Abstract
Anaplastic meningioma is a rare and aggressive brain tumor characterised by intractable recurrences and dismal outcomes. Here, we present an integrated analysis of the whole genome, transcriptome and methylation profiles of primary and recurrent anaplastic meningioma. A key finding was the delineation of distinct molecular subgroups that were associated with diametrically opposed survival outcomes. Relative to lower grade meningiomas, anaplastic tumors harbored frequent driver mutations in SWI/SNF complex genes, which were confined to the poor prognosis subgroup. Aggressive disease was further characterised by transcriptional evidence of increased PRC2 activity, stemness and epithelial-to-mesenchymal transition. Our analyses discern biologically distinct variants of anaplastic meningioma with prognostic and therapeutic significance.
Introduction
Meningiomas arise from arachnoidal cells of the meninges and are classified as grade I (80% of cases), grade II (10–20%) or grade III (1–3%). Grade III meningiomas comprise papillary, rhabdoid and anaplastic histological subtypes, with anaplastic tumors accounting for the vast majority of grade III diagnoses1,2. Nearly half of anaplastic meningiomas represent progression of a previously resected lower grade tumor, whereas the remainder arise de novo3,4. Recurrence rates are 5–20% and 20–40%, respectively, for grade I and II tumors2,5. By contrast, the majority of anaplastic meningioma patients suffer from inexorable recurrences with progressively diminishing benefit from repeated surgery and radiotherapy and 5-year overall survival of 30–60%4,6.
A recent study of 775 grade I and grade II meningiomas identified five molecular subgroups defined by driver mutation profile7. In keeping with previous smaller studies, mutually exclusive mutations in NF2 and TRAF7 were the most frequent driver events, followed by mutations affecting key mediators of PI3K and Hedgehog signalling7,8. Recurrent hotspot mutations were also identified in the catalytic unit of RNA polymerase II (POLR2A) in 6% of grade I tumors7. More recently, a study comparing benign versus de novo atypical (grade II) meningiomas found the latter to be significantly associated with NF2 and SMARCB1 mutations9. Atypical meningiomas were further defined by DNA and chromatin methylation patterns consistent with upregulated PRC2 activity, aberrant Homeobox domain methylation and transcriptional dysregulation of pathways involved in proliferation and differentiation9.
Despite the high mortality rate of anaplastic meningiomas, efforts to identify adjuvant treatment strategies have been hampered by a limited understanding of the distinctive molecular features of this aggressive subtype. A recent analysis of meningioma methylation profiles identified distinct subgroups within Grade III tumors predictive of survival outcomes, though the biology underpinning these differences and any therapeutic implications remain unknown10. Here, we present an analysis of the genomic, transcriptional and DNA methylation patterns defining anaplastic meningioma. Our results reveal molecular hallmarks of aggressive disease and suggest novel approaches to risk stratification and targeted therapy.
Results
Overview of the genomic landscape of primary and recurrent anaplastic meningioma
We performed whole genome sequencing (WGS) on a discovery set of 19 anaplastic meningiomas resected at first presentation (‘primary’). A subsequent validation cohort comprised 31 primary tumors characterised by targeted sequencing of 366 cancer genes. We integrated genomic findings with RNA sequencing and methylation array profiling in a subset of samples (Supplementary Table S1). Somatic copy number alterations and rearrangements were derived from whole genome sequencing reads, with RNA sequences providing corroborating evidence for gene fusions. Given the propensity of anaplastic meningioma to recur, we studied by whole genome sequencing 13 recurrences from 7 patients.
Excluding a hypermutated tumor (PD23359a, see Supplementary Discussion), the somatic point mutation burden of primary anaplastic meningioma was low with a median of 28 somatic coding mutations per tumor (range 11 to 71; mean sequencing coverage 66X) (Supplementary Fig. S1). Mutational signatures analysis of substitutions identified in whole genome sequences revealed the age-related, ubiquitous processes 1 and 5 as the predominant source of substitutions (Supplementary Fig. S2)11. The rearrangement landscape was also relatively quiet, with a median of 12 structural rearrangements (range 0–79) in the 18 primary tumor genomes (Supplementary Fig. S3, Table S3). Somatic retrotransposition events, a significant source of structural variants in over half of human cancers, were scarce (Supplementary Fig. S4, Table S4)12. Analysis of expressed gene fusions did not reveal any recurrent events involving putative cancer genes (Supplementary Table S5).
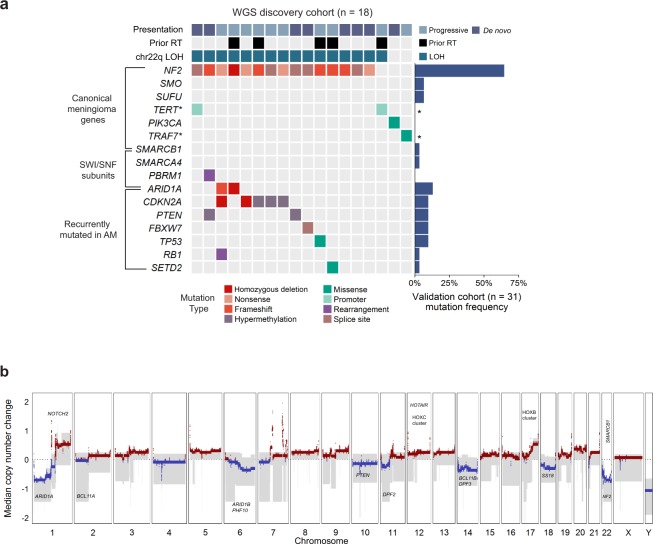
Recurrent large copy number changes were in keeping with known patterns in aggressive meningiomas, notably frequent deletions affecting chromosomes 1p, 6q, 14 and 22q (Fig. 1b, Supplementary Table S6)7,9,13.
Figure 1.
The landscape of driver mutations and copy number alterations in anaplastic meningioma. (a) The landscape of somatic driver variants in primary anaplastic meningioma. Somatic mutation and promoter methylation data is shown for a discovery cohort of 18 primary tumors characterised by whole genome sequencing. Mutations in recurrently altered genes, established meningioma genes and SWI/SNF complex subunits are included. Samples are annotated for chromosome 22q LOH, prior radiotherapy exposure, and clinical presentation (de novo verus progression from a lower grade meningioma). The bar plot to the right indicates mutation frequency in a validation cohort of 31 primary tumors sequenced with a 366 cancer gene panel. Asterisks indicate genes not included in the targeted sequencing assay. (b) Aggregate copy number profile of primary anaplastic meningioma. For the 18 tumors characterized by whole genome sequencing, the median relative copy number change was calculated across the genome in 10 kilobase segments, adjusting for ploidy. The grey shaded area indicates the first and third quantile of copy number for each genomic segment. The solid red and blue lines represent the median relative copy number gain and loss, respectively, with zero indicating no copy number change. X-axis: Chromosomal position. Y-axis: median relative copy number change. Potential target genes are noted. AM, anaplastic meningioma; LOH, loss of heterozygosity; RT, radiotherapy.
Driver genes do not delineate subgroups of anaplastic meningioma
Over 80% of low grade meningiomas segregate into 5 distinct subgroups based on driver mutation profile7,9. In anaplastic meningioma, however, we found a more uniform driver landscape dominated by deleterious mutations in NF2 (Fig. 1a). A key feature distinguishing anaplastic meningioma from its lower grade counterparts were driver events in genes of the SWI/SNF chromatin regulatory complex (Fig. 1a; Supplementary Fig. S7). The SWI/SNF (mSWI/SNF or BAF) complex is the most commonly mutated chromatin-regulatory complex in cancer14,15, and acts as a tumor suppressor in many cell types by antagonising the chromatin modifying PRC216–18. The most frequently mutated SWI/SNF component was ARID1A, which harbored at least one deleterious somatic change in 12% of our cohort of 50 primary tumors (Supplementary Table S1). ARID1A has not been implicated as a driver in grade I or grade II meningiomas7,9. Single variants in SMARCB1, SMARCA4 and PBRM1 were also detected in three tumors (Supplementary Fig. S7). In total, 16% of anaplastic meningiomas contained a damaging SWI/SNF gene mutation. By contrast, SWI/SNF genes are mutated in <5% of benign and atypical meningiomas7,9.
