Abstract
Prolyl 4-hydroxylase alpha subunit is the enzymic active site of prolyl 4-hydroxylase, which is a critical enzyme to maintain the stability of newly synthesized collagens. The expression profile and functional role of P4HA3 in gastric cancer have not been explored. In the Cancer Genome Atlas-Stomach Cancer, P4HA3 RNA is significantly upregulated in gastric cancer than in normal stomach tissues. In the Human Protein Atlas, Prolyl 4-hydroxylase alpha subunit is not detectable by immunohistochemistry staining in normal stomach tissues, but it has weak staining in 7 of 12 gastric cancer tissues. Further study showed that SNAI2 (encoding Slug) is highly coexpressed with P4HA3 (Pearson r = 0.70) in Cancer Genome Atlas-Stomach Cancer. In vitro cell assay showed that Slug could efficiently bind to the P4HA3 promoter and increase its transcription. P4HA3 exon array data in Cancer Genome Atlas-Stomach Cancer revealed that 2 exons are significantly upregulated in M1 (N = 27) cases than in M0 (N = 367) cases. In MKN-45 and AGS cells, P4HA3 upregulation could enhance cell motility and invasiveness. In Cancer Genome Atlas-Stomach Cancer, high P4HA3 exon expression is associated with significantly worse 5-year and 10-year overall survival (P = .007 and .009, respectively). Data mining in Kaplan-Meier plotter also showed that high P4HA3 expression is related to unfavorable overall survival (hazard ratio: 1.54, 95% confidence interval: 1.23-1.93, P < .001) and first progression-free survival (hazard ratio: 1.64, 95% confidence interval: 1.29-2.1, P < .001). Based on findings above, we infer that P4HA3 is epigenetically activated by Slug, and its deregulation is associated with enhanced metastasis and poor survival of gastric cancer.
Keywords: P4HA3, slug, gastric cancer, metastasis, survival
Introduction
Extracellular matrix (ECM) is a critical component of tumor microenvironment, and its remodeling is closely associated with cancer development such as cancer cell growth, survival, invasion, and motility.1 Collagens are one of the major constituents of the ECM, which constitute up to 90% of the ECM.1,2 In cancer biology, collagens are critical molecules regulating cell polarity, motility, and signaling.3 Their deposition and stiffening are associated with facilitated metastasis of many solid tumors.4
Prolyl 4-hydroxylase (P4 H) is an evolutionarily conserved enzyme, which can catalyze the formation of (2 S, 4 R)-4-hydroxyproline (Hyp).5 (2 S, 4 R)-4-hydroxyproline is a critical posttranslational modification to ensure the proper 3-dimensional folding of newly synthesized procollagen chains into triple helix.5 Therefore, P4 H is an essential enzyme to maintain collagen stability. Mammalian P4 H is a α2β2 tetramer in which the prolyl 4-hydroxylase alpha subunit (P4HA) is the enzymic substrate-binding domain.5 Three isoforms of P4HA have been identified (P4HA1, P4HA2, and P4HA3), among which P4HA1 is the most prevalent.5,6 P4HA2 is mainly expressed in osteoblasts, chondrocytes, and capillary endothelial cells.7 In comparison, the expression of P4HA3 is very low in normal fetal and adults tissues.6
As the enzymic component of P4 H, dysregulated P4HA is associated with a series of pathological processes, including tumor initiation and progression.8 P4HA1 and P4HA2 are critical for collagen deposition by breast cancer cells.4,9 Their upregulation directly increases the invasiveness potential of the cancer cells and enhances metastasis to lymph nodes and lungs.4,9 Increased expression of P4HA1 in prostate cancer can attenuate the expression of tumor suppressor FLRT3 but increase the expression of Matrix metalloproteinase 1 (MMP1) and Matrix metalloproteinase 2 (MMP2) to trigger invasion and metastasis.10 Another recent study based on the bioinformatic analysis in The Cancer Genome Atlas (TCGA) found that P4HA3 upregulation is highly correlated with genes representing ECM production in breast cancer, and higher P4HA3 expression is associated with worse prognosis.11
Extracellular matrix (ECM) remodeling also plays an essential role during the invasion and metastasis process of gastric cancer (GC).12,13 Slug, which is also known as SNAI2, is a transcription factor that is involved in regulation of ECM in GC.14,15 In this study, we found that P4HA3 is significantly upregulated in GC than in normal tissues, and its upregulation is epigenetically activated by Slug. Also, we also observed that P4HA3 upregulation is associated with GC metastasis and poor survival.
Materials and Methods
Bioinformatic Analysis Based on Data in the TCGA
P4HA3 RNA expression in gastrointestinal and some other solid tumors and in the corresponding normal tissues was examined using data from TCGA. In this database, 478 patients with GC having primary tumors, and 102 cases of normal solid tissues were included. Four hundred fifteen GC samples and 32 normal samples were subjected to RNA-seq analysis (IlluminaHiSeq UNC). The inclusion of patients and the availability of the genomic and pathological data were summarized in Supplemental Figure 1. Data analysis was performed using the FireBrowse (http://firebrowse.org/). Heatmap of P4HA3 exon expression in patients with primary GC was reviewed using the UCSC Xena browser (https://xenabrowser.net/). The correlation between P4HA3 RNA and SNAI2 RNA expression was analyzed. Kaplan-Meier plots of 5-year and 10-year OS were generated by dividing the patients into high and low P4HA1/P4HA2/P4HA3 expression groups according to median exon expression. The genes coexpressed with P4HA3 in TCGA-Stomach Cancer (STAD) were identified by using cBioPortal for Cancer Genomics.16,17
Data Mining in the Human Protein Atlas
P4HA3 expression in cancer tissues and the corresponding normal tissues were reviewed by data mining in Human Protein Atlas (http://www.proteinatlas.org/).18 The images of P4HA3 immunohistochemistry (IHC) staining in GC tissues (N = 12) and normal stomach tissues were also downloaded from this database.
