Abstract
Gene expression data using retrieved ovarian cancer (OC) samples were used to identify genes of interest and a support vector machine (SVM) classifier was subsequently established to predict the recurrence of OC. Three datasets (GSE17260, GSE44104 and GSE51088) investigating OC gene expression were downloaded from the Gene Expression Omnibus. Differentially expressed genes (DEGs) in samples from patients with non-recurrent and recurrent OC were revealed via a homogeneity test and quality control analysis. A protein-protein interaction (PPI) network was subsequently established for the DEGs using data from Biological General Repository for Interaction Datasets, Human Protein Reference Database and Database of Interacting Proteins. Degrees of interaction and betweenness centrality (BC) scores were calculated for each node in the PPI network. The top 100 genes ranked by BC scores were selected to identify feature genes via recursive feature elimination using the GSE17260 dataset. Following this, a SVM classifier was constructed and further validated using the GSE44104 and GSE51088 datasets and independent gene expression data obtained from the Cancer Genome Atlas (TCGA). A total of 639 DEGs were identified from the three gene expression datasets, and a PPI network including 249 nodes and 354 edges was constructed. A SVM classifier consisting of 39 feature genes (including cullin 3, mouse double minute 2 homolog, aurora kinase A, WW domain containing oxidoreducatase, large tumor suppressor kinase 2, sirtuin 6, staphylococcal nuclease and tudor domain containing 1, leucine rich repeats and immunoglobulin like domains 1 and aurora kinase 1 interacting protein 1) was subsequently constructed. The prediction accuracies of the SVM classifier for GSE17260, GSE44104 and GSE51088 datasets as well as data downloaded from TCGA were revealed to be 92.7, 93.3, 96.6 and 90.4%, respectively. Furthermore, the results of the present study revealed that patients with predicted non-recurrent OC survived significantly longer compared with the patients with predicted recurrent OC (P=6.598×10−6). A SVM classifier consisting of 39 feature genes was established for predicting the recurrence and prognosis of OC. Therefore, the results of the present study suggested that the 39 feature genes may serve important roles in the development of OC and may represent therapeutic biomarkers of OC.
Keywords: ovarian cancer, recurrence, gene expression data, differentially expressed genes, feature genes, support vector machine classifier
Introduction
Ovarian cancer (OC) is the seventh most commonly diagnosed cancer in women in the USA and the average five-year survival rate of patients with OC in the USA is 45% (1). OC frequently recurs following treatment (2). Furthermore, 20% of patients with stage I and II cancer experience recurrence within a 5 year period in the USA (1). Recurrence is closely associated with the prognosis of OC (1), and, therefore, there is a requirement for novel biomarkers to predict recurrence of OC in order to improve the outcome of patients with OC.
Previous studies have identified numerous relevant prognostic biomarkers (3–5). Elevated levels of serum interleukin (IL)-37 are predictive of poor prognosis in patients with epithelial OC (6). Sprouty 2 is an independent prognostic biomarker for the survival and recurrence of human epithelial OC (7). IL-8 has been revealed to represent a biomarker for prognostic prediction in patients with recurrent platinum-sensitive OC (8). In addition, upregulation of Golgi phosphoprotein 3 is associated with poor prognosis in patients with epithelial OC (9). Class III β-tubulin overexpression within the tumor microenvironment has been demonstrated to represent a prognostic biomarker for poor overall survival in patients with OC (10). Mitogen-activated protein kinase/extracellular signal-regulated kinase 1 has been reported to represent a promising candidate prognostic biomarker and to be correlated with response rates to platinum based chemotherapy in OC (11). Flap structure-specific endonuclease 1 overexpression has been revealed to be associated with the poor survival of patients exhibiting high grade and advanced stage OC (12). In addition, overexpression of fibroblast growth factor 18 (FGF18) is an independent predictive marker for poor clinical outcome in patients with OC, and FGF18 has been demonstrated to regulate OC cell migration, invasion and tumorigenicity via nuclear factor-κB activation (13). Tumor necrosis factor α-induced protein 8 overexpression is associated with epithelial OC metastasis and poor survival, and, therefore, can function as a prognostic and therapeutic biomarker for epithelial OC (14). However, biomarkers with a greater accuracy are required to predict recurrence and prognosis of OC.
In the present study, data of samples from patients with recurrent and non-recurrent OC in three gene expression datasets were analyzed to identify differentially expressed genes (DEGs). Following this, relevant feature genes were identified and subsequently used to establish a support vector machine (SVM) classifier, the results of which were further verified using independent data. The results of the present study suggested that the SVM classifier may facilitate the prediction of OC recurrence and prognosis.
Materials and methods
Gene expression data
Gene expression data were retrieved from the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) by searching for the following key words: ‘Ovarian cancer,’ ‘recurrence,’ ‘homo sapiens’ and ‘recurrence.’ Datasets were selected for further analysis if they fulfilled the following criteria: i) Included gene expression profiles of patients with OC; and ii) included gene expression profiles of patients with recurrent and non-recurrent OC. Following this, three gene expression datasets [GSE17260 (15), GSE44104 (16) and GSE51088 (17)] were downloaded for subsequent analysis (Table I).
Table I.
Summary of gene expression datasets used in the present study.
Background correction and normalization were performed using gene expression dataset GSE44104 with package affy 1.42.3 (18) of R 3.1.0 (19). Missing values were filled using the median value (20). Microarray Suite (21) was used to perform background correction. The quantile method was used for standardization.
Screening of DEGs
Prior to meta-analysis, the characteristics of the three gene expression datasets were investigated by principal component analysis (PCA) and standardized mean rank using the MetaQC package (22). The homogeneity test of gene expression profiles among datasets (internal quality control), homogeneity test of gene expression profiles with pathway database (external quality control), accuracy quality control, accuracy of feature genes and pathways, consistency quality control and consistency in the ranking of feature genes and pathways were investigated for quality control purposes using the MetaQC package.
DEGs were screened for using MetaDE.ES from the MetaDE package (23). Firstly, tests for heterogeneity of gene expression value in numerous platforms were performed using three statistical parameters: Tau2, Q value and Cochran's Q value. Values of tau2=0 and Cochran's Q value >0.05 served as the criteria for the identification of homogenous genes. Following this, the false discovery rate (<0.05) of DEGs between non-recurrent samples and recurrent samples within each dataset was investigated. Two-way clustering analysis of sample data from patients with recurrent and non-recurrent OC in each dataset was performed using selected DEGs and then visualized by a heatmap using R 3.1.0 (19).
