Abstract
Unilateral cross-incompatibility (UCI) is a unidirectional inter/intra-population reproductive barrier when both parents are self-compatible. Maize Gametophyte factor1 (Ga1) is an intraspecific UCI system and has been utilized in breeding. However, the mechanism underlying maize UCI specificity has remained mysterious for decades. Here, we report the cloning of ZmGa1P, a pollen-expressed PECTIN METHYLESTERASE (PME) gene at the Ga1 locus that can confer the male function in the maize UCI system. Homozygous transgenic plants expressing ZmGa1P in a ga1 background can fertilize Ga1-S plants and can be fertilized by pollen of ga1 plants. ZmGa1P protein is predominantly localized to the apex of growing pollen tubes and may interact with another pollen-specific PME protein, ZmPME10-1, to maintain the state of pectin methylesterification required for pollen tube growth in Ga1-S silks. Our study discloses a PME-mediated UCI mechanism and provides a tool to manipulate hybrid breeding.
Unilateral cross-incompatibility between certain varieties of maize prevents cross-fertilization and can facilitate hybrid breeding. Here the authors show that a PECTIN METHYLESTERASE gene is able to overcome this reproductive barrier and confer fertility when expressed in pollen of the male parent.
Introduction
Maize is a versatile crop grown worldwide and is used as human food, animal feed, and industrial raw materials for starch and biofuel production. World-wide corn production and consumption exceeded 38 billion bushels in 2016 and is continually increasing (2016 data, see ref. 1), benefiting from an increased maize yield that has largely resulted from harnessing heterosis as well as improved agronomic practice. To optimize heterosis in hybrid seed production, avoiding undesired pollen is required, which is also important for organic and specialty maize productions. Multiple efforts, including temporal and spatial isolations, have been made to exclude undesired pollen; however, the technique remains challenging given that maize is a cross-pollinating crop with a large volume of pollen that can travel long distances via wind and insects2,3. Unilateral cross incompatibility (UCI) may offer an effective method to manipulate pollen flow in commercial maize production4–6.
UCI is a postpollination reproductive barrier that unidirectionally prevents hybridization when male and female parents are self-compatible4,5. Since the early 1900s, a non-Mendelian segregation ratio at the sugary 1 locus has been noted in crosses between popcorn and sweet corns7, resulting from UCI controlled by one linked Gametophyte factor1 (Ga1) locus in popcorn2,4,8–10. Genetic analyses revealed that maize UCI is governed by male-expressed and female-expressed components within the Ga1 locus5,9–11, generally resembling S-locus structure, a complex locus that confers self-incompatibility (SI) in numerous plant species12–14. However, in contrast to SI, which requires a suite of polymorphic haplotypes to distinguish individuals within a population, the Ga1 system is composed of three different haplotypes: the Ga1-S haplotype possesses both male and female determinants, the Ga1-M haplotype only harbors the male determinant, and the ga1 haplotype (wild type) lacks both. During fertilization, Ga1-S plants completely reject the pollen of ga1 due to the female barrier but accept pollen of Ga1-S and Ga1-M haplotypes, though the selfing and reciprocal crosses are compatible2,3. Many efforts have been made to identify the male and female determinants3,11,15. Recently, ZmPME3 in the Ga1 region was found to be specifically expressed in the silks of Ga1-S plants, serving as a likely candidate for the female function15. However, the mechanism underlying the male function in maize UCI remains elusive.
In this study, we clone a male determinant ZmGa1P at the Ga1 locus and reveal that expression of this pollen-specific PME can overcome the barrier in the Ga1-S silks. The interaction of ZmGa1P and another pollen-specific PME, ZmPME10-1, may form a protein complex to maintain the equilibrium of the apical cell wall dynamics during pollen tube growth, offering a mechanistic view for maize UCI.
Results
Genetic cloning of a male determinant at the Ga1 locus
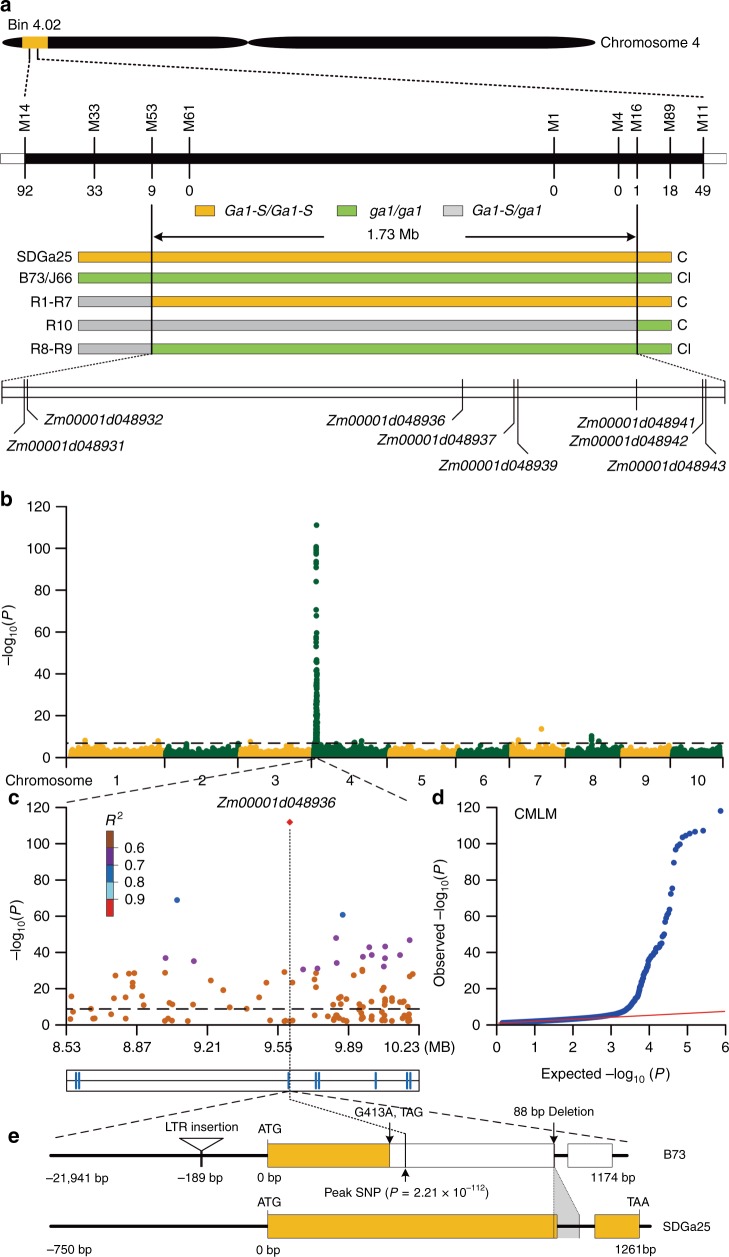
Maize Ga1 was reported in the 1900s but has remained an open issue for a century4. To address the underlying mechanism of Ga1, we utilized a map-based cloning strategy to isolate male determinant gene. In this study, approximately 21,600 individuals from two segregating populations were subjected to recombinant screening. The region containing male determinant gene was narrowed to a 1.73-Mb interval between markers M53 and M16 (Supplementary Table 1), in which eight protein-coding genes were annotated based on the B73 AGP v4 sequence16 (Fig. 1a). To identify male determinant gene, 946 inbred lines of Ga1-S, Ga1-M and ga1 haplotypes were genotyped using a 600-K SNP chip for genome-wide association analysis (Fig. 1b–d). Using a compressed mixed linear model that incorporates population structures as fixed effects and kinships as random effects to evaluate the P values (Supplementary Fig. 1), 167 of 174 male determinant associated SNPs (P < 2.15 × 10−8) were found to be intensively distributed in the mapping interval (Supplementary Table 2), and the peak SNP (P = 2.21 × 10−112) was located within Zm00001d048936 (Fig. 1b–d), suggesting this gene as the most likely candidate.
