Abstract
RAS mutations are frequent in relapsed/refractory multiple myeloma (RRMM) but functional study in primary samples is scanty. Herein, in primary myeloma plasma cells of 17 suspected RRMM, functional activation of RAS signalling was studied by Western blot of phosphorylated ERK1/2 (phospho-ERK1/2). Moreover, activating mutations in KRAS, NRAS, BRAF, and ALK were studied by PCR and bidirectional direct sequencing. Furthermore, methylation of negative RAS signalling regulator genes, RASSF1A and RASD1, were analyzed by methylation-specific PCR. As evidenced by phospho-ERK1/2 over-expression, functional RAS activation was detected in 12 (75.0%) RRMM. Of patients with functional RAS activation, sequencing data showed only seven (58.3%) patients with one each had NRAS Q61H, NRAS Q61K, KRAS G12D, KRAS G12V, KRAS G13D, KRAS Q61P, or BRAF V600E mutation, whereas five (41.7%) patients had no RAS/RAF mutation. Conversely, patients without functional RAS activation had no RAS/RAF mutation. Moreover, none of the patients with functional RAS activation had ALK mutations, or methylation of RASSF1A and RASD1. Collectively, functional activation of RAS signalling was present in majority of RRMM but only about half (58.3%) accountable by RAS/RAF mutations. If verified in larger studies, clinical investigations of MEK inhibitors are warranted regardless of RAS/RAF mutations.
Introduction
RAS signalling pathway plays a key role in the regulation of cellular proliferation1. RAS family proteins consist of HRAS, KRAS, and NRAS. In mammalian cells, binding of cytokines, growth factors or mitogens to their cognate surface receptors will lead to activation of the corresponding receptor tyrosine kinases (RTKs) in the intracellular domain, recruitment and hence formation of the cytosolic SHC/GRB2/SOS complex, whereby inactive GDP-bound RAS is converted into active GTP-bound RAS2. Activated RAS will sequentially phosphorylate and activate RAF, mitogen-activated protein kinase kinases (MEKs), and extracellular signal–regulated kinases (ERKs)3. Activated ERKs may translocate into nucleus, leading to phosphorylation and activation of multiple transcription factors, and hence gene expressions4. In cancers, activating mutations of KRAS and NRAS are frequently found and associated with over-activities of the RAS-RAF-MEK-ERK cascade, resulting in upregulation of pro-survival transcription factors involved in cell cycle progression. Other causes of constitutive activation of RAS signalling, based on phosphorylated ERK1/2 over-expression, may involve activating mutations of BRAF1 or anaplastic lymphoma kinase (ALK) receptor tyrosine kinase (localized to chromosome 2p23)5, over-expression of growth factors (such as IL-6)1, or methylation-mediated silencing of tumour-suppressive negative regulator genes, such as RASSF1A6 and RASD17, of the RAS signalling pathway.
Multiple myeloma is an incurable haematological malignancy characterized by neoplastic proliferation of clonal plasma cells in the bone marrow8. Genetic aberrations, such as t(4;14), t(14;16), deletion 17p13, and amplification of 1q21 [amp(1q21)], are associated with poor prognosis8. Activating mutations of RAS signalling pathway have been reported in approximately half of newly diagnosed myeloma and an even higher proportion of relapsed/refractory multiple myeloma (RRMM)9,10. However, evidence for functional activation of RAS signalling with over-expression of phospho-ERK1/2 by Western blot in primary myeloma plasma cells is scanty.
Previously, in a bortezomib- and lenalidomide-refractory myeloma patient, we have shown that constitutive activation of RAS signalling, as evidenced by over-expression of phospho-ERK1/2 by Western blot, was attributed by BRAF V600E but not KRAS/NRAS mutation11. Herein, we studied the activation of RAS-RAF-MEK-ERK cascade in primary myeloma plasma cells from the CD138-sorted marrow or nodal plasma cells of 17 suspected RRMM by Western blotting of phospho-ERK1/2. Status of RAS signalling activation was then correlated with activating mutations in KRAS, NRAS, BRAF, or ALK, amp(1q21), and methylation of RASSF1A and RASD1.