Construction of a protein-protein interaction (PPI) network
PPI information was downloaded from Biological General Repository for Interaction Datasets (BioGRID; thebiogrid.org), Human Protein Reference Database (HPRD; www.hprd.org) and Database of Interacting Proteins (DIP; dip.doe-mbi.ucla.edu). Using Cytoscape version 3.5.1 (http://www.cytoscape.org/) (24), DEGs were mapped into the downloaded PPIs to construct the PPI network. Gene Ontology (GO; www.geneontology.org) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis (www.kegg.jp) were performed for the genes in the PPI network using Fisher's exact test using Cytoscape version 3.5.1.
Construction of the SVM classifier
To determine which genes in the PPI network could be classified as hub genes, the degree of nodes and betweenness centrality (BC) scores were determined (25). The BC score was calculated as follows using the igraph package version 1.2.1 in R 3.1.0 (https://cran.r-project.org/web/packages/igraph/index.html).
Here, σst is the number of shortest paths from s to t; σst (ν) is the number of shortest paths from s to t that pass node v; BC score is between 0 and 1, and greater BC score indicates higher degree of hubness in the network.
The top 100 DEGs, as determined by BC scores, were selected as candidate feature genes. The dataset GSE17260 was selected as the training set because the sample is larger than the other datasets, and the difference between the number of non-recurrent samples and recurrent samples is relatively small. An optimum combination of feature genes was determined by performing recursive feature elimination using R caret_6.0–79 (https://cran.r-project.org/web/packages/caret/) (26). The SVM classifier was subsequently established to predict OC recurrence based on the expression levels of the screened feature genes.
The other two datasets (GSE44104 and GSE51088) were used to further verify the results of the SVM classifier. Sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV) and area under the receiver operating characteristic curve (AUROC) values were determined to evaluate the performance of the established SVM classifier.
Verification of results generated by the SVM classifier using independent data
A further set of microarray data from samples of patients with OC was downloaded from the Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov/) (27) and used to further verify the results of the SVM classifier. This dataset contained 222 recurrent and 173 non-recurrent OC samples. The OC samples were classified into two groups: Predicted recurrent OC samples and predicted non-recurrent OC samples. Kaplan-Meier (KM) survival curves were then plotted for the two groups to determine the reliability of the SVM classifier regarding patient prognosis.
Results
DEGs
Quality control analysis using data from the three gene expression datasets (GSE17260, GSE44104 and GSE51088) revealed that there was no significant bias among these datasets according to the SMR values (Table II) (22). In addition, PCA analysis revealed that all three datasets are distributed on the same side of the arrow, which suggest good comparability. (Fig. 1). For this reason, all three datasets were retained for subsequent analysis in the present study.
Table II.
Results of quality control measures and standardized mean rank test from data included in GSE17260, GSE44104 and GSE51088 datasets.
| Accession number | IQC | EQC | CQCg | CQCp | AQCg | AQCp | SMR |
|---|---|---|---|---|---|---|---|
| GSE17260 | 5.48 | 3.36 | 110.95 | 165.26 | 34.03 | 94.54 | 1.69 |
| GSE44104 | 4.55 | 3.29 | 66.72 | 152.42 | 27.52 | 100.64 | 2.51 |
| GSE51088 | 6.33 | 1.14 | 105.17 | 118.9 | 20.32 | 30.64 | 4.08 |
IQC, internal quality control; EQC, external quality control; AQCg, accuracy quality control of genes; AQCp, accuracy quality control of pathways; CQCg, consistency quality control of genes; CQCp, consistency quality control of pathways; SMR, standard mean rank.
Figure 1.
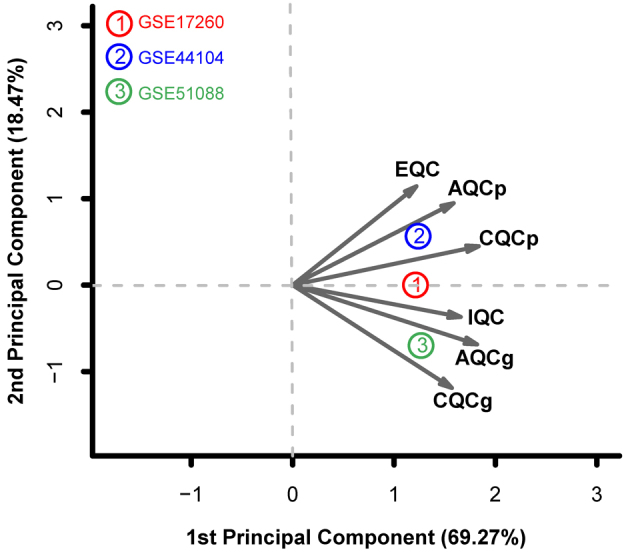
Principal component analysis of the GSE17260, GSE44104 and GSE51088 datasets. X-axis represents the first principal component and the Y-axis represents the second principal component. IQC, internal quality control; EQC, external quality control; AQCg, accuracy quality control of genes; AQCp, accuracy quality control of pathways; CQCg, consistency quality control of genes; CQCp, consistency quality control of pathways.
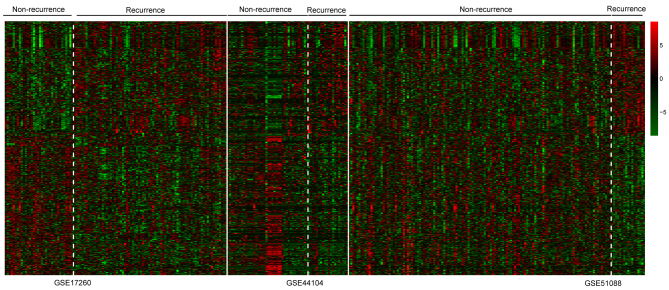
Based on the aforementioned criteria, a total of 639 DEGs were identified from the GSE17260, GSE44104 and GSE51088 datasets, including 279 upregulated DEGs and 360 downregulated DEGs. The heatmap of two-way clustering revealed marked differences in gene expression between the patient samples with recurrent and non-recurrent OC in each dataset (Fig. 2).
Figure 2.
Hierarchical clustering heatmap of samples from patients with OC in GSE17260, GSE44104 and GSE51088 datasets using the expression levels of 639 differentially expressed genes. Red panels represent high expression; green panels represent low expression. Bars represent samples from patients with non-recurrent and recurrent OC in each dataset. OC, ovarian cancer.
