) to 1.0 (deleterious). The PROVEAN score of an equal to or less than a predefined threshold of −2.5 value indicates for a “deleterious” effect. The visualisation of affected amino acid was performed on a crystal structure of the protein from the Protein Data Bank database for factor VIII protein (PDB-ID:2R7E)34 and IX protein (PDB-ID:2WPI)35 using Pymol, version 1.8.6.1 that is freely available online.
Results
Demographic and clinical data
Among the 100 HA patients in the study, 83 were severe (FVIII:C:<1%), nine were moderate (FVIII:C:1–5%), and eight were mild (FVIII:C:>5–40%) whereas, for the 15 HB patients, nine were severe (FIX:C:<1%), and six were moderate (FIX:C:1–5%) (Table 1). Fourteen of the severe HA patients developed inhibitors against factor VIII, while none of the HB patients had factor IX inhibitor. Majority of the patients were Malays (HA:54%, HB:73%), and followed by Chinese (HA:37%, HB:13%), Indians (HA:8%, HB:13%) and other (HA:1%) (Table 1).
Table 1.
Demographic data on Haemophilia A (HA) and Haemophilia B (HB) non-familial patients in a representative cohort of the Malaysian population.
Descriptive summary of the HA and HB non-familial patients included in the study. Data were expressed as patient count (n) and frequency from total patients (%).
| Description | HA, n (%) | HB, n (%) |
|---|---|---|
| Total Patients | 100 | 15 |
| Ethnicity | ||
| Malay | 54 (54) | 11 (73) |
| Chinese | 37 (37) | 2 (13) |
| Indian | 8 (8) | 2 (13) |
| Others | 1 (1) | 0 (0) |
| Disease Severity | ||
| Severe (C :< 1%) | 83 (83) | 9 (60) |
| Moderate (C: 1–5%) | 9 (9) | 6 (40) |
| Mild (C :> 5–40%) | 8 | (8) 0 (0) |
| Inhibitory Response | ||
| Yes | 14 (14) | 0 (0) |
| No | 86 (86) | 15 (100) |
F8 mutations
Out of 83 severe HA patients, 44 (53%) of them were positive for intron 22 inversion (IVS22), and three (3.6%) were positive for intron 1 inversion (IVS1). Among those 44 IVS22 positive patients, 18 of them had sporadic occurrence while 26 were familial based on the family history. For those remaining HA patients without IVS22/1 mutations, a total of 22 novel mutations were identified (Table 2) consisting of missense (8), frameshift (9), splice site (3), large deletion (1) and nonsense (1) mutations. Additionally, 21 of previously reported F8 mutations were also detected (Table 2) consisting large deletions (2), missense (10), nonsense (2), splice site (4) and frameshift (3) mutations. Excluding the IVS22 and IVS1 mutations, 41.7% of these identified mutations mainly occurred at the exon 14 of F8. In three severe HA patients, three large deletions were detected. Patient HA93 has a novel deletion which spans from exon 8 to exon 12 corresponding to A1-a1-A2 domains of factor VIII. Whereas, patient HA1 and HA41 have large deletions that span from exon 4 to exon 6 (corresponding to the A1 domain of factor VIII) and span from exon 8 to exon 9 (corresponding to A1-a1-A2 domains of factor VIII) respectively (Table 2). Unfortunately, we were unable to detect any mutation in four of the severe HA patients (HA4, HA15, HA60, and HA64).
Table 2.
The F8 mutational spectrum in Malaysian Haemophilia A (HA) patients without intron 22 and 1 inversions. The summary of the genetic alterations in the F8 gene in HA patients that were negative for intron 22 and intron 1 inversions. Nucleotide numbering (c.) is according to coding bases from A (nucleotide +1) the initiation methionine (ATG) at position −171 (F8 mRNA gene bank ref. NM_000132.3) and protein numbering (p.) follows amino acid sequences that assign the first residue Methionine as +1 in factor VIII protein sequence (NP_000123.1) according to Human Genome Variation Society guidelines.28
| Patients | Ethnicity | F8 Exon | F8 domain | Nucleotide Changes | Amino Acid Changes | Novelty | Disease Severity | Mutation Effects |
|---|---|---|---|---|---|---|---|---|
| HA5/HA36 | Malay/Malay | 14 | B | c.3610InsA | p.N1204Kfs*2 | Novel | Severe | Frameshift |
| HA9 | Malay | 4 | A1 | c.553DelAA | p.K185Rfs*14 | Novel | Severe | Frameshift |
| HA11 | Malay | Exon7/Intron7 | A1 | c.1007A>C c.1008-1009 DelTG/GT |
- | Novel | Severe | Donor splice site |
| HA17 | Chinese | 14 | B | c.49464947 DelGT | p.G1649Efs*3 | Novel | Severe | Frameshift |
| HA18 | Malay | 9 | A2 | c.1354G>T | p.D452Y | Novel | Mild | Missense |
| HA24 | Other | 14 | B | c.3625C>T | p.Q1209* | Novel | Severe | Nonsense |
| HA25 | Chinese | 8 | A1 | c.1016T>G | p.M339R | Novel | Severe | Missense |
| HA45# | Malay | 14 | B | c.3175DelA | p.V1060*fs | Novel | Severe | Frameshift |
| HA52 | Malay | 14 | B | c.4820InsA | p.T1609Nfs*3 | Novel | Severe | Frameshift |
| HA54 | Indian | Intron6 | - | c.787+1G>T | - | Novel | Severe | Donor splice site |
| HA57 | Indian | 14 | B | c.2444DelAG | p.P817Yfs*9 | Novel | Severe | Frameshift |
| HA61 | Malay | 14 | B | c.2696DelG | p.S899Ifs*6 | Novel | Moderate | Frameshift |
| HA62 | Malay | 14 | B | c.3762DelT | p.N1254Kfs*2 | Novel | Severe | Frameshift |
| HA67 | Malay | 26 | C2 | c.6986C>G | p.P2329R | Novel | Severe | Missense |
| HA70 | Chinese | 19 | A3 | c.6085A>T | p.M2029L | Novel | Mild | Missense |
| HA76 | Malay | 14 | A2 | c.2159G>T | p.G720V | Novel | Severe | Missense |
| HA77 | Malay | 17 | A3 | c.5609T>C | p.L1870P | Novel | Moderate | Missense |
| HA80 | Malay | 21 | C1 | c.6272A>C | p.K2091T | Novel | Mild | Missense |
| HA86 | Chinese | 25 | C2 | c.6857A>T | p.D2286V | Novel | Severe | Missense |
