Abstract
Since the description of the genus Arcobacter in 1991, a total of 27 species have been described, although some species have shown 16S rRNA similarities below 95%, which is the cut-off that usually separates species that belong to different genera. The objective of the present study was to reassess the taxonomy of the genus Arcobacter using information derived from the core genome (286 genes), a Multilocus Sequence Analysis (MLSA) with 13 housekeeping genes, as well as different genomic indexes like Average Nucleotide Identity (ANI), in silico DNA–DNA hybridization (isDDH), Average Amino-acid Identity (AAI), Percentage of Conserved Proteins (POCPs), and Relative Synonymous Codon Usage (RSCU). The study included a total of 39 strains that represent all the 27 species included in the genus Arcobacter together with 13 strains that are potentially new species, and the analysis of 57 genomes. The different phylogenetic analyses showed that the Arcobacter species grouped into four clusters. In addition, A. lekithochrous and the candidatus species ‘A. aquaticus’ appeared, as did A. nitrofigilis, the type species of the genus, in separate branches. Furthermore, the genomic indices ANI and isDDH not only confirmed that all the species were well-defined, but also the coherence of the clusters. The AAI and POCP values showed intra-cluster ranges above the respective cut-off values of 60% and 50% described for species belonging to the same genus. Phenotypic analysis showed that certain test combinations could allow the differentiation of the four clusters and the three orphan species established by the phylogenetic and genomic analyses. The origin of the strains showed that each of the clusters embraced species recovered from a common or related environment. The results obtained enable the division of the current genus Arcobacter in at least seven different genera, for which the names Arcobacter, Aliiarcobacter gen. nov., Pseudoarcobacter gen. nov., Haloarcobacter gen. nov., Malacobacter gen. nov., Poseidonibacter gen. nov., and Candidate ‘Arcomarinus’ gen. nov. are proposed.
Keywords: Arcobacter, Aliiarcobacter gen. nov., Pseudoarcobacter gen. nov., Haloarcobacter gen. nov., Malacobacter gen. nov., Poseidonibacter gen. nov., taxonomic criteria
Introduction
The genus Arcobacter was created by Vandamme et al. (1991) to accommodate Gram-negative, curved-shaped bacteria belonging to two species Campylobacter cryaerophila (now Arcobacter cryaerophilus) and Campylobacter nitrofigilis (now A. nitrofigilis), considered atypical campylobacters due to their ability to grow at lower temperatures (15°C–30°C) and without microaerophilic conditions (Vandamme et al., 1991). The latter species was selected as the type species for the new genus (Vandamme et al., 1991). One year later the genus was enlarged with the addition of two new species, A. skirrowii with an animal origin being isolated from aborted ovine, porcine and bovine fetuses, and from lambs with diarrhea, and A. butzleri, which was recovered from cases of human and animal diarrhea (Vandamme et al., 1992). Another two new species were incorporated into the genus in 2005. A. halophilus was isolated from water from a hypersaline lagoon in Hawaii (Donachie et al., 2005), and A. cibarius was isolated from broiled carcasses in Belgium (Houf et al., 2005). These species were assigned to the genus Arcobacter on the basis of the 16S rRNA gene similarity (94% and 95% for A. nitrofigilis with A. halophilus and A. cibarius, respectively). However, these values are equal, or even below, the cut-off of 95% for genus definition (Rosselló-Mora and Amann, 2001; Yarza et al., 2008, 2014; Tindall et al., 2010).
From 2009 onward, new species were being described year-by-year, reaching a total number of 27 in 2017. In some of these descriptions, the similarity of the 16S rRNA gene was the decisive character for taxonomic assignation at genus level, although phylogeny based on housekeeping genes (rpoB first and then gyrB and hsp60) was also included as additional, more discriminatory tools for the species (Collado et al., 2009a, 2011; De Smet et al., 2011). Using this approach, A. molluscorum, A. ellisii, A. defluvii, or A. bivalviorum were defined, among others (Collado et al., 2009a, 2011; Figueras et al., 2011a,b; Levican et al., 2012), which showed 16S rRNA similarities ranging from 91.1 to 94.7%, not supporting their common affiliation. On the other hand, the most closely related species, which showed a similarity of 99.1% were A. ellisii and A. defluvii (Collado et al., 2011), giving evidence for the first time of the poor resolution of the 16S rRNA gene for separating closely related species in the genus Arcobacter. However, the phylogenetic analysis based on the concatenated sequences of gyrB, rpoB, and cpn60 genes, together with the DNA–DNA hybridization results, clearly supported the existence of these two differentiated taxa (Figueras et al., 2011a). Also in 2011, A. trophiarum was discovered from the intestinal tract of healthy fattening pigs, which interestingly showed the closest similarities (≥97.4%) with the other species also recovered from humans or animals, i.e., A. cryaerophilus, A. thereius, A. cibarius, or A. skirrowii (De Smet et al., 2011; Figueras et al., 2014; Van den Abeele et al., 2014).
In 2013, the species A. cloacae and A. suis were described, using a Multilocus Sequence Analysis (MLSA) approach including five housekeeping genes (Levican et al., 2013) for the first time. Simultaneously, and due to the highest 16S rRNA gene similarity with A. marinus (95.5%), the species A. anaerophilus was incorporated to the genus (Sasi-Jyothsna et al., 2013). However, this species showed atypical characteristics, including lack of motility and obligate anaerobic metabolism, which led to the original description of the genus Arcobacter being emended (Sasi-Jyothsna et al., 2013). The most recently described species from shellfish are A. lekithochrous, A. haliotis, and A. canalis (Diéguez et al., 2017; Tanaka et al., 2017; Pérez-Cataluña et al., 2018a). The first one included several isolates recovered from scallop larvae and from tank seawater of a Norwegian hatchery (Diéguez et al., 2017), the second species came from an abalone of Japan (Tanaka et al., 2017) and the third from oysters submerged in a water channel contaminated with wastewater (Pérez-Cataluña et al., 2018a). However, Diéguez et al. (2018) evidenced that the species A. haliotis is a later heterotypic synonym of A. lekithochrous. Additionally, the low 16S rRNA gene similarity of A. lekithochrous with the known Arcobacter species (91.0–94.8%) found in the A. lekithochrous description made Diéguez et al. (2017) suggest that certain species might belong to other genera and recommend that a profound revision of the genus might clarify the taxonomy.
On the other hand, adding 2.5% NaCl to the enrichment medium and subculturing on marine agar, Salas-Massó et al. (2016) recognized seven potential new species from water and shellfish (mussels and/or oysters), and recovered new isolates of A. halophilus and A. marinus of which only the type strains had been known. In addition, during the characterization of the most recently described species A. canalis (Pérez-Cataluña et al., 2018a) and when trying to define the seven mentioned new species, we observed that the Arcobacter species formed several different clusters distant enough to suspect they might correspond to different genera, in agreement with Diéguez et al. (2017).
There are clear criteria for describing new bacterial species (Tindall et al., 2010; Figueras et al., 2011a,b). However, the description of a genus is usually based on a cut-off of <95% similarity in the 16S rRNA gene sequence, and a G+C (% mol) content differing by more than 10% (Rosselló-Mora and Amann, 2001; Yarza et al., 2008; Tindall et al., 2010; Yarza et al., 2014). Nowadays, genomic data like the Average Nucleotide Identity (ANI) and the in silico DNA–DNA hybridization (isDDH) are used to define bacterial species, although have not yet been fully explored for delineating genera (Konstantinidis and Tiedje, 2005; Goris et al., 2007; Richter and Rosselló-Móra, 2009; Qin et al., 2014; Chun et al., 2018).
A percentage of Average Amino-acid Identity (AAI) ranging from 60 to 80% between the compared genomes of species or strains and a Percentage of Conserved Proteins (POCPs) above 50% has been proposed if they are to belong to the same genus (Konstantinidis and Tiedje, 2005; Qin et al., 2014). Finally, the Relative Synonymous Codon Usage (RSCU) has also been used by some authors to infer evolutionary and ecological links among bacterial species (Ma et al., 2015; Farooqi et al., 2016).
Very recently, Waite et al. (2017) carried out a comparative genomic analysis of the class Epsilonproteobacteria. Using 16S and 23S rRNA, 120 single-copy marker proteins and AAI analysis they proposed its reclassification as the new phylum Epsilonbacteraeota. In that study, Waite et al. (2017) also proposed a reclassification of the genus Arcobacter as a new Family Arcobacteraceae, within the class Campylobacteria, order Campylobacterales. One weakness of this study, specifically regarding the genus Arcobacter, is that only seven validated species were included in the analysis. The new family therefore comprised only the genus Arcobacter. However, these findings also support the need for a clarification of the taxonomy of the current genus Arcobacter.
The rise of genome sequencing has dramatically changed the landscape of systematics of prokaryotes, improving different aspects such as the identification of species, the functional characterization for resolving taxonomic groups, and the resolution of the phylogeny of higher taxa (Whitman, 2015). It seems clear that the incorporation of genomics into the taxonomy will boost its credibility providing reproducible, reliable, highly informative means to infer phylogenetic relationships among prokaryotes, and avoiding unreliable methods and subjective difficult-to-replicate data (Chun and Rainey, 2014; Chun et al., 2018).
Within this modern taxonomy context, the objective of the present study was to reassess the taxonomy of the known and newly recognized Arcobacter species by using a MLSA of 13 housekeeping genes, the whole genome sequences and the derived genomic analysis. The latter analysis included ANI, isDDH, AAI, POCP, and RSCU of all Arcobacter type strains. In addition, phylogenies based on 16S and 23S rRNA gene sequences were also performed with comparative purposes. The new taxonomic criteria were stable when including whole genome sequences of a second strain of each species or of unassigned sequences obtained from the public databases.
Materials and Methods
Bacterial Strains
All 27 valid species included in the genus Arcobacter have been studied. They are represented by 39 strains, and 13 strains that are potentially new species (Table 1). Furthermore, 50 genomes of Arcobacter strains identified at species level were investigated, 39 of which were obtained in our laboratory (27 from known species and 13 from potentially new species) and the others from the public databases1,2. Five genomes that had been deposited as Arcobacter sp. in the databases were also included in the study. If there was more than one strain of a known Arcobacter species, two representative genomes for each species were included in the analysis. The only exceptions were: A. acticola (Park et al., 2016) and A. pacificus (Zhang et al., 2015), whose taxonomic positions were only inferred by the phylogenetic analysis of the 16S rRNA gene sequences published in their species descriptions, together with a MLSA of three housekeeping genes (atpA, gyrB, and rpoB) for A. pacificus (Zhang et al., 2015; Park et al., 2016). The strains considered potentially new species, and named hereafter as ‘candidate species,’ had been recognized with an MLSA analysis of five housekeeping genes (atpA, gyrA, gyrB, hsp60, and rpoB) (data not shown).
Table 1.
Strains used in this study, source of isolation and accession numbers of the available genomes.