PPI network
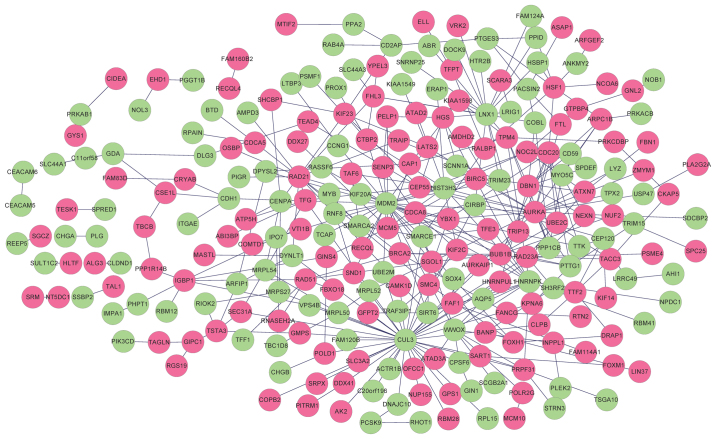
A total of 321 and 296 PPIs for selected DEGs were identified in HPRD and BioGRID, respectively. Overlapping PPIs were selected and visualized using Cytoscape (Fig. 3). The constructed PPI network contained 249 nodes (115 downregulated genes and 134 upregulated genes) and 354 edges. Functional enrichment analysis revealed the genes in the PPI network were significantly associated with 14 GO terms, including ‘cell cycle phase’, ‘M phase’, ‘mitotic cell cycle’ and ‘cell cycle process’ (Table III). Furthermore, five KEGG pathways, including ‘cell cycle’, ‘homologous recombination’, ‘purine metabolism’, ‘pathways in cancer’ and ‘DNA replication’ were revealed to be significantly enriched for the genes in the PPI network (Table IV).
Figure 3.
Protein-protein interaction network of DEGs. Red nodes indicate upregulated DEGs and green nodes indicate downregulated DEGs. DEGs, differentially expressed genes.
Table III.
Gene Ontology biological process terms significantly associated with the genes included in the protein-protein interaction network.
| Term | Count | P-value | FDR |
|---|---|---|---|
| GO:0022403, cell cycle phase | 33 | 8.56×10−15 | 1.40×10−11 |
| GO:0000279, M phase | 29 | 4.41×10−14 | 7.23×10−11 |
| GO:0000278, mitotic cell cycle | 30 | 1.24×10-13 | 2.04×10−10 |
| GO:0022402, cell cycle process | 36 | 3.14×10−13 | 5.15×10−10 |
| GO:0007067, mitosis | 23 | 1.12×10−12 | 1.83×10−9 |
| GO:0000280, nuclear division | 23 | 1.12×10−12 | 1.83×10−9 |
| GO:0000087, M phase of mitotic cell cycle | 23 | 1.61×10−12 | 2.64×10−9 |
| GO:0007049, cell cycle | 41 | 1.90×10−12 | 3.12×10−9 |
| GO:0048285, organelle fission | 23 | 2.52×10−12 | 4.13×10−9 |
| GO:0051301, cell division | 24 | 5.77×10−11 | 9.47×10−8 |
| GO:0000226, microtubule cytoskeleton organization | 13 | 1.65×10−6 | 2.71×10−3 |
| GO:0007051, spindle organization | 8 | 3.67×10−6 | 6.02×10−3 |
| GO:0007017, microtubule-based process | 16 | 4.71×10−6 | 7.72×10−3 |
| GO:0007010, cytoskeleton organization | 21 | 6.25×10−6 | 1.03×10−2 |
FDR, false discovery rate.
Table IV.
Significantly enriched Kyoto Encyclopedia of Genes and Genomes pathways for genes in the protein-protein interaction network.
| Term | Count | P-value | Genes |
|---|---|---|---|
| hsa04110:Cell cycle | 7 | 6.62×10−3 | RAD21, BUB1B, MDM2, TTK, CDC20, PTTG1, MCM5 |
| hsa03440:Homologous recombination | 3 | 5.47×10−3 | POLD1, BRCA2, RAD51 |
| hsa00230:Purine metabolism | 6 | 3.57×10−2 | POLR2G, GDA, POLD1, AK2, AMPD3, GMPS |
| hsa05200:Pathways in cancer | 9 | 4.44×10−2 | CTBP2, RALBP1, PIK3CD, TFG, MDM2, BRCA2, BIRC5, CDH1, RAD51 |
| hsa03030:DNA replication | 3 | 4.51×10−2 | POLD1, RNASEH2A, MCM5 |
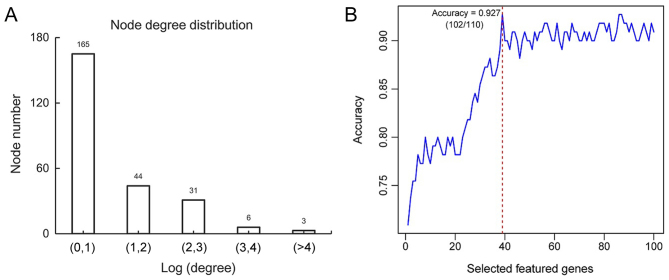
The distribution of calculated degree demonstrated that 165 genes exhibited a small degree score [Log (degree) <1]; whereas 3 genes exhibited a large degree score (Log>4; Fig. 4A). This revealed that this PPI network exhibited scale-free property similar to the majority of biological networks (25). Genes exhibiting high degrees were considered to represent hub genes and may serve important roles in the development of ovarian cancer.
Figure 4.
Distribution of the node degree of interaction and the screening of feature genes following recursive feature elimination. (A) 165 genes exhibited a small degree score. The X-axis represents the value of Log (degree of interaction) and the Y-axis represents the number of nodes in the protein-protein interaction network. (B) The accuracy is highest when the number of feature genes is 39. The X-axis indicates number of feature genes and the Y-axis represents prediction accuracy.