| HA87 | Chinese | 23 | C1 | c.6355DelC | p.Q2119Sfs*24 | Novel | Severe | Frameshift |
| HA91 | Malay | Intron22 | - | c.6429+2T>A | - | Novel | Moderate | +2 Donor splice site |
| HA93 | Malay | 8 – 12 | A1-a1-A2 | - | - | Novel | Severe | Large deletion |
| HA1 | Malay | 4–6 | A1 | - | - | Reported | Severe | Large deletion |
| HA2 | Chinese | 11 | A2 | c.1696C>T | p.L566F | Reported | Moderate | Missense |
| HA6/HA23/HA27/HA40 HA8/HA44/ | Chinese/Malay/Malay/Chinese | 14 | B | c.3629InsA | p.I1213Nfs*28 | Reported | Severe | Frameshift |
| HA50 | Malay/Malay/Malay | Intron18 | - | c.5998-1G>A | - | Reported | Severe | −1 Acceptor splice site |
| HA12 | Malay | 4 | A1 | c.524A>C | p.Y175S | Reported | Mild | Missense |
| HA26 | Chinese | 14 | B | c.4156C>T | p.Q1386* | Reported | Severe | Nonsense |
| HA32 | Chinese | 3 | A1 | c.274 G>C | p.G92R | Reported | Severe | Missense |
| HA33 | Malay | 22 | C1 | c.6317A>C | p.Q2106P | Reported | Severe | Missense |
| HA38/HA65 | Malay/Malay | 24 | C2 | c.6682C>T | p.R2228* | Reported | Severe | Nonsense |
| HA41 | Malay | 8–9 | A1-a1-A2 | - | - | Reported | Severe | Large deletion |
| HA42 | Chinese | 3 | A1 | c.274G>A | p.G92S | Reported | Mild | Missense |
| HA45# | Malay | 14 | B | c.2383A>G | p.R795G | Reported | Severe | Missense |
| HA48 | Chinese | Intron6 | - | c.787+1G>A | - | Reported | Severe | Donor splice site |
| HA55 | Malay | 18 | A3 | c.5941G>A | p.V1981M | Reported | Mild | Missense |
| HA58 | Chinese | 8 | a1 | c.1171C>T | p.R391C | Reported | Moderate | Missense |
| HA66 | Malay | 9 | A2 | c.1443G>A | p.L481L | Reported | Mild | Splice site end of Exon 9 |
| HA68 | Indian | Intron16 | - | c.5586+2T>G | - | Reported | Severe | Donor splice site |
| HA71 | Chinese | 18 | A3 | c.5879G>A | p.R1960Q | Reported | Mild | Missense |
| HA73 | Malay | 12 | A2 | c.1812G>C | p.W604C | Reported | Severe | Missense |
| HA78 | Malay | 14 | B | c.3637DelA | p.I1213Ffs*5 | Reported | Severe | Frameshift |
| HA85 | Malay | 17 | A3 | c.5689-5690 DelCT | p.L1897Vfs*6 | Reported | Severe | Frameshift |
HA, Haemophilia A,
indicates the same sample having double mutations.
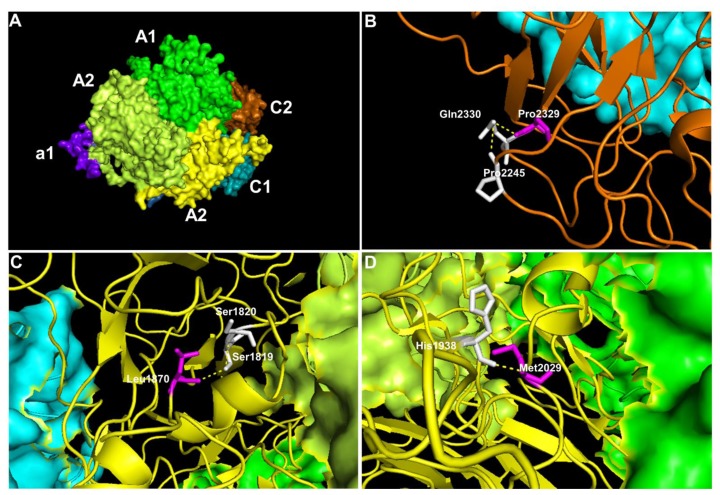
To further evaluate the impact of the novel missense mutations identified, we performed prediction analysis on the effect of these mutations on the factor VIII protein using multiple prediction software (Table 3). Except for one missense mutation in a mild HA patient, all other novel missense mutations were predicted to have damaging effects (Table 3). We also visualised the amino acid location of these novel missense mutations in factor VIII (Figure 1). For example, the HA67 patient who has a severe disease was detected to have a missense mutation of c.6986C>G that results in the substitution of proline to arginine at position 2329, and this mutation was predicted to be damaging. In the wildtype position, the large cyclic hydrophobic residue of Pro2329 lies within the C2 domain and forms hydrogen bonds with the neighbouring polar residue of glutamine (Figure 1B). Substitution of this hydrophobic proline to the positively charged arginine would disrupt this hydrogen bond. Patient HA77 who has a moderate disease was detected to have a missense mutation of c.5609T>C, which was also predicted to be damaging. For this mutation, the wildtype residue of Leu1870 forms a hydrogen bond with the polar Ser1819 residue through its hydroxyl group in the A3 domain (Figure 1C). Substitution of Leu1870 to cyclic proline would alter the hydroxyl group interaction therefore consistent with the predicted damaging score. Patient HA70 who has a mild disease also has a non-damaging missense mutation of c.6085A>T. For this mutation, the wildtype residue of Met2029 forms a hydrogen bond with the positively charged histidine residue in the A3 domain near to the core (Figure 1D). Substitution of this methionine to leucine that is similar in structure and polarity would minimally disrupt this bond, and thus consistent with the predicted score.
Table 3.
Summary of prediction values for eight novel missense mutations identified in F8 gene. Prediction of the novel missense mutation effect was performed on factor VIII protein and biological function using multiple software. SIFT score ≤ 0.05 indicates the damaging/deleterious effect and SIFT score above than 0.05 indicates a tolerated effect. A PolyPhen2 score ranges from 0.0 (tolerated) to 1.0 (deleterious), while for PROVEAN score that is equal to or less than a predefined threshold of −2.5, the variant is predicted to have a “deleterious” effect.
| Patients | Nucleotide Changes | Amino Acid Changes | Disease Severity | SIFT Score | PolyPhen2 Score | PROVEAN Score | Missense Prediction |
|---|---|---|---|---|---|---|---|
| HA18 | c.1354 G > T | D452Y | Mild | <0.05 | 1.00 | −7.71 | Damaging |
| HA25 | c.1016 T > G | M339R | Severe | <0.05 | 1.00 | −5.35 | Damaging |
| HA67 | c.6986 C > G | P2329R | Severe | <0.05 | 1.00 | −7.85 | Damaging |
| HA70 | c.6085 A > T | M2029L | Mild | 0.05 | 0.106 | −2.47 | Neutral/Benign |
| HA76 | c.2159 G > T | G720V | Severe | <0.05 | 1.00 | −7.26 | Damaging |
| HA77 | c.5609 T > C | L1870P | Moderate | <0.05 | 1.00 | −6.32 | Damaging |
| HA80 | c.6272 A > C | K2091T | Mild | 0.01 | 1.00 | −2.92 | Damaging |
| HA86 | c.6857 A > T | D2286V | Severe | <0.05 | 0.999 | −6.23 | Damaging |
HA, Haemophilia A.