In the combined cohort of 50 primary tumors, we found at least one driver mutation in NF2 in 70%, similar to the prevalence reported in atypical meningiomas and more than twice that found in grade I tumors7,9. As observed in other cancer types, it is possible that non-mutational mechanisms may contribute to NF2 loss of function in a proportion of anaplastic meningiomas19,20. We considered promoter hypermethylation as a source of additional NF2 inactivation, but found no evidence of this (Supplementary Table S7). There was no significant difference in NF2 expression between NF2 mutant and wild-type tumors (p-value 0.960; Supplementary Fig. S8), suggesting that a truncated dysfunctional protein may be expressed.
Other driver genes commonly implicated in low grade tumors were not mutated, or very infrequently (Fig. 1a). Furthermore, and consistent with the most recent reports7,9, we did not observe an increased frequency of TERT promoter mutations, previously associated with progressive or high grade tumors21. Notably13, methylation analysis revealed CDKN2A and PTEN promoter hypermethylation in 17% and 11% of primary tumors, respectively (Fig. 1a). We did not find evidence of novel cancer genes in our cohort, applying established methods to search for enrichment of non-synonymous mutations22. The full driver landscape of anaplastic meningioma, considering point mutations, structural variants with resulting copy number changes and promoter hypermethylation is presented in Supplementary Fig. S7.
The genomic landscape of recurrent tumors was largely static both with respect to driver mutations and structural variation. Driver mutations differed between primary and recurrent tumors for only two of eleven patients with serial resections available. For seven sets of recurrent tumors studied by whole genome sequencing, only two demonstrated any discrepancies in large copy number variants (PD23344 and PD23346; Supplementary Fig. S5). Similarly, matched primary and recurrent samples clustered closely together by PCA of transcriptome data, suggesting minimal phenotypic evolution (Supplementary Fig. S6).
Differential gene expression defines anaplastic meningioma subgroups with prognostic and biological significance
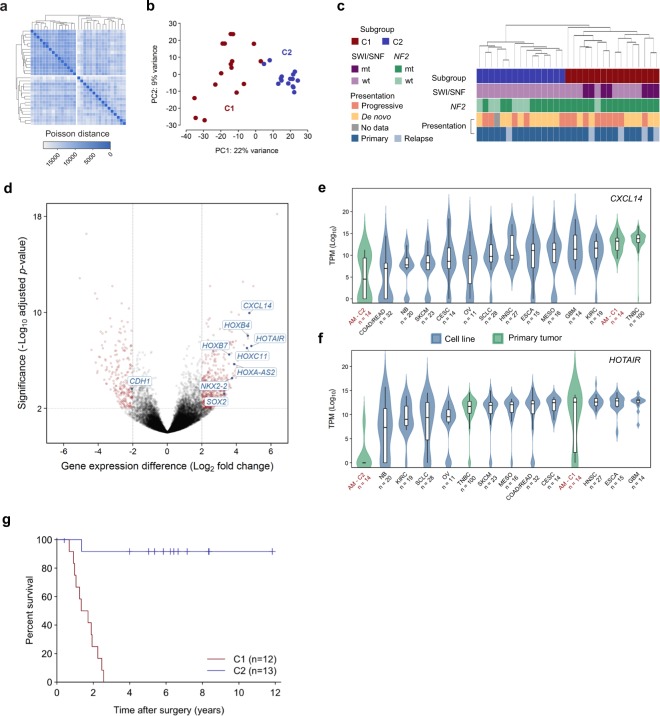
We performed messenger RNA (mRNA) sequencing of 31 anaplastic meningioma samples from a total of 28 patients (26 primary tumors and 5 recurrences). Gene expression variability within the cohort did not correlate with clinical parameters including prior radiotherapy, anatomical location or clinical presentation (de novo versus progressive tumor) (Supplementary Fig. S6). However, unsupervised hierarchical clustering demonstrated segregation of tumors into two main groups, hereafter referred to as C1 and C2 (Fig. 2a). These groups were recapitulated by principal component analysis (PCA) of normalised transcript counts (Fig. 2b), which delineated C1 as a well-demarcated cluster clearly defined by the first two principal components (PC). Of note, all SWI/SNF mutations were confined to the poor prognosis (C1) subgroup (Fig. 2c). C1 constituted a more diffuse group on PCA, distinguished from C2 mainly along the first principal component. We next retrospectively sought follow-up survival data from the time of first surgery, which was available for 25 of the 28 patients included in the transcriptome analysis (12 patients in C1, 13 in C2; mean follow-up of 1,403 days from surgery). We observed a significantly worse overall survival outcome in C1 compared to C2 (P < 0.0001; hazard ratio 17.0, 95% CI 5.2–56.0) (Fig. 2g; Supplementary Table S8). The subgroups were well balanced with respect to potential confounding features such as gender, age, radiotherapy, anatomical location and amount of residual tumor remaining after surgery (Supplementary Table S9).
Figure 2.
Transcriptomic classification of anaplastic meningioma. (a) Unsupervised hierarchical clustering and (b) principal component analysis of anaplastic meningioma gene expression revealed two subgroups (denoted C1 and C2). (c) Dendrogram obtained by unsupervised clustering annotated with clinical and genomic features. (d) Volcano plot depicting genes differentially expressed between C1 versus C2 anaplastic meningioma samples. X-axis, log2 fold change; y-axis, −log10 adjusted P-value. Genes with an adjusted P-value < 0.01 and absolute log2 fold change >2 are highlighted in red. (e,f) Box plots of (e) CXLC14 and (f) HOTAIR expression across 31 anaplastic meningomas classified into C1 and C2 subgroups, 100 primary breast tumors, and 219 cancer cell lines from 11 tumor types. Upper and lower box hinges correspond to first and third quartiles, horizontal line and whiskers indicate the median and 1.5-fold the interquartile range, respectively. Underlying violin plots show data distribution and are color-coded according to specimen source (blue, cell line; green, primary tumor). X-axis indicates tumor type and number of samples; y-axis shows log10 TPM values. (g) Kaplan-Meier curves showing overall survival for 25 anaplastic meningioma patients in C1 and C2 subgroups for whom follow-up data was available. Dashes indicate timepoints at which subjects were censored at time of last follow-up. TPM, transcripts per kilobase million; AM, anaplastic meningioma; TNBC, triple negative breast carcinoma; wt, wild-type; mt, mutated; PC, principal component.
Recent work has demonstrated that anaplastic meningiomas segregate into 2–3 prognostically significant subgroups on the basis of methylation profile10. Unsupervised hierarchical clustering using methylation data available for a subset of the cohort (n = 19) demonstrated segregation into two main groups largely overlapping the subgroups delineated on the basis of gene expression profile, though correlation with survival outcomes was less marked (Supplementary Fig. S8).