SVM classifier
Following the calculation of BC scores for each node and the subsequent ranking of the top 100 nodes, 39 feature genes [including cullin 3 (CUL3), mouse double minute 2 homolog (MDM2), aurora kinase A (AURKA), WW domain containing oxidoreducatase (WWOX), large tumor suppressor kinase (LATS)2, sirtuin 6 (SIRT6), staphylococcal nuclease and tudor domain containing 1 (SND1), leucine rich repeats and immunoglobulin like domains 1 (LRIG1) and aurora kinase 1 interacting protein 1 (AURKAIP1)] were determined by the recursive feature elimination (Table V). The highest prediction accuracy determined from analysis of training dataset GSE17260 was 92.7% [102 out of 110 samples (27 samples from patients with non-recurrent OC and 75 samples from patients with recurrent OC)] when 39 feature genes were used (Fig. 4B). The samples from patients with non-recurrent OC and recurrent OC from training dataset GSE17260 were also presented in a scatter plot, which clearly distinguished the recurrence samples from non-recurrence samples (Fig. 5). This result illustrated the effectiveness of the SVM classifier.
Table V.
Screened feature genes used for construction of support vector machine classifier as determined by recursive feature elimination.
| Gene | BC | Degree | P-value | FDR | Q value | Cochran's Q value | tau2 | Log fold change |
|---|---|---|---|---|---|---|---|---|
| CUL3 | 0.759895 | 41 | 0.009775 | 0.02304 | 0.198586 | 0.905478 | 0 | −3.08977 |
| MDM2 | 0.694803 | 25 | 0.014685 | 0.034611 | 0.513419 | 0.773593 | 0 | −1.09727 |
| AURKA | 0.558121 | 19 | 0.001087 | 0.002561 | 1.286947 | 0.525464 | 0 | 1.42154 |
| HNRNPK | 0.50414 | 13 | 0.011236 | 0.026482 | 1.021909 | 0.599923 | 0 | −0.96217 |
| RAD21 | 0.490358 | 12 | 0.014482 | 0.034133 | 0.386617 | 0.824228 | 0 | 2.596818 |
| WWOX | 0.458579 | 10 | 0.013516 | 0.031857 | 0.857721 | 0.651251 | 0 | −3.74564 |
| IGBP1 | 0.449997 | 7 | 0.011454 | 0.026997 | 0.47506 | 0.788573 | 0 | 0.321061 |
| IPO7 | 0.442128 | 5 | 0.002717 | 0.006405 | 1.598947 | 0.449566 | 0 | −1.03747 |
| RAD23A | 0.441265 | 8 | 0.012419 | 0.02927 | 0.900161 | 0.637577 | 0 | 1.194153 |
| TSTA3 | 0.436658 | 5 | 0.010425 | 0.02457 | 0.358922 | 0.835721 | 0 | 2.936405 |
| BRCA2 | 0.435695 | 5 | 0.003583 | 0.008444 | 0.100529 | 0.950978 | 0 | 1.948855 |
| FHL3 | 0.433658 | 6 | 0.001257 | 0.002963 | 0.279246 | 0.869686 | 0 | 0.987126 |
| LATS2 | 0.430752 | 4 | 0.011053 | 0.026051 | 0.09771 | 0.952319 | 0 | 0.4797 |
| NOC2L | 0.430291 | 4 | 0.013451 | 0.031703 | 0.014566 | 0.992743 | 0 | 1.077813 |
| CD2AP | 0.42926 | 4 | 0.018901 | 0.044548 | 0.026358 | 0.986908 | 0 | −1.79036 |
| TPM4 | 0.428095 | 7 | 0.013303 | 0.031355 | 1.621492 | 0.444526 | 0 | 2.932572 |
| MCM5 | 0.427881 | 7 | 0.006661 | 0.0157 | 0.397359 | 0.819813 | 0 | 0.44911 |
| CTBP2 | 0.427047 | 5 | 0.013728 | 0.032356 | 1.715255 | 0.424167 | 0 | 0.877948 |
| SIRT6 | 0.426042 | 6 | 0.009524 | 0.022448 | 0.145688 | 0.929746 | 0 | −0.81214 |
| RALBP1 | 0.42506 | 3 | 0.008046 | 0.018964 | 1.608012 | 0.447533 | 0 | 1.788163 |
| DBN1 | 0.422172 | 9 | 0.001295 | 0.003052 | 1.479727 | 0.477179 | 0 | 1.499493 |
| FAF1 | 0.420131 | 4 | 0.014442 | 0.034039 | 1.184726 | 0.553019 | 0 | 1.782133 |
| SMC4 | 0.416491 | 5 | 0.005295 | 0.01248 | 0.238612 | 0.887536 | 0 | 1.769396 |
| SND1 | 0.41646 | 3 | 0.003913 | 0.009222 | 0.920509 | 0.631123 | 0 | 1.238259 |
| TEAD4 | 0.414377 | 2 | 0.008077 | 0.019037 | 1.729928 | 0.421067 | 0 | 0.992568 |
| BANP | 0.411436 | 3 | 0.004271 | 0.010067 | 0.305887 | 0.858178 | 0 | 0.759078 |
| SART1 | 0.409053 | 3 | 0.01088 | 0.025643 | 0.068045 | 0.96655 | 0 | 1.254434 |
| INPPL1 | 0.408929 | 2 | 9.41E-05 | 0.000222 | 1.3698 | 0.504141 | 0 | 2.084808 |
| LRIG1 | 0.408929 | 2 | 0.018088 | 0.042633 | 1.901966 | 0.386361 | 0 | −1.42735 |
| LRRC49 | 0.408929 | 2 | 0.011943 | 0.028148 | 0.263645 | 0.876497 | 0 | −1.66365 |
| PCSK9 | 0.408929 | 2 | 0.000332 | 0.000782 | 0.045994 | 0.977266 | 0 | −5.13551 |
| PHPT1 | 0.408929 | 2 | 0.009973 | 0.023507 | 0.52427 | 0.769407 | 0 | −1.60935 |
| POLR2G | 0.408929 | 2 | 0.010377 | 0.024458 | 0.509447 | 0.775131 | 0 | 1.450648 |
| PPA2 | 0.408929 | 2 | 0.002415 | 0.005691 | 0.983689 | 0.611498 | 0 | −0.77017 |
| USP47 | 0.408929 | 2 | 0.016431 | 0.038727 | 1.485343 | 0.475841 | 0 | −2.70658 |
| TTK | 0.408856 | 3 | 0.019272 | 0.045422 | 0.154504 | 0.925656 | 0 | −0.23932 |
| ARFIP1 | 0.408051 | 3 | 5.30E-06 | 1.25E-05 | 1.795171 | 0.407552 | 0 | −2.91913 |
| FTL | 0.407729 | 2 | 0.00145 | 0.003417 | 1.83461 | 0.399595 | 0 | 2.012816 |
| AURKAIP1 | 0.407431 | 2 | 0.006809 | 0.016048 | 0.532797 | 0.766134 | 0 | 0.828771 |
BC, betweenness centrality score; FDR, false discovery rate.