Figure 1.
The representative model of factor VIII protein showing the affected amino acids by missense mutations. The visualisation of affected amino acids by missense mutations based on factor VIII protein (PDB:2R7E). A) The localisation of the domains in the factor VIII, B) Position of Pro2329 in the C2 domain C) Leu1870 in the A3 domain and D) Met2029 in the A3 domain. The visualisation of the whole structure of factor VIII as a surface model with colour coding that represents A1 domain (green), a1 domain (purple), A2 domain (lime), A3 domain (yellow), C1 domain (cyan) and C2 domain (orange). Except when the domain is affected, the region is visualised as a ribbon model. In this ribbon model, the affected residue (magenta), the neighbouring amino acids (white), and the hydrogen bonds (yellow dotted lines) are highlighted.
F9 mutations
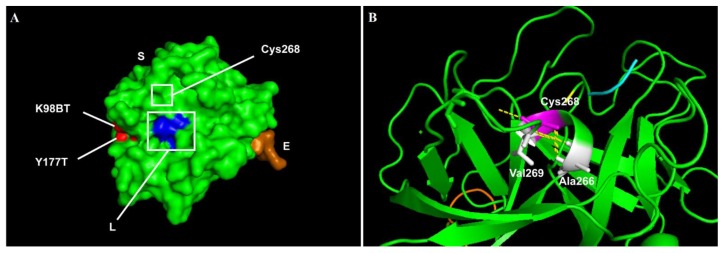
From the 15 HB patients, there were four novel mutations identified (Table 4), namely the splice site mutation (1), small deletion (1) and large deletion (1) and missense (1) mutations. A novel large deletion spans from exon 1 to exon 4, corresponding to signal-propeptide-GLA-EGF1 domains of factor IX, was detected in patient HB13 who has a severe disease. Additionally, there were ten previously reported mutations in the F9 identified among the patients (Table 4) consists of the splice site (2), missense (5), nonsense (2) and frameshift (1) mutations. A novel missense mutation (c.803G>A) in the patient HB4 was predicted to be damaging (SIFT score= <0.05, PolyPhen2 score = 1.00, PROVEAN score= −10.52). This missense mutation (c.803G>A) resulted in the substitution of cysteine to tyrosine residue at position 268. In this serine-peptidase catalytic domain, a wildtype residue of Cys268 forms strong disulphide hydrogen bonds with small hydrophobic Ala266 residue, contributing to the helical structure and folding of the factor IX (Figure 2B). Substitution of this Cys268 residue with tyrosine would affect the structure of the protein.
Table 4.
The F9 mutational spectrum in Malaysian Haemophilia B (HB) patients. The summary of the genetic alterations in the F9 gene from our Malaysian HB patients. Nucleotide numbering (c.) is according to coding bases from A (nucleotide +1) the initiation methionine (ATG) at position −29 (F9 mRNA gene bank ref. NM_000133.3) and protein numbering (p.) follows amino acid sequences that assign the first residue Methionine as +1 in factor IX protein sequence (NP_000124.1) according to Human Genome Variation Society guidelines.28
| Patient | Ethnicity | F9 Exon | F9 domain | Nucleotide Changes | Amino Acid Changes | Novelty | Disease Severity | Mutation Effects |
|---|---|---|---|---|---|---|---|---|
| HB2 | Malay | Intron2 | - | c.253-17_253-13DelTCTTT | - | Novel | Severe | Acceptor splice site |
| HB4 | Malay | 7 | Serine Protease | c.803G>A | p.C268Y | Novel | Severe | Missense |
| HB12 | Indian | 1 | Signal Peptide | c.39DelC | p.L14Sfs*7 | Novel | Moderate | Frameshift |
| HB13 | Malay | 1–4 | Signal-Pro-peptide-GLA-EGF1 | - | - | Novel | Severe | Large deletion |
| HB1 | Indian | Intron2 | - | c.252+1G>A | - | Reported | Severe | Donor splice site |
| HB3 | Malay | 8 | Serine Protease | c.1237G>A | p.G413R | Reported | Severe | Missense |
| HB5 | Malay | 8 | Serine Protease | c.1135C>T | p.R379* | Reported | Moderate | Nonsense |
| HB7/HB14 | Malay/ Malay | 2 | Pro-peptide | c.128G>A | p.R43Q | Reported | Moderate/ Severe | Missense |
| HB8 | Malay | Intron1 | - | c.88+5G>C | - | Reported | Moderate | Donor splice site |
| HB9 | Malay | 7 | Serine Protease | c.800A>G | p.H267R | Reported | Severe | Missense |
| HB10 | Malay | 5 | EGF2 | c.415G>A | p.G139S | Reported | Severe | Missense |
| HB11 | Chinese | 4 | EGF1 | c.383G>A | p.C128Y | Reported | Moderate | Missense |
| HB15 | Malay | 2 | GLA | c.223C>T | p.R75* | Reported | Severe | Nonsense |
| HB16 | Chinese | 2 | GLA | c.159_160DelAG | p.E54Vfs*7 | Reported | Moderate | Frameshift |
HB, Haemophilia B.
Figure 2.
The representative model of factor IX protein showing the affected amino acids by missense mutations. The visualisation of affected amino acids by missense mutations based on factor IX protein (PDB:2WPL) of double-mutant. A) Visualisation of the selected structure of factor IX protein as a surface model with colour-coding that represents S chain, containing Peptidase S1 domain (green), E chain containing EGF2 domain (orange) and L chain domain (blue). Original mutants’ residues from the crystal structure (2WPL) are in red. B) The affected domain of Peptidase S1 is visualised as a ribbon model, with the affected residue (Cys268, magenta), the neighbouring amino acids (white), and the hydrogen bonds (yellow dotted lines) are highlighted.
Genotype-Phenotype relationship in HA and HB patients
From those 44 severe HA patients with the IVS22 mutation, five of them developed inhibitors against factor VIII, while two of the three severe HA patients with IVS1 mutation also had factor VIII inhibitors (Table 5). Among those remaining patients negative for IVS22/1 mutations, seven patients had inhibitory presence with different types of mutations, namely patient HA38 (nonsense mutation), patient HA73 (missense mutation) and patient HA4 (undetected mutation), patients HA85 and HA87 (small deletions) and patients, HA41 and HA93 (large deletions). In contrast, none of the HB patients exhibited any inhibitory response against factor IX.
Table 5.