| Species | Strain | Source | Acc. No. Genome | Species | Strain | Source | Acc. No. Genome |
|---|---|---|---|---|---|---|---|
| A. acticola | KCTC 52212T | Seawater | NAa | A. mytili | T234 | Seawater | PDJW00b |
| A. anaerophilus | DSM 24636T | Estuarine sediment | PDKO00b | A. nitrofigilis | DSM7299T | Marshland plant | NC014166c |
| IR-1 | Utsira aquifer | NZ_JXXG00c | A. pacificus | DSM 25018T | Seawater | NAa | |
| A. aquimarinus | CECT 8442T | Mediterranean Sea | NXIJ00b | A. skirrowii | LMG 6621T | Diarrheic lamb | NXIC00b |
| A. bivalviorum | CECT 7835T | Mussels | PDKM00b | F28 | Wild pig | PDJT00b | |
| F118-4 | Mussels | PDKL00b | A. suis | CECT 7833T | Pork meat | NREO00b | |
| A. butzleri | RM4018T | Human (Clinical) | NC_009850c | A. thereius | LMG 24486T | Aborted pig foetus | LLKQ01c |
| ED1 | Microbial fuel cell | NC_017187c | DU22 | Duck cloaca | LCUJ01c | ||
| A. canalis | F138-33 | Oyster PNCe | NWVW01b | A. trophiarum | LMG 25534T | Piglet feces | PDKD00b |
| SH-4D_Col1 | Unknown | FUYO00c | CECT 7650 | Chicken cloacal swab | PDJS00b | ||
| A. cibarius | LMG 21996T | Broiler, skin | NZ_JABW00c | A. venerupis | CECT 7836T | Clams | NREP00b |
| A. cloacae | CECT 7834T | Sewage | NXII00b | Arcobacter sp. | L | Microbial fuel cell | NC_017192c |
| F26 | Mussels | PDJZ00b | AF1028 | Human feces | JART01c | ||
| A. cryaerophilus | LMG 24291T | Aborted bovine foetus | NXGK00b | CAB | Marine | Go0012496d | |
| A. defluvii | CECT 7697T | Sewage | NXIH00b | LA11 | Marine | BDIR01c | |
| A. ebronensis | CECT 8441T | Mussels | PDKK00b | LPB0137 | Environmental | CP019070c | |
| CECT 8993 | Seawater | PDKJ00b | |||||
| A. ellisii | CECT 7837T | Mussels | NXIG00b | ‘A. aquaticus’ | W112-28 | Freshwater PNCe | PDKN00b |
| A. faecis | LMG 28519T | Human septic tank | NZ_JARS00c | ‘A. caeni’ | RW17-10 | Recycled wastewater | MUXE00b |
| A. halophillus | DSM 18005T | Hypersaline lagoon | PDJY00b | ‘A. hispanicus’ | FW-54 | Wastewater | PDKI00b |
| F166-45 | Oyster PNCe | PDJY00b | ‘A. lacus’ | RW43-9 | Recycled wastewater | MUXF00b | |
| A. lanthieri | LMG 28516T | Pig manure | JARU01c | ‘A. mediterraneus’ | F156-34 | Mussels Alfacs Bay | NXIE00b |
| LMG 28517 | Dairy cattle manure | JARV01c | ‘A. miroungae’ | 9Antf | Cloaca elephant seal | PDKH00b | |
| A. lekithochrous | CECT 8942T | Great scallop larvae | NZ_MKCO00b | ‘A. neptunis’ | F146-38 | Mussels Alfacs Bay | PDKG00b |
| LMG 28652 | Abalon | PZYW00c | ‘A. porcinus’ | LMG 24487T | Aborted pig foetus | LCUH01c | |
| A. marinus | CECT 7727T | Seawater | NXAO01b | ‘A. ponticus’ | F161-33 | Cockle Alfacs Bay | PDKF00b |
| F140-37 | Clams Alfacs Bay | NWVX01b | ‘A. salis’ | F155-33 | Oyster PNCe | PDKE00b | |
| A. molluscorum | CECT 7696T | Mussels | NZ_NXFY00b | ‘A. viscosus’ | F142-34g | Mussels PNCe | PDKC00b |
| F91 | Mussels | PDJX00b | ‘A. vitoriensis’ | FW59g | Wastewater | PDKB00b | |
| A. mytili | CECT 7386T | Mussels | NXID00b | Arcobacter sp. | F2176 | Mussels | PDJV00b |
aGenome not available; bGenome sequenced in this study; cGenome obtained from NCBI database; dGenome obtained from JGI Gold atabase; ePNC means PobleNou Channel, which is a freshwater channel heavily (geometric mean of E. coli counts 4.1 × 104 c.f.u./100ml) contaminated with wastewater where shellfish were exposed for 72h (Salas-Massó et al., 2016, 2018). fThis strain was obtained from F.J. García from the Laboratorio Central de Veterinaria de Algete, MAGRAMA, Madrid, Spain; gThese strains were recovered at the Faculty of Pharmacy, University of the Basque Country (UPV-EHU), Vitoria-Gasteiz, Spain, by R. Alonso, I. Martinez-Malaxetxebarria and A. Fernández-Astorga.
Culturing for genome sequencing was carried out either on blood agar (DIFCO, Madrid, Spain) or marine agar (Scharlau, Sentmenat, Spain) at 30°C in aerobiosis for 24–72 h, depending on the requirements. DNA was extracted using Easy-DNATM gDNA Purification kit (Invitrogen, Madrid, Spain) following the manufacturer’s instructions. The integrity of the DNA was evaluated by electrophoresis of 10 μl of the sample in a 1.5% agarose gel. The total amount of DNA was quantified using QubitTM with the dsDNA Broad Range Assay kit (Invitrogen). Paired-end libraries were constructed with 50 ng of DNA using Nextera DNA Library Preparation Kit (Illumina, Lisbon, Portugal) and sequenced with MiSeq platform (Illumina). Sequencing generated 2 × 300 bp paired-end reads. Clean reads were assembled with SPAdes (Nurk et al., 2013) and the CGE assembler (Larsen et al., 2012) in order to select the better assembly. Before depositing the genomes in the NCBI database, FASTA files were screened for eukaryotic and prokaryotic sequences using BLASTn, and for adaptors with VecScreen standalone software3. The five housekeeping genes used in the first MLSA analysis (atpA, gyrA, gyrB, hsp60, and rpoB) were extracted from each genome and compared with the Sanger sequences of these genes obtained originally for the identification of the strain. The existence of a single and identical copy of these genes confirmed that the genomes were not contaminated and belonged to the correct strain. Finally, contigs were deleted if they had less than 200 bp. The genomes were deposited in the GenBank database and Table 1 lists the accession numbers.
The 55 genomes were annotated with a local installation of Prokka v1.2 (Seemann, 2014) using an e-value of 1e-06. The annotation was performed with Prokka, with the prediction tools Prodigal v2.6 (Hyatt et al., 2010) and ARAGORN v1.2 (Laslett and Canback, 2004). The prediction tool Barrnap v0.64 included in Prokka v1.2 was used for the annotation of rRNA genes. Coding sequences (CDS) were annotated, combining the Rapid Annotation Subsystems Technology (RAST) (Overbeek et al., 2014) using the classic RAST scheme and the Annotation Tools of PATRIC server (Wattam et al., 2017). The characteristics of each genome (i.e., N50, number of contigs, number of CDS, G+C content) were obtained from NCBI annotations.
Analysis of Housekeeping Genes, Ribosomal Genes, and Core Genome
Thirteen housekeeping genes (atpA, atpD, dnaA, dnaJ, dnaK, ftsZ, gyrA, hsp60, radA, recA, rpoB, rpoD, and tsf) were obtained from the genomes using BLASTn search. Sequence similarities of housekeeping genes were determined using the MegAlign program (DNASTAR®, Madison, WI, United States). Genes were aligned using ClustalW (Larkin et al., 2007) and phylogenies based on individual genes and on the concatenated sequences was constructed with MEGA version 6.0 (Tamura et al., 2013) using the Neighbor-Joining (NJ) and Maximum-Likelihood (ML) algorithms.
The phylogenetic analysis of the core genome was assessed with the Roary software (Page et al., 2015) using 80% as cut-off for the BLASTp search. The core genome alignment was extracted with the latter software and the phylogeny was inferred using SplitsTree version 4.14.2 as described in Sawabe et al. (2007) using SplitsTree version 4.14.2, with a neighbor net drawing and Jukes-Cantor correction (Bandelt and Dress, 1992; Huson and Bryant, 2005).
Furthermore, the 16S and 23S rRNA genes of each genome were obtained using RNammer (Lagesen et al., 2007). In some cases, 16S rRNA gene sequences were obtained in our laboratories by Sanger sequencing or from the GenBank. The similarity of the 16S rRNA genes was calculated using MegAlign version 7.0.0 (DNASTAR®, Madison, WI, United States). Phylogenetic trees were reconstructed with MEGA version 6.0 (Tamura et al., 2013) also using the NJ and ML algorithms. Alignments obtained for both genes were visually analyzed in order to localize signature sequences for strains or groups of strains.
Genomic Indices
In order to ensure the correct assignation at species level of each analyzed genome, the ANI and the isDDH were calculated between all the genomes (Konstantinidis and Tiedje, 2005; Richter and Rosselló-Móra, 2009; Qin et al., 2014). The ANIb was calculated using JSpeciesWS (Richter et al., 2016), the resulting matrix was clustered and visualized using ggplot2 2.2.1 package (Wickham, 2009) and the isDDH was calculated with the GGDC software using results obtained with the formula 2 (Meier-Kolthoff et al., 2013). Two other indices (AAI and POCP) described for genus classification (Konstantinidis and Tiedje, 2005; Luo et al., 2014; Qin et al., 2014) were calculated among the genomes that corresponded to the type strains of the accepted species and the reference strains of the candidate species. The AAI was calculated with the Lycoming College Newman Lab AAIr Calculator5 using the Sequence-Based Comparison Tools output file from RAST (Overbeek et al., 2014). The POCP was determined as described by Qin et al. (2014) using the following parameters to consider a peptide as a conserved protein: an e-value lower than 1e-5 and an identity percentage higher than 40% from an aligned region higher than 50%.
Finally, the RSCU was computed using the Codon Adaptation Index (CAI) developed by Sharp and Li (1987) through the CAIcal web-server (Puigbò et al., 2008). Statistical differences in the RSCU were assessed by a multinomial regression approach using the R software environment (R Core Team, 2015). The principal component analysis (PCA) was performed by the R software environment (R Core Team, 2015, and visualized using ggplot2 2.2.1 and ggfortify 0.4.4 (Wickham, 2009; Horikoshi and Tang, 2015; Tang et al., 2016) or pca3d 0.10 (Weiner, 2017) packages.
Phenotypic Analysis and Metabolic Inference
Phenotypic characterization of each described species was obtained from this study, from the original descriptions or from the summary published by On et al. (2017). For the potentially new Arcobacter species, the phenotype was characterized following the recommended minimal standards described for new taxa of the family Campylobacteraceae (Ursing et al., 1994; On et al., 2017) and with complementary tests used in the description of other Arcobacter species (Levican et al., 2013).
Inference of the metabolic routes from the genome sequences was performed with the software package Traitar (Microbial Trait Analyzer) (Weimann et al., 2016), using the protein coding genes files obtained with Prokka v1.2 (Seemann, 2014). Traitar software is based on phenotypic data extracted from the Global Infectious Disease and Epidemiology Online Network (GIDEON) and Bergey’s Systematic Bacteriology. The software uses two prediction models: the phypat classifier, which predicts the presence/absence of proteins found in the phenotype of 234 bacterial species; and the phypat+PGL classifier, which uses the same information as the phypat combined with the information of the acquisition and loss of protein families and phenotypes during evolutive events. A total of 67 traits available within the software, related to oxygen requirement, enzymatic activities, proteolysis, antibiotic resistance, morphology and motility and the use of different carbon sources, were tested and the combined results of the two predictors were analyzed using a heat map.
Results and Discussion
Strains and Genomes
All the 27 species currently included in the genus Arcobacter and 13 candidate species have been investigated in the present study, which has analyzed 55 genomes, 16 of them from the public databases and 39 sequenced in this study (Tables 1, 2). It was not possible to analyze the genomes from A. acticola and A. pacificus because we were unable to get the type strains of the species. The contigs obtained and the N50 values complied with the recently proposed minimal standards for the use of genomes in taxonomic studies (Chun et al., 2018). The genome size ranged from 1.81 Mb for A. skirrowii F28 to 3.60 Mb for A. lekithochrous CECT 8942T (Table 2). The G+C content ranged from 26.1% in A. molluscorum CECT 7696T to 34.9% in ‘A. aquaticus’ W112-28. The G+C values agree with the range from 24.6% (which corresponded to the type strain of A. anaerophilus) to 31% indicated for the genus Arcobacter in the recent emended description by Sasi-Jyothsna et al. (2013). Interestingly, 26 genomes (47.3%) showed the presence of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs) and CRISPR-associated genes, related with the immune response of the bacteria.
Table 2.
Genome characteristics and annotation results. Source of whole genome sequences as indicated in Table 1.