Figure 5.
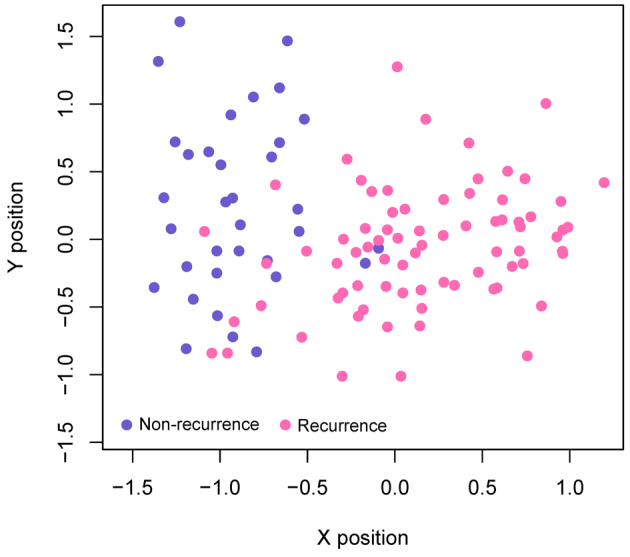
Scatter plots detailing the prediction results of GSE17260 datasets using the support vector machine classifier. Blue dots represent samples from patients with non-recurrent OC and pink dots represent samples from patients with recurrent OC. OC, ovarian cancer.
The SVM classifier was further validated using GSE44104 and GSE51088 datasets. The prediction accuracy for dataset GSE44104 was revealed to be 93.3% [56 out of 60 samples (40 samples from patients with non-recurrent OC and 16 samples from patients with recurrent OC)]. The accuracy for dataset GSE51088 was revealed to be 96.6% [142 out of 147 samples (126 non-recurrent OC samples and 16 recurrent OC samples)]. The correct rate, sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV) and area under receiver operating characteristic curve (AUROC) values were presented in Table VI. It can be observed that the SVM classifier had a good classification effect in all 3 data sets. Furthermore, the AUROC values of GSE17260, GSE44104 and GSE51088 datasets were 0.988, 0.970, and 0.967, respectively (Table VI). All values are close to 1, which means close to the perfect prediction effect.
Table VI.
Prediction results of the support vector machine classifier using sample data from GSE17260, GSE44104 and GSE51088.
| Datasets | Number of samples | Correct samples | Correct rate | Sensitivity | Specificity | PPV | NPV | AUROC |
|---|---|---|---|---|---|---|---|---|
| GSE17260 | 110 | 102 | 0.927 | 0.894 | 0.987 | 0.964 | 0.915 | 0.988 |
| GSE44104 | 60 | 56 | 0.933 | 1.000 | 0.800 | 0.909 | 1.000 | 0.970 |
| GSE51088 | 147 | 142 | 0.966 | 0.969 | 0.941 | 0.992 | 0.800 | 0.967 |
| TCGA | 395 | 357 | 0.9038 | 0.987 | 0.801 | 0.862 | 0.979 | 0.981 |
PPV, positive predictive value; NPV, negative predictive value; AUROC, area under receiver operating characteristic curve.
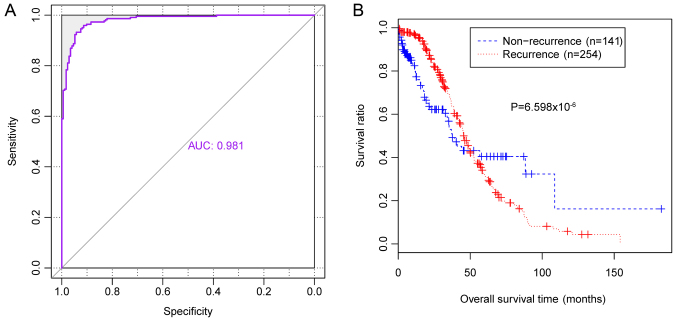
Results of validation
Prediction accuracy of independent gene expression data downloaded from TCGA was revealed to be 90.4% [357 out of 395 samples (138 samples from patients with non-recurrent OC and 219 samples from patients with recurrent OC)], with an AUROC value of 0.981 (Table VI, Fig. 6A). In addition, survival ratios were determined for the 394 patients with OC (172 patients with non-recurrent OC and 222 patients with recurrent OC). The KM survival curve revealed that survival times of patients with predicted non-recurrent OC were significantly increased compared with patients with predicted recurrent OC (P=6.598×10−6; Fig. 6B), which suggested that the classifier may accurately predict the prognosis of patients with OC.
Figure 6.
Classification efficacy of the SVM classifier using independent gene expression data from the TCGA database. (A) Receiver operating characteristic curves and (B) Kaplan-Meier survival analysis predicted by the SVM classifier for using gene expression data from the TCGA database. The blue line indicates samples from patients with non-recurrent OC and the red line indicates samples from patients with recurrent OC. TCGA, the Cancer Genome Atlas; OC, ovarian cancer; AUC, area under a curve.
Discussion
In the present study, a SVM classifier consisting of specific genes was revealed to predict the rates of non-recurrent and recurrent OC. Gene expression profiles of patients with recurrent OC were compared with patients with non-recurrent OC to identify DEGs. Homogeneity and quality control analyses using three gene expression datasets were performed to improve the prediction accuracy of the classifier. A PPI network was then constructed using identified DEGs, which included 249 nodes and 354 edges. Functional and pathway enrichment analysis demonstrated that genes in the PPI network were significantly associated with 14 GO terms, including ‘cell cycle,’ ‘homologous recombination’, ‘purine metabolism’ and ‘pathways in cancer and DNA replication’. A total of 39 genes were selected by recursive feature elimination, including CUL3, MDM2, AURKA, WWOX, LATS2, SIRT6, SND1, LRIG1 and AURKAIP1.