Fig. 1.
Map-based cloning of the pollen determinant. a Physical mapping, showing Bin 4.02 narrowed to a region between markers M53 and M16 contains 8 annotated genes. Recombinants are indicated below the markers. R1-R10, recombinants. C, cross-compatible; CI, cross-incompatible. b Genome-wide association analysis of the pollen determinant. The horizontal dashed line depicts the Bonferroni-adjusted significance threshold (P = 2.15 × 10−8). c The genome-wide association signals in the 8.53–10.23 Mb interval. The peak SNP is positioned by a vertical dashed line with a red diamond. The eight annotated genes are indicated by the blue bars at the bottom. d Quantile–quantile plot under a compressed mixed linear model. e Gene structure of Zm00001d048936 in B73 (ga1) and SDGa25 (Ga1-S)
We then constructed a bacterial artificial chromosome (BAC) library of SDGa25 (Ga1-S) to investigate the gene structure of Zm00001d048936. A 135-kb BAC clone containing two copies of Zm00001d048936 was identified. In this BAC, copy_1 is intact and expressed, but copy_2 is interrupted by a long-terminal-repeat retrotransposon (LTR) and not transcribed (Supplementary Fig. 2). In addition, 86% of the remaining BAC sequences belong to several types of transposons (Supplementary Data 1), resulting in weak collinearity between the 135-kb BAC and the B73 (ga1) reference sequence. However, alignment of the intact copy of Zm00001d048936 with B73 revealed 26 hits in the mapping region. Only one hit, the Zm00001d048936 gene in B73, exhibits a gene structure identical to that in SDGa25, whereas the other 25 hits are partial fragments (Supplementary Figs. 3). The Zm00001d048936 sequence in SDGa25 and B73 was further compared to determine genomic variations between Ga1-S and ga1 lines. In addition to synonymous and missense variations (Supplementary Figs. 4, 5), B73 has a 21,254-bp LTR insertion at the promoter region (−189-bp), an 88-bp deletion (at the junction of the 1st exon and intron), and a premature nonsense point mutation (G413A) (Fig. 1e). To investigate whether these variations are widespread in different maize lines, the 88-bp deletion was used to genotype the above 946 inbred lines. This deletion was consistently present in all ga1 lines, but absent in Ga1-S and Ga1-M lines (Supplementary Fig. 6).
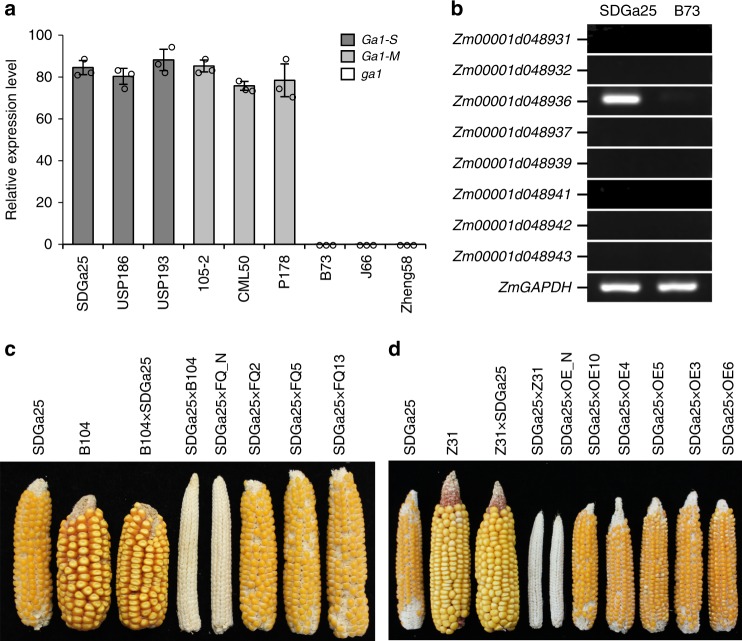
Genomic variations may disturb gene structure and/or gene expression. The transcripts of Zm00001d048936 were detected exclusively in pollen of Ga1-S and Ga1-M lines, and not in that of ga1 lines (Fig. 2a). Zm00001d048936 is also the only gene expressed in SDGa25 pollen among the eight candidates located within the mapping region (Fig. 2b). All these data demonstrated that Zm00001d048936 is a candidate for the male function.
Fig. 2.
Validation of Zm00001d048936 as a pollen determinant. a Quantitative expression analysis of Zm0001d048936 in pollen of Ga1-S, Ga1-M, and ga1 lines. Error bars represent the mean ± SD (n = 3). b Examination of the eight candidate gene expression in pollen of SDGa25 (Ga1-S) and B73 (ga1). ZmGAPDH was used as an internal control. c, d Cross-compatibility analyses, showing ears of self-pollinations and reciprocal crosses between the SDGa25 (Ga1-S) and B104 or Z31 (ga1, control plants). The SDGa25 ears pollinated with non-transgenic controls (FQ_N and OE_N) and transgenic plants expressing ZmGa1P. c Transgenic plants made with the construct shown in Supplementary Fig 7a, d T2 generation of transgenic plants made with the construct shown in Supplementary Fig 8a
Expressing Zm00001d048936 endows ga1 with male function
To verify Zm00001d048936 as a male determinant, a 6,287-bp genomic fragment of Zm00001d048936 cloned from SDGa25 was introduced into the inbred line B104 (ga1) (Supplementary Fig. 7a). Pollen from three independent T1 transgenic (FQ2, FQ5, and FQ13) and nontransgenic plants were used to fertilize detasseled SDGa25 (Ga1-S) plants, separately. All the SDGa25 plants fertilized with pollen from the transgenic plants exhibited a full seed-set (Fig. 2c), but those fertilized with pollen from nontransgenic B104 had no seeds, providing genetic evidence for the conclusion that Zm00001d048936 is a pollen determinant. In addition, a construct harboring full-length cDNA of Zm00001d048936 from SDGa25 (Supplementary Fig. 8a) was introduced into maize inbred line Z31 (ga1) to generate over-expression plants (OE). We thereby pollinated detasseled SDGa25 plants with pollen from Z31 and five independent transgenic lines (OE3, OE4, OE5, OE6, and OE10) at T0, T1, and T2 generations, separately. A full seed-set was observed using transgenic pollen, but no seed-set was obtained using pollen from Z31 (Fig. 2d and Supplementary Fig. 8b). PCR analyses further confirmed the heterozygotic genotype in the seeds resulting from cross-pollination (Supplementary Figs. 7b and 8c, d) and corroborated the expression of Zm00001d048936 in the pollen of plants expressing the genomic fragment and of the OE lines (Supplementary Figs. 7c, 8e). Therefore, expressing Ga1-type Zm00001d048936 endows ga1 pollen with the ability to penetrate the female barrier of Ga1-S.
To determine whether Zm00001d048936 has female function, the five T2 OE lines that exhibited high level of Zm00001d048936 transcripts in silks were pollinated with B73 (ga1) pollen. The full seed-set suggested that Zm00001d048936 does not possess the function of the female barrier (Supplementary Fig. 9). We thus conclude that Zm00001d048936, which is designated ZmGa1P, is a pollen/male determinant of the Ga1 locus.