Data Mining in Kaplan-Meier Plotter
Association between P4HA3 expression and overall survival (OS) or first progression-free survival (FPS) was examined using Kaplan-Meier plotter, which is an online database containing gene expression data and survival information of 1065 patients with GC.19 The patients were grouped by median P4HA3 expression. The hazard ratio (HR) with 95% confidence intervals (CIs) and log-rank P value were calculated. The number at risk is indicated below the survival curves.
Cell Culture and Transfection
Poorly differentiated gastric adenocarcinoma cancer cell line MKN-45 and AGS cells were obtained from the Institute of Basic Medical Sciences of the Chinese Academy of Medical Sciences (Beijing, China). MKN-45 cells were cultured in a standard RPMI-1640, and AGS cells were grown in a standard Dulbecco's Modified Eagle Medium (DMEM) medium, each supplemented with 10% (vol/vol) fetal bovine serum (FBS) and 1% (vol/vol) penicillin and streptomycin (10000 µg/mL) in a humidified chamber at 37°C in the presence of 5% CO2.
Lentiviral Slug and P4HA3 expression particle and the corresponding negative controls were purchased from GeneCopoeia (Rockville, Maryland). Lentiviral Slug short hairpin RNA (shRNA; sc-38393-V) and P4HA3 shRNA (sc-97000-V) and the corresponding negative controls were purchased from Santa Cruz Biotechnology (Santa Cruz, California). Cells were infected with the lentiviral particles in the presence of Polybrene.
Real-Time Quantitative Reverse Transcription PCR
Briefly, total RNA in the cell samples were extracted using the Trizol Reagent (Invitrogen, Carlsbad, California, USA). Then, the complementary DNA (cDNA) was obtained by reverse transcription using the iScript cDNA Synthesis kit (Bio-Rad, Carlsbad, California) following the manufacturer’s instructions. To quantify the expression of P4HA3 messenger RNA (mRNA), qPCR was conducted using the gene-specific primers (forward, 5′-AAGTGGAGTACCGCATCAGC-3′ and reverse, 5′-TTGGTGACGTAGCATGGTCAA-3′) and the SYBR Select Master Mix (Applied Biosystems) in an ABI 7900HT Fast Real-Time PCR System (Applied Biosystems, Carlsbad, California). RNA expression was normalized to the housekeeping gene GAPDH and was calculated used the 2−ΔΔCT method.
Western Blotting
Cells were harvested and lysed in an Radioimmunoprecipitation assay (RIPA) buffer. Protein lysates of 30 µg were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis followed by western blot analysis. The antibodies used were as follows: rabbit monoclonal anti-Slug (1:200, sc-166476, Santa Cruz, Cambridge, UK), anti-P4HA3 (1:1000, ab101657, Abcam), and anti-β-actin (1:1000, ab3280, Abcam, Cambridge, UK). Antigen–antibody complexes were detected by electrochemiluminiscence Western blotting detection reagent (GE Healthcare, Marlborough, Massachusetts). Band densitometry was performed using ImageJ software (version 1.48, Bio-Rad, Hercules, California, USA).
Transwell Assay
Transwell assay was conducted using a Matrigel invasion chamber (BD Bioscience, San Jose, California) in a 24-well cell culture plate following the manufacturer’s instruction. Briefly, MKN-45 and AGS cells with P4HA3 overexpression or knockdown were seeded into chamber inserts containing an 8-μm pore size membrane with a thin layer matrigel matrix, with 500 μL serum-free RPMI-1640 or DMEM medium. The bottom of the well was filled with 700 μL RPMI-1640 or DMEM medium with 20% FBS. Forty-eight hours later, cells invaded to the lower surface of the membrane were fixed, while the noninvading cells on the upper surface were removed. The invaded cells were stained with 0.1% Crystal violet, and the number was then determined for 3 independent fields under a microscope.
Wound Healing Assay
Briefly, MKN-45 and AGS cells were cultured in 6-well plates and were infected with lentiviral P4HA3 shRNA particles or the negative controls. Twenty-four hours later, confluent cell monolayers were manually wounded by scraping the cells with a 200-μL pipette tip. Wound images were taken at 0 and 24 hours after the scratch. The wound areas were measured using the ImageJ software (n = 3).
ChIP-qPCR
The promoter sequence of P4HA3 was obtained from GeneCopoeia (>HPRM41923, NM_001288748, Supplemental Figure). The possible Slug-binding sites in the promoter region were predicted using the JASPAR Database (http://jaspar.genereg.net/). ChIP assay was conducted using the Upstate-ChIP Assay Kit (Upstate, Lake Placid, New York) according to the manufacturer’s instructions. Briefly, equal aliquots of chromatin supernatants from MKN-45 or AGS cells were subjected to immunoprecipitation with 1 μg of normal goat immunoglobulin G (IgG) or anti-Slug overnight at 4°C with rotation. After reverse cross-link of protein/DNA complexes to free DNA, the ChIP-enriched DNA was analyzed by quantitative polymerase chain reaction (qPCR) using the ABI 7900HT sequence detection system and SYBR green master mix. Primers used for P4HA3 promoter were left, 5′-CCATTGCATATTGCACAACC-3′and right, 5′-CAGGCAATGTTCATTCATGC-3′. The location of the primers was indicated in Figure 1F. The validated nonspecific primers (left, 5′-TTTTACGGGGCAACTACGGC-3′ and right, 5′-CAGTGGCATCCATTAGCAGGTC-3′) were used as a negative control.