Factor VIII inhibitory response distribution across the mutational spectrum of F8 in a representative cohort of Malaysians. Factor VIII inhibitory response distribution across the mutation types in Haemophilia A patients (n=14) within the F8 gene. Data were expressed as count (n) and frequency (%). Numbers of mutations are not equal to the number of patients, due to some patients share the same mutations. IVS, Intron Inversion.
| Mutation type | No. of mutations n (%) |
No. of patients with inhibitors, n (% of total mutations of same type) | |||
|---|---|---|---|---|---|
|
| |||||
| Malay, n (%) | Chinese, n (%) | Indian, n (%) | Other, n (%) | ||
| IVS22 | 44 (44) | 2 (4.54) | 2 (4.54) | 1 (2.27) | - |
| IVS1 | 3 (3) | 1 (33.3) | - | 1 (33.3) | - |
| Missense | 18 (18) | 1 (5.56) | - | - | - |
| Nonsense | 3 (3) | 1 (33.3) | - | - | - |
| Large deletion | 3 (3) | 2 (66.6) | - | - | - |
| Small deletion | 9 (9) | 1 (11.1) | 1 (11.1) | - | - |
| Small Insertion | 3 (3) | - | - | - | - |
| Splice donor site | 7 (5) | - | - | - | - |
| No mutation detected | 4 (4) | 1 (25.0) | - | - | - |
Discussion
This study is the first to report a comprehensive mutational spectrum of F8 in 100 HA patients from Malaysia, together with the mutational profile of F9 in 15 HB patients. Among the 83 severe and non-familial HA patients, 53% of them have IVS22 mutation which is slightly higher than other Asian populations.36–40 We also identified 22 novel mutations in F8 and four novel mutations in F9. Among those with novel mutations, we found one HA patient and one HB patient each with a novel large deletion in the F8 and F9 respectively, and both patients have severe disease. Unfortunately, we were unable to detect any mutation in F8 in four HA patients with severe disease. Similarly, previous studies also reported that in a small number of HA patients, no mutation was detected despite the use of multiple techniques.38,41,42 This is probably due to the location of the mutations which lie deep within the introns or outside the analysable region of F8 using the current techniques.43,44 Given the associations between F8 and F9 mutational status and the disease severity as well as the development of inhibitors following the treatment, our findings suggest that detection of the mutational spectrum of F8 and F9 can improve the disease management and treatment outcome in HA and HB patients in Malaysia.
Three of the severe HA patients have large deletions, including one novel deletion. Patient HA93 with the novel deletion affecting A1-a1-A2 domains of factor VIII showed a high inhibitor level at the age of 12 years. Likewise, patient HA41 with a large deletion of A1-a1-A2 domains also had a high inhibitor level since the age of 4 years. Large deletions and nonsense mutations identified in factor VIII are shown to associate with higher risk of inhibitory response,16,36,45–47 particularly at the A2 and C2 domains.48 This evidence is in agreement with our findings, as our patient HA1 who has a large deletion affecting only the A1 domain did not develop the inhibitory response. Thus, depending on which factor VIII domains that are being affected by the large deletions, a differential outcome in the inhibitory response may be observed. Therefore, further investigations are needed to evaluate the prognostic value of these large deletions in predicting the inhibitory response.
Excluding IVS22 and IVS1 mutations, 41.7% of the identified mutations in the present study mainly occurred in exon 14 that is corresponding to the B domain of factor VIII. B domain has no homology sequence to any other known genes and has been shown to participate in the intracellular processing and trafficking of factor VIII.49 The role of the B domain in the pro-coagulant activity is minimal, as this domain is cleaved off during the activation.50,51 Here, we reported that among our eight severe HA patients, seven novel nonsense/frameshift mutations identified were in exon 14. Previous studies have reported that only some of these nonsense/frameshift mutations were causative mutations,52 despite that they could result in premature termination or frameshift codon. Therefore, further works should be pursued to elucidate the pathogenic impact of these mutations on factor VIII activity and production. Due to the limitation of the budget, we did not perform a functional study to assess the effects of these novel exon 14 mutations. However, four of these identified mutations are near to the ‘hotspot’ region at codon position 1210–1213 that is associated with a severe phenotype.53 Further functional studies are needed to elucidate the mechanism of how these mutations can affect factor VIII activity.
Missense mutations may represent polymorphisms,54 thus may require further evaluation. In the present study, seven novel missense mutations in F8 were predicted to be damaging. The structural visualisations of the affected amino acids in factor VIII (Figure 1) were consistent with the prediction scores, therefore suggesting that these mutations are more likely to be disease-causing. For example, we identified that patient HA67 has the strongest prediction of disease-causing mutation (Pro2326Arg) in the C2 domain, and this was consistent with the severe disease phenotype exhibited by the patient. Any mutation that lies within the C2 domain is likely to be causative as the C1/2 domains are essential for the binding of factor VIII to the von Willebrand factor55 and membrane-binding motif of tenase complex.56,57
In comparison to previous studies, we found 21 recurrent mutations in F8. Among them, a nonsense mutation (c.6682C>T) was reported in various populations,6,7 including two cases from the Asian populations.58,59 Another recurrent missense mutation (c.1171C>T, Arg391Cys) was also reported before with differential disease outcomes,6,7 even though this mutation lies within the thrombin activation site.60–62 HA disease severity can vary upon which substitution of the amino acid at Arg391/Arg372 (mature protein) that can influence the rate of thrombin cleavage.63 Histidine residue substitution at Arg372 position resulted in lower activation and thrombin cleavage, though no effect on factor VIII pro-coagulation activity,63 thus consistent with the mild disease phenotype. Whereas, Arg372 substitutions to cysteine, leucine, and proline residues were reported in moderate to severe HA patients,6,7 due to impairments in thrombin cleavage and activity.64,65 Interestingly, our severe patient HA45 has double mutations in F8, namely one novel frameshift deletion (c.3175DelA) and a previously reported missense mutation (c.2383A>G) in one Taiwanese woman with a severe disease phenotype.66 As this recurrent missense mutation (c.2383A>G) lies within the B domain, therefore, it may not be a disease-causing mutation. However, a presence of frameshift deletion would affect the factor VIII synthesis and function.