ZmGa1P encodes a pollen PME and modulates pollen tube growth
Alignment of the ZmGa1P sequence revealed that this gene encodes a putative PME, a class of critical factors affecting pollen tube growth17. Fertilization in plants involves a series of physiological processes, including hydration and germination of pollen grains, growth of pollen tubes for penetration and navigation in the transmitting tract to deliver sperm, and recognition between sperm nuclei and egg cells18. To dissect the cause of UCI regulated by Ga1, we analyzed pollen tube growth in silks. Although both pollen types can germinate on Ga1-S stigma, growth of ga1 pollen tubes was significantly inhibited compared with Ga1-S (Supplementary Fig. 10a, b). Our results, together with the results from other groups3,11,19, revealed that the cross-incompatibility of the Ga1 locus is caused by abnormal pollen tube growth.
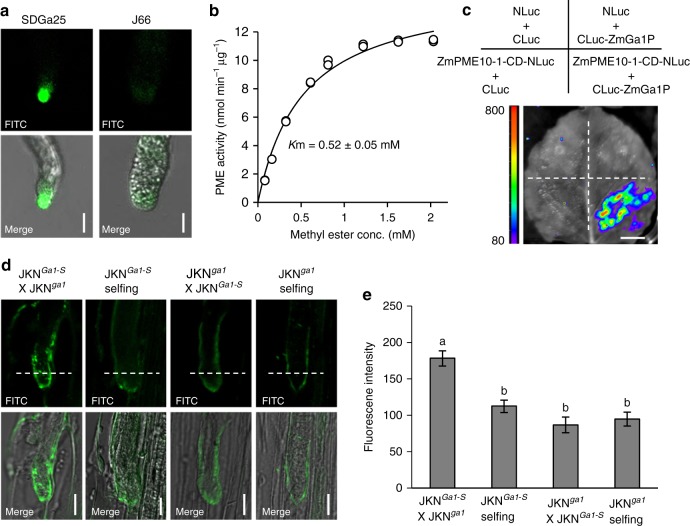
Among multiple factors that affect pollen tube growth, the mechanical strength and plasticity of the pollen tube wall controlled by the equilibrium of pectin esterification-deesterification at the apical region of pollen tubes is crucial20,21. ZmGa1P was therefore proposed to function in pollen tubes and affect their wall properties. To verify this hypothesis, we examined ZmGa1P expression in various tissues of Ga1-S and ga1 lines. Quantitative PCR analysis revealed that ZmGa1P is specifically expressed in Ga1-S pollen (Supplementary Fig. 10c). The encoded proteins were further found to be mainly localized at the pollen tube apex of Ga1-S plants, as revealed by immunolabeling the germinated pollen tubes with a specific anti-peptide antibody (Fig. 3a and Supplementary Fig. 10d, e). These findings are consistent with the view that PMEs function at the pollen tube apex20. Therefore, ZmGa1P is a PME candidate that specifically functions in the apex region of pollen tubes.
Fig. 3.
Characterization of ZmGa1P biochemical properties. a Immuno-staining the pollen tubes of SDGa25 (Ga1-S) and J66 (ga1) using the anti-ZmGa1P antibody. Scale bar = 10 μm. b Kinetic analysis of PME activity of secretory proteins in the fraction 8 of SDGa25. Error bars represent the mean ± SD of three replicates. c Split-luciferase complementation assay showing the interaction between ZmGa1P and the PME domain (CD) of ZmPME10-1 in N. benthamiana leaves infiltrated with the construct combinations shown above. Scale bar = 1 cm. d Immuno-detecting highly methylesterified pectins in the longitudinally sectioned pollen tubes of near isogenic lines with LM20 antibody. Scale bars = 10 μm. FITC, Fluorescein isothiocyanate. e Quantification of the fluorescent signal intensity of immune-examinations, to show the varied pectin DM on ga1 pollen tube walls growing in Ga1-S silks. Error bars represent the mean ± SE (n > 15 pollen tubes). a and b indicate that the means differ according to analysis of variance and Tukey’s test (P < 0.01)
Identifying ZmGa1P and ZmPME10-1 in Ga1-S pollen secretome
PME often functions in the apoplast20,21. To determine the biochemical property of ZmGa1P, we fractionated proteins secreted by pollen during germination. Comparing the fractionation profile of secretory proteins between Ga1-S and ga1 pollen, an ~42-kD band was clearly detected in fractions 8–10 of Ga1-S (Supplementary Fig. 11a) but absent in ga1 (Supplementary Fig. 11b). This band was recognized as ZmGa1P by the anti-ZmGa1P antibody (Supplementary Fig. 11a), indicating that Ga1-S pollen secreted large amounts of ZmGa1P. The pectoplate assay further revealed obvious PME activity in fractions 6–10 (Supplementary Fig. 11), suggesting that these fractions likely include active PMEs. We then subjected fractions 6–10 of Ga1-S and ga1 pollen to LC-MS/MS analyses. Secretome analysis identified approximately one hundred proteins in each fraction (Supplementary Data 2). ZmGa1P and Zm00001d026422 (designated as ZmPME10-1) were two major annotated PMEs in Ga1-S pollen, whereas ZmPME10-1 was the most abundant PME in ga1 pollen (Supplementary Figs. 12, 13 and Supplementary Table 3). Therefore, ZmGa1P plus ZmPME10-1 and ZmPME10-1 appeared to be the dominant PMEs in Ga1-S and ga1 pollen, respectively. We selected fraction 8 as a representative to analyze PME activity. The fraction of Ga1-S line exhibited saturable kinetics with a Km of 0.52 mM (Fig. 3b); the value was 0.44 mM in ga1 lines (Supplementary Fig. 14). Hence, the PME activity in Ga1-S and ga1 lines may mainly result from the presence of ZmGa1P plus ZmPME10-1 and ZmPME10-1 in the pollen tubes, respectively.
A PME complex controls pectin esterification of pollen tubes
Although the detected PMEs in Ga1-S and ga1 pollen tubes exhibited obvious PME activity in vitro, only the pollen harboring ZmGa1P can penetrate the silk barrier of the Ga1 locus, suggesting that the in planta PME activity mediated by ZmGa1P may differ from that in ga1 lines. The presence of ZmGa1P and ZmPME10-1 together in the Ga1-S line prompted us to hypothesize that the two proteins may interact. We treated the secretory proteins of fraction 8 with disuccinimidyl suberate (DSS), a widely used chemical for protein cross-linking analysis. The 42-kD ZmGa1P shifted to a higher molecular weight ~100 kD (Supplementary Fig. 15a), indicating the existence of a protein complex. By subjecting the ~100-kD cross-linked band to LC-MS/MS analysis, ZmGa1P and ZmPME10-1 were identified as the most likely components in this complex (Supplementary Fig. 15b and Supplementary Data 3). To obtain in planta evidence, we crosslinked the protein extracts from unpollinated and pollinated Ga1-S silks using DSS. The ~100-kD protein complex was also found in the self-pollinated Ga1-S silks (Supplementary Fig. 15c). Application of the split-luciferase complementary assay in the transiently transformed tobacco leaves further corroborated the interaction between ZmGa1P and the PME domain or signal peptide-depleted version of ZmPME10-1 (Fig. 3c and Supplementary Fig. 15d).