Results
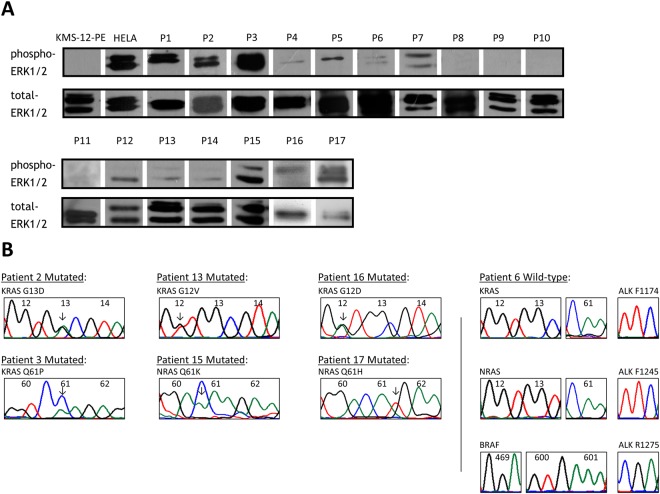
Of the 17 patients, one patient (P5) had bone marrow examination for suspected myeloma relapse due to onset of pancytopenia, which revealed Philadelphia chromosome-negative acute lymphoblastic leukaemia but no evidence of myeloma, hence was excluded from analysis. By Western blot analysis on primary myeloma plasma cells, 12/16 (75.0%) RRMM patients showed phospho-ERK1/2, indicating constitutive activation of the RAS-RAF-MEK-ERK signalling pathway (Fig. 1A and Table 1). Of the 12 patients with functional ERK activation, seven (58.3%) showed RAS/RAF mutations with one each had NRAS Q61H, NRAS Q61K, KRAS G12D, KRAS G12V, KRAS G13D, KRAS Q61P, or BRAF V600E mutation [this case was reported in Chim et al.11] (Fig. 1B and Table 1). However, the other five patients with functional ERK activation had no RAS/RAF mutation at the selected mutation hotspots, including KRAS/NRAS (codon 12, 13 and 61) and BRAF (codon 469 and 600), which account for almost all RAS activation in cancers1. Moreover, in the remaining four patients without functional ERK activation, no RAS/RAF mutation was detected.
Figure 1.
RAS signalling activation in patients with relapsed and/or refractory myeloma. (A) Western blot analysis of CD138-sorted bone marrow plasma cells showed ERK1/2 activation. KMS-12-BM served as negative and HeLa cells as positive control for RAS signalling activation based on phospho-ERK1/2 expression. Cropped blots were shown, whereas full-length blots are available upon request. (B) Sequencing analysis of NRAS, KRAS, and ALK showed KRAS G13D (patient 2), KRAS Q61P (patient 3), KRAS G12V (patient 13), NRAS Q61K (patient 15), KRAS G12D (patient 16), and NRAS Q61H (patient 17) mutations, whereas wild-type was also illustrated (patient 6).
Table 1.
RAS-RAF-MEK-ERK activation, RAS/RAF mutation, ALK mutation, 1q21 amplification, and RASSF1A/RASD1 methylation in patients with RRMM.