We identified four novel mutations in F9, including in the severe patient HB13 who had a novel large deletion affecting the signal-pro-peptide-GLA-EGF1 domains of factor IX. This finding is consistent with the previous findings that 90% of the large deletions identified were in severe HB patients48,51,60 and associated with higher risk of inhibitor development. 67 One HB2 patient with severe disease has a novel small 5bp deletion (c.253-17_253-13delTCTTT) at the acceptor splice site in intron 2. This novel small deletion is similar to a previously reported 5bp deletion (c.253-18_253-14delTTCTT) in two Malaysian siblings with moderate disease.19 Despite a difference of a single nucleotide position, the two siblings19 and our patient exhibited differential disease outcomes, in which our 5bp deletion is more detrimental due to being nearer to the intron-exon boundary. As this novel 5bp deletion may interfere with the acceptor-binding site and causing an exon skipping event,68 therefore it could explain such differential disease phenotypes. We also found a novel frameshift mutation in the signal-peptide domain (patient HB12) in which this deletion of C nucleotide is consistent with previous findings that any mutation lies within the early pre-pro leader sequences of factor IX is detrimental.69 In our severe HB4 patient, the novel missense mutation (c.803G>A) is likely a disease-causing mutation as it lays within the serine-protease domain and is also predicted to disrupt the helix structure of factor IX (Figure 2B), consistent with the vital role of the serine-protease domain in factor IX activity.33
In comparison to previous studies of F9, we found ten previously reported mutations. Both nonsense mutations (c.1135C>T and c.223C>T) have been reported in various populations,5 including two Malaysian patients (c.1135C>T only).19 Similarly, a frameshift mutation (c.159_160DelAG), the missense mutations (c.415G>A and c.128G>A) and a splice site mutation c.252+1G>A were also reported before in Malaysian patients.19 As for the remaining recurrent missense mutations, these have been reported in non-Malaysian populations. The missense mutation of c.383G>A was reported before in German70 and Indian patients71 though, the latter had a severe disease phenotype. The recurrence splice site mutation, c.88+5G>A was reported in a Chinese patient with the same moderate disease.72 Two recurrence missense mutations (c.1237G>A and c.800A>G) in our patients are possibly the disease-causing missense mutations because they are within the serine-peptidase domain.73 Intriguingly, a missense mutation (c.800A>G) in our severe HB9 patient was reported before with differential disease phenotypes across two different populations namely, in a French patient with a moderate phenotype74 and an Indian patient with a severe phenotype. 75
Conclusions
This study is the first to comprehensively analyse the mutational spectrum of F8 in HA patients in Malaysia. The 53% prevalence of the IVS22 mutation in our severe HA patients is slightly higher than other Asian populations. A total of 22 and four novel mutations were identified in F8 and F9 respectively, thus suggesting a high heterogeneity of molecular changes in factor VIII and IX in our local patients. How these mutations can affect the disease severity and the inhibitor development, is worth exploring further to provide a better understanding of the genotype-phenotype association in our patients. These mutational profiles of our Malaysian HA and HB patients can provide a useful reference database in the detection of carrier status and the diagnosis of HA and HB in the Malaysian population.
Acknowledgements
The authors would like to acknowledge the Haematology Department of Singapore General Hospital for generously sharing their procedures of PCR amplification and direct sequencing.
Footnotes
Competing interests: The authors have declared that no competing interests exist.
References
- 1.Bowen DJ. Haemophilia A and haemophilia B: Molecular insights. Mol Pathol. 2002;55(2):127–44. doi: 10.1136/mp.55.2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Brasi C, El-Maarri O, Perry DJ, Oldenburg J, Pezeshkpoor B, Goodeve A. Genetic testing in bleeding disorders. Haemophilia. 2014;20(0 4):54–8. doi: 10.1111/hae.12409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lakich D, Kazazian HH, Antonarakis SE, Gitschier J. Inversions disrupting the factor VIII gene are a common cause of severe haemophilia A. Nat Genet. 1993;5(3):236–41. doi: 10.1038/ng1193-236. [DOI] [PubMed] [Google Scholar]
- 4.Bagnall RD, Waseem N, Green PM, Giannelli F. Recurrent inversion breaking intron 1 of the factor VIII gene is a frequent cause of severe hemophilia A. Blood. 2002;99(1):168–74. doi: 10.1182/blood.V99.1.168. doi: 10.1182/blood.V99.1.168. [DOI] [PubMed] [Google Scholar]
- 5.Rallapalli PM, Kemball-Cook G, Tuddenham EG, Gomez K, Perkins SJ. An interactive mutation database for human coagulation factor IX provides novel insights into the phenotypes and genetics of hemophilia B. J Thromb Haemost. 2013;11(7):1329–40. doi: 10.1111/jth.12276. doi: 10.1111/jth.12276. [DOI] [PubMed] [Google Scholar]
- 6.EAHAD Coagulation Factor Variant Databases. 2017. [cited 13th March 2017]. Available from: http://www.factorviii-db.org/index.php.
- 7.Factor 8 Variant Database. 2014. Available from: http://factorviii-db.org/
- 8.Franchini M. The modern treatment of haemophilia: a narrative review. Blood Transfusion. 2013;11(2):178–82. doi: 10.2450/2012.0166-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walsh CE, Soucie JM, Miller CH the United States Hemophilia Treatment Center N. Impact of inhibitors on hemophilia a mortality in the United States. Am J Hematol. 2015;90(5):400–5. doi: 10.1002/ajh.23957. doi: 10.1002/ajh.23957. [DOI] [PubMed] [Google Scholar]
- 10.Wight J, Paisley S. The epidemiology of inhibitors in haemophilia A: A systematic review. Haemophilia. 2003;9(4):418–35. doi: 10.1046/j.1365-2516.2003.00780.x. doi: 10.1046/j.1365-2516.2003.00780.x. [DOI] [PubMed] [Google Scholar]