| Species | No. Contigs | N50 (Kb) | CDS (Total) | CDS (Coding) | RNA Genes | tRNAs | ncRNAs | CRISPR Arrays | G+C (%) | Size (Mb) |
|---|---|---|---|---|---|---|---|---|---|---|
| A. anaerophilus DSM 24636T | 40 | 186 | 2,938 | 2,922 | 45 | 40 | 2 | 1 | 29.9 | 2.98 |
| A. anaerophilus IR1 | 7 | 1,179 | 3,360 | 3,024 | 61 | 47 | 2 | 3 | 30.2 | 3.25 |
| ‘A. aquaticus’ W112-28T | 20 | 370 | 2,500 | 2,487 | 55 | 45 | 3 | 0 | 34.9 | 2.53 |
| A. aquimarinus CECT 8442T | 68 | 75 | 2,473 | 2,463 | 46 | 42 | 2 | 0 | 26.6 | 2.46 |
| A. bivalviorum CECT 7835T | 179 | 461 | 2,786 | 2,728 | 50 | 41 | 3 | 0 | 28.2 | 2.75 |
| A. bivalviorum F118-4 | 26 | 209 | 2,652 | 2,652 | 47 | 38 | 3 | 0 | 28.1 | 2.71 |
| A. butzleri RM4018T | 1 | – | 2,261 | 2,256 | 71 | 54 | 2 | 0 | 27.0 | 2.34 |
| A. butzleri ED1 | 1 | – | 2,151 | 2,145 | 71 | 54 | 2 | 0 | 27.1 | 2.26 |
| ‘A. caeni RW17-10T | 59 | 123 | 2,357 | 2,337 | 58 | 51 | 3 | 0 | 27.1 | 2.42 |
| A. canalis CECT8984T | 50 | 166 | 2,733 | 2,720 | 53 | 48 | 2 | 1 | 27.3 | 2.78 |
| A. canalis SH-4D_Col1 | 69 | 72 | 2,716 | 2,663 | 63 | 52 | 2 | 1 | 27.1 | 2.82 |
| A. cibarius LMG 21996T | 44 | 119 | 2,156 | 2,110 | 68 | 46 | 2 | 0 | 27.1 | 2.20 |
| A. cloacae CECT 7834T | 135 | 135 | 2,826 | 2,795 | 58 | 51 | 2 | 3 | 26.8 | 2.78 |
| A. cloacae F26 | 40 | 218 | 2,470 | 2,459 | 53 | 44 | 2 | 1 | 26.9 | 2.51 |
| A. cryaerophilus LMG 24291T | 91 | 54 | 2,092 | 2,081 | 49 | 40 | 3 | 0 | 27.2 | 2.06 |
| A. defluvii CECT 7697T | 80 | 166 | 2,921 | 2,894 | 57 | 49 | 2 | 2 | 26.3 | 2.94 |
| A. ebronensis CECT 8441T | 103 | 188 | 3,089 | 3,072 | 47 | 39 | 3 | 1 | 29.2 | 3.15 |
| A. ebronensis W129-34 | 126 | 217 | 3,206 | 3,171 | 46 | 40 | 3 | 2 | 29.2 | 3.23 |
| A. ellisii CECT 7837T | 135 | 177 | 2,875 | 2,840 | 64 | 52 | 2 | 1 | 26.9 | 2.80 |
| A. faecis LMG 28519T | 55 | 127 | 2,429 | 2,376 | 76 | 53 | 2 | 1 | 27.2 | 2.50 |
| A. halophilus DSM 18005T | 111 | 56 | 2,677 | 2,660 | 54 | 46 | 3 | 2 | 27.4 | 2.75 |
| A. halophilus F166-45 | 90 | 56 | 2,879 | 2,864 | 59 | 51 | 2 | 2 | 27.0 | 2.96 |
| ‘A. hispanicus’ FW54T | 76 | 148 | 2,228 | 2,207 | 46 | 40 | 3 | 1 | 26.4 | 2.21 |
| ‘A. lacus’ RW43-9T | 24 | 295 | 2,194 | 2,182 | 47 | 40 | 2 | 0 | 26.8 | 2.22 |
| A. lanthieri LMG 28516T | 29 | 466 | 2,223 | 2,190 | 73 | 52 | 3 | 1 | 26.7 | 2.29 |
| A. lanthieri AF1581 | 24 | 353 | 2,199 | 2,186 | 88 | 57 | 3 | 0 | 26.8 | 2.26 |
| A. lekithochrous CECT 8942T | 436 | 343 | 3,628 | 3,316 | 88 | 75 | 3 | 0 | 28.6 | 3.61 |
| A. lekithochrous LMG 28652 | 82 | 343 | 3,499 | 3,330 | 61 | 55 | 3 | 0 | 28.2 | 3.50 |
| A. marinus CECT 7727T | 162 | 54 | 2,809 | 2,781 | 55 | 50 | 2 | 0 | 27.0 | 2.87 |
| A. marinus F140-37 | 76 | 67 | 2,725 | 2,652 | 59 | 48 | 2 | 0 | 27.0 | 2.78 |
| ‘A. mediterraneus’ F156-34T | 29 | 689 | 2,769 | 2,750 | 47 | 41 | 3 | 1 | 27.3 | 2.83 |
| ‘A. miroungae’ 9AntT | 35 | 363 | 1,868 | 1,847 | 46 | 41 | 2 | 1 | 28.1 | 1.84 |
| A. molluscorum CECT 7696T | 117 | 121 | 2,746 | 2,736 | 58 | 49 | 3 | 6 | 26.1 | 2.76 |
| A. molluscorum F91 | 240 | 150 | 2,951 | 2,889 | 71 | 58 | 3 | 2 | 26.3 | 2.89 |
| A. mytili CECT 7386T | 126 | 70 | 2,950 | 2,934 | 58 | 48 | 3 | 1 | 26.3 | 2.97 |
| A. mytili T234 | 145 | 37 | 2,735 | 2,723 | 54 | 48 | 3 | 0 | 26.4 | 2.77 |
| ‘A. neptunis’ F146-38T | 36 | 267 | 2,627 | 2,614 | 57 | 45 | 3 | 0 | 27.1 | 2.65 |
| A. nitrofigilis DSM 7299T | 1 | – | 3,101 | 3,086 | 69 | 55 | 2 | 1 | 28.4 | 3.19 |
| ‘A. ponticus’ F161-33 | 24 | 597 | 2,632 | 2,621 | 46 | 36 | 3 | 0 | 28.1 | 2.74 |
| ‘A. porcinus’ LMG 24487T | 70 | 123 | 2,186 | 2,112 | 47 | 41 | 2 | 0 | 27.0 | 2.14 |
| ‘A. salis’ F155-33T | 153 | 169 | 2,932 | 2,904 | 50 | 43 | 3 | 0 | 29.0 | 2.93 |
| A. skirrowii LMG 6621T | 62 | 306 | 2,029 | 2,006 | 48 | 42 | 2 | 2 | 27.7 | 1.97 |
| A. skirrowii F28 | 110 | 40 | 1,911 | 1,897 | 46 | 41 | 2 | 0 | 27.8 | 1.81 |
| A. suis CECT 7833T | 122 | 142 | 2,646 | 2,613 | 57 | 52 | 2 | 0 | 27.3 | 2.62 |
| A. thereius LMG 24486T | 2 | 1,039 | 1,896 | 1,883 | 57 | 46 | 2 | 3 | 27.0 | 1.91 |
| A. thereius DU22 | 19 | 252 | 2,006 | 1,983 | 47 | 42 | 2 | 1 | 26.8 | 2.01 |
| A. trophiarum CECT 7650 | 37 | 152 | 1,911 | 1,894 | 48 | 37 | 3 | 0 | 28.0 | 1.90 |
| A. trophiarum LMG 25534T | 266 | 86 | 2,167 | 2,071 | 49 | 41 | 3 | 0 | 29.4 | 2.00 |
| A. venerupis CECT 7836T | 234 | 182 | 3,319 | 3,267 | 64 | 52 | 2 | 0 | 28.0 | 3.28 |
| ‘A. viscosus’ F142-34T | 82 | 65 | 2,772 | 2,756 | 55 | 48 | 3 | 1 | 26.6 | 2.79 |
| ‘A. vitoriensis’ FW59T | 144 | 179 | 2,617 | 2,570 | 53 | 46 | 2 | 0 | 27.4 | 2.58 |
| Arcobacter sp. CAB | 367 | 20 | 3,596 | 3,392 | NA | 31 | NA | NA | 28.2 | 3.48 |
| Arcobacter sp. F2176 | 99 | 178 | 3,212 | 3,186 | 67 | 57 | 2 | 0 | 28.1 | 3.27 |
| Arcobacter sp. LA11 | 53 | 229 | 3,006 | 2,961 | 49 | 43 | 3 | 0 | 27.9 | 3.10 |
| Arcobacter sp. LPB0137 | 1 | – | 2,731 | 2,698 | 85 | 64 | 2 | 0 | 27.7 | 2.87 |
| Arcobacter sp. La | 1 | – | 2,847 | 2,834 | 73 | 56 | 2 | 1 | 26.6 | 2.95 |
| Arcobacter sp. AF1028b | 46 | 148 | 2,336 | 2,285 | 71 | 51 | 2 | 1 | 27.2 | 2.41 |
aGenome sequenced in this study; bGenome obtained from NCBI database; cGenome obtained from JGI Gold database. Our results show that these strains belong to the species. dA. defluvii and eA. faecis.
Taxonomic and Phylogenetic Analysis
Similarities in the 16S rRNA gene sequences among type and representative strains of the different Arcobacter species (all the 27 species currently included in the genus and the 13 new candidate species) showed a wide range of values (Supplementary Tables S1, S4). They ranged from 90.8% (observed between A. anaerophilus and A. faecis) to 99.9% (between A. butzleri and ‘A. lacus’). The lower range of similarity (90.8%) is due to the fact that those species, as occurred with others, were assigned within the genus based on the premise that 16S rRNA gene similarity was higher with any type strain of Arcobacter than with other taxa. However, in some cases being below the 95% cut-off value for genus delimitation (Rosselló-Mora and Amann, 2001; Yarza et al., 2008; Tindall et al., 2010; Figueras et al., 2011a,b). It is interesting to point out that 16S rRNA gene sequence similarities among A. nitrofigilis, the type species of the genus, and the other described species ranged from 93.2% (with A. thereius) to 95.9% (with A. venerupis). Furthermore, A. nitrofigilis showed higher similarities than the threshold value of 95% with only seven species (A. acticola, ‘A. caeni,’ A. cloacae, A. defluvii, A. ellisii, A. suis, and A. venerupis) out of the 27 accepted species. In any case, from the analysis of the similarities in the 16S rRNA gene sequences among the Arcobacter species it is clear that this gene has limited value and that other approaches available in the genomic era of taxonomy are needed for their study.
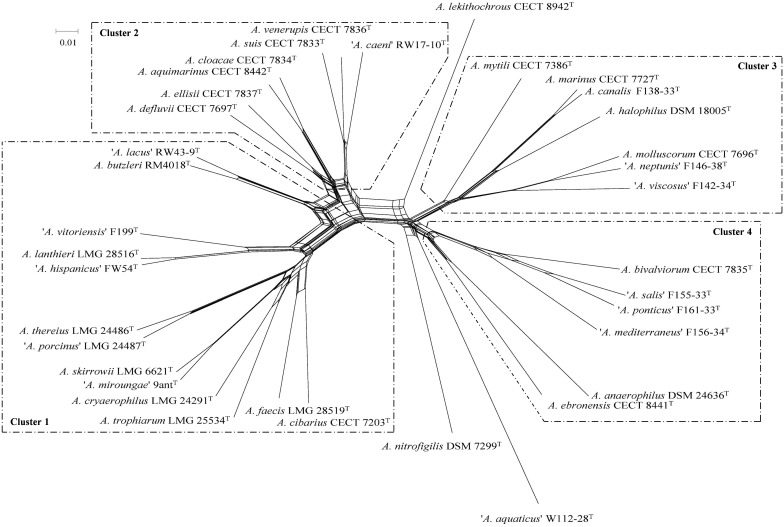
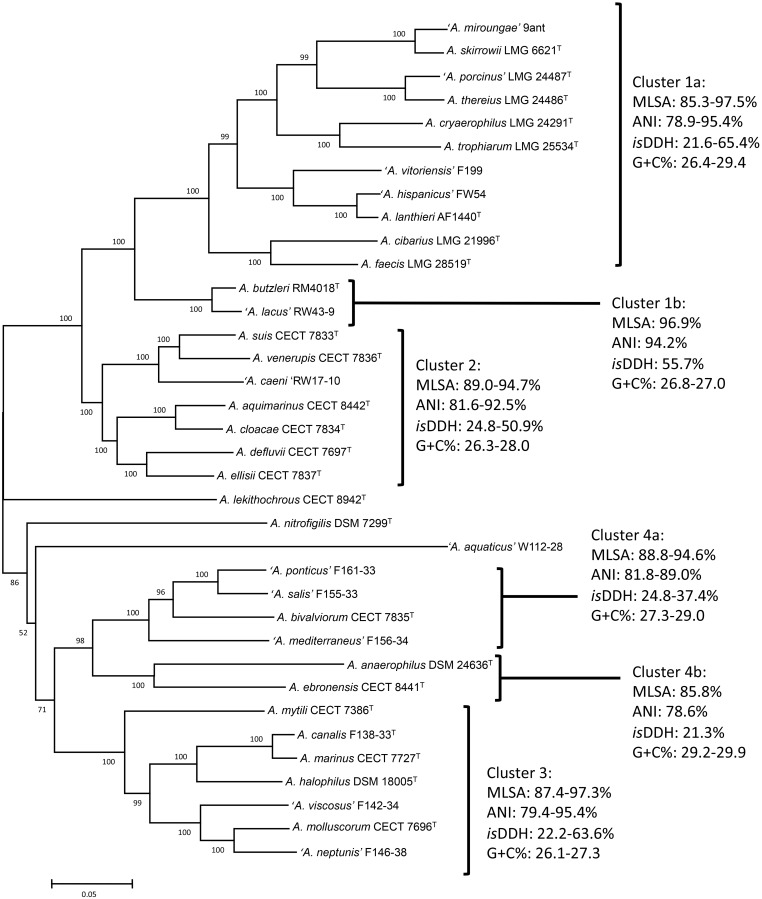
Phylogenetic analysis based on the core genome made up of 286 genes (Figure 1 and Supplementary Table S5) and also on the concatenated sequences of 13 housekeeping genes of the representative Arcobacter strains (Figure 2) revealed that the Arcobacter species could be grouped into 4 major monophyletic clusters. Cluster 1, comprised seven validated species: A. butzleri, A. cibarius, A. cryaerophilus, A. lanthieri, A. skirrowii, A. thereius, and A. trophiarum, together with A. faecis (species described but not validated yet) and five candidate taxa ‘A. hispanicus,’ ‘A. lacus,’ ‘A. miroungae,’ ‘A. porcinus,’ and ‘A. vitoriensis’ (Figure 1). Cluster 2 embraced the species A. aquimarinus, A. cloacae, A. defluvii, A. ellisii, A. suis, and A. venerupis, as well as the non-validated A. acticola and the candidatus ‘A. caeni.’ Cluster 3 included five species, A. canalis, A. halophilus, A. marinus, A. molluscorum, and A. mytili, together with two candidates, ‘A. neptunis’ and ‘A. viscosus.’ Finally, Cluster 4 included the species A. anaerophilus, A. bivalviorum, and A. ebronensis, as well as the candidates ‘A. mediterraneus,’ ‘A. ponticus,’ and ‘A. salis.’ The split decomposition network analysis of the core genome showed that the species A. lekithochrous CECT 8942T and A. nitrofigilis DSM 7299T appeared as orphan species. Furthermore, with this analysis the candidatus ‘A. aquaticus’ W112-28 also appeared in a separate branch near to A. nitrofigilis DSM 7299T. On the other hand, both analyses, MLSA and core genome, confirmed the existence of two sub-clusters in Cluster 1 (again A. butzleri and ‘A. lacus’ were located in the most distant branch within the cluster), and also two subgroups could be observed in Cluster 4, one comprising the species A. anaerophilus and A. ebronensis, and the other including the rest of species within this cluster (Figures 1, 2). All the clusters and sub-clusters showed a similarity in the concatenated sequences of the 13 housekeeping genes higher than 85% (Figure 2).