Figure 1.
P4HA3 is epigenetically activated by Slug in gastric cancer. A, Regression analysis of the expression between P4HA3 and SNAI2 in Cancer Genome Atlas-Stomach Cancer (TCGA-STAD). B-C, Quantitative polymerase chain reaction (qPCR) analysis of P4HA3 messenger RNA (mRNA) expression in MKN-45 (B) and AGS (C) cells 36 hours after infection of lentiviral Slug expression particles (B) or lentiviral Slug short hairpin RNA (shRNA) (C). D-E, Western blot analysis of Slug and P4HA3 expression in MKN-45 (D) and AGS (E) cells 48 hours after infection of lentiviral Slug expression particles (D) or lentiviral Slug shRNA (E). F, Predicted high score Slug binding sites in the P4HA3 promoter region. G-H, Fold-enrichment of Slug binding at the P4HA3 promoter relative to the background in MKN-45 (G) and AGS (H) cells was measured by ChIP-qPCR. The primer set for chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) is indicated in figure F. Upon normalization to GAPDH, results were expressed as n-fold compared to IgG. I, The luciferase reporter constructs carrying intact or truncated P4HA3 promoter sequences were introduced into HEK-293 cells preinfected with lentiviral Slug expression particles. Luciferase activity was measured 24 hours posttransfection. PWM indicates position weight matrix. **, P < .01.
Dual Luciferase Assay
P4HA3 promoter has 1318 nucleotides, including 1254 nucleotides upstream and 63 nucleotides downstream of the transcriptional start site site (Supplemental Figure). To further verify the predicted binding sites and the effect of Slug on transcription activity, the intact promoter sequence (−1253/+63) and 4 truncated promoter sequences, including −800/+63, −600/+63, −400/+63, and −400/+63, were PCR amplified from the P4HA3 promoter plasmid. Then, the fragments were cloned into the site between XhoI and Hind III of the pGL3-basic luciferase reporter vector, respectively. HEK-293 cells preinfected with lentiviral Slug expression particles were plated into 12-well plates (1×105 cells/well) and were cotransfected the with 1.5 μg reconstructed luciferase plasmids or the empty control and 0.05 μg phRL-TK by using Superfectin (Qiagen, Valencia, California). Twenty-four hours after transfection, cells were lysed, and the luciferase activity was assessed using the dual-luciferase reporter assay system (Promega, Madison, Wisconsin) with a luminometer (Promega). The luciferase activity was normalized to the activity of renilla luciferase.
Statistical Analysis
Data were reported as means (standard deviation) . The group difference was performed by 2-tailed Student t test or analysis of variance with Student-Newman-Keuls test as a post hoc test. Log-rank test was performed to detect the difference between the survival curves. Regression analysis was performed to identify the correlation between the expression of P4HA3 and SNAI2 and among P4HA1/P4HA2/P4HA3. P < .05 was considered statistically significant.
Results
P4HA3 is Significantly Upregulated in Gastric Cancer Tissues than in Normal Controls
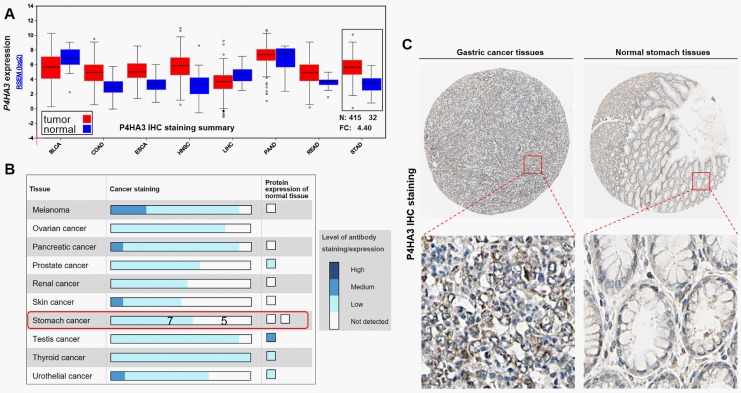
By data mining in TCGA, we first characterized P4HA3 expression in some solid tumors (Figure 2A). Results showed that P4HA3 expression is significantly higher in GC tissues (N = 415) than in normal controls (N = 32; Figure 2A). Then, we examined P4HA3 expression at the protein level by data mining in the Human Protein Atlas. In normal gastric tissues, the P4HA3 expression is usually not detectable by IHC staining (Figure 2B and C). However, among 12 cases of GC examined, 7 cases have detectable P4HA3 expression (Figure 2B), mainly in cytoplasma and membrane (Figure 2C).
Figure 2.