- 11.Lacroix-Desmazes S, Scott DW, Goudemand J, van den Berg M, Makris M, van Velzen AS, Santagostino E, Lillicrap D, Rosendaal FR, Hilger A, Sauna ZE, Oldenburg J, Mantovani L, Mancuso ME, Kessler C, Hay CRM, Knoebl P, Di Minno G, Hoots K, Bok A, Brooker M, Buoso E, Mannucci PM, Peyvandi F. Summary report of the First International Conference on inhibitors in haemophilia A. Blood Transfusion. 2017;15(6):568–76. doi: 10.2450/2016.0252-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamiya T, Takahashi I, Saito H. Retrospective study of inhibitor formation in Japanese hemophiliacs. Int J Hematol. 1995;62(3):175–81. doi: 10.1016/0925-5710(95)00405-H. [DOI] [PubMed] [Google Scholar]
- 13.Ljung R. Gene mutations and inhibitor formation in patients with hemophilia B. Acta Haematol. 1995;94(Suppl 1):49–52. doi: 10.1159/000204029. [DOI] [PubMed] [Google Scholar]
- 14.Aledort LM, Dimichele DM. Inhibitors occur more frequently in African-American and Latino haemophiliacs. Haemophilia. 1998;4(1):68. doi: 10.1046/j.1365-2516.1998.0146c.x. [DOI] [PubMed] [Google Scholar]
- 15.Carpenter SL, Michael Soucie J, Sterner S, Presley R Hemophilia Treatment Center Network I. Increased prevalence of inhibitors in Hispanic patients with severe haemophilia A enrolled in the Universal Data Collection database. Haemophilia. 2012;18(3):e260–e5. doi: 10.1111/j.1365-2516.2011.02739.x. doi: 10.1111/j.1365-2516.2011.02739.x. [DOI] [PubMed] [Google Scholar]
- 16.Gouw SC, van den Berg HM, Oldenburg J, Astermark J, de Groot PG, Margaglione M, Thompson AR, van Heerde W, Boekhorst J, Miller CH, le Cessie S, van der Bom JG. F8 gene mutation type and inhibitor development in patients with severe hemophilia A: Systematic review and meta-analysis. Blood. 2012;119(12):2922–34. doi: 10.1182/blood-2011-09-379453. doi: 10.1182/blood-2011-09-379453. [DOI] [PubMed] [Google Scholar]
- 17.Malaysian Ministry of Health M. Health technology assessment report: Management of haemophilia. Kuala Lumpur, Malaysia: Ministry of Health; 2012. Contract No.: MOH/P/PAK/258.12(TR) [Google Scholar]
- 18.Moses EJ, Ling SP, Al-Hassan FM, Karim FA, Yusoff NM. Identification of novel mutations in exon 14 of the F8 gene in Malaysian patients with severe Hemophilia A. Indian J Clin Biochem. 2012;27(2):207–8. doi: 10.1007/s12291-011-0161-z. doi: 10.1007/s12291-011-0161-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balraj P, Ahmad M, Khoo AS, Ayob Y. Factor IX mutations in haemophilia B patients in Malaysia: a preliminary study. Malays J Pathol. 2012;34(1):67–9. [PubMed] [Google Scholar]
- 20.Ishak R, Zakaria Z. Detection of carrier status of hemophilia B using DNA markers. Southeast Asian J Trop Med Public Health. 1997;28(3):629–30. [PubMed] [Google Scholar]
- 21.White GC, Rosendaal F, Aledort LM, Lusher JM, Rothschild C, Ingerslev J on behalf of the Factor VIII and Factor IX Subcommittee. Definitions in Hemophilia. Recommendation of the scientific subcommittee on factor VIII and factor IX of the scientific and standardization committee of the International Society on thrombosis and haemostasis. Thromb Haemost. 2001;85(3):560. doi: 10.1055/s-0037-1615621. [DOI] [PubMed] [Google Scholar]
- 22.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Q, Nozari G, Sommer SS. Single-tube polymerase chain reaction for rapid diagnosis of the inversion hotspot of mutation in Hemophilia A. Blood. 1998;92(4):1458–9. [PubMed] [Google Scholar]
- 24.Liu Q, Sommer SS. Subcycling-PCR for multiplex long-distance amplification of regions with high and low GC content: Application to the inversion hotspot in the factor VIII gene. Biotechniques. 1998;25(6):1022–8. doi: 10.2144/98256rr01. [DOI] [PubMed] [Google Scholar]
- 25.Vidal F, Farssac E, Altisent C, Puig L, Gallardo D. Rapid Hemophilia A molecular diagnosis by a simple DNA sequencing procedure: Identification of 14 novel mutations. Thromb Haemost. 2001;85(4):580–3. doi: 10.1055/s-0037-1615637. [DOI] [PubMed] [Google Scholar]
- 26.Hinks JL, Winship PR, Makris M, Preston FE, Peake IR, Goodeve AC. A rapid method for haemophilia B mutation detection using conformation sensitive gel electrophoresis. Br J Haematol. 1999;104(4):915–8. doi: 10.1046/j.1365-2141.1999.01274.x. doi: 10.1046/j.1365-2141.1999.01274.x. [DOI] [PubMed] [Google Scholar]
- 27.Vidal F, Farssac E, Altisent C, Puig L, Gallardo D. Factor IX gene sequencing by a simple and sensitive 15-hour procedure for haemophilia B diagnosis: Identification of two novel mutations. Br J Haematol. 2000;111(2):549–51. doi: 10.1111/j.1365-2141.2000.02389.x. doi: 10.1111/j.1365-2141.2000.02389.x. [DOI] [PubMed] [Google Scholar]
- 28.den Dunnen JT, Dalgleish R, Maglott DR, Hart RK, Greenblatt MS, McGowan-Jordan J, Roux A-F, Smith T, Antonarakis SE, Taschner PEM on behalf of the Human Genome Variation Society, the Human Variome Project, and the Human Genome Organisation. HGVS Recommendations for the description of sequence variants: 2016 Update. Hum Mutat. 2016;37(6):564–9. doi: 10.1002/humu.22981. doi: 10.1002/humu.22981. [DOI] [PubMed] [Google Scholar]
- 29.Li T, Miller CH, Payne AB, Craig Hooper W. The CDC Hemophilia B mutation project mutation list: A new online resource. Mol Genet Genomic Med. 2013;1(4):238–45. doi: 10.1002/mgg3.30. doi: 10.1002/mgg3.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flanagan SE, Patch AM, Ellard S. Using SIFT and PolyPhen to predict loss-of-function and gain-of-function mutations. Genet Test Mol Biomarkers. 2010;14(4):533–7. doi: 10.1089/gtmb.2010.0036. doi: 10.1089/gtmb.2010.0036. [DOI] [PubMed] [Google Scholar]