Transcriptional programs segregating indolent and aggressive anaplastic meningioma
Nineteen hundred genes underpinned the differentiation of anaplastic meningioma into subgroups C1 and C2, which could be reduced to only 6 transcripts selected on the basis of PCA coefficient and differential expression analysis (see Methods; Supplementary Tables S10 and S11, Fig. S9). Pathway enrichment analysis was most significant for evidence of epithelial-mesenchymal transition (EMT) in the C1 tumors, with concordant loss of E-cadherin (CDH1) and upregulation of CXCL14, both prognostic biomarkers in diverse other cancers (Supplementary Table S12, Fig. 2d–f)23–25. EMT, which involves reprogramming of adherent epithelial cells into migratory mesenchymal cells, is critical for embryogenesis and tissue plasticity, and can play an important role in malignant progression, metastasis and therapy resistance24,26. Interestingly, NF2 and the closely related cytoskeletal protein ezrin normally help maintain E-cadherin expression at adherence junctions, whereas HOXB7 and HOXB9, both overexpressed in C1 tumors, suppress CDH1 expression27–29. It is increasingly recognised that CXCL14 and other EMT mediators are often derived from cancer-associated fibroblasts (CAFs) and function in a paracrine manner25,30,31. It is hence possible that some of the gene expression patterns we observed may reflect differences in the tumor stromal compartment, itself an increasingly recognised therapeutic target30,32,33.
The C1 tumors were further characterised by upregulation of transcriptional programs associated with increased proliferation, PRC2 activity and stem cell phenotype (Supplementary Table S13). Hox genes constituted a notable proportion of the transcripts distinguishing the two anaplastic meningioma subgroups, largely underpinning the significance of pathways involved in tissue morphogenesis. Furthermore, differentially methylated genes were also significantly enriched for Hox genes, with pathway analysis results corroborating the main biological themes apparent from the transcriptome (Supplementary Tables S14 and S15). Given the transcriptional evidence of increased PRC2 activity in the C1 subgroup, is noteworthy that SWI/SNF gene mutations occurred exclusively in C1 tumors (P = 0.016, Fisher’s exact test).
Comparison of the anaplastic and benign meningioma transcriptome
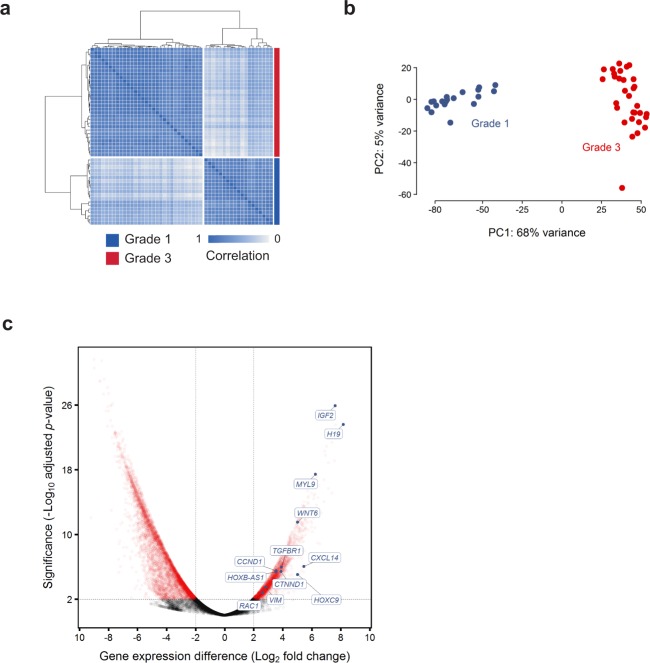
Previous studies investigating the relationship between meningioma WHO grade and gene expression profiles have included few anaplastic tumors34,35. We therefore extended our analysis to include published RNA sequences from 19 benign grade I meningiomas. External data was processed using our in-house pipeline with additional measures taken to minimise batch effects (Methods, Supplementary Tables S16 and S17). Unsupervised hierarchical clustering and principal component analysis demonstrated clear tumor segregation by histological grade (Fig. 3a,b). In keeping with previous reports, the anaplastic tumors demonstrated marked upregulation of major growth factor receptor and kinase circuits implicated in meningioma pathogenesis, notably epidermal growth factor receptor (EGFR), insulin-like growth factor (IGFR), vascular endothelial growth factor receptor (VEGFR) and mTOR complex 1 (mTORC1) kinase complex36–41.
Figure 3.
Differences in gene expression profile between grade I and anaplastic meningomas. (a,b) Normalised transcript counts from grade I and anaplastic meningioma samples clustered by (a) Pearson’s correlation coefficient and (b) principal component analysis. (c) Volcano plot illustrating differences in gene expression between anaplastic versus grade I meningiomas with selected genes indicated. The horizontal axis shows the log2 fold change and the vertical axis indicates the −log10 adjusted P-value. Genes with an adjusted P-value < 0.01 and absolute log2 fold change >2 are highlighted in red. PC, principal component.
Consistent with there being a coherent biological trend across histological grades and anaplastic meningioma subgroups, we noted significant overlap between genes differentially expressed between grades and between C1 and C2 tumors (hypergeometric distribution P = 5.08 × 10−9). In keeping with this finding, formal pathway analysis identified significant dysregulation of stemness, proliferation, EMT and PRC2 activity (Supplementary Tables S18 and S19). The most significantly dysregulated pathways also included TGF-beta, Wnt and integrin signalling, mediators of invasion and mesenchymal differentiation that are normally in part controlled by NF2 and other Hippo pathway members20,24,42. Yes-associated protein 1 (Yap1), a cornerstone of oncogenic Hippo signalling, is frequently overexpressed in cancer and synergises with Wnt signalling to induce EMT43,44. YAP1 was upregulated in anaplastic tumors along with MYL9, a key downstream effector essential for Yap1-mediated stromal reprogramming (Fig. 3c)43.
Discussion
Meningiomas constitute a common, yet diverse tumor type with few therapeutic options6,7,9,45. Efforts to improve clinical outcomes have been hampered by limited understanding of the molecular determinants of aggressive disease. Here, we explored genomic, epigenetic and transcriptional features of anaplastic meningioma, the most lethal meningioma subtype4.
Frequent somatic changes in SWI/SNF complex genes, predominantly ARID1A, constitute the main genomic distinction between anaplastic and lower grade meningiomas7,9. SWI/SNF inactivation is associated with aberrant PRC2 activation, stem cell-like phenotype and poor outcomes in diverse cancer types46–48.
Although anaplastic tumors resist comprehensive classification based on driver mutation patterns, transcriptional profiling revealed two biologically distinct subgroups with dramatically divergent survival outcomes. This finding is emblematic of the limitations of histopathological grading as a risk stratification system for meningioma2,4,10,45,49. All SWI/SNF mutations were confined to the poor prognosis (C1) subgroup, which was further characterised by transcriptional signatures of PRC2 target activation, stemness, proliferation and mesenchymal differentiation. These findings were in part underpinned by differential expression of Hox genes. Acquisition of invasive capacity and stem cell traits are frequently co-ordinately dysregulated in cancer, often through subversion of Hox gene programs integral to normal tissue morphogenesis50–52. Hox genes have a central role in orchestrating vertebrate development and act as highly context-dependent oncogenes and tumor suppressors in cancer51,53. Several of the most starkly upregulated Hox genes in the C1 tumors consistently function as oncogenes across a range of solid and haematological malignancies, including HOTAIR, HOXB7, HOXA4, HOXA-AS2, HOXC11, and NKX2-228,29,51,54–62. Like many other long non-coding RNAs (lncRNA), HOTAIR and HOXA-AS2 modulate gene expression primarily by interacting directly with chromatin remodelling complexes, exerting oncogenic activity by recruiting PRC2 to target genes54,56,61–65. HOXA-AS2 has been shown to mediate transcriptional repression of the tumor suppressor gene CDKN2A (p16INK4A), deletion of which is associated with poor meningioma survival54,61,62,66,67. Given the antagonistic relationship between the SWI/SNF and PRC2 chromatin regulators, deleterious SWI/SNF mutations and overexpression of lncRNAs known to mediate PRC2 activity emerge as potentially convergent mechanisms underpinning the differences between C1 and C2 tumors68. Further endorsing a link between transcriptional subgroups and chromatin dysregulation, 15 of the differentially expressed transcripts delineating C1 and C2 subgroups (absolute log2 fold change >2 and FDR < 0.01) are among the 50 genes most often associated with frequently bivalent chromatin segments (FBS) in cancer, including 11 transcripts from the HOXB cluster on chromosome 1769. This overlap was highly statistically significant (hypergeometric distribution P = 1.98 × 10−11). Bivalent, or epigenetically ‘poised’, chromatin is characterised by finely balanced activating (H3K4me1/H3K4me3) and repressive (H3K27me3) histone marks and pre-loaded DNA polymerase II poised to transcribe in response to modest epigenetic changes70. Bivalent chromatin most often marks genes involved in developmental reprogramming, in particular Hox cluster genes and homeotic non-coding transcripts, and is a frequent target of aberrant chromatin modification in cancer65,69,71.