P4HA3 is significantly upregulated in gastric cancer tissues than in normal controls. A, P4HA3 messenger RNA (mRNA) expression in some solid tumors. Box plots were obtained by data mining in FireBrowse. B, P4HA3 IHC staining summary in some cancers and the corresponding normal tissues. C, Representative images of P4HA3 IHC staining in gastric cancer tissues and normal stomach tissues. Data and images were obtained from the Human Protein Atlas (http://www.proteinatlas.org/ENSG00000149380-P4HA3/tissue/stomach and http://www.proteinatlas.org/ENSG00000149380-P4HA3/pathology/tissue/stomach+cancer).
P4HA3 is Epigenetically Activated by Slug in Gastric Cancer
To explore the mechanism of elevated P4HA3 expression in GC, we screened the genes coexpressed with P4HA3 in TCGA-STAD and observed that SNAI2 (encoding Slug) is highly coexpressed with P4HA3 (Pearson r = 0.70, Figure 1A). Since Slug is a transcription factor that can modulate multiple genes in GC, we further studied whether it has a regulative effect on P4HA3 expression. In both MKN-45 and AGS cells, Slug overexpression significantly increased P4HA3 expression at the mRNA and protein level (Figure 1B and D). Slug shRNA reduced around 54% Slug expression in MKN-45 cells while inhibited about 72% Slug expression in AGS cells (Figure 1E). Consecutively, Slug shRNA resulted in over 70% of P4HA3 mRNA inhibition (Figure 1C) and over 35% of P4HA3 protein inhibition in both MKN-45 and AGS cells (Figure 1E). To further explore the underlying mechanisms, the promoter sequence of P4HA3 (Supplemental Figure 2) was subjected to the prediction of Slug-binding sites. Results showed that there are 2 possible Slug-binding sites in the promoter of P4HA3 (Figure 1F). To verify the prediction, ChIP-qPCR was first performed with primers designed near the predicted binding sites. Results confirmed that the DNA samples immunoprecipitated by Slug antibody from MKN-45 and AGS cells had a significantly higher enrichment of P4HA3 promoter fragments than the controls (Figure 1G and H). To further validate the direct binding and associated activating effect, luciferase reporter constructs carrying truncated P4HA3 promoter sequences were generated. The dual luciferase assay showed that Slug overexpression significantly increased the luciferase activity of the reporter carrying intact P4HA3 promoter sequence (Figure 1I). In addition, truncating anyone of the 2 predicted Slug-binding sites resulted in significant decrease in luciferase activity in HEK-293 cells preinfected with lentiviral Slug expression particles (Figure 1I). These findings suggest that Slug can efficiently bind to the P4HA3 promoter and increase its transcription.
P4HA3 Upregulation is Associated With Gastric Cancer Metastasis
By using the cBioPortal for Cancer Genomics, we found that MMP2, MMP14, and MMP19 are highly coexpressed with P4HA3 in TCGA-STAD (Table 1). Some collagen genes, such as COL6A3, COL1A2, COL3A1, COL5A2, COL5A1, COL8A1, COL1A1, COL12A1, and COL15A1, are also highly coexpressed with P4HA3 (Table 1). These findings imply that P4HA3 might be related to ECM remodeling in GC. By comparing the expression of P4HA3 between lymph nodal positive and negative cases, we failed to identify significant association (Figure 3A). However, by comparing its expression between the intestinal and diffuse histological subtypes, we found that the more malignant diffuse subtype had significantly higher P4HA3 expression compared to the intestinal subtype (Figure 3B). Then, we further assessed the association between P4HA3 and GC metastasis. By comparing P4HA3 exon expression in patients with metastasis (M1, N = 27) and without metastasis (M0, N = 367), we observed that 2 exons are significantly upregulated in M1 cases than in M0 cases (Figure 4A and B). Then, we tried to further identify the functional role of P4HA3 in cell motility and invasion. Both MKN-45 and AGS cells were transfected with PH4A3 lentiviral expression particles or shRNA (Figure 4C). Results of wound healing assay and transwell assay showed that P4HA3 overexpression significantly enhanced motility and invasion of the cancer cells (Figure 4D and E), while P4HA3 inhibition substantially reduced their motility and invasion potential (Figure 4F and G).
Table 1.
MMP and Collagen Genes Coexpressed With P4HA3 in TCGA-STAD.
| Gene Symbol | Cytoband | Pearson Score | Spearman Score |
|---|---|---|---|
| MMP2 | 16q12.2 | 0.82 | 0.76 |
| MMP19 | 12q14 | 0.66 | 0.64 |
| MMP14 | 14q11.2 | 0.65 | 0.77 |
| COL6A3 | 2q37 | 0.8 | 0.79 |
| COL1A2 | 7q22.1 | 0.74 | 0.88 |
| COL3A1 | 2q31 | 0.73 | 0.86 |
| COL5A2 | 2q14-q32 | 0.73 | 0.84 |
| COL5A1 | 9q34.2-q34.3 | 0.69 | 0.84 |
| COL8A1 | 3q12.3 | 0.69 | 0.79 |
| COL1A1 | 17q21.33 | 0.64 | 0.84 |
| COL12A1 | 6q12-q13 | 0.62 | 0.76 |
| COL15A1 | 9q21-q22 | 0.62 | 0.58 |
| COL14A1 | 8q23 | 0.56 | 0.49 |
| COL18A1 | 21q22.3 | 0.56 | 0.65 |
| COL6A1 | 21q22.3 | 0.55 | 0.58 |
| COL6A2 | 21q22.3 | 0.54 | 0.6 |
| COL16A1 | 1p35-p34 | 0.5 | 0.63 |
Abbreviations: MMP: Matrix metallopeptidase; TCGA-STAD, Cancer Genome Atlas-Stomach Cancer.
a Results were obtained from cBioPortal for Cancer Genomics.