| Patient | pERK1/2 | Amp(1q21) | KRAS codon | NRAS codon | BRAF codon | ALK codon | RASSF1A | RASD1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 12 | 13 | 61 | 12 | 13 | 61 | 469 | 600 | 601 | 1174 | 1245 | 1275 | Methylation | Methylation | |||
| 1 | Pos | Pos | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | UU | UU |
| 2 | Pos | Pos | WT | G(G/A)C | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | UU | UU |
| 3 | Pos | Pos | WT | WT | C(A/C)A | WT | WT | WT | WT | WT | WT | WT | WT | WT | UU | UU |
| 4 | Pos | Pos | WT | WT | WT | WT | WT | WT | WT | G(T/A)G | WT | WT | WT | WT | UU | UU |
| 5^ | Pos | Neg | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | UU | UU |
| 6 | Pos | Neg | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | UU | UU |
| 7 | Pos | Pos | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | UU | UU |
| 8 | Neg | Pos | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | ND | ND | UU | UU |
| 9 | Neg | Neg | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | UU | UU |
| 10 | Neg | Pos | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | UU | UU |
| 11 | Neg | N/A | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | UU | UU |
| 12 | Pos | N/A | WT | WT | WT | WT | WT | WT | WT | WT | WT | ND | ND | ND | ND | ND |
| 13 | Pos | N/A | G(G/T)T | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | UU | UU |
| 14 | Pos | Pos | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | ND | ND |
| 15 | Pos | Pos | WT | WT | WT | WT | WT | (C/A)AA | WT | WT | WT | WT | WT | WT | UU | UU |
| 16 | Pos | N/A | G(G/A)T | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | UU | UU |
| 17 | Pos | N/A | WT | WT | WT | WT | WT | CAT | WT | WT | WT | WT | WT | WT | UU | UU |
Keys: RRMM, relapsed/refractory multiple myeloma; Pos, positive; Neg, negative; N/A, not applicable; WT, wild-type; ND, not done due to insufficient DNA; ALK codon 1174/1245/1275 WT, TTC/TTC/CGA; KRAS codon 12/13/61 WT, GGT/GGC/CAA; NRAS codon 12/13/61 WT, GGT/GGT/CAA; BRAF codon 469/600/601 WT, GGA/GTG/AAA; UU, completely unmethylated; ^patient in very good partial remission but had bone marrow examination because of pancytopenia, which revealed Philadelphia chromosome-negative acute lymphoblastic leukaemia with no bone marrow plasmacytosis.
Furthermore, alternative mechanism of functional activation of RAS signalling by activating mutations of ALK (F1174, F1245, or R1275), which accounted for ERK activation in diagnostic and relapsed neuroblastoma5, was not found in our patients with functional activation of RAS (Fig. 1B). In addition, as amp(1q21) is an adverse cytogenetic aberration that may be acquired in myeloma patients at relapse12 with overexpression of CKS1B13, association of amp(1q21) with activation of RAS signalling in myeloma was investigated (Supplementary Fig. S1). However, by FISH in marrow samples at relapse, amp(1q21) was present in seven with and two without phospho-ERK1/2 (P = 0.700; Table 1), hence not associated with functional activation of RAS.
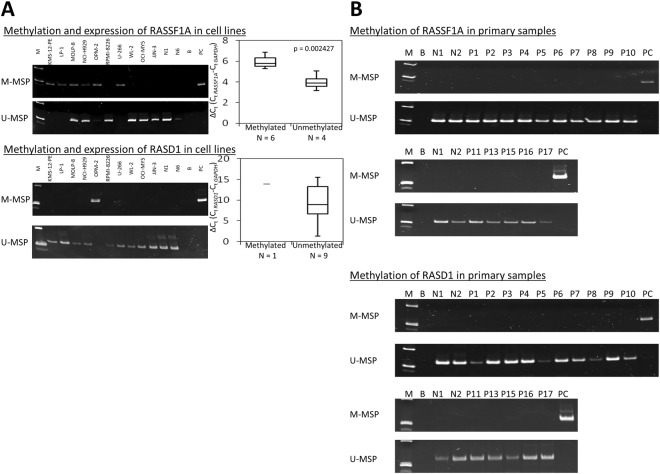
Finally, to investigate if methylation-mediated silencing of tumour suppressor genes negatively regulating RAS signalling pathway may account for functional activation of RAS, DNA methylation of RASSF1A and RASD1 promoters were studied by methylation-specific PCR (MSP). Similar to previous reports6,7, methylation of RASSF1A and RASD1 were absent in normal controls, including CD138-sorted normal bone marrow plasma cells (n = 8) and normal peripheral blood buffy coats (n = 10) (Supplementary Fig. S2), but detected in six (60%) and one (10%) human myeloma cell lines respectively, hence tumour-specific (Fig. 2A). Moreover, methylation of RASSF1A and RASD1 was associated with low expression in human myeloma cell lines (Fig. 2A), thereby demonstrating methylation-mediated gene silencing. However, in primary samples, neither RASSF1A nor RASD1 methylation was detected (Fig. 2B).
Figure 2.
Methylation-mediated silencing of tumour-suppressive negative regulators of the RAS signalling pathway. (A) Methylation-specific PCR indicated methylation of RASSF1A and RASD1 in human myeloma cell lines. Quantitative real-time RT-PCR showed an inverse correlation between methylation and expression of each of RASSF1A and RASD1. (B) Methylation-specific PCR showed absence of methylation of RASSF1A and RASD1 in primary samples of patients with relapsed and/or refractory myeloma. M: marker; B: reagent blank; N: normal control; P: patient; PC: positive control with methylated DNA.