PMEs catalyze pectin deesterification20. To visualize the in vivo difference in the degree of methyl esterification (DM) at the pollen-tube apexes, we developed near isogenic lines JKNGa1-S and JKNga1 that differ at the Ga1 locus and compared the DM of the pollen tube walls between selfing and reciprocal crosses. The pollinated silk sections were immunolabeled by LM2022, an antibody that preferentially recognizes methylesterified pectin. Compared with the apical region of ga1 pollen tubes growing in the JKNGa1-S silks that exhibited significantly increased fluorescent signal intensity, those grown in the selfed JKNga1 silks as well as the Ga1-S pollen tubes penetrating in JKNGa1-S and JKNga1 silks exhibited weak intensity (Fig. 3d, e). This result suggested high DM pectin presenting in the ga1 pollen tubes upon growing in Ga1-S silks, which may result from compromised PME activity. These findings also implied the impact of the female determinant on modifying the methylesterified pectin levels in the pollen tube walls. Hence, assembly of a PME complex that contains ZmGa1P and ZmPME10-1 is essential for modulating pectin properties to overcome the silk barrier of Ga1.
Application of the Ga1 locus in manipulation of pollen flow
UCI may provide a selective biological barrier to manipulate pollen flow among different categories of maize cultivars. Unlike popcorn and teosinte, the majority of dent and flint maize varieties carry the ga1 haplotype5. Introduction of the Ga1 locus into ga1 lines may offer an effective method to isolate undesired pollen in commercial maize production. To validate the commercial value of the Ga1 locus in dent and flint corn production, we developed a near isogenic white kernel hybrid KYN3 carrying Ga1-S. Based on field tests at two different locations for two years, this line rejected ga1 pollen from adjacent plants without physical isolation as indicated by kernel colors (Fig. 4), demonstrating that the Ga1 locus represents a feasible commercial carrier with significant market potential.
Fig. 4.
Field testing the isolation of ga1 pollen from adjacent plants by Ga1 locus. a KYN3ga1(left) and its near isogenic line KYN3Ga1-S (right) grown surrounded by a ga1 line with purple corn in Beijing, China. b KYN3Ga1-S (white ears) was mingled with a ga1 line with yellow corn in Jilin, China
Discussion
Incompatibility in flowering plants has a great impact on species evolution and resource exploitation. The maize Ga1 locus has been studied for approximately one century, but low sequence homology between popcorn and conventional maize and difficulties in phenotyping and genotyping made cloning the determinants a daunting challenge11. In this study, despite more than 20,000 individuals were screened for recombinants, the mapping region was unable to be reduced less than 1.73 Mb, indicating genomic complexity at the Ga1 locus even though it is not near the centromere. Combining with genome-wide association analysis, we cloned a ZmGa1P gene that performs the male function of the Ga1 locus and demonstrated that ZmGa1P interacts with ZmPME10-1 to form a complex, which is essential for maintenance of the pectin methylesterification degree on the pollen tube walls for navigation through Ga1-S silks. Moreover, cloning of genes involved in UCI provides a tool for the control of pollen flow in commercial maize production and heterosis manipulation. Because Ga1-M can overcome Ga1-S barrier and this haplotype has been found at a much higher frequency in maize lines than previously thought23, its risk to Ga1-S hybrids should be aware.
Pectin deesterification catalyzed by PMEs can free carboxyl groups on pectin chains, promoting pectin crosslinking with Ca2+ and stiffening the cell wall24. PME activity can therefore modulate the pectin esterification-deesterification degree at the apical region of pollen tube walls and thereby affect the balance of plasticity and mechanical strength that is essential for pollen tube growth17,20,21. The role of PMEs in pollen tube growth regulation is well known; however, how PMEs control cross-incompatibility (CI) has not been reported. This study identified a pollen tube-specific PME, a type-II/group 1 PME (lacking a pro-region)25,26, as a pollen determinant of the Ga1 locus in maize UCI. Another type-II PME, ZmPME3, has been reported to function as a female determinant candidate15. Maize potentially evolved a previously undescribed PME-mediated mechanism to control UCI, which is distinct from the known mechanisms that govern SI and CI in higher plants, such as the RNase-mediated system in Solanaceae, Rosaceae, and Plantaginaceae12,13,27 and the peptide ligand-kinase system in Brassicaceae and Papaveraceae28–30.
PME activity should be tightly controlled to fine-tune the pectin properties in specific regions of the walls, e.g., the pollen tube apex, to fit their roles in plant growth and development20. However, how male- and female-PME regulates pollen tube growth in UCI remains unknown. Differential expression patterns of PMEs and environmental conditions are key factors affecting PME activity. Here, we demonstrated that a pollen-specific PME is a male determinant, whereas the female determinant candidate appears to be a silk-specific PME15, providing crucial spatial and temporal information to unravel the intriguing action of both PMEs. However, other components involved in pectin configuration control cannot be excluded. In this study, biochemical analyses identified a PME complex, in which ZmPME10-1, a pro-region harboring type-I/group 2 PME23,24, represents one of the key components and interacts with ZmGa1P. Of note, the assembly of a PME complex found here is a previously undescribed mode for PME action. As an abundant pollen-specific PME, the function of ZmPME10-1 regulating pollen tube growth is expected because a mutation in VANGUARD1, a homolog isoform in Arabidopsis, exhibited abnormal pollen tube growth and male sterility17. Although overt PME activity has been detected in the pollen secretory proteins containing ZmPME10-1 or the ZmGa1P-ZmPME10-1 complex, an increase in methylesterified pectin that may result from compromised PME activity was only discovered in ga1 pollen tubes upon growth in Ga1-S silks. We hypothesize that the pollen PME complex may interact with the female component to coordinate the final state of pectin methylation in pollen tube cell walls. However, further studies are required to clearly explore the intrinsic mechanism of Ga1-mediated UCI in maize.
Taken together, our findings open a door for understanding the molecular mechanisms underlying UCI in maize and likely other monocot plants31,32, which will contribute to crop molecular breeding.
Methods
Plant materials
The maize lines used in this study, including the association and mapping populations, were planted in the experimental fields of the Institute of Genetic and Developmental Biology (IGDB), Chinese Academy of Sciences (CAS) in Beijing and San Ya, Hainan province. The transgenic plants were grown in the fields and greenhouse with a 16-h light (25 °C)/8-h dark (18 °C) cycle. All plants were planted in plots at 0.6-m intervals in the horizontal direction and 0.4-m intervals in the longitudinal direction.
Determination of the haplotype of Ga1
To determine the haplotype of Ga1 in 1,299 inbred lines, SDGa25 (Ga1-S) was used as a female tester and planted in batches at 7-day intervals. Pollen from each of the 1,299 inbred lines was harvested and used to manually pollinate a set of three SDGa25 plants that were detasseled before flowering19,33. The resulting maize ear-florets (ears)34 with a full seed-set indicated bearing male function, whereas those with less than five seeds indicated a lack of male function. The identified inbred lines containing the male determinant gene were also used as females and were pollinated with B73 (ga1) pollen to test female function. The resulting ears with the full seed-set indicated a lack of female function, whereas those with less than five seeds indicated harboring female function. These tests were replicated at two different locations for two years. In total, 20 Ga1-S lines, 54 Ga1-M lines, and 872 ga1 lines were determined (Supplementary Data 4). Genotyping ZmGa1P in the indicated Ga1-S, Ga1-M, and ga1 lines was shown in Supplementary Figure 17.