FIGURE 1.
Split decomposition network constructed with the concatenated sequences of 284 core genes from the genomes of 36 type and representative strains of Arcobacter. Scale bar, base substitutions per site.
FIGURE 2.
Phylogenetic tree constructed with 36 type and representative strains of Arcobacter species based on concatenated sequences of 13 housekeeping genes by the Maximum-Likelihood algorithm (model GTR+G+I). Numbers at nodes denote the level of bootstrap based on 1,000 replicates; only values greater than 50% are shown. Scale bar, base substitutions per site.
Phylogenies based on the 16S and 23S rRNA gene sequences, undertaken with the NJ and ML approacheserealso constructed with comparative purposes. 16S rRNA based tree showed also the four major clusters although less defined (Supplementary Figure S1A). Species within Cluster 1, showed 16S rRNA gene sequence similarities ranging from 96.1 to 99.9%. Cluster 2 yielded similarities among species for the 16S rRNA gene between 96.7 and 99.6%, whereas within Cluster 3 ranged between 93.0 and 99.1%. Finally, Cluster 4 included species with a range of 16S rRNA sequence similarity from 94.0 to 99.5%. With the exception of Cluster 3, similarity values within the clusters (>94–95%) were within the classical boundaries for genus assignation in bacterial taxonomy (Rosselló-Mora and Amann, 2001; Yarza et al., 2008, 2014; Tindall et al., 2010; Figueras et al., 2011a,b). Our results agree with those from a recent study by Yarza et al. (2014), who investigated 568 taxa and described a threshold in 16S rRNA sequence identity of 94.5% for genus delineation.
Similar groups and topology, with only minor differences, were obtained when the 23S rRNA gene sequences were used to analyze the phylogeny of the genus (Supplementary Figure S2). In this analysis, the recently described species A. acticola, and A. pacificus could not be included because of the unavailability of the type strains and/or whole genome sequences. The same four major clusters formed in the 23S rRNA gene phylogenetic tree, and the species A. lekithochrous and A. nitrofigilis appeared also as orphan species (Supplementary Figure S2). Within Cluster 1 two subgroups could also be obtained, differentiating the species A. butzleri and ‘A. lacus’ from the rest of the species. Similarly, the species A. anaerophilus and A. ebronensis formed a differentiated subgroup in Cluster 4.
The visual analysis of the alignments obtained with the sequences of the 16S and 23S rRNA genes allowed the localization of signature motifs, especially in the 16S rRNA gene, for the different clusters established in the phylogenetic analysis. In these sequences, a total of 16 locations were found, presenting nucleotide combinations characteristic for the clusters (Supplementary Figure S3). Some of these motifs were located in helix regions as interactions with proteins of the ribosomal 30S subunit, such as helix 21 (region V4) or helix 28/44 (region V9), and therefore had a considerable level of protection against mutations (Adilakshmi et al., 2008; Kitahara et al., 2012). There are some studies on the presence of signature regions with taxonomic/phylogenetic implications in the ribosomal genes (Martínez-Murcia et al., 1992, 2007; Ue et al., 2011;Řeháková et al., 2014; Martínez-Murcia and Lamy, 2015). Some regions with signature motifs detected in the present study have also shown implications for phylogenetic analysis in cyanobacteria, including regions H15, H17, H21, H22-H23, H41, and H44 (Řeháková et al., 2014). A tree was also constructed weighting such positions (Supplementary Figure S1B), which allowed a better definition of the main clusters observed with the whole 16S rRNA sequences although, as expected, differentiation among species within each cluster was lower. Two sub-clusters were observed in Cluster 1, where the species A. butzleri and ‘A. lacus’ grouped into a well-differentiated branch with respect to the other species in the cluster (Supplementary Figure S1B). In this analysis, A. pacificus was clearly located in the Cluster 3, whereas in Cluster 4, A anaerophilus was the borderline species, while A. ebronensis and ‘A. mediterraneus’ were located in an independent branch (Supplementary Figure S1B). Therefore, the signature motifs described here might be a new tool for identification of the different clusters and/or genus.
Genomic Indices
The results of the calculations of the ANI and the isDDH among the 36 studied genomes are given in the Supplementary Table S2 and Supplementary Figure S4. The results of the ANI and isDDH calculations showed that the genomes grouped into the same clusters observed by the analyses of the MLSA of the 13 housekeeping and core genes (Figures 1, 2). Ranges of ANI within each cluster were from 75.2 to 95.4%, whereas isDDH values were between 19.5 and 65.4% (Figure 2 and Table 3). These results confirm the phylogenetic analysis for the 13 new candidate species because all of them showed ANI and isDDH values of <96% and <70%, respectively, which are the cut-off values proposed for the delineation of new species (Konstantinidis and Tiedje, 2005; Goris et al., 2007; Richter and Rosselló-Móra, 2009; Figueras et al., 2017). As discussed in other studies, the ANI and isDDH indices provided reliable information for the delineation of Arcobacter species and are also included in the minimal guidelines to define species using genomes (Whiteduck-Léveillée et al., 2015, 2016; Figueras et al., 2017; Chun et al., 2018). Although those indices are not considered useful for delimiting genera, each of the four clusters showed values that ranged between 75.2 and 81.8% as their lowest ANI, which might be the suitable range for separating different, closely related genera. These values are relatively similar to those reported by Qin et al. (2014) that found 68–82% interspecies ANI values among the genera that they studied. Values of ANI obtained for the candidate species ‘A. aquaticus’ were lower than the other results, from 70.0% with A. cryaerophilus LMG 24291T to 71.9% with A. bivalviorum CECT 7835T and more in line with the Qin et al. (2014) results of 68% (Supplementary Table S2). In the case of the isDDH the lower values among species in the same cluster ranged between 19.5 and 24.8%, and again these might be the levels associated to different genera.
Table 3.
Intra-cluster similarities (%) obtained for the 16S rRNA gene and for the different genomic indexes analyzed.
| 16S RNA gene | MLSA | ANI | isDDH | AAI | POCP | G+C% (mol) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cluster 1a | 96.8–99.8 | 85.3–97.5 | 78.9–95.4 | 21.6–65.4 | 72.5–95.0 | 68.2–95.6 | 26.4–29.4 | 26.4–29.4 | ||||||
| 96.1–99.9 | 85.3–97.5 | 77.7–95.4 | 20.5–65.4 | 71.5–95.0 | 67.0–95.6 | |||||||||
| Cluster 1b | 99.9 | 96.9 | 94.2 | 55.7 | 93.7 | 84.3 | 26.8–27.0 | |||||||
| Cluster 2 | 96.7–99.6 | 89.0–94.7 | 81.6–92.5 | 24.8–50.9 | 73.1–93.5 | 71.7–87.5 | 26.3–28.0 | |||||||
| Cluster 3 | 94.2–99.1 | 87.4–97.3 | 79.4–95.4 | 22.2–63.6 | 67.6–95.7 | 75.4–91.4 | 26.1–27.3 | |||||||
| Cluster 4a | 96.6–99.5 | 88.8–94.6 | 81.8–89.0 | 24.8–37.4 | 80.3–83.4 | 74.4–90.7 | 27.3–29.0 | 27.3–29.9 | ||||||
| 94.0–99.5 | 85.8–94.6 | 75.2–89.0 | 19.5–37.4 | 68.7–83.4 | 71.6–90.7 | |||||||||
| Cluster 4b | 96.9 | 85.8 | 78.6 | 21.3 | 78.4 | 77.9 | 29.2–29.9 | |||||||
With the aim of confirming if the clusters observed might represent different genera, as suggested by the phylogenetic analyses, the similarity indices AAI and POCP were also calculated (Supplementary Table S3). In agreement with the 60–80% AAI that have been described for species belonging to the same genus (Konstantinidis and Tiedje, 2005) all our clusters showed lower ranges of between 67.6 to 80.3% (Table 3). All the clusters also complied with the POCP proposed for genus separation above 50% (Luo et al., 2014; Qin et al., 2014) because as shown in Table 3 all clusters showed the lowest values from 67.0 to 75.4%.
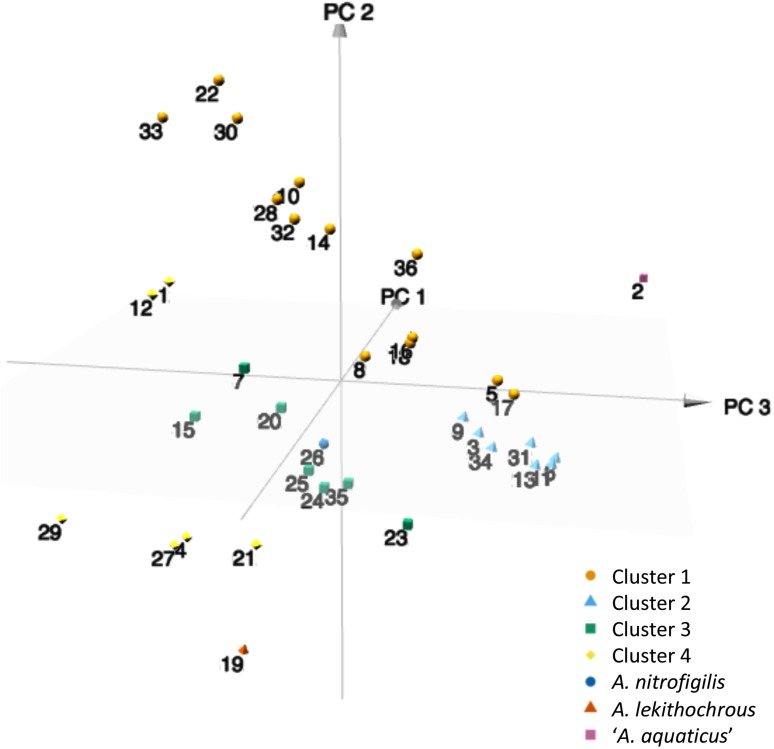
It is widely known that synonymous codon usage varies among organisms and that it is related to differences in G+C content, replication strand skew, or gene expression (Suzuki et al., 2008; Farooqi et al., 2016). The interaction of these factors may vary among species depending on their evolutionary process (Ma et al., 2015). It has also been suggested that the extent of codon usage bias plays a role in the adaptation of prokaryotic organisms to their environments and lifestyles (Botzman and Margalit, 2011). To analyze the overall codon usage trends of the Arcobacter species, the frequencies of the different codons were obtained from the whole genomes and the RSCU was computed using the CAI, which is a useful tool for estimating codon usage bias (Ma et al., 2015; Farooqi et al., 2016). A first finding was that all the Arcobacter species presented a preferential use of the codons finishing in A or T (Supplementary Figure S5), which might be expected due to their low G+C% content. The characteristic pattern showed by A. aquaticus is noteworthy (Supplementary Figure S5), which supports its differentiation from the other species in Cluster 3 as well as its unique taxonomy. Such difference was the only statistically significant (p < 0.05) in the multinomial regression analysis carried out.
Next, the codon usage trends were analyzed by PCA to reveal possible evolutionary relationships. Interestingly, different groups of strains could be observed in the three-dimensional graphic (Figure 3), which correlated with those clusters established in the different phylogenetic analyses, as shown above. As reported previously for different species of Mycoplasma (Marenda et al., 2005; Ma et al., 2015), PCA provides an additional pathway to investigate the evolutionary direction of the Arcobacter species. In addition, similarities in the synonymous codon usage patterns might reflect similar lifestyles (pathogenic vs. non-pathogenic) and adaptation to certain environments (marine water, shellfish, etc.).
FIGURE 3.