Figure 3.
P4HA3 expression is higher in diffuse gastric cancer compared to intestinal gastric cancer. A-B, Comparison of P4HA3 expression between lymph nodal positive/negative cases (A) and between 2 histological subtypes (diffuse and intestinal) of gastric cancer (B), according to the Lauren classification.
Figure 4.
P4HA3 upregulation is associated with gastric cancer metastasis. A, Heatmap of P4HA3 exon expression in Cancer Genome Atlas-Stomach Cancer (TCGA-STAD). B, P4HA3 exon expression in patients with metastasis (M1, N = 27) and without metastasis (M0, N = 367). C, Western blot analysis of P4HA3 expression in MKN-45 and AGS cells 48 hours after infection of lentiviral P4HA3 expression particles or lentiviral shRNA. D-G, Quantitative results of wound healing assay (D and F) and transwell assay (E and G) that were conducted 48 hours after indicating infection in MKN-45 and AGS cells. The relative wound areas at 24 hours compared to 0 hours after scratch were calculated to reflect the speed of wound healing. The relative proportion of invaded cells in P4HA3 overexpression/shRNA groups compared to Vector/shNC groups was calculated to reflect the capability of cell invasion. **, P < .01.
P4HA3 but not P4HA1/P4HA2 Upregulation is Associated with Poor Survival
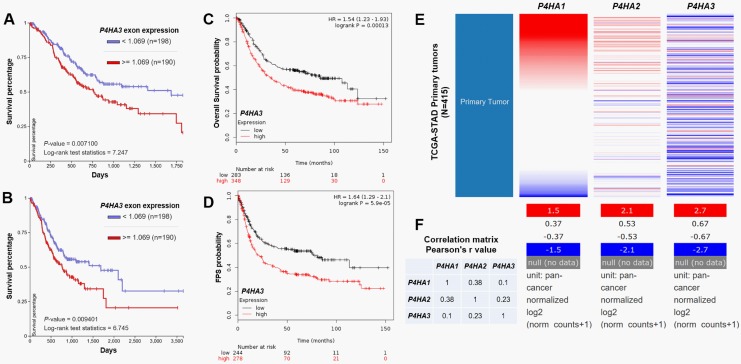
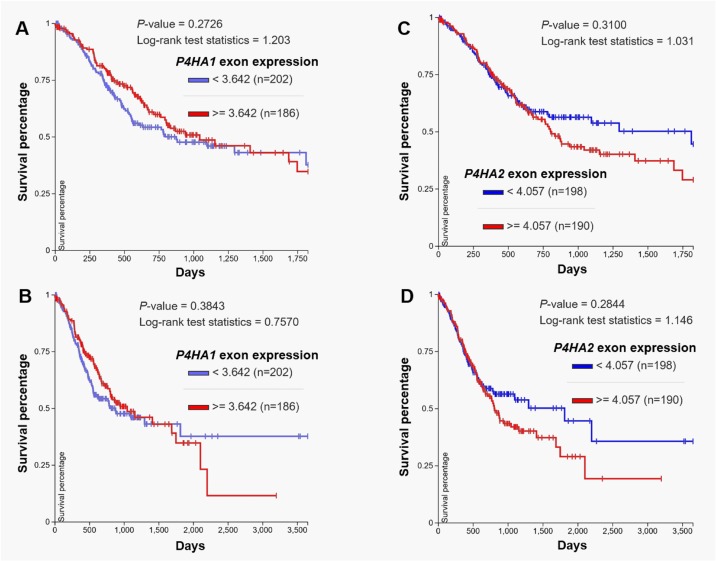
Via analyzing the survival outcomes of patients with GC in TCGA-STAD, we observed that high P4HA3 exon expression is associated with significantly worse 5-year and 10-year OS (P = .007 and .009, respectively; Figure 5A and B). Following data mining in Kaplan-Meier plotter also showed that high P4HA3 expression is associated with unfavorable OS (hazard ratio [HR]: 1.54, 95% confidence interval [CI]: 1.23-1.93, P < .001) and FPS (HR: 1.64, 95% CI: 1.29-2.1, P < .001; Figure 5C and D). Since P4HA1, P4HA2, and P4HA3 have similar functions, we also examined the association between P4HA1/P4HA2 expression and OS in patients with GC. In TCGA-STAD, no significant correlation was observed among P4HA1/P4HA2/P4HA3 (Figure 5E and F). Log-rank test of Kaplan-Meier curves also indicated that there is no significant difference in survival between high and low P4HA1 or P4HA2 exon expression groups (Figure 6A-D).
Figure 5.
P4HA3 upregulation is associated with poor survival of patients with gastric cancer. A-B, Kaplan-Meier curves of 3-year (A) and 5-year (B) overall survival of patients with gastric cancer grouped by high or low P4HA3 expression (median splitting) in Cancer Genome Atlas-Stomach Cancer (TCGA-STAD). Survival curves were generated by using the UCSC Xena. C-D, Kaplan-Meier curves of overall survival (C) and FPS (D) of patients with gastric cancer grouped by high or low P4HA3 expression (median splitting) in Kaplan-Meier Plotter. E-F, Heatmap of P4HA1/P4HA2/P4HA3 expression in Cancer Genome Atlas-Stomach Cancer (TCGA-STAD) (E) and correlation matrix (F) showing their correlations.