Fine-mapping of ZmGa1P
Two different BC1F1 segregating populations, ga1 (B73 or J66)//ga1 (B73 or J66)/Ga1-S (SDGa25) and Ga1-S (SDGa25)//ga1 (B73 or J66)/Ga1-S (SDGa25), were used for mapping. The first population was developed using B73 or J66 as female and the F1 (B73 or J66 crossing SDGa25) plants as male. The resulting individuals in this BC1F1 population generated two types of gametes (Ga1-S or ga1). Additional phenotyping is required. To save labor, we produced the second population, in which SDGa25 (Ga1-S) lines were used as female and pollinated with pollen from the F1 (B73 or J66 crossing SDGa25) plants. Given the gametophytic nature of the male determinant, the resulting individuals in this BC1F1 population only had Ga1-S type gametes; thus, phenotyping is unnecessary3. In total, we used 21,600 individuals, including 5,700 from the first population and 13,943 from the second population, to screen the recombinants with the molecular markers (Supplementary Tables 4 and 5) designed according to the B73 sequence (RefGen_V4, www.maizegdb.org). Consequently, a total of 9 recombinants nearby the M53 marker on the distal side and 1 recombinant at the M16 marker on the proximal side were identified. The male determinant was ultimately mapped within a 1.73-Mb region.
Genome-wide association analysis
More than 946 inbred lines containing Ga1-S, Ga1-M, and ga1 haplotypes were employed to constitute a GWAS panel. DNA was extracted from five seedlings of each line and genotyped using the Axiom Maize Genotyping Array consisting of sequence information from 616,201 variants. The high-density SNP chip has been successfully applied for association mapping and comparative genomics35. Quality control was performed by limiting the MAF (minor allele frequency) value per SNP to less than 0.05, the missing rate per SNP to less than 0.1 and the missing rate per individual to less than 0.1. After filtering, 465,112 polymorphic SNPs and 875 individuals (19 Ga1-S, 42 Ga1-M, and 814 ga1 lines) qualified for association analysis. The phenotype values of each inbred line in the association test were assigned 1 or 0 to represent cross-compatible and incompatible, respectively. To examine the association between SNPs and the traits, Tassel 3.0 with a compressed mixed linear model (CMLM) was employed for GWAS36–38 as this model considers the population structure (Q) and familial relationship (K). The analysis was conducted with the GAPIT package in R39. The population structure was analyzed by principle component analysis (PCA). The first four principal components of PCA were used as a covariate matrix for the CMLM as fixed effects. The relative K was incorporated as a random effect in the CMLM, calculated by the VanRaden method40. The threshold for significant association (P < 2.15 × 10−8) was corrected using the Bonferroni correction method (P = 0.01/n, 0.01 was the type I error rate, and n was the total number of markers used for the association test)41. To identify the candidate genes underlying the association signals, we performed linkage disequilibrium (LD) analysis on each chromosome and plotted the whole genome LD decay against physical distance. The average distance over which LD decayed to half of its maximum value in our association panel was approximately 100 kb. Among the identified unique association signals, the annotated genes within 200 kb of the lead SNP in the mapping interval were validated and regarded as the candidate gene. The corresponding positions of each SNP were mapped based on the reference sequence B73 AGP_v416.
Construction and screening of the SDGa25 BAC library
To construct a BAC library of SDGa25, high-molecular weight genomic DNA was extracted from leaf tissues, partially digested by the restriction enzyme HindIII, and ligated into the vector pIndigoBAC536-S42,43. The ligation products were transformed into ElectroMAX DH10B T1 phage-resistant cells (Invitrogene) and screened on LB medium with chloramphenicol (12.5 mg/L), 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (80 mg/L) and isopropyl β-d-1-thiogalactoside (100 mg/L). White colonies were individually picked into 384-well microtiter plates. In total, 207,260 BAC clones were arranged in 540 384-well plates, representing a 10-fold coverage of the maize genome. We developed a PCR-based method to screen clones harboring the pollen determinant. A 384-well stock was duplicated and pooled to extract a primary DNA pool using the ZR BAC DNA Miniprep Kit (Zymo Research Corporation, Irvine, CA, USA). In total, 540 primary DNA pools were prepared and screened using the primers listed in Supplementary Table 6.
BAC DNA sequencing was performed using PacBio SMRT sequencing technology44 by the Nextomics Biosciences Company (Wuhan, China). In short, SMRTbell DNA template libraries with an average ~17-kb insert size for the BAC sample were prepared from sheared DNA by G-tubes (Covaris) according to the manufacturer’s recommendations. The library was bound with P6 polymerase, and complexes were then loaded onto SMRT cells (version V3). SMRT sequencing was performed on PacBio RSII using C4 sequencing reagents according to standard protocols. Movie lengths were 240 min for the SMRT cell. In total, one SMRT cell yielded output data of 1,376,923,513 bp with an average coverage of ∼4653× and an average length of 7688 bp subreads. To assemble the BAC sequence, clean reads obtained by removing adapters and low-quality reads (reads length <500 bp, quality <0.8) were assembled de novo using the RS hierarchical genome assembly process (HGAP) protocol version 2.3.045. Finally, a 154,617-bp gapless completed circular sequence was assembled and trimmed into clean data of 135,485 bp with 44.5% GC content. Annotation was performed using the FGENESH gene annotation program (www.softberry.com)46. Interspersed repeats and low-complexity DNA sequences were screened using the RepeatMasker program (http://www.repeatmasker.org/).
Generation of maize transformation plants
For the genomic complementary assay, the 6,287-bp Zm00001d048936 genomic fragment containing the coding region and the 2,980-bp promoter and 2,046-bp terminator regions from SDGa25 was inserted into pCAMBIA3300 to generate the genomic construct. In addition, the ZmGa1P coding sequence was amplified from SDGa25 and inserted into the pCAMBIA2300 vector between the maize ubiquitin promoter and the nopaline synthase terminator to generate an OE construct. The resulting constructs were introduced into the wild-type inbred line B104 (the genomic construct) and Z31 (the OE construct) through Agrobacterium tumefaciens infection, separately. Positive transgenic events were identified using the primers presented in Supplementary Table 6. Pollen from T0 to T2 transgenic plants was collected to fertilize SDGa25 plants to test cross-compatibility. At least five individuals from each transgenic event and the corresponding nontransgenic sibling plants were selected to provide pollen, and maize ears from at least three SDGa25 plants were used as the female testers. To examine the function of ZmGa1P in the female barrier, the transgenic plants were used as the female to pollinate B73 pollen. These crosses were evaluated by examining the seed-set on the resulting ears. Genotyping results on the seeds of parents and the indicated cross-pollinations were shown in Supplementary Figure 17, 18.
Gene expression analyses
To examine the expression pattern of the candidate genes located within the mapping region, roots, stems, leaves, anthers, and silks were harvested from B73 and SDGa25 plants at the proper growth stages. Total RNA was isolated from the different maize tissues using TRIzol (Invitrogen) and treated with RNase-free DNase I (Thermo Fisher Scientific) to remove DNA according to the manufacturer. Complementary DNA (cDNA) was synthesized from 2 μg of total RNA using RevertAid reverse transcriptase (Thermo Fisher Scientific) with the oligoT18 primer. For PCR analysis, cDNAs were used as template and amplified with the primers shown in Supplementary Table 6 (26 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 15 s). RT-PCR examinations of the candidate gene expression in pollen were shown in Supplementary Figure 16. The expression profile of ZmPME10-1 was also analyzed based on online public RNAseq data47. Three biological replicates were tested.