3D plot of the three major axes generated by principal component analysis (PCA) of the RSCU values computed for the 36 type and representative strains of Arcobacter species. 1, A. anaerophilus DSM 24636T; 2, ‘A. aquaticus’ W112-28; 3, A. aquimarinus CECT 8442T; 4, A. bivalviorum; 5, A. butzleri RM4018T; 6, ‘A. caeni’ RW17-10; 7, A. canalis CECT 8984T; 8 A. cibarius LMG 21996T; 9, A. cloacae CECT 7834T; 10, A. cryaerophilus LMG 24291T; 11, A. defluvii CECT 7697T; 12, A. ebronensis F128-2T; 13, A. ellisii CECT 7837T; 14, A. faecis AF1078T; 15, A. halophilus DSM 18005T; 16, ‘A. hispanicus’ FW54; 17, ‘A. lacus’ RW43-9; 18, A. lanthieri AF1440T; 19, A. lekithochrous LFT1.7T; 20, A. marinus CECT 7727T; 21, ‘A. mediterraneus’ F156-34; 22, ‘A. miroungae’ 9Ant; 23, A. molluscorum CECT 7696T; 24, A. mytili W112-28; 25, ‘A. neptunis’ F146-38; 26, A. nitrofrigilis DSM 7299T; 27, ‘A. ponticus’ F161-33; 28, ‘A. porcinus’ LMG 24487; 29, ‘A. salis’ F155-33; 30, A. skirrowii LMG 6621T; 31, A. suis CECT 7833T; 32, A. thereius LMG 24486T; 33, A. trophiarum LMG 25534T; 34, A. venerupis CECT 7836T; 35, ‘A. viscosus’ F142-34; 36, ‘A. vitoriensis’ F199.
Metabolic Inference and Phenotypic Analysis
Phylogenetic and genomic analysis confirmed the existence of four clusters among the validated and candidate Arcobacter species, which comply with the cut-off values established for the differentiation of independent genera. A thorough phenotypic analysis was therefore carried out to determine if the description of new taxa at genus level was possible or if such clusters were only clades or genomovars within the genus Arcobacter. In fact, this is what has occurred in a recent polyphasic study of 52 A. cryaerophilus strains (including genome information) in which, despite four different genomospecies being recognized, the phenotypic characterization did not allow their differentiation into separate species and were therefore considered genomovars (Pérez-Cataluña et al., 2018a).
Phenotypic inference using Traitar confirmed the lack of reaction of Arcobacter species to most of the tests commonly used for bacterial identification (Supplementary Figure S6). Thus, all the type and representative strains rendered negative results, regardless of the predictor employed, for use as the sole carbon source of sugars (D-Mannitol, D-Mannose, Salicin, or Trehalose, among others) and carboxylic acids (Citrate or Malonate). Such results have been previously reported in the original descriptions of the species (see review of On et al., 2017). On the other hand, there was some incongruence between results from Traitar and those obtained by classical characterization for some tests, including growth on MacConkey agar or urea hydrolysis (data not shown). A possible explanation is related with the macro-accuracy of the predictors employed in the Traitar analysis (82.6–85.5%), as reported in the original description of the microbial trait analyzer (Weimann et al., 2016). The fact that some of the Arcobacter species studied are halophilic cannot be ignored, since some of the media usually employed in the wet-lab characterization are developed for non-halophilic microorganisms.
The heat maps built from the combined results of both predictors in the Traitar analysis revealed the existence of similarity groups regarding the metabolic characteristics of the Arcobacter type strains (Supplementary Figure S6). In most case, clustering of strains supported the groups obtained with genomic tools, although some incongruence was also observed, such as for A. butzleri (better related here to A. defluvii, A. ellisii or A. cloacae), A. mytili (closest Traitar species ‘A. caeni’) or A. venerupis (forming a branch with A. ebronensis and ‘A. ponticus’). In any case, Traitar might be helpful as a first-step method for phenotypic inference, although further verification should be made, especially in environmental bacterial species with special growth requirements (i.e., halophilic conditions).
A deep review of the characteristics reported in the original descriptions of the Arcobacter species, together with results obtained in our respective laboratories, allowing phenotypic traits to differentiate the clusters established by the phylogenetic and genomic analyses (Table 4). Growth at 37°C in microaerophilic condition, the halophilic character, the ability to grow in presence of glycine, safranin, oxgall, or triphenyltetrazolium chloride (TTC), the presence of some enzymatic activities, such as catalase, urease or indoxyl acetate hydrolysis, and resistance to cefoperazone among others, were the main differentiating traits. Most of these characters are included in the minimal standards for describing new species in the families Campylobacteraceae and Helicobacteraceae (On et al., 2017), and they should, therefore, also be maintained for the new family Arcobacteraceae proposed by Waite et al. (2017), once this taxonomical change is validated. The phenotypic differentiation proposed in Table 4 enabled to further describe the new genera that corresponded to the different clusters of Arcobacter species determined in the present study.
Table 4.
Differential phenotypic traits among the different clusters of Arcobacter species obtained on the basis of the characteristics of the type and representative strains of the species included in each group.
| Test | A. nitrofigilis | Cluster 1 | Cluster 2 | Cluster 3 | Cluster 4 | A. lekithochrous | A. aquaticus |
|---|---|---|---|---|---|---|---|
| Growth at/on | |||||||
| CO2 37°C | – | V | V | + | V | – | + |
| 0.5% NaCl | – | +a | + | V | –b | –c | + |
| 4% NaCl | + | – | – | + | + | – | – |
| 1% Glycine | – | V | – | V | V | – | – |
| 0.05% Safranin | – | + | V | V | V | + | + |
| 0.04 TTC | – | V | – | – | – | + | – |
| 1% Oxgall | – | V | V | – | –d | – | – |
| CCDA | – | V | V | –e | – | + | + |
| Enzymatic activities | |||||||
| Catalase | – | +f | + | V | V | + | – |
| Urease | + | – | V | – | – d | – | – |
| Indoxyl acetate hydrolysis | + | +f | + | V | V | – | – |
| Nitrate reduction | + | V | + | –g | V | – | – |
| Resistance to cefoperazone (64 mg/l) | ND | V | – | V | – | – | + |
+, positive result; -, negative result; V, variable result in all the species of the cluster; aWith the exception of A. skirrowii; bWith the exception of A. pacificus; cA. lekithochrous needs sea salts to grow; dWith the exception of A. ebronensis; eWith the exception of A. molluscorum; fWith the exception of A. cibarius; gWith the exception of A. anaerophilus. ND, not determined.
Stability of the Genomic-Based Clustering
In order to test the stability of the new taxonomical scheme proposed, we analyzed the whole genome sequences using second strains from each species or from unassigned sequences obtained from the public databases. That analysis is shown in Supplementary Figure S7 and included 55 genomes. These new phylogenetic analyses of the core genome also using a Split network showed that the four clusters were maintained, but the two clusters (Clusters 3 and 4) that include species able to grow in media containing 2.5% NaCl appeared in the right place (Supplementary Figure S7). The genome of Arcobacter sp. LPB0137 obtained from the NCBI database grouped with the species A. lekithochrous CECT 8942T, while the genomes Arcobacter sp. LA11 and CAB grouped together in a separate branch near to Cluster 4. Interestingly, the ANI and isDDH values of 91.4% and 45.8% between strain F2176, previously identified as A. nitrofigilis (Figueras et al., 2008), and the type strains of this species along with the phylogenetic position (Supplementary Figure S7), revealed that this strain belonged to another potentially new species. Furthermore, strains L and AF1028, deposited at the NCBI database as Arcobacter sp. were identified as A. defluvii and A. faecis, respectively, because they clustered with the type strains of those species (Supplementary Figure S7). This was also confirmed by the ANI and isDDH results being above 96% and 70%, respectively.
Collado and Figueras (2011), in their review about the epidemiology and clinical significance of the genus Arcobacter, reported that these bacteria should be considered quite atypical within the class Epsilonproteobacteria because of the great diversity of hosts and habitats from which they have been isolated. In order to show if the clusters obtained have a relationship with their ecological habitat, the origin of each strain is also given in Supplementary Figure S7. Despite the fact that only two strains from each species were included in the analysis, each of the clusters embraced species that had been recovered from common or related origins. Cluster 1 included by strains isolated from humans and animals, from wastewater and from broiler skin (A. cibarius CECT 7203T). The fact that some strains isolated from wastewater that was contaminated by humans or animal excreta, gives evidence of the relationship of these sources. This finding agrees with the high abundance of Arcobacter in wastewater and in water contaminated with fecal pollution (Collado et al., 2008, 2010). Among the species of Cluster 1, both by metagenomics analysis or direct plating without enrichment (Fisher et al., 2014; Levican et al., 2016), the species A. cryaerophilus was the prevalent species in wastewater, while the species A. butzleri is normally predominant in studies that investigate water and food samples of animal origin, such as different types of meats using an enrichment step (Collado et al., 2009b; Collado and Figueras, 2011; Hsu and Lee, 2015; and references therein). So far, only the species A. cryaerophilus, A. thereius, A. trophiarum, A. cibarius or A. skirrowii have been recovered from humans or animals (De Smet et al., 2011; Figueras et al., 2014; Van den Abeele et al., 2014) and all these species are as commented in the same cluster.
Cluster 2 included strains from different origins but was dominated by species that came from wastewater, shellfish or food products. In this sense, A. defluvii CECT 7697T and ‘A. caeni’ RW17-10 were isolated from wastewater, while the strain A. defluvii L was recovered from a microbial fuel cell. Strains of A. defluvii have also been recovered from shellfish in other studies (Levican et al., 2014; Salas-Massó et al., 2016). The strain A. suis CECT 7833T was isolated from pork meat, but other isolates have also been obtained from buffalo milk in Italy (Levican et al., 2013; Giacometti et al., 2015). The other five strains in the cluster were isolated from shellfish, wastewater and seawater (Table 1 and Supplementary Figure S7). The other two clusters (Clusters 3 and 4) included strains isolated from seawater shellfish giving evidence of the marine origin of these clusters. The orphan species (A. nitrofigilis DSM7299T, A. lekithochrous CECT 8942T, and ‘A. aquaticus’ W112-28) also corresponded to strains isolated from marine environments and their phylogenetic position was close to the two marine clusters (3 and 4).
As indicated in the review by Collado and Figueras (2011), there are many uncultured or not-yet-described species of Arcobacter, which have been recognized on the basis of nearly full-length 16S rRNA gene sequences, and which probably outnumber those species that were already known at that time. Their hosts and/or habitats are very diverse and include cod larvae, cyanobacterial mats, activated sludge, tidal and marine sediments, estuarine and river water, plankton, coral, tubeworms, snails, etc. (Collado et al., 2011; and references therein). In the near future new species can be expected to emerge that will reinforce the value of the different genera proposed in this study.
Conclusion
Genomic information obtained through next-generation sequencing leads to great advances in the systematics of prokaryotes (Whitman, 2015), not only to the general understanding of prokaryotic biology but also for the resolution of the phylogeny of taxa higher than species. Single gene phylogeny, including 16S rRNA gene, has often limitations that analysis of complete genome sequences can overcome. The study aims to use this modern taxonomy approach to clarify the relationships of the diverse Arcobacter species.
The results obtained in the present study confirmed the opinion of some authors on the need for a clarification of the taxonomy of the genus Arcobacter. The phylogenetic analyses derived from the MLSA of 13 genes and of the core genome as well as the existence of signature regions in the 16S rRNA gene have shown, together with the genomic indexes ANI (75.2–81.8%), isDDH (19.5–24.8%), AAI (67.6–80.3%), and POCP (67.0–75.4%), to be useful tools for delimiting several genomic and phylogenetic groups within this genus. The intra-genus ranges and cut-off values established here might also be helpful for future taxonomic studies in other bacterial groups.
Such genomic variability, together with the determination of combinations of differentiating phenotypic traits allowed the division of the current genus Arcobacter in at least six different genera for which the names Aliiarcobacter gen. nov., Pseudoarcobacter gen. nov., Haloarcobacter gen. nov., Malacobacter gen. nov., and Poseidonibacter gen. nov. are proposed. In addition, the candidate species ‘A. aquaticus’ also constitutes a new genus for which the name Candidate ‘Arcomarinus’ gen. nov. is proposed, although such proposal should be formulated in parallel to the formal description of the species.
According to Tindall et al. (2010) “the type strain of a genus is the most important reference organism to which a novel species has to be compared.” In the case of the genus Arcobacter, the type species has rarely been isolated (Collado et al., 2009b; Toh et al., 2011; Levican et al., 2016; Salas-Massó et al., 2016) and in fact, all the analyses show that A. nitrofigilis is an orphan species and the only representative of the genus Arcobacter, for which an emended description is provided.
The other genera are described here while taking into account the species validated at the time of writing but with the confidence that the formal description of the candidate species would fit in such descriptions. Thus, the genus Aliiarcobacter gen. nov. is described comprising seven species Aliiarcobacter cryaerophilus comb. nov., A. butzleri comb. nov., A. skirrowii comb. nov., A. cibarius comb. nov., A. thereius comb. nov., A. trophiarum comb. nov., A. lanthieri comb. nov., and A. faecis comb. nov. On the other hand, the genus Pseudoarcobacter gen. nov. includes the species Pseudoarcobacter defluvii comb. nov., P. ellisii comb. nov., P. venerupis comb. nov., P. cloacae comb. nov., P. suis comb. nov., P. aquimarinus comb. nov., and P. acticola comb. nov. Four species, Malacobacter halophilus comb. nov., M. mytili comb. nov., M. marinus comb. nov., M. molluscorum comb. nov., and M. pacificus comb. nov. are compiled in the new genus Malacobacter gen. nov., whereas the genus Haloarcobacter gen. nov. comprises three species Haloarcobacter bivalviorum comb. nov., H. anaerophilus comb. nov., and H. ebronensis comb. nov. Finally, the genus Poseidonibacter gen. nov. has a unique species Poseidonibacter lekithochrous comb. nov.