- 31.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protocols. 2009;4(8):1073–81. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 32.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7(10):e46688. doi: 10.1371/journal.pone.0046688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: Mutation prediction for the deep-sequencing age. Nat Meth. 2014;11(4):361–2. doi: 10.1038/nmeth.2890. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 34.Shen BW, Spiegel PC, Chang C-H, Huh J-W, Lee J-S, Kim J, Kim Y-H, Stoddard BL. The tertiary structure and domain organization of coagulation factor VIII. Blood. 2008;111(3):1240–7. doi: 10.1182/blood-2007-08-109918. doi: 10.1182/blood-2007-08-109918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zögg T, Brandstetter H. Structural Basis of the Cofactor- and Substrate-Assisted Activation of Human Coagulation Factor IXa. Structure. 2009;17(12):1669–78. doi: 10.1016/j.str.2009.10.011. doi: 10.1016/j.str.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 36.Pinto P, Ghosh K, Shetty S. F8 gene mutation profile in Indian hemophilia A patients: Identification of 23 novel mutations and factor VIII inhibitor risk association. Mutat Res. 2016;786:27–33. doi: 10.1016/j.mrfmmm.2016.02.002. doi: 10.1016/j.mrfmmm.2016.02.002. https://doi.org/10.1016/j.mrfmmm.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 37.Xue F, Zhang L, Sui T, Ge J, Gu D, Du W, Zhao H, Yang R. Factor VIII gene mutations profile in 148 Chinese hemophilia A subjects. Eur J Haematol. 2010;85(3):264–72. doi: 10.1111/j.1600-0609.2010.01481.x. doi: 10.1111/j.1600-0609.2010.01481.x. [DOI] [PubMed] [Google Scholar]
- 38.Ahmed R, Ivaskevicius V, Kannan M, Seifried E, Oldenburg J, Saxena R. Identification of 32 novel mutations in the factor VIII gene in Indian patients with hemophilia A. Haematologica. 2005;90(2):283–4. [PubMed] [Google Scholar]
- 39.Shekari Khaniani M, Ebrahimi A, Daraei S, Derakhshan SM. Genotyping of Intron Inversions and Point Mutations in Exon 14 of the FVIII Gene in Iranian Azeri Turkish Families with Hemophilia A. Indian J Hematol Blood Transfus. 2016;32(4):475–80. doi: 10.1007/s12288-016-0699-2. doi: 10.1007/s12288-016-0699-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mousavi SH, Mesbah-Namin SA, Rezaie N, Zeinali S. Frequencies of intron 1 and 22 inversions of factor VIII gene: A first report in Afghan patients with severe haemophilia A. Haemophilia. 2018;0(0) doi: 10.1111/hae.13491. doi: 10.1111/hae.13491. [DOI] [PubMed] [Google Scholar]
- 41.Citron M, Godmilow L, Ganguly T, Ganguly A. High throughput mutation screening of the factor VIII gene (F8C) in hemophilia A: 37 novel mutations and genotype–phenotype correlation. Hum Mutat. 2002;20(4):267–74. doi: 10.1002/humu.10119. doi: 10.1002/humu.10119. [DOI] [PubMed] [Google Scholar]
- 42.Guo Z, Yang L, Qin X, Liu X, Zhang Y. Spectrum of Molecular Defects in 216 Chinese Families With Hemophilia A: Identification of Noninversion Mutation Hot Spots and 42 Novel Mutations. Clin Appl Thromb Hemost. 2017;24(1):70–8. doi: 10.1177/1076029616687848. doi: 10.1177/1076029616687848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lyu C, Xue F, Liu X, Liu W, Fu R, Sun T, Wu R, Zhang L, Li H, Zhang D, Yang R, Zhang L. Identification of mutations in the F8 and F9 gene in families with haemophilia using targeted high-throughput sequencing. Haemophilia. 2016;22(5):e427–e34. doi: 10.1111/hae.12924. doi: 10.1111/hae.12924. [DOI] [PubMed] [Google Scholar]
- 44.Pezeshkpoor B, Zimmer N, Marquardt N, Nanda I, Haaf T, Budde U, Oldenburg J, El-Maarri O. Deep intronic ‘mutations’ cause hemophilia A: Application of next generation sequencing in patients without detectable mutation in F8 cDNA. J Thromb Haemost. 2013;11(9):1679–87. doi: 10.1111/jth.12339. doi: 10.1111/jth.12339. [DOI] [PubMed] [Google Scholar]
- 45.Repessé Y, Slaoui M, Ferrandiz D, Gautier P, Costa C, Costa JM, Lavergne JM, Borel-Derlon A. Factor VIII (FVIII) gene mutations in 120 patients with hemophilia A: Detection of 26 novel mutations and correlation with FVIII inhibitor development. J Thromb Haemost. 2007;5(7):1469–76. doi: 10.1111/j.1538-7836.2007.02591.x. doi: 10.1111/j.1538-7836.2007.02591.x. [DOI] [PubMed] [Google Scholar]
- 46.Oldenburg J, El-Maarri O, Schwaab R. Inhibitor development in correlation to factor VIII genotypes. Haemophilia. 2002;8:23–9. doi: 10.1046/j.1351-8216.2001.00134.x. doi: 10.1046/j.1351-8216.2001.00134.x. [DOI] [PubMed] [Google Scholar]
- 47.Miller CH, Benson J, Ellingsen D, Driggers J, Payne A, Kelly FM, Soucie JM, Craig Hooper W The Hemophilia Inhibitor Research Study Investigators. F8 and F9 mutations in US haemophilia patients: Correlation with history of inhibitor and race/ethnicity. Haemophilia. 2012;18(3):375–82. doi: 10.1111/j.1365-2516.2011.02700.x. doi: 10.1111/j.1365-2516.2011.02700.x. [DOI] [PubMed] [Google Scholar]
- 48.Prescott R, Nakai H, Saenko EL, Scharrer I, Nilsson IM, Humphries JE, Hurst D, Bray G, Scandella D. the inhibitor antibody response is more complex in Hemophilia A patients than in most nonhemophiliacs with factor VIII autoantibodies. Blood. 1997;89(10):3663–71. [PubMed] [Google Scholar]
- 49.Pipe SW. Functional roles of the factor VIII B domain. Haemophilia. 2009;15(6):1187–96. doi: 10.1111/j.1365-2516.2009.02026.x. doi: 10.1111/j.1365-2516.2009.02026.x. [DOI] [PubMed] [Google Scholar]
- 50.Burke RL, Pachl C, Quiroga M, Rosenberg S, Haigwood N, Nordfang O, Ezban M. The functional domains of coagulation factor VIII:C. J Biol Chem. 1986;261(27):12574–8. [PubMed] [Google Scholar]
- 51.Pittman D, Alderman E, Tomkinson K, Wang J, Giles A, Kaufman R. Biochemical, immunological, and in vivo functional characterization of B-domain-deleted factor VIII. Blood. 1993;81(11):2925–35. [PubMed] [Google Scholar]
- 52.Shelley N, Miao-Liang L, RTA Some factor VIII exon 14 frameshift mutations cause moderately severe haemophilia A. Br J Haematol. 2001;115(4):977–82. doi: 10.1046/j.1365-2141.2001.03173.x. doi: 10.1046/j.1365-2141.2001.03173.x. [DOI] [PubMed] [Google Scholar]
- 53.Oldenburg J, Ananyeva NM, Saenko EL. Molecular basis of haemophilia A. Haemophilia. 2004;10:133–9. doi: 10.1111/j.1365-2516.2004.01005.x. doi: 10.1111/j.1365-2516.2004.01005.x. [DOI] [PubMed] [Google Scholar]