Figure 6.
P4HA1 or P4HA2 expression is not associated with the survival of gastric cancer. A-D, Kaplan-Meier curves of 3-year (A and C) and 5-year (B and D) overall survival of patients with gastric cancer grouped by high and low P4HA1 (A-B) or P4HA2 (C-D) expression (median splitting) in Cancer Genome Atlas-Stomach Cancer (TCGA-STAD). Survival curves were generated by using the UCSC Xena.
Discussion
As the enzymic active site of P4 H, the expression profile and functional role of P4HA3 in GC have not been explored. Although P4HA3 expression is usually low in normal human tissues, its catalytic properties are very similar to those of P4HA1 and P4HA2.6 In this study, via data mining in TCGA-STAD, we found that P4HA3 RNA is significantly upregulated in GC than in normal stomach tissues. In the Human Protein Atlas, we observed that P4HA3 is not detectable by IHC staining in normal stomach tissues, but it has weak staining in 7 of 12 GC tissues. These findings suggest that P4HA3 might be activated in some GC cases.
Via bioinformatic prediction, we found that Slug is coexpressed with P4HA3 and has 2 possible binding sites in the P4HA3 promoter. In fact, Slug is usually upregulated in GC20 and confers increased potential of motility and invasion to the cancer cells.21 In cancer biology, Slug can be either transcription suppressor or activator, depending on specific cancers and environment. Traditionally, Slug can negatively regulate the transcription of multiple genes by binding to E-box motifs in their promoters, such as E-cadherin22 and Phosphatase and tensin homolog (PTEN).23 Recent studies found that it can activate the transcription of hexokinase-2 and MMP1 by binding to their respective promoters in breast cancer24,25 and can also activate ZEB1 expression in melanoma via promoter binding.26 In GC, increased Slug expression has well-characterized oncogenic properties and also confers malignant behaviors to the cancers. Slug-induced epithelial-to-mesenchymal transition results in enhanced GC cell invasion and metastasis.27,28 Slug-positive gastric tumors are at a worse stage and also associated with a higher risk of lymph node metastasis, lymphatic invasion, and venous invasion than the tumors with negative Slug expression.20,29 However, its downstream regulation in GC has not been fully understood. In this study, we found that Slug can transactivate P4HA3 expression in GC, which is a novel epigenetic regulative effect of Slug in GC.
In this current study, via bioinformatic analysis, we observed that 2 P4HA3 exons are significantly upregulated in M1 cases than in M0 cases. However, we failed to identify any significant association between P4HA3 and lymphatic invasion. Very limited studies reported the functional role of P4HA3 in cancer,11 and no studies have explored the role of P4HA3 in colorectal cancer. The only known clue is that P4HA3 is a collagen-modifying enzyme causing 4-hydroxylation of prolines,6 and it is the only P4HA gene that seems to be selectively expressed in the stroma of breast tumors.11 Since P4HA3 is a gene modulating collagen fibres, we then tried to evaluate its contribution to collagen deposition in GC and association with tumor metastasis. Among P4HA3 coexpressed MMPs, MMP2 is able to degrade type IV collagen, and its upregulation is associated with ECM remodeling, vascular invasion, and metastasis of gastric cancer.30–32 MMP14 also had a significant correlation with lymph node metastasis and tumor stage33 and can also promote vascular invasion via enhancing the expression of vascular endothelial growth factor .34 In fact, extramural vascular invasion independently predicts metastases and poor survival in patients with GC.35,36 Therefore, we hypothesized that P4HA3 upregulation might be associated with distant metastasis via enhancing extramural vascular invasion of GC. Among P4HA3 coexpressed collagens, COL6A3, COL1A1, COL1A2, COL3A1, COL5A1 and COL5A2 are known upregulated genes in GC.37,38 COL1A2 expression level is significantly associated with the histological type and lymph node status.39,40 COL5A1 is among the signature genes associated with prognosis of GC.38 In in vitro studies using MKN-45 and AGS cells, we confirmed that P4HA3 upregulation could enhance cell motility and invasiveness. Therefore, we infer that P4HA3 is among the network regulating metastasis of GC by modulating ECM remodeling and vascular invasion. In the future, it is meaningful to further investigate the detailed functional role of P4HA3 in ECM remodeling, especially in ECM production or desmoplastic reaction and extramural vascular invasion.
P4HA3 upregulation might predict poor prognosis in breast cancer.11 In this study, we further examined the association between P4HA3 expression and survival of patients with GC. Based on data mining in 2 large databases, we observed that high P4HA3 expression is associated with unfavorable survival. However, no significant difference in survival was found between high and low P4HA1 or P4HA2 exon expression groups. These findings suggest that deregulated P4HA3 might be a specific indicator of poor survival of GC.
Conclusion
Based on findings above, we infer that P4HA3 is significantly upregulated in GC than in normal tissues and its upregulation is epigenetically activated by Slug. P4HA3 deregulation is associated with enhanced metastasis and poor survival of GC.