To determine the expression level of ZmGa1P in various inbred lines or transgenic and nontransgenic plants, mature anthers were collected from 20 of the Ga1-S, Ga1-M, and ga1 lines and the transgenic plants. Total RNA was extracted from anthers as described above. After synthesizing cDNA, quantitative PCR (qPCR) was performed on a LightCycler 480 Real-Time PCR System (Roche Diagnostics) with SYBR Green Master mix (45 cycles of 95 °C for 10 s, 60 °C for 30 s, and 72 °C for 10 s). ZmGAPDH was used as an internal control and the 2−ΔΔCt method was used to calculate the relative gene expression level. At least three biological replicates were performed.
Pollen isolation
To harvest the pollen used in this study, tassels were vigorously shook and covered with paper bags in the evening before pollen harvest33. Pollen grains were collected by shaking the tassels in paper bags at 9:30 a.m. and 2:00 p.m. The fresh pollen grains were used for pollination and in vitro germination.
In vivo pollen-tube growth assay
B73 and SDGa25 used for in vivo pollen-tube growth analysis were grown in experimental fields for self-crossing and reciprocal crossing11. Six hours after pollination, silks were collected and fixed in 4% paraformaldehyde in 50 mM sodium phosphate, pH 7.0 (PBS) for at least one day. The fixed silks were washed with the above buffer and immersed in 1 M NaOH overnight for softening. The pistil was then stained with 0.1% aniline blue (Amresco) in 0.1 M potassium phosphate (pH 8.0) for 6 h. The samples were observed under an Axio Skop2 microscope (Zeiss) equipped with an ultraviolet filter. The length of pollen tubes was evaluated between the pollen entry place and the tip of pollen tubes using ImageJ software (http://rsb.info.nih.gov/ij/). Twenty-four pollen tubes from at least three plants were examined in each cross.
Detection of ZmGa1P in the in vitro germinated pollen tubes
Fresh pollen harvested from the Ga1-S plants and wild-type lines was sprayed onto the germination medium (10% [w/v] sucrose, 0.0005% [w/v] H3BO3, 10 mM CaCl2, 0.05 mM KH2PO4, and 6% [w/v] polyethylene glycol 4000) in petri dishes immediately48. After germination at room temperature for 20–60 min, pollen tubes were collected by precipitation and fixed with 4% paraformaldehyde in PBS. For immunolabeling of ZmGa1P at pollen tubes, a rabbit antibody was generated using a polypeptide (70–83 aa) of ZmGa1P as an immunogen. The fixed pollen tubes were extensively rinsed in PBS and then probed with the purified primary antibody of ZmGa1P at a 1:1000 dilution. Fluorescein isothiocyanate (FITC) coupled with anti-rabbit IgG (Sigma, F0382) was used as a secondary antibody at a 1:500 dilution. Fluorescence signals were recorded using a confocal laser scanning microscope (Axio imager Z2, Zeiss). Western blot of secretory proteins from pollen of the maize lines was shown in Supplementary Figure 19.
Scretome assay
Supernatant of the medium after germination of SDGa25 and J66 pollen was collected by centrifugation at 200×g at room temperature to remove pollen. The supernatant samples that contained secretory proteins were loaded onto a HiTrap Q HP column (GE) equilibrated with buffer A (20 mM Tris-HCl, pH 7.5) and fractionated with a linear gradient of 0–1 M NaCl in buffer A after extensive washing in the FPLC system (ÄKTA pure, GE). Proper amounts of each eluent fraction were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) for staining with Coomassie Brilliant Blue G-250 (CBB) or for western blotting by probing with anti-ZmGa1P antibody (at 1:500 dilution) to monitor the presence of ZmGa1P proteins. The CBB stained gels of fraction 6–10 were further subjected to liquid chromatography tandem-mass spectrometry (LC-MS/MS) analyses49. In brief, the gel slices were subjected to an in-gel digestion with trypsin. After extraction with 60% acetonitrile, the resultant peptides were separated on a reverse phase C18 column and detected with a linear ion trap mass spectrometer (Thermo Finnigan). The generated mass spectra data were analyzed with Proteome Discoverer (Thermo Fischer Scientific) and were summarized in the Supplementary Table 3 and Supplementary Data 2 if the peptide spectrum match (PSM) was greater than 3. Three biological replicates were tested. Western blots and CBB stained SDS-PAGE gels of secretory proteins from pollen of maize lines were shown in Supplementary Figure 20.
Pectoplate assay
Pectin methyl esterase activity was analyzed based on the report50. In brief, the agarose plate was prepared by pouring 50 mL of media containing 0.1% (w/v) citrus pectin (Sigma), 1% (w/v) LE agarose (USB), 12.5 mM citric acid, and 50 mM Na2HPO4, pH 6.5, into 12-cm2 petri dishes. After solidification, the plates were punched using a capillary tube at equal distance. Equal volumes of the eluent fractions were loaded into the punched wells and incubated at 30 °C for 10–16 h. The plates were then stained with 0.05% (w/v) ruthenium red (R2751, Sigma) for 30 min and destained by rinsing with distilled water. The stained circle size indicated PME activity. Three biological replicates were tested. Pectoplate assays were shown in Supplementary Figure 20.
Quantification of PME activity
To quantify PME activity, different amounts of fractions were added to the reaction solution (0.01 U/μL alcohol oxidase, 0.5% citrus pectin, 200 mM sodium phosphate, pH 6.2) and incubated at room temperature for 30 min. Fluoral-P (Sigma) was then added to a final concentration of 4 mg/mL. After incubation at room temperature for 5 min, the fluorescent intensity was measured in an Enspire reader (PerkinElmer) at 510 nm upon excitation at 405 nm. The released methanol amounts that represent PME activity were quantified based on the standard curve established using a methanol gradient. Three biological replicates were tested.
DSS crosslink assay
To determine the relationship between ZmGa1P and ZmPME10-1, fraction 8 of pollen secretory proteins of SDGa25 and J66 pollen was desalted with a PD MiniTrap G-10 column (GE) in 50 mM hydroxyethyl piperazineethanesulfonic acid buffer (pH 8.0) and concentrated in a filtrate column (Amicon, Millipore). The conditioned samples were cross-linked by adding disuccinimidyl suberate (DSS, Sigma) at a 75-fold molar excess. After incubation for 1 h at room temperature, the reaction was quenched by adding 1 M Tris (pH 8.0) to a final concentration of 50 mM. The samples were separated on a NuPAGE 4–12% Bis-Tris gel (Invitrogen) and subjected to western blotting using the antibody against ZmGa1P at 1:500 dilution. For mass spectrometry analyses, the ~100-kD bands separated by SDS-PAGE were stained with CBB and excised for LC-MS/MS analyses as described above. This experiment was also performed using the total proteins extracted from unpollinated and pollinated SDGa25 silks. Three biological replicates were tested. Western blots for crosslinking proteins were shown in Supplementary Figure 21.
Firefly luciferase complementation assay
To examine protein-protein interactions, a split luciferase complementation assay was employed51. The coding sequence of ZmGa1P without a signal peptide (82−1,137 nt) was amplified from SDGa25 and fused to the pCAMBIA-35S-CLuc vector. Simultaneously, the coding region without a signal peptide (88−2,184 nt) and the PME domain (1210−2187 nt) of ZmPME10-1 were amplified from pollen of SDGa25, and inserted into vector pCAMBIA-35S-NLuc, respectively. The resulting constructs were transformed into A. tumefaciens and cultivated overnight. The Agrobacterium samples were resuspended in the infiltration medium containing 10 mM MgCl2, 10 mM 2-(N-morpholino) ethanesulfonic acid (MES), and 200 mM acetosyringone, and incubated for 1–3 h at room temperature. For co-infiltration, equal volumes of Agrobacterium containing the NLuc- or CLuc-fused construct were pairwise mixed at OD600 of 1.6 to 1.8 and infiltrated the abaxial side of 4- to 6-week-old N. benthamiana leaves. After infiltration, plants were cultivated in the dark for 12 h and then moved into a greenhouse with a 16-h light/8-h dark cycle. The infiltrated leaves were cut approximately 48 h later and sprayed with 1 mM luciferin. Images were captured using a low-light cooled CCD imaging apparatus (CHEMIPROHT 1300B/LND, 16 bits; Roper Scientific). The signal peptide cleavage sites of the above proteins were predicted using the SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP/)52. Three biological replicates were tested.