Constitutive activation of nuclear factor erythroid 2 like 2 (NRF2) is associated with acquisition of malignant features in OC (28,29). Markedly increased frequencies of DNA and mRNA alterations compared with healthy controls affect components of the kelch like ECH associated protein 1 (KEAP1)/CUL3/ring-box 1 (RBX1) E3-ubiquitin ligase complex, which regulates NRF2 expression, have been revealed via sequencing of KEAP1, CUL3 and RBX1 in a cohort of 568 samples obtained from patients with OC detailed in TCGA (30). MDM2 is a nuclear-localized E3 ubiquitin ligase that promotes tumor formation by targeting tumor suppressor proteins, including p53, and has an important role in the development of OC (31). It has been previously demonstrated that overexpression of MDM2 can increase cisplatin cytotoxicity in human ovarian cell lines (32). Furthermore, it has been demonstrated that antagonists of MDM2 can induce apoptosis in human ovarian cancer cells and synergize with cisplatin to attenuate the chemoresistance of patients exhibiting wild-type tumor protein p53 (33). AURKA expression has been revealed to be closely correlated with prognosis of endometrioid OC in a study including 51 tumor samples (34), which may result from its role in the regulation of OC cell migration and adhesion (35). The predominant full-length transcript (variant 1) of WWOX functions as a suppressor of ovarian tumorigenesis (36) by inducing apoptosis in detached cells, and regulating the interaction between tumor cells and the extracellular matrix (37). WWOX can regulate the cell cycle and apoptosis of OC stem cells (38), which suggests that WWOX may represent an important molecular target for the treatment of OC. Numerous studies have reported that miR-25 and miR-181b can promote OC by targeting LATS2, which is a serine/threonine protein kinase belonging to the LATS tumor suppressor family and is involved in the proliferation, migration and invasion of OC cells (39,40). SIRT6, a member of NAD+ dependent class III deacetylase sirtuin family, has been revealed to inhibit the proliferation of OC cells by downregulating Notch 3 expression (41). Decreased expression of SIRT6 has been revealed to promote tumor cell growth and is closely correlated with poor prognosis of OC (42). Therefore, SIRT6 may represent a therapeutic target for the prevention and treatment of OC. LRIG1 is a tumor suppressor used in clinical practice (43). Decreased LRIG1 expression has been demonstrated to propagate chemoresistance in etoposide-resistant human OC cells by downregulating multidrug resistance-associated protein 1 and apoptosis (44). In addition, AURKAIP1 promotes the degradation of the Aurora A oncogene via an alternative ubiquitin-independent pathway (45). Therefore, AURKAIP1 may be involved in the development and recurrence of OC. SND1, a transcriptional co-activator, has been demonstrated to promote breast cancer metastasis via the tumor growth factor β1/mad (smad) mothers against dpp pathway (46), which has been previously used for the prediction of colon cancer prognosis (47), and to promote prostate cancer via interaction with KH domain-containing RNA-binding signal transduction-associated protein 1 (48). However, the role of SND1 in OC remains unclear. Studies on the aforementioned feature genes may help to determine the complex molecular mechanisms underlying the recurrence of OC.
In the present study, a SVM classifier consisting of 39 specific genes was constructed and verified for the prediction of the recurrence of OC. The prediction accuracy of the SVM classifier for GSE17260, GSE44104 and GSE51088 datasets was 92.7, 93.3 and 96.6%, respectively. The prediction accuracy of the SVM classifier using independent gene expression data downloaded from TCGA demonstrated an accuracy of 90.4%. Furthermore, the patients with predicted non-recurrent OC exhibited a significantly longer survival time compared with patients with predicted recurrent OC (P=6.598×10−6); therefore suggesting that the SVM classifier has the potential for use in the prognostic prediction of patients with OC. Unlike sequencing technology, the SVM classifier only requires the expression levels of 39 genes for prognostic prediction. Therefore, application of the established SVM classifier is more economical and efficient compared with sequencing for the prognostic prediction of patients with OC.
In conclusion, a SVM classifier consisting of 39 genes was established in the present study for the accurate prediction of the recurrence of OC. The 39 included genes serve roles in the development of OC and may represent novel therapeutic targets for the treatment of OC. Furthermore, the established SVM classifier may be used for prognostic prediction in patients with OC. However, further studies investigating an independent cohort of patients with non-recurrent and recurrent OC are required to further validate the results of the present study.
Acknowledgements
Not applicable.
Funding
This study was supported by Hubei Province's Outstanding Medical Academic Leader Program and Hubei Province Health and Family Planning Scientific Research project (grant no. WJ2015MA024) and the general project of Natural Science Foundation of Hubei Province (grant no. 2017CFB335).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JZ and LL performed data analyses and wrote the manuscript. LW, XL and HX contributed significantly in data analyses. LC conceived and designed the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
In the original article of the datasets, the trials were approved by the local institutional review boards of all participating centers, and informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Howlader N, Noone AM, Krapcho M, Garshell J, Miller D, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z, et al., editors. SEER Cancer Statistics Review 1975–2012. National Cancer Institute; Bethesda, MD: 2015. [Nov 18;2015 ]. [Google Scholar]