Discussion
We have demonstrated frequent functional activation of RAS signalling pathway in 75% of RRMM, but only about half (58.3%) accountable by RAS/RAF mutations. In majority of previous studies, RAS activation, defined and hence inferred by the presence of RAS/RAF DNA mutations, was present in about 25–55% of newly diagnosed myeloma, and about 45–81% of RRMM9,10,14–17. However, our study is the first functional study of RAS activation in primary myeloma samples by Western blot of phospho-ERK1/2.
There is a recent study, in which, functional RAS activation was assessed by immunohistochemistry (IHC) for phospho-ERK1/2, and revealed functional RAS activation in 41.7% RRMM, in which RAS/RAF mutations were found in 80%18. However, the discrepancy in the frequency of functional RAS activation between of both studies might be partly explained by the different definitions of ERK activation between IHC and Western blot. In the IHC study18, ERK activation was defined by the presence of median/strong IHC signal for phospho-ERK1/2 (intensity score >1) in ≥30% of tumour cells. On the contrary, in our study, ERK activation was defined by the presence of Western blot signal for phospho-ERK1/2 in protein lysate of CD138-sorted myeloma plasma cells of the patients’ marrow and the positive (HeLa cells) but not negative (KMS-12-PE cells) controls. However, given that the incidence of myeloma is much higher in the Western countries with an incidence of 6.6/100,000/year in the US19,20, as compared to about 1.7/100,000/year in the Hong Kong Chinese21, a genuine difference in the frequency of RAS activation remains possible. On the other hand, in contrast to the IHC study showing the presence of NRAS G13R in cases without functional RAS activation, our data showed absence of RAS/RAF mutation in all RRMM patients without functional RAS activation, suggesting that functional RAS signaling activation defined by Western blotting of phosphor-ERK1/2 may be more specific. Collectively, our data showed that functional RAS activation was prevalent in RRMM that would otherwise be underestimated by the frequency of RAS/RAF DNA mutations.
Regarding the mechanism of RAS activation, RAS/RAF mutation is the most important cause. Indeed, various studies had shown frequent RAS/RAF mutations in particular in RRMM9,15. Other mechanisms of RAS activation include gain-of-function mutation of oncogenes including ALK22–24. In contrast to frequent ALK mutations accounting for RAS activation in neuroblastoma5,25, our study did not demonstrate any ALK mutation.
On the other hand, RAS activation may result from loss-of-function of tumour suppressor genes, either by loss of function mutation or promoter DNA methylation of negative regulators of RAS signalling. DNA methylation is an alternative mechanism of gene silencing mediated by addition of methyl groups to the cytosine rings of the CpG dinucleotides in the promoter-associated CpG islands of tumor suppressor genes26. In myeloma, multiple tumour suppressor genes and miRNAs have been shown to be associated with methylation-mediated gene silencing27. For instance, RASSF1A and RASD1 promoter DNA methylation have been shown in myeloma to be associated with activation of RAS signalling6,7. Herein, we showed tumour-specific methylation of both RASSF1A and RASD1 in myeloma cell lines, as evidenced by the presence of methylated MSP signals in myeloma cell lines but not normal control DNA. Moreover, the inverse correlation between RASD1 methylation and expression was consistent with methylation-mediated gene silencing. Indeed, in a genome-wide methylation study of 115 primary myeloma samples using Illumina 27 K28, RASD1 methylation was inversely correlated with expression, hence further testifying the role of methylation-mediated silencing for RASD1 in myeloma patients. However, in contrast to the previous reports of RASSF1A and RASD1 hypermethylation in primary myeloma samples6,7, RASSF1A or RASD1 methylation was absent in all samples, including those with functional RAS activation. Therefore, methylation of either RASSF1A or RASD1 may not be important for the activation of RAS in RRMM. However, in view of the limited number of samples herein, further studies with larger number of patients are required. Moreover, investigation into additional mechanisms of activation of RAS signalling is warranted. In addition to promoter DNA methylation, RASSF1A inactivation had been shown to be associated with a repressive chromatin configuration with histone deacetylation and H3K9 dimethylation in prostate cancer cell lines29.Nonetheless, in primary lung, liver and renal cancer tissues, RASSF1A inactivation has been consistently shown to be mediated by promoter DNA methylation, hence an important role in the regulation of RASSF1A expression30.