Immunolabeling analysis
To visualize the difference of pectin esterification in pollen tubes grown in silks, two near isogenic lines that vary in Ga1 locus, JKNGa1-S and JKNga1, were developed by a series of backcrosses. The silks of JKNGa1-S and JKNga1 that pollinated with themselves or with reciprocal pollen were fixed in 4% paraformaldehyde. After dehydration with an ethanol gradient, the silks were paraffin embedded. Ten-micrometer thick longitudinal sections were prepared using a microtome (RM2235, Leica). For immunolabeling, the silk sections were dewaxed, rinsed in PBS and probed with LM2022, an antibody against highly esterified homogalacturonan (University of Leeds, UK), at a 1:20 dilution. FITC-coupled anti-rat IgG (Sigma, F1763) was used as a secondary antibody at a 1:500 dilution. Fluorescence signals were recorded with a confocal laser scanning microscope (Axio imager Z2, Zeiss). In this experiment, silks from at least three individual plants were employed for analysis.
Field tests of the Ga1 locus in pollen isolation
A marker-assisted backcrossing (MAB) strategy was employed to introgress the Ga1 locus from SDGa25 into the elite parental lines of the KYN3 hybrid with white kernels. KYN3 parental lines were crossed with SDGa25 to produce the first generation, which was further backcrossed with KYN3 parental lines eight times using a single-seed descent approach. Three polymorphic markers M14, M1, and M10 (Supplementary Table 4) located in the ZmGa1P interval were used to select the Ga1 locus during backcrosses. After nine rounds of backcrossing, individuals carrying the Ga1 locus were selfed. The near isogenic hybrids carrying homozygous Ga1 locus JKN3Ga1-S and wild-type JKN3ga1 were subjected to field tests in the farms located in Beijing and Jilin, China, respectively. The near isogenic hybrids grown in the fields were surrounded by or mingled with ga1 hybrids that produce yellow or purple kernels without physical isolation. The outcrossing with the pollen from adjacent plants was evaluated by the kernel color on the resulting ears.
Electronic supplementary material
Description of Additional Supplementary Files
Acknowledgements
We thank Prof. Weicai Yang and Dr. Hongju Li for critical discussion, and Prof. Staffan Persson and Prof. Scott NeSmith for comments on the manuscript. This work was supported by the National Transgenic Science and Technology Program, Ministry of Agriculture of China (2016ZX08010-002), the National Key Research and Development Program of China (2016YFD0101803 and 2013BAD05B01), the National Natural Science Foundation of China (91535105 and 31271729), and Youth Innovation Promotion Association CAS (2016094 and 2017136).
Author contributions
H.C., Y.Z., and Y.X. designed and supervised the project. Z.Z., Z.C., J.L., S.X., Y.H., Jie L., M.W. and X.L. performed mapping, gene expression, and transformation analyses. B.Z., D.Z., H.W., and Lanjun Z. performed biochemical and immune labeling assays and examined PME activity. D.C. detected the pollen tube length growing in silks. H.Z. performed GWAS analysis. Yu’e Z. contributed to the BAC library construction and screening. Z.Z., B.Z., Y.Z., L.Z., and H.C. analyzed the data and interpreted the results. Y.Z., H.C., Z.Z. and Z.B. wrote the paper with the inputs from all authors. H.C., Y.Z., L.Z., and Y.X. contributed equally to this work.
Data availability
The authors declare that all other data supporting the findings of this study, as well as vectors and plant seeds generated as part of this study, are available from the corresponding authors upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zhaogui Zhang, Baocai Zhang.
Contributor Information
Li Zhao, Email: zhaoli@genetics.ac.cn.
Yihua Zhou, Email: yhzhou@genetics.ac.cn.
Huabang Chen, Email: hbchen@genetics.ac.cn.
Electronic supplementary material
Supplementary Information accompanies this paper at 10.1038/s41467-018-06139-8.
References
- 1.United States Department of Agriculture Foreign Agricultural Service. https://www.fas.usda.gov/data/grain-world-markets-and-trade 2016.
- 2.Nelson, O. E. in The Maize Handbook, M. Freeling, V. Walbot, Eds. (Springer New York, 1994), 496–503.
- 3.Liu X, et al. Fine mapping of the maize cross-incompatibility locus using a homogeneous population. Crop Sci. 2014;54:873–881. doi: 10.2135/cropsci2013.09.0598. [DOI] [Google Scholar]
- 4.Jones DF. Selective fertilization among the gametes from the same individuals. Proc. Natl Acad. Sci. USA. 1924;10:218–221. doi: 10.1073/pnas.10.6.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kermicle JL. A selfish gene governing pollen-pistil compatibility confers reproductive isolation between maize relatives. Genetics. 2006;172:499–506. doi: 10.1534/genetics.105.048645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kermicle JL, Evans MM. The Zea mays sexual compatibility gene ga2: naturally occurring alleles, their distribution, and role in reproductive isolation. J. Hered. 2010;101:737–749. doi: 10.1093/jhered/esq090. [DOI] [PubMed] [Google Scholar]
- 7.Correns C. Scheinbare ausnahmen von der Mendel’shen spaltungsregel fur bastarde. Ber. Dtsch. Bot. Ges. 1902;20:159–172. [Google Scholar]
- 8.Mangelsdorf PC, Jones DF. The expression of Mendelian factors in the gametophyte of maize. Genetics. 1926;11:423–455. doi: 10.1093/genetics/11.5.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jimenez JR, Nelson OE. A new fourth chromosome gametophyte locus in maize. J. Hered. 1965;56:259–263. doi: 10.1093/oxfordjournals.jhered.a107433. [DOI] [Google Scholar]
- 10.Kermicle JL, Evans MM. Pollen–pistil barriers to crossing in maize and teosinte result from incongruity rather than active rejection. Sex. Plant. Reprod. 2005;18:187–194. doi: 10.1007/s00497-005-0012-2. [DOI] [Google Scholar]
- 11.Zhang H, et al. Genetic analysis and fine mapping of the Ga1-S gene region conferring cross-incompatibility in maize. Theor. Appl. Genet. 2012;124:459–465. doi: 10.1007/s00122-011-1720-7. [DOI] [PubMed] [Google Scholar]
- 12.McClure BA, et al. Style self-incompatibility gene products of Nicotiana alata are ribonucleases. Nature. 1989;342:955–957. doi: 10.1038/342955a0. [DOI] [PubMed] [Google Scholar]
- 13.Li W, Chetelat RT. A pollen factor linking inter- and intraspecific pollen rejection in tomato. Science. 2010;330:1827–1830. doi: 10.1126/science.1197908. [DOI] [PubMed] [Google Scholar]
- 14.Fujii S, Kubo K, Takayama S. Non-self- and self-recognition models in plant self-incompatibility. Nat. Plants. 2016;2:16130. doi: 10.1038/nplants.2016.130. [DOI] [PubMed] [Google Scholar]