In the context of recent studies of lower grade meningiomas, our findings raise the possibility that the balance between PRC2 and SWI/SNF activity may have broader relevance to meningioma pathogenesis. Compared to grade I tumors, atypical meningiomas are more likely to harbor SMARCB1 mutations and large deletions encompassing chromosomes 1q, 6q and 14q. Notably, these genomic regions encompass ARID1A and several other SWI/SNF subunit genes. Both SMARCB1 mutations and the aforementioned copy number changes were associated with epigenetic evidence of increased PRC2 activity, differential Homeobox domain methylation, and upregulation of proliferation and stemness programs in atypical grade II meningiomas9.
The extent to which SWI/SNF depletion plays a role in meningioma development may be therapeutically relevant. Diverse SWI/SNF mutated cancers exhibit dependence on both catalytic and non-catalytic functions of EZH2, a core subunit of PRC272–74. Several EZH2 inhibitors are in development with promising initial clinical results75. Other modulators of PRC2 activity, including HOTAIR, may also be relevant therapeutic targets76,77. Furthermore, growing recognition of the relationship between EMT and resistance to conventional and targeted anti-cancer agents has profound implications for rational integration of treatment approaches32,33. Notably, EGFR inhibition has yielded disappointing response rates in meningioma despite high EGFR expression37,78. A mesenchymal phenotype is strongly associated with resistance to EGFR inhibitors in lung and colorectal cancer32,33,79–81. Combining agents that abrogate EMT with other therapies is a promising strategy for addressing cell-autonomous and extrinsic determinants of disease progression and may warrant further investigation in meningioma32,33.
This study has revealed biologically and prognostically significant anaplastic meningioma subgroups and identified potentially actionable alternations in SWI/SNF genes, PRC2 activity and EMT regulatory networks. However, a substantially larger series of tumors, ideally nested in a prospective multicentre observational study, will be required to expand upon our main findings and explore mechanistic and therapeutic ramifications of meningioma diversity.
Methods
Sample selection
DNA was extracted from 70 anaplastic meningiomas; 51 samples at first resection (‘primary’) and 19 from subsequent recurrences. Matched normal DNA was derived from peripheral blood lymphocytes. Written informed consent was obtained for sample collection and DNA sequencing from all patients in accordance with the Declaration of Helsinki and protocols approved by the NREC/Health Research Authority (REC reference 7/YH/0101) and Ethics Committee at University Hospital Carl Gustav Carus, Technische Universität Dresden, Germany (EK 323122008). Samples underwent independent specialist pathology review (V.P.C and K.A). DNA extracted from fresh-frozen material was submitted for whole genome sequencing whereas that derived from formalin-fixed paraffin-embedded (FFPE) material underwent deep targeted sequencing of 366 cancer genes.
One tumor sample PD23348 (and two subsequent recurrences) separated from the main study samples in a principal components analysis of transcriptomic data (Supplementary Fig. S10). Analysis of WGS and RNA sequencing data identified an expressed gene fusion, NAB2-STAT6. This fusion is pathognomonic of meningeal hemangiopericytoma, now classified as a separate entity, solitary fibrous tumors82–84. We therefore excluded three samples from this tumor from further study. A second sample (PD23354a), diagnosed as an anaplastic meningioma with papillary features, was found to have a strong APOBEC mutational signature as well as an EML4-ALK gene fusion (exon 6 EML4, exon 19 ALK) (Supplementary Fig. S11)85. Therefore this sample was also removed as a likely metastasis from a primary lung adenocarcinoma. The hypermutator sample PD23359a underwent additional pathological review to confirm the diagnosis of anaplastic meningioma (K.A., Department of Histopathology, Cambridge University Hospital, Cambridge, UK).
RNA was extracted from fresh-frozen material from 34 primary and recurrent tumors, 3 of which were from PD23348 and were subsequently excluded from final analyses (Supplementary Table S1).
Whole genome sequencing
Short insert 500 bp genomic libraries were constructed, flowcells prepared and sequencing clusters generated according to Illumina library protocols86. 108 base/100 base (genomic), or 75 base (transcriptomic) paired-end sequencing were performed on Illumina X10 genome analyzers in accordance with the Illumina Genome Analyzer operating manual. The average sequence coverage was 65.8X for tumor samples and 33.8X for matched normal samples (Supplementary Table S1).
Targeted genomic sequencing
For targeted sequencing we used a custom cRNA bait set (Agilent) to enrich for all coding exons of 366 cancer genes (Supplementary Table S20). Short insert libraries (150 bp) were prepared and sequenced on the Illumina HiSeq 2000 using 75 base paired-end sequencing as per Illumina protocol. The average sequence coverage was 469X for the tumor samples.
RNA sequencing and data processing
For transcriptome sequencing, 350 bp poly-A selected RNA libraries were prepared on the Agilent Bravo platform using the Stranded mRNA library prep kit from KAPA Biosystems. Processing steps were unchanged from those specified in the KAPA manual except for use of an in-house indexing set. Reads were mapped to the GRCh37 reference genome using STAR (v2.5.0c)87. Mean sequence coverage was 128X. Read counts per gene, based on the union of all exons from all possible transcripts, were then extracted BAM files using HTseq (v0.6.1)88. Transcripts Per kilobase per Million reads (TPM) were generated using an in-house python script (https://github.com/TravisCG/SI_scripts/blob/master/tpm.py)87,88. We downloaded archived RNA sequencing FASTQ files for 19 grade I meningioma samples representing the major mutational groups (NF2/chr22 loss, POLR2A, KLF4/TRAF7, PI3K mutant) (ArrayExpress: GSE85133)7. Reads were then processed using STAR and HTseq as described above. Cancer cell line (n = 252) and triple-negative breast cancer (n = 100) RNA sequencing data was generated in-house by the aforementioned sequencing and bioinformatic pipeline.
Expressed gene fusions were sought using an in-house pipeline incorporating three algorithms: TopHat-Fusion (v2.1.0), STAR-Fusion (v0.1.1) and deFuse (v0.7.0) (https://github.com/cancerit/cgpRna)87,89,90. Fusions identified by one or two algorithms or also detected in the matched normal sample were flagged as likely artefacts. Fusions were further annotated according to whether they involved a kinase or known oncogene and whether they occurred near known fragile sites or rearrangement break points91 (Supplementary Table S5).
The C1 and C2 subgroups were defined by unsupervised hierarchical clustering using Poisson distance between samples92,93. Poisson distance was calculated using the PoissonDistance function implemented in the ‘PoiClaClu’ R package92 and unsupervised hierarchical clustering performed with the stats::hclust() function using the 250 transcripts with the most variable expression across tumors. Silhouette information was computed using the cluster::silhouette() function. The highest mean silhouette score was consistently achieved with two clusters.
Differential gene expression and pathway enrichment analysis
The DESeq2 R package was used for all differential gene expression analyses94,95. DESeq2 uses shrinkage estimation of dispersion for the sample-specific count normalization and subsequently applies a linear regression method to identify differentially expressed genes (DEGs)94,95.