Finally, as our study showed functional activation of RAS in the majority (75%) of RRMM regardless of the presence of RAS/RAF mutation, activated RAS signaling poses a potential therapeutic target in RRMM. Moreover, as functional activation of RAS may occur with either RAS or RAF mutations, inhibition of RAS signalling appears more effective by the use of inhibitors to downstream signalling effectors such as MEK or ERK1/2. Indeed, there are ongoing clinical trials using MEK inhibitor, trametinib, in RRMM, which appeared promising31,32.
Conclusions
Collectively, functional activation of RAS signalling, as evidenced by ERK activation, was present in the majority of RRMM, but only accountable by known RAS/RAF mutations in half. In view of prevalent functional RAS activation regardless of RAS/RAF mutations, a clinical trial of MEK inhibitors is warranted in RRMM.
Methods
Patients
Seventeen patients with suspected RRMM, including those with “relapsed” or “relapsed-and-refractory myeloma” were included for functional study of activation of RAS-RAF-MEK-ERK cascade by Western blot of phospho-ERK1/2 in primary myeloma plasma cells from the CD138-sorted marrow or nodal plasma cells.
Relapsed myeloma is defined as previously treated myeloma that progresses and requires the initiation of salvage therapy but does not meet criteria for either “primary refractory myeloma” or “relapsed-and-refractory myeloma” categories33. Relapsed and refractory myeloma is defined as disease that is nonresponsive while on salvage therapy, or progresses within 60 days of last therapy in patients who have achieved minimal response (MR) or better at some point previously before then progressing in their disease course33.
Patient demographics were described in Supplementary Table S1. This study has been approved by the Institutional Review Board of Queen Mary Hospital with informed consent in accordance with the Declaration of Helsinki. All study methods were performed in accordance with relevant guidelines and regulations.
Cell cultures
Human myeloma cell lines LP-1 and RPMI-8226 were kindly provided by Prof. Robert Orlowski (Department of Lymphoma/Myeloma, Division of Cancer Medicine, The University of Texas MD Anderson Cancer Center, Houston, TX, USA), JJN-3 and OCI-MY5 by Prof. Wee Joo Chng (Department of Medicine, Yong Loo Lin School of Medicine, National University of Singapore), and WL-2 by Prof. Andrew Zannettino (Myeloma Research Programme, The University of Adelaide, Australia). NCI-H929 was purchased from American Type Culture Collection (Manassas, VA, USA). Other myeloma cell lines (KMS-12-PE, MOLP-8, OPM-2 and U-266) were purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) (Braunschweig, Germany). Cells were cultured in RPMI-1640 medium (IMDM for LP-1), supplemented with 10% fetal bovine serum, 50 U/ml of penicillin and 50 ug/ml streptomycin, and incubated in a humidified atmosphere of 5% CO2 at 37 °C. All cell culture reagents were purchased from Invitrogen (Carlsbad, CA, USA).
Western blotting for ERK1/2 activation
Purified CD138+ plasma cells obtained from magnetic-activated cell sorting (Miltenyi Biotec, Cologne, Germany) were lysed in RIPA buffer supplemented with phosphatase inhibitor cocktail and 1 mM PMSF. Cell debris was removed by centrifugation at 10,000 × g for 5 min at 4 °C. Protein lysate was heated in an equal volume of blue loading buffer at 95 °C for 5 min, and separated on 10% SDS-PAGE. Separated samples were then transferred to a 0.45 μm PVDF membrane (Amersham Biosciences, Buckinghamshire, UK). The membrane was blocked at room temperature for 1 hour in 5% skim milk diluted in PBS-Tween 20 (0.5% v/v). The membrane was then incubated with specific primary antibody (1:1000) (Cell signalling, Danvers, MA, USA) at 4 °C overnight with shaking. After washing 3 times of 15 minutes each in PBS-Tween 20 (0.5% v/v), the membrane was incubated with specific horseradish peroxidase conjugate secondary antibody (1:1000) (Bio-Rad) at room temperature for 1 hour. After washing 3 times of 15 minutes each in PBS-Tween 20 (0.5% v/v), signals were detected by ECL Western blotting detection reagents (Amersham Biosciences, Buckinghamshire, UK) and exposed to X-ray film.