- 2.Davidson B, Tropé CG. Ovarian cancer: Diagnostic, biological and prognostic aspects. Womens Health (Lond) 2014;10:519–533. doi: 10.2217/WHE.14.37. [DOI] [PubMed] [Google Scholar]
- 3.Gloss BS, Samimi G. Epigenetic biomarkers in epithelial ovarian cancer. Cancer Lett. 2014;342:257–263. doi: 10.1016/j.canlet.2011.12.036. [DOI] [PubMed] [Google Scholar]
- 4.Leung F, Diamandis EP, Kulasingam V. Ovarian cancer biomarkers: Current state and future implications from high-throughput technologies. Adv Clin Chem. 2014;66:25–77. doi: 10.1016/B978-0-12-801401-1.00002-5. [DOI] [PubMed] [Google Scholar]
- 5.Au KK, Josahkian JA, Francis JA, Squire JA, Koti M. Current state of biomarkers in ovarian cancer prognosis. Future Oncol. 2015;11:3187–3195. doi: 10.2217/fon.15.251. [DOI] [PubMed] [Google Scholar]
- 6.Huo J, Hu J, Liu G, Cui Y, Ju Y. Elevated serum interleukin-37 level is a predictive biomarker of poor prognosis in epithelial ovarian cancer patients. Arch Gynecol Obstet. 2017;295:459–465. doi: 10.1007/s00404-016-4258-8. [DOI] [PubMed] [Google Scholar]
- 7.Masoumi-Moghaddam S, Amini A, Wei AQ, Robertson G, Morris DL. Sprouty 2 protein, but not Sprouty 4, is an independent prognostic biomarker for human epithelial ovarian cancer. Int J Cancer. 2015;137:560–570. doi: 10.1002/ijc.29425. [DOI] [PubMed] [Google Scholar]
- 8.Lee JM, Trepel JB, Choyke P, Cao L, Sissung T, Houston N, Yu M, Figg WD, Turkbey IB, Steinberg SM, et al. CECs and IL-8 have prognostic and predictive utility in patients with recurrent platinum-sensitive ovarian cancer: Biomarker correlates from the randomized phase-2 trial of olaparib and cediranib compared with olaparib in recurrent platinum-sensitive ovarian cancer. Front Oncol. 2015;5:123. doi: 10.3389/fonc.2015.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng Y, He F, Wu H, Huang H, Zhang L, Han X, Liu J. GOLPH3L is a novel prognostic biomarker for epithelial ovarian cancer. J Cancer. 2015;6:893–900. doi: 10.7150/jca.11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roque DM, Buza N, Glasgow M, Bellone S, Bortolomai I, Gasparrini S, Cocco E, Ratner E, Silasi DA, Azodi M, et al. Class III β-tubulin overexpression within the tumor microenvironment is a prognostic biomarker for poor overall survival in ovarian cancer patients treated with neoadjuvant carboplatin/paclitaxel. Clin Exp Metastasis. 2014;31:101–110. doi: 10.1007/s10585-013-9614-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Penzvalto Z, Lanczky A, Lenart J, Meggyesházi N, Krenács T, Szoboszlai N, Denkert C, Pete I, Győrffy B. MEK1 is associated with carboplatin resistance and is a prognostic biomarker in epithelial ovarian cancer. BMC Cancer. 2014;14:837. doi: 10.1186/1471-2407-14-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdel-Fatah TM, Russell R, Albarakati N, Maloney DJ, Dorjsuren D, Rueda OM, Moseley P, Mohan V, Sun H, Abbotts R, et al. Genomic and protein expression analysis reveals flap endonuclease 1 (FEN1) as a key biomarker in breast and ovarian cancer. Mol Oncol. 2014;8:1326–1338. doi: 10.1016/j.molonc.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei W, Mok SC, Oliva E, Kim SH, Mohapatra G, Birrer MJ. FGF18 as a prognostic and therapeutic biomarker in ovarian cancer. J Clin Invest. 2013;123:4435–4448. doi: 10.1172/JCI70625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu T, Gao H, Chen X, Lou G, Gu L, Yang M, Xia B, Yin H. TNFAIP8 as a predictor of metastasis and a novel prognostic biomarker in patients with epithelial ovarian cancer. Br J Cancer. 2013;109:1685–1692. doi: 10.1038/bjc.2013.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshihara K, Tajima A, Yahata T, Kodama S, Fujiwara H, Suzuki M, Onishi Y, Hatae M, Sueyoshi K, Fujiwara H, et al. Gene expression profile for predicting survival in advanced-stage serous ovarian cancer across two independent datasets. PLoS One. 2010;5:e9615. doi: 10.1371/journal.pone.0009615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu YH, Chang TH, Huang YF, Huang HD, Chou CY. COL11A1 promotes tumor progression and predicts poor clinical outcome in ovarian cancer. Oncogene. 2014;33:3432–3440. doi: 10.1038/onc.2013.307. [DOI] [PubMed] [Google Scholar]
- 17.Karlan BY, Dering J, Walsh C, Orsulic S, Lester J, Anderson LA, Ginther CL, Fejzo M, Slamon D. POSTN/TGFBI-associated stromal signature predicts poor prognosis in serous epithelial ovarian cancer. Gynecol Oncol. 2014;132:334–342. doi: 10.1016/j.ygyno.2013.12.021. [DOI] [PubMed] [Google Scholar]
- 18.Liu WM, Mei R, Di X, Ryder TB, Hubbell E, Dee S, Webster TA, Harrington CA, Ho MH, Baid J, Smeekens SP. Analysis of high density expression microarrays with signed-rank call algorithms. Bioinformatics. 2002;18:1593–1599. doi: 10.1093/bioinformatics/18.12.1593. [DOI] [PubMed] [Google Scholar]
- 19.R Development Core Team, corp-author. R: a language and environment for statistical computing. the R Foundation for Statistical Computing; Vienna: 2016. [Google Scholar]
- 20.de Souto MC, Jaskowiak PA, Costa IG. Impact of missing data imputation methods on gene expression clustering and classification. BMC Bioinformatics. 2015;16:64. doi: 10.1186/s12859-015-0494-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Affymetrix® Microarray Suite. User's Guide. Version 5.0. Affymetrix, Inc.; Santa Clara: 2001. [Google Scholar]