Preliminary comparison of anaplastic and externally-generated grade I meningioma data revealed evidence of laboratory batch effects, which we mitigated with two batch-correction methods: RUVg and PEER96,97. RUVg estimates the factor attributed to spurious variation using control genes that are assumed to have constant expression across samples98–100. We selected control genes (RPL37A, EIF2B1, CASC3, IPO8, MRPL19, PGK1 and POP4) on the basis of previous studies of suitable control genes for transcript-based assays in meningioma101. PEER (‘probabilistic estimation of expression residuals’) is based on factor analysis methods that infer broad variance components in the measurements. PEER can find hidden factors that are orthogonal to the known covariates. We applied this feature of PEER to remove additional hidden effect biases. The final fitted linear regression model consists of the factor identified by RUVg method that represents the unwanted laboratory batch effect and 13 additional hidden factors found by PEER that are orthogonal to the estimated laboratory batch effect. Using this approach we were able to reduce the number of DEGs from more than 18000 to 8930, of which <4,000 are predicted to be protein-coding.
To identify biological pathways differentially expressed between meningioma grades and anaplastic meningioma subgroups we applied a functional class scoring algorithm using a collection of 461 published gene sets mapped to 10 canonical cancer hallmarks (Supplementary Table S21)50,102–106. We further corroborated these findings with a more general Gene Ontology (GO) pathway analysis107.
Identification of 6 transcripts recapitulating anaplastic meningioma clusters
Mapped RNA sequencing reads were normalised using the regularised logarithm (rlog) function implemented by the DESeq2 package94,95. PCA was performed using the top 500 most variably expressed transcripts and the R stats::prcomp function108. Given that primary component 1 (PC1) was the vector most clearly distinguishing the closely clustered C2 subgroup from the more diffusely clustered C1 (Fig. 3a), we extracted the top 50 transcripts with the highest absolute PC1 coefficients. We then identified the subset that overlapped with the most significantly differentially expressed genes (absolute log2 fold change >4 and adjust p-value < 0.0001) between i) the C1 and C2 anaplastic meningioma subgroups and ii) the C1 anaplastic meningiomas and the 19 grade I tumors (Supplementary Tables S10 and S17). Iteratively reducing the number of PC1 components identified the minimum number of transcripts that recapitulated segregation of C1 and C2 tumors upon unsupervised hierarchical clustering and PCA (Supplementary Table S11, Fig. S9).
Processing of genomic sequencing data
Genomic reads were aligned to the reference human genome (GRCh37) using the Burrows-Wheeler Aligner, BWA (v0.5.9)109. CaVEMan (Cancer Variants Through Expectation Maximization: http://cancerit.github.io/CaVEMan/) was used for calling somatic substitutions. Small insertions and deletions (indels) in tumor and normal reads were called using a modified Pindel version 2.0. (http://cancerit.github.io/cgpPindel/) on the NCBI37 genome build110,111. Annotation was according to ENSEMBL version 58. Structural variants were called using a bespoke algorithm, BRASS (BReakpoint AnalySiS) (https://github.com/cancerit/BRASS) as previously described112.
The ascatNGS algorithm was used to estimate tumor purity and ploidy and to construct copy number profiles from whole genome data113.
Identification of cancer genes based on the impact of coding mutations
To identify recurrently mutated driver genes, we applied an established dN/dS method that considers the mutation spectrum, the sequence of each gene, the impact of coding substitutions (synonymous, missense, nonsense, splice site) and the variation of the mutation rate across genes22.
Identification of driver mutations in known cancer genes
Non-synonymous coding variants detected by Caveman and Pindel algorithms were flagged as putative driver mutations according to set criteria and further curated following manual inspection in the Jbrowse genome browser114. Variants were screened against lists of somatic mutations identified by a recent study of 11,119 human tumors encompassing 41 cancer types and also against a database of validated somatic drivers identified in cancer sequencing studies at the Wellcome Trust Sanger Institute (Supplementary Tables S22 and S23)115.
Copy number data was analysed for homozygous deletions encompassing tumor suppressor genes and for oncogene amplifications exceeding 5 or 9 copies for diploid and tetraploid genomes, respectively. Only focal (<1 Mb) copy number variants meeting these criteria were considered potential drivers. Additional truncating events (disruptive rearrangement break points, nonsense point mutations, essential splice site mutations and frameshift indels) in established tumor suppressors were also flagged as potential drivers. Only rearrangements with breakpoints able to be reassembled at base pair resolution are included in this dataset.
TraFiC pipeline for retrotransposon integration detection
For the identification of putative solo-L1 and L1-transduction integration sites, we used the TraFiC (Transposome Finder in Cancer) algorithm12. TraFiC uses paired-end sequencing data for the detection of somatic insertions of transposable elements (TEs) and exogenous viruses. The identification of somatic TEs (solo-L1, Alu, SINE, and ERV) is performed in three steps: (i) selection of candidate reads, (ii) transposable element masking, (iii) clustering and prediction of TE integration sites and (iv) filtering of germline events12.
Methylation arrays and analysis
We performed quantitative methylation analysis of 850,000 CpG sites in 25 anaplastic meningiomas. Bisulfite-converted DNA (bs-DNA) was hybridized on the Ilumina Infinium HumanMethylationEPIC BeadChip array following the manufacturer’s instructions. All patient DNA samples were assessed for integrity, quantity and purity by electrophoresis in a 1.3% agarose gel, picogreen quantification and Nanodrop measurements. Bisulfite conversion of 500 ng of genomic DNA was done using the EZ DNA Methylation Kit (Zymo Research), following the manufacturer’s instructions. Resulting raw intensity data (IDATs) were normalized using the Illumina normalization method developed under the minfi R package (v1.19.10). Normalized intensities were then used to calculate DNA methylation levels (beta values). We then excluded from the analysis the positions with background signal levels in methylated and unmethylated channels (p > 0.01). Finally we removed probes with one or more single nucleotide polymorphisms (SNPs) with a minor allele frequency (MAF) >1% in the first 10 bp of the interrogated CpG, as well as the probes related to X and Y chromosomes. From the filtered positions, we selected only CpG sites present both in promoter regions (TSS, 5′UTR and 1st exon) and CpG islands (UCSC database, genome version hg19).
For the supervised analysis of the probes, CpG sites were selected by applying an ANOVA test to identify statistically significant CpG positions (FDR adjusted p-value < 0.01) that were differentially methylated among the compared groups (Δβ > 0.2). Selected CpG sites were later clustered based on the Manhattan distances aggregated by ward’s linkage. Finally, the genes corresponding to the selected CpGs were used to perform a Gene Set Enrichment Analysis (GSEA) with curated gene sets in the Molecular Signatures Database116. The gene sets used were: H: hallmark gene sets, BP: GO biological process, CC: GO cellular component, MF: GO molecular function and C3: motif gene sets (http://software.broadinstitute.org/gsea/msigdb/collections.jsp). The gene clusters resulting from the hypergeometric test with a FDR adjusted p-value < 0.05 were finally considered. We observed high levels of methylation for CREBBP in the majority of tumor samples, however, similar patterns were manifest in normal tissue controls, hence CREBBP hypermethyation does not appear to be a feature of oncogenesis in these samples.
For principal component analysis, we used the R function prcomp to calculate the Singular Value Decomposition of the beta value matrix after removing the CpGs without methylation information. We plotted the first two principal components which contain most variation by using the ggbiplot R package (http://github.com/vqv/ggbiplot). For each group we plotted a normal data ellipse with size defined as a normal probability equal to 0.68. Unsupervised hierarchical clustering was performed with the stats::hclust() function using the 75 probes with the highest variance in methylation beta values.
Mutational signature analysis
Mutational signature extraction was performed using the nonnegative matrix factorization (NNMF) algorithm11. Briefly, the algorithm identifies a minimal set of mutational signatures that optimally explains the proportions of mutation types found across a given mutational catalogue and then estimates the contribution of each identified signature to the mutation spectra of each sample.