Supplemental Material
Supplementary_figure_1 for P4HA3 is Epigenetically Activated by Slug in Gastric Cancer and its Deregulation is Associated With Enhanced Metastasis and Poor Survival by Hu Song, Lingling Liu, Zhaoquan Song, Yongqiang Ren, Chao Li, and Jiege Huo in Technology in Cancer Research & Treatment
Supplemental Material
supplementary_figure_2 for P4HA3 is Epigenetically Activated by Slug in Gastric Cancer and its Deregulation is Associated With Enhanced Metastasis and Poor Survival by Hu Song, Lingling Liu, Zhaoquan Song, Yongqiang Ren, Chao Li, and Jiege Huo in Technology in Cancer Research & Treatment
Abbreviations
- cDNA
complementary DNA
- CI
confidence intervals
- ECM
extracellular matrix
- FPS
first progression-free survival
- FBS
fetal bovine serum
- HR
hazard ratio
- IHC
immunohistochemistry
- mRNA
messenger RNA
- OS
overall survival
- P4H
Prolyl 4-hydroxylase
- Hyp
(2 S, 4 R)-4-hydroxyproline
- P4HA
prolyl 4-hydroxylase alpha
- qPCR
quantitative polymerase chain reaction
- STAD
Stomach Cancer
- shRNA
short hairpin RNA
- TCGA
Cancer Genome Atlas
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by the Project of Nanjing Science and Technology Commission of Jiangsu Province (No. 201605064).
ORCID iD: Chao Li, MD http://orcid.org/0000-0001-8349-0911
http://orcid.org/0000-0001-8349-0911
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Gilkes DM, Semenza GL, Wirtz D. Hypoxia and the extracellular matrix: drivers of tumour metastasis. Nat Rev Cancer. 2014;14(6):430–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shoulders MD, Raines RT. Collagen structure and stability. Annu Rev Biochem. 2009;78:929–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kwon MJ, Kim Y, Choi Y, et al. The extracellular domain of syndecan-2 regulates the interaction of HCT116 human colon carcinoma cells with fibronectin. Biochem Biophys Res Commun. 15 2013;431(3):415–420. [DOI] [PubMed] [Google Scholar]
- 4. Gilkes DM, Chaturvedi P, Bajpai S, et al. Collagen prolyl hydroxylases are essential for breast cancer metastasis. Cancer Res. 2013;73(11):3285–3296. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5. Gorres KL, Raines RT. Prolyl 4-hydroxylase. Crit Rev Biochem Mol Biol. 2010;45(2):106–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kukkola L, Hieta R, Kivirikko KI, Myllyharju J. Identification and characterization of a third human, rat, and mouse collagen prolyl 4-hydroxylase isoenzyme. J Biol Chem. 2003;278(48):47685–47693. [DOI] [PubMed] [Google Scholar]
- 7. Vonk LA, Doulabi BZ, Huang CL, Helder MN, Everts V, Bank RA. Endoplasmic reticulum stress inhibits collagen synthesis independent of collagen-modifying enzymes in different chondrocyte populations and dermal fibroblasts. Biochem Cell Biol. 2010;88(3):539–552. [DOI] [PubMed] [Google Scholar]
- 8. Zhou T, Erber L, Liu B, Gao Y, Ruan HB, Chen Y. Proteomic analysis reveals diverse proline hydroxylation-mediated oxygen-sensing cellular pathways in cancer cells. Oncotarget. 2016;7(48):79154–79169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xiong G, Deng L, Zhu J, Rychahou PG, Xu R. Prolyl-4-hydroxylase alpha subunit 2 promotes breast cancer progression and metastasis by regulating collagen deposition. BMC Cancer. 2014;14:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chakravarthi BV, Pathi SS, Goswami MT, et al. The miR-124-prolyl hydroxylase P4HA1-MMP1 axis plays a critical role in prostate cancer progression. Oncotarget. 2014;5(16):6654–6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Winslow S, Lindquist KE, Edsjo A, Larsson C. The expression pattern of matrix-producing tumor stroma is of prognostic importance in breast cancer. BMC Cancer. 2016;16(1):841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yu S, Yu Y, Zhang W, et al. FOXO3a promotes gastric cancer cell migration and invasion through the induction of cathepsin L. Oncotarget. 2016;7(23):34773–34784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zuo QF, Cao LY, Yu T, et al. MicroRNA-22 inhibits tumor growth and metastasis in gastric cancer by directly targeting MMP14 and Snail. Cell Death Dis. 2015;6(11):e2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ji J, Jia S, Jia Y, Ji K, Hargest R, Jiang WG. WISP-2 in human gastric cancer and its potential metastatic suppressor role in gastric cancer cells mediated by JNK and PLC-gamma pathways. Br J Cancer. 2015;113(6):921–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lin SY, Lee YX, Yu SL, Chang GC, Chen JJ. Phosphatase of regenerating liver-3 inhibits invasiveness and proliferation in non-small cell lung cancer by regulating the epithelial-mesenchymal transition. Oncotarget. 2016;7(16):21799–21811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lindskog C. The potential clinical impact of the tissue-based map of the human proteome. Expert Rev Proteomics. 2015;12(3):213–215. [DOI] [PubMed] [Google Scholar]
- 19. Szasz AM, Lanczky A, Nagy A, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7(31):49322–49333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Uchikado Y, Okumura H, Ishigami S, et al. Increased slug and decreased E-cadherin expression is related to poor prognosis in patients with gastric cancer. Gastric Cancer. 2011;14(1):41–49. [DOI] [PubMed] [Google Scholar]