- 22.Kang DD, Sibille E, Kaminski N, Tseng GC. MetaQC: Objective quality control and inclusion/exclusion criteria for genomic meta-analysis. Nucleic Acids Res. 2012;40:e15. doi: 10.1093/nar/gkr1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi C, Hong L, Cheng Z, Yin Q. Identification of metastasis-associated genes in colorectal cancer using metaDE and survival analysis. Oncol Lett. 2016;11:568–574. doi: 10.3892/ol.2015.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shannon PI, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goh KI, Oh E, Jeong H, Kahng B, Kim D. Classification of scale-free networks; Proc Natl Acad Sci USA; 2002; pp. 12583–12588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qureshi MN, Min B, Jo HJ, Lee B. Multiclass classification for the differential diagnosis on the ADHD subtypes using recursive feature elimination and hierarchical extreme learning machine: Structural MRI study. PLoS One. 2016;11:e0160697. doi: 10.1371/journal.pone.0160697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomczak K, Czerwińska P, Wiznerowicz M. The cancer genome atlas (TCGA): An immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sporn MB, Liby KT. NRF2 and cancer: The good, the bad and the importance of context. Nat Rev Cancer. 2012;12:564–571. doi: 10.1038/nrc3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao H, Zhou Q, Zhang Z, Wang Q, Sun Y, Yi X, Feng Y. NRF2 is overexpressed in ovarian epithelial carcinoma and is regulated by gonadotrophin and sex-steroid hormones. Oncol Rep. 2012;27:1918–1924. doi: 10.3892/or.2012.1700. [DOI] [PubMed] [Google Scholar]
- 30.Martinez VD, Vucic EA, Thu KL, Pikor LA, Hubaux R, Lam WL. Unique pattern of component gene disruption in the NRF2 inhibitor KEAP1/CUL3/RBX1 E3-ubiquitin ligase complex in serous ovarian cancer. Biomed Res Int. 2014;2014:159459. doi: 10.1155/2014/159459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ginath S, Menczer J, Friedmann Y, Aingorn H, Aviv A, Tajima K, Dantes A, Glezerman M, Vlodavsky I, Amsterdam A. Expression of heparanase, Mdm2, and erbB2 in ovarian cancer. Int J Oncol. 2001;18:1133–1144. doi: 10.3892/ijo.18.6.1133. [DOI] [PubMed] [Google Scholar]
- 32.Mi RR, Ni H. MDM2 sensitizes a human ovarian cancer cell line. Gynecol Oncol. 2003;90:238–244. doi: 10.1016/S0090-8258(03)00282-8. [DOI] [PubMed] [Google Scholar]
- 33.Mir R, Tortosa A, Martinez-soler F, Vidal A, Condom E, Pérez-Perarnau A, Ruiz-Larroya T, Gil J, Giménez-Bonafé P. Mdm2 antagonists induce apoptosis and synergize with cisplatin overcoming chemoresistance in TP53 wild-type ovarian cancer cells. Int J Cancer. 2013;132:1525–1536. doi: 10.1002/ijc.27832. [DOI] [PubMed] [Google Scholar]
- 34.Yang F, Guo X, Yang G, Rosen DG, Liu J. AURKA and BRCA2 expression highly correlate with prognosis of endometriofid ovarian carcinoma. Mod Pathol. 2011;24:836–845. doi: 10.1038/modpathol.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Do TV, Xiao F, Bickel LE, Klein-Szanto AJ, Pathak HB, Hua X, Howe C, O'Brien SW, Maglaty M, Ecsedy JA, et al. Aurora kinase A mediates epithelial ovarian cancer cell migration and adhesion. Oncogene. 2014;33:539–549. doi: 10.1038/onc.2012.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gourley C, Paige AJW, Taylor KJ, Scott D, Francis NJ, Rush R, Aldaz CM, Smyth JF, Gabra H. WWOX mRNA expression profile in epithelial ovarian cancer supports the role of WWOX variant 1 as a tumour suppressor, although the role of variant 4 remains unclear. Int J Oncol. 2005;26:1681–1689. doi: 10.3892/ijo.26.6.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gourley C, Paige AJ, Taylor KJ, Ward C, Kuske B, Zhang J, Sun M, Janczar S, Harrison DJ, Muir M, et al. WWOX gene expression abolishes ovarian cancer tumorigenicity in vivo and decreases attachment to fibronectin via integrin alpha3. Cancer Res. 2009;69:4835–4842. doi: 10.1158/0008-5472.CAN-08-2974. [DOI] [PubMed] [Google Scholar]
- 38.Yan H, Tong J, Lin X, Han Q, Huang H. Effect of the WWOX gene on the regulation of the cell cycle and apoptosis in human ovarian cancer stem cells. Mol Med Rep. 2015;12:1783–1788. doi: 10.3892/mmr.2015.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng S, Pan W, Jin Y, Zheng J. MiR-25 promotes ovarian cancer proliferation and motility by targeting LATS2. Tumour Biol. 2014;35:12339–12344. doi: 10.1007/s13277-014-2546-0. [DOI] [PubMed] [Google Scholar]
- 40.Xia Y, Gao Y. MicroRNA-181b promotes ovarian cancer cell growth and invasion by targeting LATS2. Biochem Biophys Res Commun. 2014;447:446–451. doi: 10.1016/j.bbrc.2014.04.027. [DOI] [PubMed] [Google Scholar]
- 41.Zhang J, Yin XJ, Xu CJ, Ning YX, Chen M, Zhang H, Chen SF, Yao LQ. The histone deacetylase SIRT6 inhibits ovarian cancer cell proliferation via down-regulation of Notch 3 expression. Eur Rev Med Pharmacol Sci. 2015;19:818–824. [PubMed] [Google Scholar]
- 42.Zhang G, Liu Z, Qin S, Li K. Decreased expression of SIRT6 promotes tumor cell growth correlates closely with poor prognosis of ovarian cancer. Eur J Gynaecol Oncol. 2015;36:629–632. doi: 10.1097/IGC.0000000000000350. [DOI] [PubMed] [Google Scholar]
- 43.Lindquist D, Kvarnbrink S, Henriksson R, Hedman H. LRIG and cancer prognosis. Acta Oncol. 2014;53:1135–1142. doi: 10.3109/0284186X.2014.953258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang H, Yao J, Yin J, Wei X. Decreased LRIG1 in human ovarian cancer cell SKOV3 upregulates MRP-1 and contributes to the chemoresistance of VP16. Cancer Biother Radiopharm. 2016;31:125–132. doi: 10.1089/cbr.2015.1970. [DOI] [PubMed] [Google Scholar]
- 45.Lim SK, Gopalan G. Aurora-A kinase interacting protein 1 (AURKAIP1) promotes Aurora-A degradation through an alternative ubiquitin-independent pathway. Biochem J. 2007;403:119–127. doi: 10.1042/BJ20061272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu L, Liu X, Cui K, Di Y, Xin L, Sun X, Zhang W, Yang X, Wei M, Yao Z, Yang J. SND1 acts downstream of TGFβ1 and upstream of Smurf1 to promote breast cancer metastasis. Cancer Res. 2015;75:1275–1286. doi: 10.1158/0008-5472.CAN-14-2387. [DOI] [PubMed] [Google Scholar]
- 47.Wang N, Du X, Zang L, Song N, Yang T, Dong R, Wu T, He X, Lu J. Prognostic impact of metadherin–SND1 interaction in colon cancer. Mol Biol Rep. 2012;39:10497–10504. doi: 10.1007/s11033-012-1933-0. [DOI] [PubMed] [Google Scholar]
- 48.Cappellari M, Bielli P, Paronetto MP, Ciccosanti F, Fimia GM, Saarikettu J, Silvennoinen O, Sette C. The transcriptional co-activator SND1 is a novel regulator of alternative splicing in prostate cancer cells. Oncogene. 2014;33:3794–3802. doi: 10.1038/onc.2013.360. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.