Patient survival analysis
The Kaplan-Meier method was used to analyze survival outcomes by the log-rank Mantel-Cox test, with hazard ratio and two-sided 95% confidence intervals calculated using the Mantel_Haenszel test (GraphPad Prism, ver 7.02). Overall survival data from time of first surgery for each anaplastic meningioma within gene-expression defined subgroups C1 and C2 was collected and used to plot a Kaplan-Meier survival curve.
Supplementary Discussion
A hypermutator anaplastic meningioma with a haploid genome
One primary anaplastic meningioma resected from an 85-year old female (PD23359a) had a hypermutator phenotype, with 27,332 point mutations and LOH across nearly its entire genome (Supplementary Fig. S12, Table S24). Independent pathological review confirmed the original diagnosis of anaplastic meningioma, and transcriptome analysis demonstrated that this tumor clustered appropriately with the rest of the cohort (Fig. 3a,b). The majority of the mutations were substitutions, 72% of which were C > T transitions. We identified two deleterious mutations in DNA damage repair mediators: a TP53 p.R248Q missense mutation and a homozygous truncating variant in the mismatch repair gene MSH6 (p.L1330Vfs*9). Despite the latter finding, mutational signatures analysis was dominated by signature 1, with no evidence of signatures typically associated with defects in homologous recombination, mismatch repair or POLE activity (signatures 3, 6, 10, 15, 20 or 26). The copy number profile is most consistent with this tumor having first undergone haploidization of its genome, with the exception of chromosomes 7, 19 and 20, followed by whole genome duplication (Supplementary Fig. S12). Of note, several important oncogenes are located on chromosome 7, including EGFR, MET and BRAF. Widespread LOH has been described in a significant proportion of oncocytic follicular thyroid cancers where preservation of chromosome 7 heterozygosity has also been observed117.
Electronic supplementary material
Acknowledgements
This work was supported by the Wellcome Trust, Cancer Research UK, Meningioma UK and Tadhg and Marie-Louise Flood. U.M. was personally supported by a Cancer Research UK Clinician Scientist Fellowship; G.C. by a Wellcome Trust Clinical PhD Fellowship (WT098051); F.M. by A.I.L. (Associazione Italiana Contro le Leucemie-Linfomi e Mieloma ONLUS) and by S.I.E.S. (Società Italiana di Ematologia Sperimentale); S.B. was funded by a Wellcome Trust Intermediate Clinical Research Fellowship and a St. Baldrick’s Foundation Robert J. Arceci Innovation Award. J.M.C.T. is supported by ERC Starting Grant StG-2016_716290_SCUBA CANCERS and by MINECO Grants RYC 2014 14999 and SAF2015-66368-P. J.B. was funded by the charity Brain Tumour Research. The samples were received from the tissue banks from Cambridge (UK), Dresden (Germany), Liverpool (UK), Plymouth (UK) and Tel Aviv (Israel). The Human Research Tissue Bank is supported by the NIHR Cambridge Biomedical Research Centre. We are grateful to the patients who enabled this study and to the clinical teams coordinating their care.
Author Contributions
G.C. and N.K. performed mRNA expression analysis. G.C. and P.T. analysed whole genome and targeted sequencing data. I.M. performed statistical analyses to detect novel driver mutations. S.M. analysed methylation array data. F.M. generated mutational signatures analysis. J.M.C.T. and M.C. performed retrotransposon analysis. C.O.H. and J.D. performed protein expression analysis. A.B., S.B. and M.Y. contributed to data analysis strategy. A.Y., T.N., G.R.B. and J.T. provided informatic support. T.S., R.W.K., M.K., G.S., D.P., A.D., C.E.M., A.Y., I.N., S.J.P., C.W., Z.R., M.D.J., R.Z. and K. S. provided samples and clinical data. S.B., G.S.V., I.N. and M.W.M. provided conceptual advice. V.P.C. and K.A. carried out central pathology review. U.M. and T.S. devised and supervised the project. G.C. wrote the manuscript with input from U.M., S.B., T.S., P.T., and G.S.V. All authors approved the manuscript.
Data Availability
All sequencing data that support the findings of this study have been deposited in the European Genome-Phenome Archive and are accessible through the accession numbers EGAS00001000377, EGAS00001000828, EGAS00001000859, EGAS00001001155 and EGAS00001001873. All other relevant data are available from the corresponding author on request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Grace Collord and Patrick Tarpey contributed equally.
Contributor Information
Thomas Santarius, Email: ts381@cam.ac.uk.
Ultan McDermott, Email: um1@sanger.ac.uk.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-31659-0.
References
- 1.Mawrin C, Perry A. Pathological classification and molecular genetics of meningiomas. J Neurooncol. 2010;99:379–391. doi: 10.1007/s11060-010-0342-2. [DOI] [PubMed] [Google Scholar]
- 2.Rogers CL, et al. Pathology concordance levels for meningioma classification and grading in NRG Oncology RTOGTrial 0539. Neuro Oncol. 2016;18:565–574. doi: 10.1093/neuonc/nov247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moliterno J, et al. Survival in patients treated for anaplastic meningioma. Journal of neurosurgery. 2015;123:23–30. doi: 10.3171/2014.10.JNS14502. [DOI] [PubMed] [Google Scholar]
- 4.Champeaux C, Wilson E, Brandner S, Shieff C, Thorne L, World Health Organization grade III meningiomas. A retrospective study for outcome and prognostic factors assessment. Br J Neurosurg. 2015;29:693–698. doi: 10.3109/02688697.2015.1054350. [DOI] [PubMed] [Google Scholar]
- 5.Durand A, et al. WHO grade II and III meningiomas: a study of prognostic factors. J Neurooncol. 2009;95:367–375. doi: 10.1007/s11060-009-9934-0. [DOI] [PubMed] [Google Scholar]
- 6.Buttrick S, Shah AH, Komotar RJ, Ivan ME. Management of Atypical and Anaplastic Meningiomas. Neurosurg Clin N Am. 2016;27:239–247. doi: 10.1016/j.nec.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Clark, V. E. et al. Recurrent somatic mutations in POLR2A define a distinct subset of meningiomas. Nat Genet48, 1253–1259, 10.1038/ng.3651http://www.nature.com/ng/journal/vaop/ncurrent/abs/ng.3651.html - supplementary-information (2016). [DOI] [PMC free article] [PubMed]
- 8.Clark VE, et al. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science. 2013;339:1077–1080. doi: 10.1126/science.1233009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harmanci AS, et al. Integrated genomic analyses of de novo pathways underlying atypical meningiomas. Nat Commun. 2017;8:14433. doi: 10.1038/ncomms14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sahm F, et al. DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol. 2017;18:682–694. doi: 10.1016/S1470-2045(17)30155-9. [DOI] [PubMed] [Google Scholar]
- 11.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tubio JM, et al. Mobile DNA in cancer. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science. 2014;345:1251343. doi: 10.1126/science.1251343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galani V, et al. Genetic and epigenetic alterations in meningiomas. Clinical neurology and neurosurgery. 2017;158:119–125. doi: 10.1016/j.clineuro.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Kadoch C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shain AH, Pollack JR. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS One. 2013;8:e55119. doi: 10.1371/journal.pone.0055119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell. 2009;136:200–206. doi: 10.1016/j.cell.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol Cell Biol. 2008;28:3457–3464. doi: 10.1128/MCB.02019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 19.Morales FC, Molina JR, Hayashi Y, Georgescu MM. Overexpression of ezrin inactivates NF2 tumor suppressor in glioblastoma. Neuro Oncol. 2010;12:528–539. doi: 10.1093/neuonc/nop060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petrilli AM, Fernandez-Valle C. Role of Merlin/NF2 inactivation in tumor biology. Oncogene. 2016;35:537–548. doi: 10.1038/onc.2015.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goutagny S, et al. High incidence of activating TERT promoter mutations in meningiomas undergoing malignant progression. Brain Pathol. 2014;24:184–189. doi: 10.1111/bpa.12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martincorena I, et al. Universal Patterns of Selection in Cancer and Somatic Tissues. Cell. 2017;171:1029–1041.e1021. doi: 10.1016/j.cell.2017.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berx G, van Roy F. Involvement of members of the cadherin superfamily in cancer. Cold Spring Harbor perspectives in biology. 2009;1:a003129. doi: 10.1101/cshperspect.a003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 25.Benarafa C, Wolf M. CXCL14: the Swiss army knife chemokine. Oncotarget. 2015;6:34065–34066. doi: 10.18632/oncotarget.6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 27.Pujuguet P, Del Maestro L, Gautreau A, Louvard D, Arpin M. Ezrin regulates E-cadherin-dependent adherens junction assembly through Rac1 activation. Mol Biol Cell. 2003;14:2181–2191. doi: 10.1091/mbc.e02-07-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayashida T, et al. HOXB9, a gene overexpressed in breast cancer, promotes tumorigenicity and lung metastasis. Proc Natl Acad Sci USA. 2010;107:1100–1105. doi: 10.1073/pnas.0912710107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu X, et al. HOXB7, a homeodomain protein, is overexpressed in breast cancer and confers epithelial-mesenchymal transition. Cancer Res. 2006;66:9527–9534. doi: 10.1158/0008-5472.CAN-05-4470. [DOI] [PubMed] [Google Scholar]