Mutation analysis of KRAS, NRAS, BRAF, and ALK
Direct sequencing analysis was employed to study codons 12, 13, and 61 of KRAS and NRAS, which encompass more than 96% of all RAS mutations in human cancers34; to examine codons 469, 600 and 601 of BRAF, which represent more than 85% of all BRAF mutations in human cancers35; and to study codons 1174, 1245, 1275 of ALK, which account for 85% of ALK mutations in diagnostic neuroblastoma36. In brief, genomic DNA extracted from CD138-sorted plasma cells was amplified by PCR, followed by bidirectional direct sequencing. Primer sets specific to these regions adopted from literatures were shown in Supplementary Table S2 5,35,37. Results were compared to reference sequences NM_004304 (ALK), NM_004985 (KRAS), NM_002524 (NRAS) and NM_004333 (BRAF).
Fluorescence in situ hybridization (FISH)
Detection of cytogenetic abnormalities was performed on myeloma cells in the bone marrow by FISH, as previously described38. Interphase FISH was performed on slides for the examination of high-risk (HR) karyotypes, including t(4;14), t(14;16), del(17p), and amp(1q21), by IGH/FGFR3 DF FISH Probe kit (Vysis, USA), IGH/MAF DF FISH Probe kit (Vysis, USA), TP53/CEP17 FISH Probe kit (Vysis, USA), and CKS1B/CDKN2C (P18) amplification/deletion probe (Cytocell) respectively, in accordance with the International Myeloma Workshop Consensus recommendation. At least 200 nuclei were analyzed and scored independently by two persons. The cutoff for positivity was above 5% or at least 10 positive nuclei based on test validation data.
Methylation-specific PCR (MSP) for RASSF1A and RASD1
Genomic DNA of CD138-sorted marrow plasma cells was extracted by QIAamp DNA Blood Mini Kit (Qiagen, Germany), followed by bisulfite conversion of unmethylated cytosine to uracil (but unaffecting methylated cytosine) using EpiTect Bisulfite Kit (Qiagen, Germany). Healthy bone marrow donor DNA and enzymatically methylated control DNA (CpGenome Universal Methylated DNA, Chemicon) served as negative control and positive control respectively in all PCR. Primers used for the methylated MSP (M-MSP) and unmethylated MSP (U-MSP) are shown in Supplementary Table S2. MSP was performed in a Veriti thermal cycler (Applied Biosystems, Foster City, CA). The MSP mixture contained 50 ng of bisulfite-treated DNA, 0.2 mM dNTPs, MgCl2, 10 pmol of each primer, 1 X PCR buffer, and 2.5 units of AmpliTaq Gold DNA Polymerase (ABI, Foster City, CA) in a final volume of 25 μl. Five microliters of PCR products were loaded onto 6% non-denaturing polyacrylamide gels, electrophoresed, and visualized under ultraviolet light after staining with ethidium bromide.
Quantitative real-time reverse transcription-PCR (qRT-PCR)
In myeloma cell lines, expression of RASSF1A or RASD1 was studied by SYBR Green-based qRT-PCR. In brief, total RNA was isolated using mirVana™ miRNA Isolation Kit (Ambion, Austin, TX, USA), followed by reverse transcription to cDNA using QuantiTect Reverse Transcription Kit (Qiagen), according to the manufacturers’ instructions. The resulting cDNA was used as template for qRT-PCR (iQ SYBR Green Supermix, Bio-Rad), with GAPDH as endogenous control. Primer sequences were listed in Supplementary Table S2. Correlation between methylation and expression was studied by Student’s t-test (two-tailed), whereas P < 0.05 was regarded as statistical significant.
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).
Ethics approval and informed consent
The study has been approved by Institutional Review Board of Queen Mary Hosital (UW 05-269T/932), and written informed consent was obtained from patient for publication of this study and any accompanying data or images.