Emended Description of the Genus Arcobacter Vandamme et al., 1991 emend. Vandamme et al., 1992 and Sasi-Jyothsna et al., 2013
Arcobacter (Ar’co.bac.ter. L. n. arcus, bow; Gr. n. bacter, rod; M. L. masc. n. Arcobacter, bow-shaped rod).
Cells are Gram-negative, curved rods 0.2–0.9 μm in diameter and 1–3 μm long. Coccoid bodies are found in old cultures but are not rapidly produced under aerobic conditions. Motile with a rapid corkscrew motion. Each cell possesses a single polar flagellum. Does not swarm. Chemoorganotrophic. Utilizes organic and amino acids as carbon sources, but not carbohydrates. Respiratory metabolism with oxygen as the terminal electron acceptor; anaerobic growth with aspartate and fumarate, but not with nitrate. Nitrate usually reduced to nitrite. Requires NaCl for growth. Grows at temperatures of 10°C–35°C but not at 42°C. Catalase, oxidase, urease, and nitrogenase positive. Phosphatase, sulfatase and indole negative. Does not hydrolyze esculin, casein, DNA, gelatine, hippurate or starch. Fluorescent pigments are not produced. Unable to grow with glycine (1% wt/vol), safranin (0.05% wt/vol), oxgall (1% wt/vol), or 2,3,5-triphenyltetrazolium chloride (0.04%, wt/vol). Positive for the hydrolysis of indoxyl acetate. Poly-β-hydroxybutyrate not produced.
The base composition of the DNA is 28.1–28.4% G+C as determined from the genomes.
The type species is Arcobacter nitrofigilis.
Description of Aliiarcobacter gen. nov.
Aliiarcobacter (A.li.i.ar.co.bac’ter, L. pronoun alius other, another; N.L. masc. n. Arcobacter a bacterial generic name; N.L. masc. n. Aliiarcobacter the other Arcobacter).
Cells are Gram-negative, curved rods 0.2–0.5 μm in diameter and 1–3 μm long. Motile by single polar flagellum. Does not swarm. Chemoorganotrophic. Oxidase and catalase positive. No growth occur at 4% NaCl. Growth occurs at 15°C–42°C. Carbohydrates are not fermented. Nitrate usually reduced to nitrite. Positive for the hydrolysis of indoxyl acetate and negative for urease. Growth does not occur in the presence 2,3,5-triphenyltetrazolium chloride (0.04%, wt/vol) or glycine (1% wt/vol). Some species may grow in the presence of safranin (0.05% wt/vol) or oxgall (1% wt/vol). Fluorescent pigments are not produced. Some species are sensitive to cefoperazone (64 mg/l). Range of DNA G+C content is 26.4–29.4 mol%.
The type species is Aliiarcobacter cryaerophilus.
Description of Aliiarcobacter cryaerophilus comb. nov.
Basonym: Campylobacter cryaerophila Neill et al., 1985.
Other synonym: Arcobacter cryaerophilus Vandamme et al., 1991.
The description is the same given by Neill et al. (1985). The type strain is A169/BT (= NCTC 1185T = ATCC 43158T).
Description of Aliiarcobacter butzleri comb. nov.
Basonym: Campylobacter butzleri Kiehlbauch et al., 1991.
Other synonym: Arcobacter butzleri Vandamme et al., 1992.
The description is the same given by Vandamme et al. (1992). The type strain is LMG 10828T (= CDC D2686T = ATCC 49616T).
Description of Aliiarcobacter skirrowii comb. nov.
Basonym: Arcobacter skirrowii Vandamme et al., 1992.
The description is the same given by Vandamme et al. (1992). The type strain is Skirrow 449/80T (= LMG 6621T = CCUG 10374T).
Description of Aliiarcobacter cibarius comb. nov.
Basonym: Arcobacter cibarius Houf et al., 2005.
The description is the same given by Houf et al. (2005). The type strain is LMG 21996T (= CCUG 48482T).
Description of Aliiarcobacter thereius comb. nov.
Basonym: Arcobacter thereius Houf et al., 2009.
The description is the same given by Houf et al. (2009). The type strain is LMG 24486T (= CCUG 56902T).
Description of Aliiarcobacter trophiarum comb. nov.
Basonym: Arcobacter trophiarum De Smet et al., 2011.
The description is the same given by De Smet et al. (2011). The type strain is 64T (= LMG 25534T = CCUG 59229T).
Description of Aliiarcobacter lanthieri comb. nov.
Basonym: Arcobacter lanthieri Whiteduck-Léveillée et al., 2015.
The description is the same given by Whiteduck-Léveillée et al. (2015). The type strain is AF1440T (= LMG 28516T = CCUG 66485T).
Description of Aliiarcobacter faecis comb. nov.
Basonym: Arcobacter faecis Whiteduck-Léveillée et al., 2016.
The description is the same given by Whiteduck-Léveillée et al. (2016). The type strain is AF1078T (= LMG 28519T = CCUG 66484T).
Description of Pseudoarcobacter gen. nov.
Pseudoarcobacter (Pseu.do.ar.co.bac’ter, Gr. adj. pseudes, false; N.L. masc. n. Arcobacter a bacterial generic name; N.L. masc. n. Pseudoarcobacter, false Arcobacter).
Gram-negative, cells are rod shaped and motile. Cell size 0.2–0.9 μm in diameter and 0.4–2.2 μm long. Some species may present cells up to 10 μm in length. Oxidase and catalase positive. No growth occurs at 4% NaCl. Growth occurs at 15–37°C, but not at 42°C. Carbohydrates are not fermented. Reduce nitrate to nitrite. Positive for the hydrolysis of indoxyl acetate. Some species may hydrolyze urea. Growth does not occur in the presence 2,3,5-triphenyltetrazolium chloride (0.04%, wt/vol) or glycine (1% wt/vol). Some species may grow in the presence of safranin (0.05% wt/vol) or oxgall (1% wt/vol). Sensitive to cefoperazone (64 mg/l). Range of DNA G+C content is 26.3–28.0 mol%.
The type species is Pseudoarcobacter defluvii.
Description of Pseudoarcobacter defluvii comb. nov.
Basonym: Arcobacter defluvii Collado et al., 2011.
The description is the same given by Collado et al. (2011). The type strain is SW28-11T (= CECT 7697T = LMG 25694T).
Description of Pseudoarcobacter ellisii comb. nov.
Basonym: Arcobacter ellisii Figueras et al., 2011b.
The description is the same given by Figueras et al. (2011b). The type strain is F79-6T (= CECT 7837T = LMG 26155T).
Description of Pseudoarcobacter venerupis comb. nov.
Basonym: Arcobacter venerupis Levican et al., 2012.
The description is the same given by Levican et al. (2012). The type strain is F67-11T (= CECT 7836T = LMG 26156T).
Description of Pseudoarcobacter cloacae comb. nov.
Basonym: Arcobacter cloacae Levican et al., 2013.
The description is the same given by Levican et al. (2013). The type strain is SW28-13T (= CECT 7834T = LMG 26153T)
Description of Pseudoarcobacter suis comb. nov.
Basonym: Arcobacter suis Levican et al., 2013.
The description is the same given by Levican et al. (2013). The type strain is F41T (= CECT 7833T = LMG 26152T).
Description of Pseudoarcobacter aquimarinus comb. nov.
Basonym: Arcobacter aquimarinus Levican et al., 2015.
The description is the same given by Levican et al. (2015). The type strain is W63T (= CECT 8442T = LMG 27923T).
Description of Pseudoarcobacter acticola comb. nov.
Basonym: Arcobacter acticola Park et al., 2016.
The description is the same given by Park et al. (2016). The type strain is AR-13T (= KCTC 52212T = NBRC 112272T).
Description of Malacobacter gen. nov.
Malacobacter (Ma.la.co.bac’ter; Gr. n. malaco, soft, with soft boy, mollusc; Gr. n. bacter, rod; N.L. masc. n. Malacobacter, bacteria isolated from molluscs).
Gram-negative, cells are rod shaped and motile. Cell size 0.1–0.6 μm wide and 0.5–3.6 μm long. Oxidase positive and catalase variable among species. Halophilic, no growth can be obtained without NaCl and capable to grow up to 4% NaCl. Growth occurs at 15°C–37°C. Does not grow at 37°C in microaerophilic conditions nor at 42°C in anaerobiosis. Carbohydrates are not fermented. Does not reduce nitrate to nitrite. Negative for the hydrolysis of urea. Some species may hydrolyze indoxyl acetate. Growth does not occur in the presence of oxgall (1% wt/vol) or 2,3,5-triphenyltetrazolium chloride (0.04%, wt/vol). Some species may grow in the presence of glycine (1% wt/vol) or safranin (0.05% wt/vol). Sensitive to cefoperazone (64 mg/l) variable among species. Range of DNA G+C content is 26.1–27.3 mol%.
The type species is Malacobacter halophilus.
Description of Malacobacter halophilus comb. nov.
Basonym: Arcobacter halophilus Donachie et al., 2005.
The description is the same given by Donachie et al. (2005). The type strain is LA31BT (= ATCC BAA-1022T = CIP 108450T).
Description of Malacobacter mytili comb. nov.
Basonym: Arcobacter mytili Collado et al., 2009a.
The description is the same given by Collado et al. (2009a). The type strain is F2075T (= CECT 7386T = LMG 24559T).
Description of Malacobacter marinus comb. nov.
Basonym: Arcobacter marinus Kim et al., 2010.
The description is the same given by Kim et al. (2010), with the exception of variable result among strains for the hydrolysis of the indoxyl-acetate under microaerobic conditions (Salas-Massó et al., 2016). The type strain is CL-S1T (= KCCM 90072T = JCM 15502T).
Description of Malacobacter canalis comb. nov.
Basonym: Arcobacter canalis Pérez-Cataluña et al., 2018b.
The description is the same given by Pérez-Cataluña et al. (2018b). The type strain is F138-33T (= CECT 8984T = LMG 29148T).
Description of Malacobacter molluscorum comb. nov.
Basonym: Arcobacter molluscorum Figueras et al., 2011a.
The description is the same given by Figueras et al. (2011a). The type strain is F98-3T (= CECT 7696T = LMG 25693T).
Description of Malacobacter pacificus comb. nov.
Basonym: Arcobacter pacificus Zhang et al., 2015.
The description is the same given by Zhang et al. (2015). The type strain is SW028T (= DSM 25018T = JCM 17857T = LMG 26638T).
Description of Haloarcobacter gen. nov.
Haloarcobacter (Ha.lo.ar.co.bac’ter, Gr. n. halo, salt; N.L. masc. n. Arcobacter, a bacterial generic name; N.L. masc. n. Haloarcobacter, Arcobacter salt loving).
Gram-negative, cells are rod shaped and motile. Cell size 0.1–0.5 μm in diameter and 0.9–2.5 μm in length. Oxidase positive and catalase variable among species. Halophilic, growth can be obtained within the range of 0.5% (variable among species) and up to 4% NaCl. Growth occurs at 15–42°C. Growth at 37°C in microaerophilic conditions or at 42°C in anaerobiosis variable among species. Carbohydrates are not fermented. Some species may reduce nitrate to nitrite. Negative for the hydrolysis of urea (with the exception of H. ebronensis). Some species may hydrolyze indoxyl acetate. Growth does not occur in the presence of oxgall (1% wt/vol) (with the exception of H. molluscorum) or 2,3,5-triphenyltetrazolium chloride (0.04%, wt/vol). No growth on CCDA. Some species may grow in the presence of glycine (1% wt/vol) or safranin (0.05% wt/vol). Sensitive to cefoperazone (64 mg/l). Range of DNA G+C content is 27.3–29.9 mol%.
The type species is Haloarcobacter bivalviorum.
Description of Haloarcobacter bivalviorum comb. nov.
Basonym: Arcobacter bivalviorum Levican et al., 2012.
The description is the same given by Levican et al. (2012). The type strain is F4T (= CECT 7835T = LMG 26154T).
Description of Haloarcobacter anaerophilus comb. nov.
Basonym: Arcobacter anaerophilus Sasi-Jyothsna et al., 2013.
The description is the same given by Sasi-Jyothsna et al. (2013). The type strain is JC84T (= KCTC 15071T = MTCC 10956T = DSM 24636T).
Description of Haloarcobacter ebronensis comb. nov.
Basonym: Arcobacter ebronensis Levican et al., 2015.
The description is the same given by Levican et al. (2015). The type strain is F128-2T (= CECT 8441T = LMG 27922T).
Description of Poseidonibacter gen. nov.
Poseidonibacter (Po.se.i.do.ni.bac’ter, Gr. n. Poseidon, God of the sea; Gr. n. bacter, rod; N.L. masc. n. Poseidonibacter referring to the marine habitat of this bacteria).