- 30.Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer. 2016;16:582–598. doi: 10.1038/nrc.2016.73. [DOI] [PubMed] [Google Scholar]
- 31.Sjoberg E, Augsten M, Bergh J, Jirstrom K, Ostman A. Expression of the chemokine CXCL14 in the tumour stroma is an independent marker of survival in breast cancer. Br J Cancer. 2016;114:1117–1124. doi: 10.1038/bjc.2016.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gotwals, P. et al. Prospects for combining targeted and conventional cancer therapy with immunotherapy. Nat Rev Cancer advance online publication, 10.1038/nrc.2017.17 (2017). [DOI] [PubMed]
- 33.Marcucci F, Stassi G, De Maria R. Epithelial-mesenchymal transition: a new target in anticancer drug discovery. Nat Rev Drug Discov. 2016;15:311–325. doi: 10.1038/nrd.2015.13. [DOI] [PubMed] [Google Scholar]
- 34.Watson MA, et al. Molecular characterization of human meningiomas by gene expression profiling using high-density oligonucleotide microarrays. Am J Pathol. 2002;161:665–672. doi: 10.1016/S0002-9440(10)64222-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wrobel G, et al. Microarray-based gene expression profiling of benign, atypical and anaplastic meningiomas identifies novel genes associated with meningioma progression. Int J Cancer. 2005;114:249–256. doi: 10.1002/ijc.20733. [DOI] [PubMed] [Google Scholar]
- 36.Mawrin C, et al. Different activation of mitogen-activated protein kinase and Akt signaling is associated with aggressive phenotype of human meningiomas. Clin Cancer Res. 2005;11:4074–4082. doi: 10.1158/1078-0432.CCR-04-2550. [DOI] [PubMed] [Google Scholar]
- 37.Mawrin, C., Chung, C. & Preusser, M. Biology and clinical management challenges in meningioma. American Society of Clinical Oncology educational book. American Society of Clinical Oncology. Meeting, e106–115, 10.14694/EdBook_AM.2015.35.e106 (2015). [DOI] [PubMed]
- 38.Lopez-Lago MA, Okada T, Murillo MM, Socci N, Giancotti FG. Loss of the tumor suppressor gene NF2, encoding merlin, constitutively activates integrin-dependent mTORC1 signaling. Mol Cell Biol. 2009;29:4235–4249. doi: 10.1128/MCB.01578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.James MF, et al. NF2/merlin is a novel negative regulator of mTOR complex 1, and activation of mTORC1 is associated with meningioma and schwannoma growth. Mol Cell Biol. 2009;29:4250–4261. doi: 10.1128/MCB.01581-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson MD, Okedli E, Woodard A, Toms SA, Allen GS. Evidence for phosphatidylinositol 3-kinase-Akt-p7S6K pathway activation and transduction of mitogenic signals by platelet-derived growth factor in meningioma cells. Journal of neurosurgery. 2002;97:668–675. doi: 10.3171/jns.2002.97.3.0668. [DOI] [PubMed] [Google Scholar]
- 41.Weisman AS, Raguet SS, Kelly PA. Characterization of the epidermal growth factor receptor in human meningioma. Cancer Res. 1987;47:2172–2176. [PubMed] [Google Scholar]
- 42.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 43.Calvo F, et al. Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat Cell Biol. 2013;15:637–646. doi: 10.1038/ncb2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenbluh J, et al. beta-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell. 2012;151:1457–1473. doi: 10.1016/j.cell.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Louis DN, et al. The2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 46.Le Loarer F, et al. SMARCA4 inactivation defines a group of undifferentiated thoracic malignancies transcriptionally related to BAF-deficient sarcomas. Nat Genet. 2015;47:1200–1205. doi: 10.1038/ng.3399. [DOI] [PubMed] [Google Scholar]
- 47.Luchini C, et al. Prognostic role and implications of mutation status of tumor suppressor gene ARID1A in cancer: a systematic review and meta-analysis. Oncotarget. 2015;6:39088–39097. doi: 10.18632/oncotarget.5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu C, Allis CD. SWI/SNF complex in cancer. Nat Genet. 2017;49:178–179. doi: 10.1038/ng.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goldbrunner R, et al. EANO guidelines for the diagnosis and treatment of meningiomas. Lancet Oncol. 2016;17:e383–391. doi: 10.1016/S1470-2045(16)30321-7. [DOI] [PubMed] [Google Scholar]
- 50.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 51.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10:361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 52.Xu Q, et al. Long non-coding RNA regulation of epithelial-mesenchymal transition in cancer metastasis. Cell Death Dis. 2016;7:e2254. doi: 10.1038/cddis.2016.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 54.Xie M, et al. Long noncoding RNA HOXA-AS2 promotes gastric cancer proliferation by epigenetically silencing P21/PLK3/DDIT3 expression. Oncotarget. 2015;6:33587–33601. doi: 10.18632/oncotarget.5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bao X, et al. Knockdown of long non-coding RNA HOTAIR increases miR-454-3p by targeting Stat3 and Atg12 to inhibit chondrosarcoma growth. Cell Death Dis. 2017;8:e2605. doi: 10.1038/cddis.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim K, et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li X, et al. Long non-coding RNA HOTAIR, a driver of malignancy, predicts negative prognosis and exhibits oncogenic activity in oesophageal squamous cell carcinoma. Br J Cancer. 2013;109:2266–2278. doi: 10.1038/bjc.2013.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ozes AR, et al. NF-kappaB-HOTAIR axis links DNA damage response, chemoresistance and cellular senescence in ovarian cancer. Oncogene. 2016;35:5350–5361. doi: 10.1038/onc.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shi J, et al. Long non-coding RNA in glioma: signaling pathways. Oncotarget. 2017 doi: 10.18632/oncotarget.15175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ding J, et al. Long noncoding RNA HOXA-AS2 represses P21 and KLF2 expression transcription by binding with EZH2, LSD1 in colorectal cancer. Oncogenesis. 2017;6:e288. doi: 10.1038/oncsis.2016.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao H, Zhang X, Frazao JB, Condino-Neto A, Newburger PE. HOX antisense lincRNA HOXA-AS2 is an apoptosis repressor in all trans retinoic acid treated NB4 promyelocytic leukemia cells. J Cell Biochem. 2013;114:2375–2383. doi: 10.1002/jcb.24586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 64.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goutagny S, et al. Genomic profiling reveals alternative genetic pathways of meningioma malignant progression dependent on the underlying NF2 status. Clin Cancer Res. 2010;16:4155–4164. doi: 10.1158/1078-0432.CCR-10-0891. [DOI] [PubMed] [Google Scholar]