Electronic supplementary material
Acknowledgements
This work was supported by Hong Kong Blood Cancer Foundation and NSFC (81470369) awarded to C.S.C. The funders had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author Contributions
C.S.C. contributed to conception, design, analysis and interpretation of data, and drafting the manuscript of the study. K.Y.W. contributed to design, experiments, analysis and interpretation of data, and drafting the manuscript of the study. Q.Y., L.Q.Y., Z.L., E.S.K.M. involved in experiments, and analysis and interpretation of data. All authors read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Kwan Yeung Wong and Qiumei Yao contributed equally.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-31820-9.
References
- 1.Downward J. Targeting RAS signalling pathways in cancer therapy. Nature Reviews Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 2.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26:3291. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 3.Steelman L, et al. Contributions of the Raf/MEK/ERK, PI3K/PTEN/Akt/mTOR and Jak/STAT pathways to leukemia. Leukemia. 2008;22:686. doi: 10.1038/leu.2008.26. [DOI] [PubMed] [Google Scholar]
- 4.Little A, Smith P, Cook S. Mechanisms of acquired resistance to ERK1/2 pathway inhibitors. Oncogene. 2013;32:1207. doi: 10.1038/onc.2012.160. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y, et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008;455:971. doi: 10.1038/nature07399. [DOI] [PubMed] [Google Scholar]
- 6.Ng MH, et al. Alterations of RAS signalling in Chinese multiple myeloma patients: absent BRAF and rare RAS mutations, but frequent inactivation of RASSF1A by transcriptional silencing or expression of a non‐functional variant transcript. British Journal of Haematology. 2003;123:637–645. doi: 10.1046/j.1365-2141.2003.04664.x. [DOI] [PubMed] [Google Scholar]
- 7.Nojima M, et al. Genomic screening for genes silenced by DNA methylation revealed an association between RASD1 inactivation and dexamethasone resistance in multiple myeloma. Clinical Cancer Research. 2009;15:4356–4364. doi: 10.1158/1078-0432.CCR-08-3336. [DOI] [PubMed] [Google Scholar]
- 8.Chng W, et al. IMWG consensus on risk stratification in multiple myeloma. Leukemia. 2014;28:269–277. doi: 10.1038/leu.2013.247. [DOI] [PubMed] [Google Scholar]
- 9.Bezieau S, et al. High incidence of N and K Ras activating mutations in multiple myeloma and primary plasma cell leukemia at diagnosis. Human Mutation. 2001;18:212–224. doi: 10.1002/humu.1177. [DOI] [PubMed] [Google Scholar]
- 10.Walker BA, et al. Mutational spectrum, copy number changes, and outcome: results of a sequencing study of patients with newly diagnosed myeloma. Journal of Clinical Oncology. 2015;33:3911–3920. doi: 10.1200/JCO.2014.59.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chim CS, Wong KY. Bortezomib/bendamustine/dexamethasone induced good PR in refractory relapse post auto-SCT with constitutive RAS activation due to V600E BRAF mutation. Bone marrow transplantation. 2014;49:1545–1547. doi: 10.1038/bmt.2014.192. [DOI] [PubMed] [Google Scholar]
- 12.Hanamura I, et al. Frequent gain of chromosome band 1q21 in plasma-cell dyscrasias detected by fluorescence in situ hybridization: incidence increases from MGUS to relapsed myeloma and is related to prognosis and disease progression following tandem stem-cell transplantation. Blood. 2006;108:1724–1732. doi: 10.1182/blood-2006-03-009910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi L, et al. Over-expression of CKS1B activates both MEK/ERK and JAK/STAT3 signaling pathways and promotes myeloma cell drug-resistance. Oncotarget. 2010;1:22. doi: 10.18632/oncotarget.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat Rev Cancer. 2012;12:335–348. doi: 10.1038/nrc3257. [DOI] [PubMed] [Google Scholar]
- 15.Chng WJ, et al. Clinical and biological significance of RAS mutations in multiple myeloma. Leukemia. 2008;22:2280–2284. doi: 10.1038/leu.2008.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mulligan G, et al. Mutation of NRAS but not KRAS significantly reduces myeloma sensitivity to single-agent bortezomib therapy. Blood. 2014;123:632–639. doi: 10.1182/blood-2013-05-504340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith D, et al. RAS mutation status and bortezomib therapy for relapsed multiple myeloma. British journal of haematology. 2015;169:905–908. doi: 10.1111/bjh.13258. [DOI] [PubMed] [Google Scholar]
- 18.Xu J, et al. Molecular signaling in multiple myeloma: association of RAS/RAF mutations and MEK/ERK pathway activation. Oncogenesis. 2017;6:e337. doi: 10.1038/oncsis.2017.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA: a cancer journal for clinicians. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 20.Waxman AJ, et al. Racial disparities in incidence and outcome in multiple myeloma: a population-based study. Blood. 2010;116:5501–5506. doi: 10.1182/blood-2010-07-298760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong Kong Hospital Authority. Hong Kong Cancer Registry, Hospital Authority, http://www3.ha.org.hk/cancereg/ (2018).