Gram-negative, cells are rod shaped and motile. Oxidase and catalase positive. Halophilic, no growth can be obtained without seawater or the addition of combined marine salts to the medium. Growth occurs at 15°C–25°C, but not at 37°C or 42°C. Range of pH for growth is 6–8. Carbohydrates are not fermented. Reduce nitrate to nitrite. Negative for the hydrolysis of indoxyl acetate and urea. Growth occurs in the presence of safranin (0.05% wt/vol), and 2,3,5-triphenyltetrazolium chloride (0.04%, wt/vol), but not in the presence of glycine (1% wt/vol) sensitive to cefoperazone (30 μg). Possess ubiquinone MK-6 as a respiratory quinone. DNA G+C content is 28.7 mol%.
The type species is Poseidonibacter lekithochrous.
Description of Poseidonibacter lekithochrous comb. nov.
Basonym: Arcobacter lekithochrous Diéguez et al., 2017.
The description is the same given by Diéguez et al. (2017). The type strain is LFT1.7T (= CECT 8942T = DSM 100870T).
Author Contributions
MF and JR designed the work. AP-C, NS-M, and AD performed the phenotypic and phylogenetic experiments. AP-C and SB carried out the genome sequencing and analysis. AP-C, AL, and JR performed the bioinformatic work. JR, MF, AP-C, and AD wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. F. J. García (Laboratorio Central de Veterinaria de Algete, MAGRAMA, Madrid, Spain) and Drs. R. Alonso, I. Martinez-Malaxetxebarria, and A. Fernandez-Astorga [Faculty of Pharmacy, University of the Basque Country (UPV-EHU), Vitoria-Gasteiz, Spain], for kindly providing some of the Arcobacter strains. AP-C thanks Institut d’Investigació Sanitària Pere Virgili (IISPV) for her Ph.D. fellowship and NS-M thanks the Universitat Rovira i Virgili (URV), the Institut de Recerca i Tecnologia Agroalimentària (IRTA) and the Banco Santander for her Ph.D. fellowship.
Funding. This work was supported in part by Grants JPIW2013-69095-C03-03 from the Ministerio de Economía y Competitividad (MINECO), AQUAVALENS of the Seventh Framework Program (FP7/2007-2013) grant agreement 311846 from the European Union and AGL2013-42628-R and AGL2016-77539-R (AEI/FEDER UE) from the Agencia Estatal de Investigación (Spain).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02077/full#supplementary-material
References
- Adilakshmi T., Bellur D. L., Woodson S. A. (2008). Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature 455 1268–1272. 10.1038/nature07298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt H. J., Dress A. W. M. (1992). Split decomposition: a new and useful approach to phylogenetic analysis of distance data. Mol. Phylogenet. Evol. 1 242–252. 10.1016/1055-7903(92)90021-8 [DOI] [PubMed] [Google Scholar]
- Botzman M., Margalit H. (2011). Variation in global codon usage bias among prokaryotic organisms is associated with their lifestyles. Genome Biol. 12:R109. 10.1186/gb-2011-12-10-r109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun J., Oren A., Ventosa A., Christensen H., Arahal D. R., da Costa M. S., et al. (2018). Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 68 461–466. 10.1099/ijsem.0.002516 [DOI] [PubMed] [Google Scholar]
- Chun J., Rainey F. A. (2014). Integrating genomics into the taxonomy and systematics of the Bacteria and Archaea. Int. J. Syst. Evol. Microbiol. 64 316–324. 10.1099/ijs.0.054171-0 [DOI] [PubMed] [Google Scholar]
- Collado L., Cleenwerck I., Van Trappen S., De Vos P., Figueras M. J. (2009a). Arcobacter mytili sp. nov., an indoxyl acetate –hydrolysis- negative bacterium isolated from mussels. Int. J. Syst. Evol. Microbiol. 59 1391–1396. 10.1099/ijs.0.003749-0 [DOI] [PubMed] [Google Scholar]
- Collado L., Guarro J., Figueras M. J. (2009b). Prevalence of Arcobacter in meat and shellfish. J. Food Prot. 72 1102–1106. [DOI] [PubMed] [Google Scholar]
- Collado L., Figueras M. J. (2011). Taxonomy, epidemiology, and clinical relevance of the genus Arcobacter. Clin. Microbiol. Rev. 24 174–192. 10.1128/CMR.00034-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado L., Inza I., Guarro J., Figueras M. J. (2008). Presence of Arcobacter spp. in environmental waters correlates with high levels of fecal pollution. Environ. Microbiol. 10 1635–1640. 10.1111/j.1462-2920.2007.01555.x [DOI] [PubMed] [Google Scholar]
- Collado L., Kasimir G., Perez U., Bosch A., Pinto R., Saucedo G., et al. (2010). Occurrence and diversity of Arcobacter spp. along the Llobregat River catchment, at sewage effluents and in a drinking water treatment plant. Water Res. 44 3696–3702. 10.1016/j.watres.2010.04.002 [DOI] [PubMed] [Google Scholar]
- Collado L., Levican A., Pérez J., Figueras M. J. (2011). Arcobacter defluvii sp. nov., isolated from sewage samples. Int. J. Syst. Evol. Microbiol. 61 2155–2161. 10.1099/ijs.0.025668-0 [DOI] [PubMed] [Google Scholar]
- De Smet S., Vandamme P., De Zutter L., On S. L. W., Douidah L., Houf K. (2011). Arcobacter trophiarum sp. nov., isolated from fattening pigs. Int. J. Syst. Evol. Microbiol. 61 356–361. 10.1099/ijs.0.022665-0 [DOI] [PubMed] [Google Scholar]
- Diéguez A. L., Balboa S., Magnesen T., Romalde J. L. (2017). Arcobacter lekithochrous sp. nov., isolated from a molluscan hatchery. Int. J. Syst. Evol. Microbiol. 67 1327–1332. 10.1099/ijsem.0.001809 [DOI] [PubMed] [Google Scholar]
- Diéguez A. L., Pérez-Cataluña A., Figueras M. J., Romalde J. L. (2018). Arcobacter haliotis Tanaka et al., 2017 is a later heterotypic synonym of Arcobacter lekithochrous Diéguez et al., 2017. Int. J. Syst. Evol. Microbiol. 10.1099/ijsem.0.002909 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Donachie S. P., Bowman J. P., On S. L. W., Alam M. (2005). Arcobacter halophilus sp. nov., the first obligate halophile in the genus Arcobacter. Int. J. Syst. Evol. Microbiol. 55 1271–1277. 10.1099/ijs.0.63581-0 [DOI] [PubMed] [Google Scholar]
- Farooqi M. S., Mishra D., Rai N., Singh D., Rai A., Chaturvedi K., et al. (2016). Genome-wide relative analysis of codon usage bias and codon context pattern in the bacteria Salinibacter ruber, Chromohalobacter salexigens and Rhizobium etli. Biochem. Anal. Biochem. 5:257 10.4172/2161-1009.1000257 [DOI] [Google Scholar]
- Figueras M. J., Collado L., Guarro J. (2008). A new 16S rDNA-RFLP method for the discrimination of the accepted species of Arcobacter. Diagn. Microbiol. Infect. Dis. 62 11–15. 10.1016/j.diagmicrobio.2007.09.019 [DOI] [PubMed] [Google Scholar]
- Figueras M. J., Collado L., Levican A., Pérez J., Solsona M. J., Yustes C. (2011a). Arcobacter molluscorum sp. nov., a new species isolated from shellfish. Syst. Appl. Microbiol. 34 105–109. 10.1016/j.syapm.2010.10.001 [DOI] [PubMed] [Google Scholar]
- Figueras M. J., Levican A., Collado L., Inza M. I., Yustes C. (2011b). Arcobacter ellisii sp. nov., isolated from mussels. Syst. Appl. Microbiol. 34 414–418. 10.1016/j.syapm.2011.04.004 [DOI] [PubMed] [Google Scholar]
- Figueras M. J., Levican A., Pujol I., Ballester F., Rabada Quilez M. J., Gomez-Bertomeu F. (2014). A severe case of persistent diarrhoea associated with Arcobacter cryaerophilus but attributed to Campylobacter sp. and a review of the clinical incidence of Arcobacter spp. New Microbes New Infect. 2 31–37. 10.1002/2052-2975.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueras M. J., Pérez-Cataluña A., Salas-Massó N., Levican A., Collado L. (2017). ‘Arcobacter porcinus’ sp. nov., a novel Arcobacter species uncovered by Arcobacter thereius. New Microbe New Infect. 15 104–106. 10.1016/j.nmni.2016.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher J. C., Levican A., Figueras M. J., McLellan S. L. (2014). Population dynamics and ecology of Arcobacter in sewage. Front. Microbiol. 5:525 10.3389/fmicb.2014.00525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacometti F., Salas-Massó N., Serraino A., Figueras M. J. (2015). Characterization of Arcobacter suis isolated from water buffalo (Bubalus bubalis) milk. Food Microbiol. 51 186–191. 10.1016/j.fm.2015.06.004 [DOI] [PubMed] [Google Scholar]
- Goris J., Konstantinidis K. T., Klappenbach J. A., Coenye T., Vandamme P., Tiedje J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57 81–91. 10.1099/ijs.0.64483r-0 [DOI] [PubMed] [Google Scholar]
- Horikoshi M., Tang Y. (2015). ggfortify: Data Visualization Tools for Statistical Analysis Results. Available at: https://CRAN.R-project.org/package=ggfortify [Google Scholar]
- Houf K., On S. L. W., Coenye T., Debruyne L., De Smet S., Vandamme P. (2009). Arcobacter thereius sp. nov., isolated from pigs and ducks. Int. J. Syst. Evol. Microbiol. 59 2599–2604. 10.1099/ijs.0.006650-0 [DOI] [PubMed] [Google Scholar]
- Houf K., On S. L. W., Coenye T., Mast J., Van Hoof J., Vandamme P. (2005). Arcobacter cibarius sp. nov., isolated from boiled carcasses. Int. J. Syst. Evol. Microbiol. 55 713–717. 10.1099/ijs.0.63103-0 [DOI] [PubMed] [Google Scholar]
- Hsu T. T. D., Lee J. (2015). Global distribution and prevalence of Arcobacter in food and water. Zoonoses Public Health 62 579–589. 10.1111/zph.12215 [DOI] [PubMed] [Google Scholar]
- Huson D. H., Bryant D. (2005). Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23 254–267. 10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- Hyatt D., Chen G. L., Locascio P. F., Land M. L., Larimer F. W., Hauser L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehlbauch A., Brenner D. J., Nicholson M. A., Baker C. N., Patton C. M., Steigerwalt A. G., et al. (1991). Campylobacter butzleri sp. nov., isolated from humans and diarrheal illness. J. Clin. Microbiol. 29 376–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. M., Hwang C. Y., Cho B. C. (2010). Arcobacter marinus sp. nov. Int. J. Syst. Evol. Microbiol. 60 531–536. 10.1099/ijs.0.007740-0 [DOI] [PubMed] [Google Scholar]
- Kitahara K., Yasutake Y., Miyazaki K. (2012). Mutational robustness of 16S ribosomal RNA, shown by experimental horizontal gene transfer in Escherichia coli. PNAS 109 19220–19225. 10.1073/pnas.1213609109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis K. T., Tiedje J. M. (2005). Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. U.S.A. 102 2567–2572. 10.1073/pnas.0409727102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagesen K., Hallin P., Rødland E. A., Stærfeldt H.-H., Rognes T., Ussery D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35 3100–3108. 10.1093/nar/gkm160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Larsen M. V., Cosentino S., Rasmussen S., Friis C., Hasman H., Marvig R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50 1355–1361. 10.1128/JCM.06094-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslett D., Canback B. (2004). ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32 11–16. 10.1093/nar/gkh152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levican A., Collado L., Aguilar C., Yustes C., Diéguez A. L., Romalde J. L., et al. (2012). Arcobacter bivalviorum sp. nov. and Arcobacter venerupis sp. nov., new species isolated from shellfish. Syst. Appl. Microbiol. 35 133–138. 10.1016/j.syapm.2012.01.002 [DOI] [PubMed] [Google Scholar]
- Levican A., Collado L., Figueras M. J. (2013). Arcobacter cloacae sp. nov. and Arcobacter suis sp. nov., two new species isolated from food and sewage. Syst. Appl. Microbiol. 36 22–27. 10.1016/j.syapm.2012.11.003 [DOI] [PubMed] [Google Scholar]
- Levican A., Collado L., Figueras M. J. (2016). The use of two culturing methods in parallel reveals a high prevalence and diversity of Arcobacter spp. in a wastewater treatment plant. Biomed Res. Int. 2016:8132058. 10.1155/2016/8132058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levican A., Collado L., Yustes C., Aguilar C., Figueras M. J. (2014). Higher water temperature and incubation under aerobic and microaerobic conditions increase the recovery and diversity of Arcobacter spp. from shellfish. Appl. Environ. Microbiol. 80 385–391. 10.1128/AEM.03014-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levican A., Rubio-Arcos S., Martínez-Murcia A., Collado L., Figueras M. J. (2015). Arcobacter ebronensis sp. nov. and Arcobacter aquimarinus sp. nov., two new species isolated from marine environment. Syst. Appl. Microbiol. 38 30–35. 10.1016/j.syapm.2014.10.011 [DOI] [PubMed] [Google Scholar]