- 21. Yang L, Liang H, Wang Y, et al. MiRNA-203 suppresses tumor cell proliferation, migration and invasion by targeting slug in gastric cancer. Protein Cell. 2016;7(5):383–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Adhikary A, Chakraborty S, Mazumdar M, et al. Inhibition of epithelial to mesenchymal transition by E-cadherin up-regulation via repression of slug transcription and inhibition of E-cadherin degradation: dual role of scaffold/matrix attachment region-binding protein 1 (SMAR1) in breast cancer cells. J Biol Chem. 2014;289(37):25431–25444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Uygur B, Abramo K, Leikina E, Vary C, Liaw L, Wu WS. SLUG is a direct transcriptional repressor of PTEN tumor suppressor. Prostate. 2015;75(9):907–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Geng C, Li J, Ding F, et al. Curcumin suppresses 4-hydroxytamoxifen resistance in breast cancer cells by targeting SLUG/Hexokinase 2 pathway. Biochem Biophys Res Commun. 2016;473(1):147–153. [DOI] [PubMed] [Google Scholar]
- 25. Shen CJ, Kuo YL, Chen CC, Chen MJ, Cheng YM. MMP1 expression is activated by Slug and enhances multi-drug resistance (MDR) in breast cancer. PLoS One. 2017;12(3):e0174487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wels C, Joshi S, Koefinger P, Bergler H, Schaider H. Transcriptional activation of ZEB1 by Slug leads to cooperative regulation of the epithelial-mesenchymal transition-like phenotype in melanoma. J Invest Dermatol. 2011;131(9):1877–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen D, Zhou H, Liu G, Zhao Y, Cao G, Liu Q. SPOCK1 promotes the invasion and metastasis of gastric cancer through Slug-induced epithelial-mesenchymal transition. J Cell Mol Med. 2017;22(7):797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li Z, Mou H, Wang T, et al. A non-secretory form of FAM3B promotes invasion and metastasis of human colon cancer cells by upregulating Slug expression. Cancer Lett. 2013;328(2):278–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee HH, Lee SH, Song KY, et al. Evaluation of Slug expression is useful for predicting lymph node metastasis and survival in patients with gastric cancer. BMC Cancer. 2017;17(1):670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mrena J, Wiksten JP, Nordling S, Kokkola A, Ristimaki A, Haglund C. MMP-2 but not MMP-9 associated with COX-2 and survival in gastric cancer. J Clin Pathol. 2006;59(6):618–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu CY, Wu MS, Chen YJ, et al. Clinicopathological significance of MMP-2 and TIMP-2 genotypes in gastric cancer. Eur J Cancer. 2007;43(4):799–808. [DOI] [PubMed] [Google Scholar]
- 32. Tsai CY, Wang CS, Tsai MM, et al. Interleukin-32 increases human gastric cancer cell invasion associated with tumor progression and metastasis. Clin Cancer Res. 2014;20(9):2276–2288. [DOI] [PubMed] [Google Scholar]
- 33. Shim KN, Jung SA, Joo YH, Yoo K. Clinical significance of tissue levels of matrix metalloproteinases and tissue inhibitors of metalloproteinases in gastric cancer. J Gastroenterol. 2007;42(2):120–128. [DOI] [PubMed] [Google Scholar]
- 34. Zheng L, Li D, Xiang X, et al. Methyl jasmonate abolishes the migration, invasion and angiogenesis of gastric cancer cells through down-regulation of matrix metalloproteinase 14. BMC Cancer. 2013;13:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cheng J, Wu J, Ye Y, Zhang C, Zhang Y, Wang Y. The prognostic significance of extramural venous invasion detected by multiple-row detector computed tomography in stage III gastric cancer. Abdom Radiol (NY). 2016;41(7):1219–1226. [DOI] [PubMed] [Google Scholar]
- 36. Cheng J, Wu J, Ye Y, Zhang C, Zhang Y, Wang Y. Extramural venous invasion detected by MDCT as an adverse imaging feature for predicting synchronous metastases in T4 gastric cancer. Acta Radiol. 2017;58(4):387–393. [DOI] [PubMed] [Google Scholar]
- 37. Sun H. Identification of key genes associated with gastric cancer based on DNA microarray data. Oncol Lett. 2016;11(1):525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen Z, Soutto M, Rahman B, et al. Integrated expression analysis identifies transcription networks in mouse and human gastric neoplasia. Genes Chromosomes Cancer. 2017;56(7):535–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rong L, Huang W, Tian S, Chi X, Zhao P, Liu F. COL1A2 is a Novel biomarker to improve clinical prediction in human gastric cancer: integrating bioinformatics and meta-analysis. Pathol Oncol Res. 2017;24(1):129–134. [DOI] [PubMed] [Google Scholar]
- 40. Li J, Ding Y, Li A. Identification of COL1A1 and COL1A2 as candidate prognostic factors in gastric cancer. World J Surg Oncol. 2016;14(1):297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary_figure_1 for P4HA3 is Epigenetically Activated by Slug in Gastric Cancer and its Deregulation is Associated With Enhanced Metastasis and Poor Survival by Hu Song, Lingling Liu, Zhaoquan Song, Yongqiang Ren, Chao Li, and Jiege Huo in Technology in Cancer Research & Treatment
supplementary_figure_2 for P4HA3 is Epigenetically Activated by Slug in Gastric Cancer and its Deregulation is Associated With Enhanced Metastasis and Poor Survival by Hu Song, Lingling Liu, Zhaoquan Song, Yongqiang Ren, Chao Li, and Jiege Huo in Technology in Cancer Research & Treatment