- 22.Chiarle R, Voena C, Ambrogio C, Piva R, Inghirami G. The anaplastic lymphoma kinase in the pathogenesis of cancer. Nature Reviews Cancer. 2008;8:11–23. doi: 10.1038/nrc2291. [DOI] [PubMed] [Google Scholar]
- 23.Dance M, Montagner A, Salles J-P, Yart A, Raynal P. The molecular functions of Shp2 in the Ras/Mitogen-activated protein kinase (ERK1/2) pathway. Cellular signalling. 2008;20:453–459. doi: 10.1016/j.cellsig.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 24.Wandzioch E, Edling CE, Palmer RH, Carlsson L, Hallberg B. Activation of the MAP kinase pathway by c-Kit is PI-3 kinase dependent in hematopoietic progenitor/stem cell lines. Blood. 2004;104:51–57. doi: 10.1182/blood-2003-07-2554. [DOI] [PubMed] [Google Scholar]
- 25.Eleveld TF, et al. Relapsed neuroblastomas show frequent RAS-MAPK pathway mutations. Nature genetics. 2015;47:864. doi: 10.1038/ng.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baylin SB. DNA methylation and gene silencing in cancer. Nature clinical practice Oncology. 2005;2:S4–S11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 27.Wong, K. Y. & Chim, C. S. DNA methylation of tumor suppressor protein-coding and non-coding genes in multiple myeloma (2015). [DOI] [PubMed]
- 28.Kaiser MF, et al. Global methylation analysis identifies prognostically important epigenetically inactivated tumor suppressor genes in multiple myeloma. Blood. 2013;122:219–226. doi: 10.1182/blood-2013-03-487884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawamoto K, et al. Epigenetic modifications of RASSF1A gene through chromatin remodeling in prostate cancer. Clinical Cancer Research. 2007;13:2541–2548. doi: 10.1158/1078-0432.CCR-06-2225. [DOI] [PubMed] [Google Scholar]
- 30.Donninger H, Vos MD, Clark GJ. The RASSF1A tumor suppressor. Journal of cell science. 2007;120:3163–3172. doi: 10.1242/jcs.010389. [DOI] [PubMed] [Google Scholar]
- 31.Heuck CJ, et al. Inhibiting MEK in MAPK pathway-activated myeloma. Leukemia. 2016;30:976–980. doi: 10.1038/leu.2015.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trudel, S. et al. (Am Soc Hematology, 2016).
- 33.Rajkumar SV, et al. Consensus recommendations for the uniform reporting of clinical trials: report of the International Myeloma Workshop Consensus Panel 1. Blood. 2011;117:4691–4695. doi: 10.1182/blood-2010-10-299487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Research. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 36.Bresler SC, et al. ALK mutations confer differential oncogenic activation and sensitivity to ALK inhibition therapy in neuroblastoma. Cancer cell. 2014;26:682–694. doi: 10.1016/j.ccell.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neri A, Knowles DM, Greco A, McCormick F, Dalla-Favera R. Analysis of RAS oncogene mutations in human lymphoid malignancies. Proceedings of the National Academy of Sciences. 1988;85:9268–9272. doi: 10.1073/pnas.85.23.9268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma ES, Wang CL, Wong AT, Choy G, Chan TL. Target fluorescence in-situ hybridization (Target FISH) for plasma cell enrichment in myeloma. Molecular cytogenetics. 2016;9:63. doi: 10.1186/s13039-016-0263-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.