- 67.Bostrom J, et al. Alterations of the tumor suppressor genes CDKN2A (p16(INK4a)), p14(ARF), CDKN2B (p15(INK4b)), and CDKN2C (p18(INK4c)) in atypical and anaplastic meningiomas. Am J Pathol. 2001;159:661–669. doi: 10.1016/S0002-9440(10)61737-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kadoch C, Crabtree GR. Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Sci Adv. 2015;1:e1500447. doi: 10.1126/sciadv.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bernhart SH, et al. Changes of bivalent chromatin coincide with increased expression of developmental genes in cancer. Sci Rep. 2016;6:37393. doi: 10.1038/srep37393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 72.Helming KC, Wang X, Roberts CW. Vulnerabilities of mutant SWI/SNF complexes in cancer. Cancer Cell. 2014;26:309–317. doi: 10.1016/j.ccr.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim KH, et al. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nat Med. 2015;21:1491–1496. doi: 10.1038/nm.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bitler, B. G. et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med21, 231–238, 10.1038/nm.3799http://www.nature.com/nm/journal/v21/n3/abs/nm.3799.html - supplementary-information (2015). [DOI] [PMC free article] [PubMed]
- 75.Kim KH, Roberts CW. Targeting EZH2 in cancer. Nat Med. 2016;22:128–134. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ozes AR, et al. Therapeutic targeting using tumor specific peptides inhibits long non-coding RNA HOTAIR activity in ovarian and breast cancer. Scientific reports. 2017;7:894. doi: 10.1038/s41598-017-00966-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pfister SX, Ashworth A. Marked for death: targeting epigenetic changes in cancer. Nature reviews. Drug discovery. 2017;16:241–263. doi: 10.1038/nrd.2016.256. [DOI] [PubMed] [Google Scholar]
- 78.Norden AD, et al. Phase II trials of erlotinib or gefitinib in patients with recurrent meningioma. J Neurooncol. 2010;96:211–217. doi: 10.1007/s11060-009-9948-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Byers LA, et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19:279–290. doi: 10.1158/1078-0432.CCR-12-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Buonato JM, Lazzara MJ. ERK1/2 blockade prevents epithelial-mesenchymal transition in lung cancer cells and promotes their sensitivity to EGFR inhibition. Cancer Res. 2014;74:309–319. doi: 10.1158/0008-5472.CAN-12-4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thomson S, Petti F, Sujka-Kwok I, Epstein D, Haley JD. Kinase switching in mesenchymal-like non-small cell lung cancer lines contributes to EGFR inhibitor resistance through pathway redundancy. Clinical & experimental metastasis. 2008;25:843–854. doi: 10.1007/s10585-008-9200-4. [DOI] [PubMed] [Google Scholar]
- 82.Chmielecki J, et al. Whole-exome sequencing identifies a recurrent NAB2-STAT6 fusion in solitary fibrous tumors. Nat Genet. 2013;45:131–132. doi: 10.1038/ng.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gao F, et al. Inversion-mediated gene fusions involving NAB2-STAT6 in an unusual malignant meningioma. Br J Cancer. 2013;109:1051–1055. doi: 10.1038/bjc.2013.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schweizer L, et al. Meningeal hemangiopericytoma and solitary fibrous tumors carry the NAB2-STAT6 fusion and can be diagnosed by nuclear expression of STAT6 protein. Acta Neuropathol. 2013;125:651–658. doi: 10.1007/s00401-013-1117-6. [DOI] [PubMed] [Google Scholar]
- 85.Soda M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 86.Kozarewa I, et al. Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes. Nature methods. 2009;6:291–295. doi: 10.1038/nmeth.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics (Oxford, England) 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McPherson A, et al. deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS computational biology. 2011;7:e1001138. doi: 10.1371/journal.pcbi.1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim D, Salzberg SL. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome biology. 2011;12:R72. doi: 10.1186/gb-2011-12-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bignell GR, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463:893–898. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.M. Witten, D. Witten, D. M.: Classification and clustering of sequencing data using a Poisson model. Ann. Appl. Stat. 5(4), 2493–2518 Vol. 5 (2012).
- 93.Reeb PD, Bramardi SJ, Steibel JP. Assessing Dissimilarity Measures for Sample-Based Hierarchical Clustering of RNA Sequencing Data Using Plasmode Datasets. PLoS One. 2015;10:e0132310. doi: 10.1371/journal.pone.0132310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Anders S, Huber W. Differential expression analysis for sequence count data. Genome biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Peixoto L, et al. How data analysis affects power, reproducibility and biological insight of RNA-seq studies in complex datasets. Nucleic acids research. 2015;43:7664–7674. doi: 10.1093/nar/gkv736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stegle O, Parts L, Piipari M, Winn J, Durbin R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat Protoc. 2012;7:500–507. doi: 10.1038/nprot.2011.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang X, et al. Analysis of gene expression profiling in meningioma: deregulated signaling pathways associated with meningioma and EGFL6 overexpression in benign meningioma tissue and serum. PLoS One. 2012;7:e52707. doi: 10.1371/journal.pone.0052707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Savvidis C, Koutsilieris M. Circadian rhythm disruption in cancer biology. Mol Med. 2012;18:1249–1260. doi: 10.2119/molmed.2012.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sharma S, Ray S, Moiyadi A, Sridhar E, Srivastava S. Quantitative proteomic analysis of meningiomas for the identification of surrogate protein markers. Sci Rep. 2014;4:7140. doi: 10.1038/srep07140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pfister C, Tatabiga MS, Roser F. Selection of suitable reference genes for quantitative real-time polymerase chain reaction in human meningiomas and arachnoidea. BMC Res Notes. 2011;4:275. doi: 10.1186/1756-0500-4-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Iorio F, et al. Population-level characterization of pathway alterations with SLAPenrich dissects heterogeneity of cancer hallmark acquisition. bioRxiv. 2016 doi: 10.1101/077701. [DOI] [Google Scholar]
- 104.Ben-Porath I, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sarrio D, et al. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008;68:989–997. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- 106.Wong DJ, et al. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell stem cell. 2008;2:333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.The Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic acids research43, D1049–D1056, 10.1093/nar/gku1179 (2015). [DOI] [PMC free article] [PubMed]
- 108.R: A language and environment for statistical computing (R Foundation for Statistical Computing, Vienna, Austria, 2016).
- 109.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England) 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics (Oxford, England) 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Raine KM, et al. cgpPindel: Identifying Somatically Acquired Insertion and Deletion Events from Paired End Sequencing. Current protocols in bioinformatics. 2015;52:15.17.11–12. doi: 10.1002/0471250953.bi1507s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nik-Zainal S, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Raine KM, et al. ascatNgs: Identifying Somatically Acquired Copy-Number Alterations from Whole-Genome Sequencing Data. Current protocols in bioinformatics. 2016;56:15.19.11–15.19.17. doi: 10.1002/cpbi.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Buels R, et al. JBrowse: a dynamic web platform for genome visualization and analysis. Genome biology. 2016;17:66. doi: 10.1186/s13059-016-0924-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chang MT, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. 2016;34:155–163. doi: 10.1038/nbt.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Corver WE, et al. Genome haploidisation with chromosome 7 retention in oncocytic follicular thyroid carcinoma. PLoS One. 2012;7:e38287. doi: 10.1371/journal.pone.0038287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data that support the findings of this study have been deposited in the European Genome-Phenome Archive and are accessible through the accession numbers EGAS00001000377, EGAS00001000828, EGAS00001000859, EGAS00001001155 and EGAS00001001873. All other relevant data are available from the corresponding author on request.