- 15.Moran Lauter AN, Muszynski MG, Huffman RD, Scott MP. A pectin metethylesterase ZmPme3 is expressed in gametophyte factor1-s (Ga1-s) slks and maps to that locus in maize (Zea mays L.) Front. Plant Sci. 2017;8:1926. doi: 10.3389/fpls.2017.01926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiao Y, et al. Improved maize reference genome with single-molecule technologies. Nature. 2017;546:524. doi: 10.1038/nature22971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang L, et al. VANGUARD1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. Plant Cell. 2005;17:584–596. doi: 10.1105/tpc.104.027631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilhelmi LK, Preuss D. Self-sterility in Arabidopsis due to defective pollen tube guidance. Science. 1996;274:1535–1537. doi: 10.1126/science.274.5292.1535. [DOI] [PubMed] [Google Scholar]
- 19.Lausser A, Kliwer I, Srilunchang KO, Dresselhaus T. Sporophytic control of pollen tube growth and guidance in maize. J. Exp. Bot. 2010;61:673–682. doi: 10.1093/jxb/erp330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bosch M, Hepler PK. Pectin methylesterases and pectin dynamics in pollen tubes. Plant Cell. 2005;17:3219–3226. doi: 10.1105/tpc.105.037473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian GW, Chen MH, Zaltsman A, Citovsky V. Pollen-specific pectin methylesterase involved in pollen tube growth. Dev. Biol. 2006;294:83–91. doi: 10.1016/j.ydbio.2006.02.026. [DOI] [PubMed] [Google Scholar]
- 22.Verhertbruggen Y, Marcus SE, Haeger A, Ordaz-Ortiz JJ, Knox JP. An extended set of monoclonal antibodies to pectic homogalacturonan. Carbohydr. Res. 2009;344:1858–1862. doi: 10.1016/j.carres.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 23.Zachary GJ, Major MG. Identification of M-type gametophyte factors in maize genetic resources. Crop Sci. 2018;58:719–727. doi: 10.2135/cropsci2017.09.0560. [DOI] [Google Scholar]
- 24.Catoire L, Pierron M, Morvan C, du Penhoat CH, Goldberg R. Investigation of the action patterns of pectinmethylesterase isoforms through kinetic analyses and NMR spectroscopy. Implications in cell wall expansion. J. Biol. Chem. 1998;273:33150–33156. doi: 10.1074/jbc.273.50.33150. [DOI] [PubMed] [Google Scholar]
- 25.Micheli F. Pectin methylesterases: cell wall enzymes with important roles in plant physiology. Trends Plant Sci. 2001;6:414–419. doi: 10.1016/S1360-1385(01)02045-3. [DOI] [PubMed] [Google Scholar]
- 26.Pelloux J, Rusterucci C, Mellerowicz EJ. New insights into pectin methylesterase structure and function. Trends Plant Sci. 2007;12:267–277. doi: 10.1016/j.tplants.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Sijacic P, et al. Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature. 2004;429:302–305. doi: 10.1038/nature02523. [DOI] [PubMed] [Google Scholar]
- 28.Schopfer CR, Nasrallah ME, Nasrallah JB. The male determinant of self-incompatibility in Brassica. Science. 1999;286:1697–1700. doi: 10.1126/science.286.5445.1697. [DOI] [PubMed] [Google Scholar]
- 29.Takasaki T, et al. The S receptor kinase determines self-incompatibility in Brassica stigma. Nature. 2000;403:913–916. doi: 10.1038/35002628. [DOI] [PubMed] [Google Scholar]
- 30.Zachary GJ, Major MG. Self-incompatibility in Papaver. advances in integrating the signalling network. Biochem. Soc. Trans. 2014;42:370–376. doi: 10.1042/BST20130248. [DOI] [PubMed] [Google Scholar]
- 31.Kakeda K, et al. Molecular and genetic characterization of the S locus in Hordeum bulbosum L., a wild self-incompatible species related to cultivated barley. Mol. Genet. Genom. 2008;280:509–519. doi: 10.1007/s00438-008-0383-9. [DOI] [PubMed] [Google Scholar]
- 32.Hackauf B, Wehling P. Approaching the self-incompatibility locus Z in rye (Secale cereale L.) via comparative genetics. Theor. Appl. Genet. 2005;110:832–845. doi: 10.1007/s00122-004-1869-4. [DOI] [PubMed] [Google Scholar]
- 33.Baltazar BM, de Jesus Sanchez-Gonzalez J, de la Cruz-Larios L, Schoper JB. Pollination between maize and teosinte: an important determinant of gene flow in Mexico. Theor. Appl. Genet. 2005;110:519–526. doi: 10.1007/s00122-004-1859-6. [DOI] [PubMed] [Google Scholar]
- 34.Nelson OE. Non-reciprocal cross-sterility in maize. Genetics. 1952;37:101–124. doi: 10.1093/genetics/37.2.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Unterseer S, et al. A powerful tool for genome analysis in maize: development and evaluation of the high density 600 k SNP genotyping array. BMC Genom. 2014;15:823. doi: 10.1186/1471-2164-15-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bradbury PJ, et al. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- 37.Yu J, et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006;38:203–208. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Z, et al. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 2010;42:355–360. doi: 10.1038/ng.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lipka AE, et al. GAPIT: genome association and prediction integrated tool. Bioinformatics. 2012;28:2397–2399. doi: 10.1093/bioinformatics/bts444. [DOI] [PubMed] [Google Scholar]
- 40.VanRaden PM. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008;91:4414–4423. doi: 10.3168/jds.2007-0980. [DOI] [PubMed] [Google Scholar]
- 41.Li MX, Yeung JM, Cherny SS, Sham PC. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum. Genet. 2012;131:747–756. doi: 10.1007/s00439-011-1118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo M, Wing RA. An improved method for plant BAC library construction. Methods Mol. Biol. 2003;236:3–20. doi: 10.1385/1-59259-413-1:3. [DOI] [PubMed] [Google Scholar]
- 43.Shi X, Zeng H, Xue Y, Luo M. A pair of new BAC and BIBAC vectors that facilitate BAC/BIBAC library construction and intact large genomic DNA insert exchange. Plant. Methods. 2011;7:33. doi: 10.1186/1746-4811-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 45.Chin CS, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 46.Solovyev V, Kosarev P, Seledsov I, Vorobyev D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 2006;7(Suppl 1):S10. doi: 10.1186/gb-2006-7-s1-s10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sekhon RS, et al. Genome-wide atlas of transcription during maize development. Plant J. 2011;66:553–563. doi: 10.1111/j.1365-313X.2011.04527.x. [DOI] [PubMed] [Google Scholar]
- 48.Schreiber DN, Dresselhaus T. In vitro pollen germination and transient transformation of Zea mays and other plant species. Plant. Mol. Biol. Rep. 2003;21:31–41. doi: 10.1007/BF02773394. [DOI] [Google Scholar]
- 49.Hafidh S, et al. Quantitative proteomics of the tobacco pollen tube secretome identifies novel pollen tube guidance proteins important for fertilization. Genome Biol. 2016;17:81. doi: 10.1186/s13059-016-0928-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lionetti V. PECTOPLATE: the simultaneous phenotyping of pectin methylesterases, pectinases, and oligogalacturonides in plants during biotic stresses. Front. Plant Sci. 2015;6:331. doi: 10.3389/fpls.2015.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen H, et al. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008;146:368–376. doi: 10.1104/pp.107.111740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The authors declare that all other data supporting the findings of this study, as well as vectors and plant seeds generated as part of this study, are available from the corresponding authors upon request.