- 54.Ogata K, Selvaraj SR, Miao HZ, Pipe SW. Most factor VIII B domain missense mutations are unlikely to be causative mutations for severe Hemophilia A: Implications for genotyping. J Thromb Haemost. 2011;9(6):1183–90. doi: 10.1111/j.1538-7836.2011.04268.x. doi: 10.1111/j.1538-7836.2011.04268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jacquemin M, Lavend’homme R, Benhida A, Vanzieleghem B, d’Oiron R, Lavergne J-M, Brackmann HH, Schwaab R, VandenDriessche T, Chuah MKL, Hoylaerts M, Gilles JGG, Peerlinck K, Vermylen J, Saint-Remy J-MR. A novel cause of mild/moderate hemophilia A: Mutations scattered in the factor VIII C1 domain reduce factor VIII binding to von Willebrand factor. Blood. 2000;96(3):958–65. [PubMed] [Google Scholar]
- 56.Liu Z, Lin L, Yuan C, Nicolaes GAF, Chen L, Meehan EJ, Furie B, Furie B, Huang M. Trp(2313)-His(2315) of factor VIII C2 domain is involved in membrane binding: Structure of a complex between the C2 domain and an inhibitor of membrane binding. J Biol Chem. 2010;285(12):8824–9. doi: 10.1074/jbc.M109.080168. doi: 10.1074/jbc.M109.080168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Foster P, Fulcher C, Houghten R, Zimmerman T. Synthetic factor VIII peptides with amino acid sequences contained within the C2 domain of factor VIII inhibit factor VIII binding to phosphatidylserine. Blood. 1990;75(10):1999–2004. [PubMed] [Google Scholar]
- 58.Liu J, Zhang Y, Wang H, Huang W, Cao W, Wang X, Qu B, Wang H, Shao H, Wang Z, Chen L, Huang W. [Molecular characterization of genetic defects in hemophilia in Shanghai]. Zhonghua Xue Ye Xue Za Zhi. 1997;18(9):464–7. [PubMed] [Google Scholar]
- 59.Fidanci ID, Kavakli K, Uçar C, Timur Ç, Meral A, Klnç Y, Saylan H, Kazanc E, Çaglayan SH. Factor 8 (F8) gene mutation profile of Turkish hemophilia A patients with inhibitors. Blood Coagul Fibrinolysis. 2008;19(5):383–8. doi: 10.1097/MBC.0b013e3282f9b193. doi: 10.1097/MBC.0b013e3282f9b193. [DOI] [PubMed] [Google Scholar]
- 60.Pipe SW, Eickhorst AN, McKinley SH, Saenko EL, Kaufman RJ. Mild Hemophilia A caused by increased rate of factor VIII A2 subunit dissociation: Evidence for nonproteolytic inactivation of factor VIIIa in vivo. Blood. 1999;93(1):176–83. [PubMed] [Google Scholar]
- 61.Celie PHN, Van Stempvoort G, Jorieux S, Mazurier C, Van Mourik JA, Mertens K. Substitution of Arg527 and Arg531 in factor VIII associated with mild haemophilia A: Characterization in terms of subunit interaction and cofactor function. Br J Haematol. 1999;106(3):792–800. doi: 10.1046/j.1365-2141.1999.01590.x. doi: 10.1046/j.1365-2141.1999.01590.x. [DOI] [PubMed] [Google Scholar]
- 62.Pieters J, Lindhout T, Hemker H. In situ-generated thrombin is the only enzyme that effectively activates factor VIII and factor V in thromboplastin-activated plasma. Blood. 1989;74(3):1021–4. [PubMed] [Google Scholar]
- 63.Nogami K, Zhou Q, Wakabayashi H, Fay PJ. Thrombin-catalyzed activation of factor VIII with His substituted for Arg372 at the P(1) site. Blood. 2005;105(11):4362–8. doi: 10.1182/blood-2004-10-3939. doi: 10.1182/blood-2004-10-3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shima M, Ware J, Yoshioka A, Fukui H, Fulcher C. An arginine to cysteine amino acid substitution at a critical thrombin cleavage site in a dysfunctional factor VIII molecule. Blood. 1989;74(5):1612–7. [PubMed] [Google Scholar]
- 65.Pittman DD, Kaufman RJ. Proteolytic requirements for thrombin activation of anti-hemophilic factor (factor VIII) Proc Natl Acad Sci U S A. 1988;85(8):2429–33. doi: 10.1073/pnas.85.8.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ma GC, Chang SP, Chen M, Kuo SJ, Chang CS, Shen MC. The spectrum of the factor 8 (F8) defects in Taiwanese patients with haemophilia A. Haemophilia. 2008;14(4):787–95. doi: 10.1111/j.1365-2516.2008.01687.x. doi: 10.1111/j.1365-2516.2008.01687.x. [DOI] [PubMed] [Google Scholar]
- 67.Radic CP, Rossetti LC, Abelleyro MM, Candela M, Bianco RP, Pinto MdT, Larripa IB, Goodeve A, De Brasi CD. Assessment of the F9 genotype-specific FIX inhibitor risks and characterization of 10 novel severe F9 defects in the first molecular series of Argentine patients with haemophilia B. Thromb Haemost. 2013;109(1):24–33. doi: 10.1160/TH12-05-0302. doi: 10.1160/TH12-05-0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baralle D, Baralle M. Splicing in action: Assessing disease causing sequence changes. J Med Genet. 2005;42(10):737–48. doi: 10.1136/jmg.2004.029538. doi: 10.1136/jmg.2004.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hamasaki-Katagiri N, Salari R, Simhadri VL, Tseng SC, Needlman E, Edwards NC, Sauna ZE, Grigoryan V, Komar AA, Przytycka TM, Kimchi-Sarfaty C. Analysis of F9 point mutations and their correlation to severity of haemophilia B disease. Haemophilia. 2012;18(6):933–40. doi: 10.1111/j.1365-2516.2012.02848.x. doi: 10.1111/j.1365-2516.2012.02848.x. [DOI] [PubMed] [Google Scholar]
- 70.Wulff K, Bykowska K, Lopaciuk S, Herrmann FH. Molecular analysis of hemophilia B in Poland: 12 novel mutations of the factor IX gene. Acta Biochim Pol. 1999;46(3):721–6. [PubMed] [Google Scholar]
- 71.Li X, Drost JB, Roberts S, Kasper C, Sommer SS. Factor IX mutations in South Africans and African Americans are compatible with primarily endogenous influences upon recent germline mutations. Hum Mutat. 2000;16(4):371. doi: 10.1002/1098-1004(200010)16:4<371::AID-HUMU11>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 72.Yu T, Dai J, Liu H, Ding Q, Lu Y, Wang H, Wang X, Fu Q. Spectrum of F9 mutations in Chinese haemophilia B patients: Identification of 20 novel mutations. Pathology. 2012;44(4):342–7. doi: 10.1097/PAT.0b013e328353443d. doi: 10.1097/PAT.0b013e328353443d. [DOI] [PubMed] [Google Scholar]
- 73.Di Scipio RG, Kurachi K, Davie EW. Activation of human factor IX (Christmas factor) J Clin Invest. 1978;61(6):1528–38. doi: 10.1172/JCI109073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Attali O, Vinciguerra C, Trzeciak MC, Durin A, Pernod G, Gay V, Ménart C, Sobas F, Dechavanne M, Négrier C. Factor IX gene analysis in 70 unrelated patients with Haemophilia B: Description of 13 new mutations. Thromb Haemost. 1999;82(5):1437–42. [PubMed] [Google Scholar]
- 75.Jayandharan GR, Shaji RV, Baidya S, Nair SC, Chandy M, Srivastava A. Molecular characterization of factor IX gene mutations in 53 patients with haemophilia B in India. Thromb Haemost. 2005;94(4):883–6. [PubMed] [Google Scholar]