- Luo C., Rodriguez-R L. M., Konstantinidis K. T. (2014). MyTaxa: an advanced taxonomic classifier for genomic and metagenomic sequences. Nucleic Acids Res. 42:e73. 10.1093/nar/gku169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X. X., Feng Y. P., Bai J. L., Zhang D. R., Lin X. S., Ma Z. R. (2015). Nucleotide composition bias and codon usage trends of gene populations in Mycoplasma capriolum subsp. capriolum and M. agalactiae. J. Genetics 94 251–260. 10.1007/s12041-015-0512-2 [DOI] [PubMed] [Google Scholar]
- Marenda M. S., Sagne E., Poumarat F., Citti C. (2005). Suppression subtractive hybridization as a basis to assess Mycoplasma agalactiae and Mycoplasma bovis genomic diversity and species-specific sequences. Microbiology 151 475–489. 10.1099/mic.0.27590-0 [DOI] [PubMed] [Google Scholar]
- Martínez-Murcia A., Lamy B. (2015). “Molecular diagnostics by genetic methods,” in Aeromonas, ed. Graf J. (Poole: Caister Academic Press; ), 155–200. [Google Scholar]
- Martínez-Murcia A. J., Benlloch S., Collins M. D. (1992). Phylogenetic interrelationships of members of the genera Aeromonas and Plesiomonas as determined by 16S ribosomal DNA sequencing: lack of congruence with results of DNA-DNA hybridizations. Int. J. Syst. Bacteriol. 42 412–421. 10.1099/00207713-42-3-412 [DOI] [PubMed] [Google Scholar]
- Martínez-Murcia A. J., Figueras M. J., Saavedra M. J., Stackebrandt E. (2007). The recently proposed species Aeromonas sharmana sp. nov., isolate GPTSA-6T, is not a member of the genus Aeromonas. Int. Microbiol. 10 61–64. [PubMed] [Google Scholar]
- Meier-Kolthoff J. P., Auch A. F., Klenk H. P., Göker M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. 10.1186/1471-2105-14-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill S. D., Campbell J. N., O’Brien J. J., Weatherup S. T. C., Ellis W. A. (1985). Taxonomic position of Campylobacter cryaerophila sp. nov. Int. J. Syst. Bacteriol. 35 342–356. 10.1099/00207713-35-3-342 [DOI] [Google Scholar]
- Nurk S., Bankevich A., Antipov D., Gurevich A. A., Korobeynikov A., Lapidus A., et al. (2013). Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 20 714–737. 10.1089/cmb.2013.0084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- On S. L. W., Miller W. G., Houf K., Fox J. G., Vandamme P. (2017). Minimal standards for describing new species belonging to the families Campylobacteraceae and Helicobacteraceae: campylobacter, Arcobacter, Helicobacter and Wolinella spp. Int. J. Syst. Evol. Microbiol. 67 5296–5311. 10.1099/ijsem.0.002255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R., Olson R., Pusch G. D., Olsen G. J., Davis J. J., Disz T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology ( RAST ). Nucleic Acids Res 42 D206–D214. 10.1093/nar/gkt1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page A. J., Cummins C. A., Hunt M., Wong V. K., Reuter S., Holden M. T. G., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31 3691–3693. 10.1093/bioinformatics/btv421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Jung Y. T., Kim S., Yoon J. H. (2016). Arcobacter acticola sp. nov., isolated from seawater of the East Sea in South Korea. J. Microbiol. 54 655–659. 10.1007/s12275-016-6268-4 [DOI] [PubMed] [Google Scholar]
- Pérez-Cataluña A., Collado L., Salgado O., Lefiñanco V., Figueras M. J. (2018a). A polyphasic and taxogenomic evaluation uncovers Arcobacter cryaerophilus as a species complex that embraces four genomovars. Front. Microbiol 9:805. 10.3389/fmicb.2018.00805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Cataluña A., Salas-Massó N., Figueras M. J. (2018b). Arcobacter canalis sp. nov., isolated from a water canal contaminated with urban sewage. Int. J. Syst. Evol. Microbiol. 68 1258–1264. 10.1099/ijsem.0.002662 [DOI] [PubMed] [Google Scholar]
- Puigbò P., Bravo I. G., Garcia-Vallve S. (2008). CAIcal: a combined set of tools to assess codon usage adaptation. Biol. Dir. 3:38. 10.1186/1745-6150-3-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Q.-L., Xie B. B., Zhang X. Y., Chen X. L., Zhou B. C., Zhou J., et al. (2014). A proposed genus boundary for the prokaryotes based on genomic insights. J. Bacteriol. 196 2210–2215. 10.1128/JB.01688-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2015). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Řeháková K., Johansen J. R., Bowen M. B., Martin M. P., Sheil C. A. (2014). Variation in secondary structure of the 16S rRNA molecule in Cyanobacteria with implications for phylogenetic analysis. Fottea 14 161–178. 10.5507/fot.2014.013 [DOI] [Google Scholar]
- Richter M., Rosselló-Móra R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106 19126–19131. 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M., Rosselló-Móra R., Oliver Glöckner F., Peplies J. (2016). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32 929–931. 10.1093/bioinformatics/btv681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosselló-Mora R., Amann R. (2001). The species concept for prokaryotes. FEMS Microbiol. Rev. 25 39–67. 10.1111/j.1574-6976.2001.tb00571.x [DOI] [PubMed] [Google Scholar]
- Salas-Massó N., Andree K. B., Furones M. D., Figueras M. J. (2016). Enhanced recovery of Arcobacter spp. using NaCl in culture media and re-assessment of the traits of Arcobacter marinus and Arcobacter halophilus isolated from marine water and shellfish. Sci. Total Environ. 56 1355–1361. 10.1016/j.scitotenv.2016.05.197 [DOI] [PubMed] [Google Scholar]
- Salas-Massó N., Pérez-Cataluña A., Collado L., Levican A., Figueras M. J. (2018). “Arcobacter”, in Handbook of Foodborne Diseases, ed. Liu D. (Boca Raton, FL: CRC Press, Taylor & Francis Group; ). [Google Scholar]
- Sasi-Jyothsna T. S., Rahul K., Ramaprasad E. V. V., Sasikala C., Ramana C. V. (2013). Arcobacter anaerophilus sp. nov., isolated from an estuarine sediment and emended description of the genus Arcobacter. Int. J. Syst. Evol. Microbiol. 63 4619–4625. 10.1099/ijs.0.054155-0 [DOI] [PubMed] [Google Scholar]
- Sawabe T., Kita-Tsukamoto K., Thompson F. L. (2007). Inferring the evolutionary history of vibrios by means of multilocus sequence analysis. J. Bacteriol. 189 7932–7936. 10.1128/JB.00693-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. (1987). The codon adaptation index – a mesure of directional synonymous codon usage bias, and its potential applications. Nucleic Acid Res. 15 1281–1295. 10.1093/nar/15.3.1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Brown C. J., Fornay L. J., Top E. M. (2008). Comparison of correspondence analysis methods for synonymous codon usage in bacteria. DNA Res. 15 357–365. 10.1093/dnares/dsn028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R., Cleenwerck I., Mizutani Y., Iehata S., Bossier P., Vandamme P. (2017). Arcobacter haliotis sp. nov., isolated from abalone species Haliotis gigantea. Int. J. Syst. Evol. Microbiol 67 3050–3056. 10.1099/ijsem.0.002080 [DOI] [PubMed] [Google Scholar]
- Tang Y., Horikoshi M., Li W. (2016). Ggfortify: unified interface to visualize statistical result of popular R Packages. The R J. 8 474–485. [Google Scholar]
- Tindall B. J., Rosselló-Mora R., Busse H. J., Ludwig W., Kämpfer P. (2010). Notes on the characterization of prokaryote strains for taxonomic purposes. Int. J. Syst. Evol. Microbiol. 60 249–266. 10.1099/ijs.0.016949-0 [DOI] [PubMed] [Google Scholar]
- Toh H., Sharma V. K., Oshima K., Kondo S., Hattori M., Ward F. B., et al. (2011). Complete genome sequences of Arcobacter butzleri ED-1 and Arcobacter sp. strain L, both isolated from a microbial fuel cell. J. Bacteriol. 193 6411–6412. 10.1128/JB.06084-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ue H., Matsuo Y., Kasai H., Yokota A. (2011). Demequina globuliformis sp. nov., Demequina oxidasica sp. nov. and Demequina aurantiaca sp. nov., actinobacteria isolated from marine environments, and proposal of Demequinaceae fam. nov. Int. J. Syst. Evol. Microbiol. 61 1322–1329. 10.1099/ijs.0.024299-0 [DOI] [PubMed] [Google Scholar]
- Ursing J. B., Lior H., Owen R. J. (1994). Proposal of minimal standards for describing new species of the family Campylobacteraceae. Int. J. Syst. Bacteriol. 44 842–845. 10.1099/00207713-44-4-842 [DOI] [PubMed] [Google Scholar]
- Van den Abeele A. M., Vogelaers D., Van Hende J., Houf K. (2014). Prevalence of Arcobacter species among humans, Belgium, 2008-2013. Emerg. Infect. Dis. 20 1731–1734. 10.3201/eid2010.140433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme P., Falsen E., Rossau R., Hoste B., Segers P., Tytgat R., et al. (1991). Revision of Campylobacter, Helicobacter and Wolinella taxonomy: emendation of generic descriptions and proposal of Arcobacter gen. nov. Int. J. Syst. Bacteriol. 41 88–103. 10.1099/00207713-41-1-88 [DOI] [PubMed] [Google Scholar]
- Vandamme P., Vancanneyt M., Pot B., Mels L., Hoste B., Dewettinck D., et al. (1992). Polyphasic taxonomic study of the emended genus Arcobacter with Arcobacter butzleri comb. nov. and Arcobacter skirrowii sp. nov., an aerotolerant bacterium isolated from veterinary specimens. Int. J. Syst. Bacteriol. 42 344–356. 10.1099/00207713-42-3-344 [DOI] [PubMed] [Google Scholar]
- Waite D. W., Vanwonterghem I., Rinke C., Parks D. H., Zhang Y., Takai K., et al. (2017). Comparative genomic analysis of the class Epsilonproteobacteria and proposed reclassification to Epsilonbacteraeota (phyl. nov.). Front. Microbiol. 8:682 10.3389/fmicb.2017.00682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattam A. R., Davis J. J., Assaf R., Boisvert S., Brettin T., Bun C., et al. (2017). Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 45 D535–D542. 10.1093/nar/gkw1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimann A., Mooren K., Frank J., Pope P. B., Bremges A., McHardy A. C. (2016). From genomes to phenotypes: traitar, the microbial trait analyzer. mSystems 1 e101–e116. 10.1128/mSystems.00101-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner J. (2017). Pca3d: Three Dimensional PCA Plots. Available at: https://CRAN.R-project.org/package=pca3d. [Google Scholar]
- Whiteduck-Léveillée K., Whiteduck-Léveillée J., Cloutier M., Tambong J. T., Xu R., Topp E., et al. (2015). Arcobacter lanthieri sp. nov., isolated from pig and dairy cattle manure. Int. J. Syst. Evol. Microbiol. 65 2709–2716. 10.1099/ijs.0.000318 [DOI] [PubMed] [Google Scholar]
- Whiteduck-Léveillée K., Whiteduck-Léveillée J., Cloutier M., Tambong J. T., Xu R., Topp E., et al. (2016). Identification, characterization and description of Arcobacter faecis sp. nov., isolated from a human waste septic tank. Syst. Appl. Microbiol. 39 93–99. 10.1016/j.syapm.2015.12.002 [DOI] [PubMed] [Google Scholar]
- Whitman W. B. (2015). Genome sequences as the type material for taxonomic descriptions of prokaryotes. Syst. Appl. Microbiol. 38 217–222. 10.1016/j.syapm.2015.02.003 [DOI] [PubMed] [Google Scholar]
- Wickham H. (2009). Ggplot2: Elegant Graphics for Data Analysis. New York, NY: Springer-Verlag; 10.1007/978-0-387-98141-3 [DOI] [Google Scholar]
- Yarza P., Richter M., Peplies J., Euzeby J., Amann R., Schleifer K. H., et al. (2008). The All-Species Living Tree project: a 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst. Appl. Microbiol. 31 241–250. 10.1016/j.syapm.2008.07.001 [DOI] [PubMed] [Google Scholar]
- Yarza P., Yilmaz P., Pruesse E., Glöckner F. O., Ludwig W., Schleifer K. H., et al. (2014). Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 12 635–645. 10.1038/nrmicro3330 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Yu C., Wang X., Yu S., Zhang X. H. (2015). Arcobacter pacificus sp. nov., isolated from seawater of the South Pacific Gyre. Int. J. Syst. Evol. Microbiol. 66 542–547. 10.1099/ijsem.0.000751 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.