Abstract
Brain-type glycogen phosphorylase (PYGB) is an enzyme that metabolizes glycogen, whose function is to provide energy for an organism in an emergency state. The present study purposed to investigate the role and mechanism of PYGB silencing on the growth and apoptosis of prostate cancer cells. A cell counting kit-8 assay and flow cytometry were performed to determine the cell viability, apoptosis and reactive oxygen species (ROS) content, respectively. Colorimetry was performed to analyze the activity of caspase-3. Western blotting and reverse transcription-quantitative polymerase chain reaction were used to evaluate the associated mRNA and protein expression levels. The results revealed that PYGB was upregulated in prostate cancer tissues and was associated with disease progression. In addition, PYGB silencing suppressed the cell viability of PC3 cells. PYGB silencing promoted apoptosis of PC3 cells via the regulation of the expression levels of cleaved-poly (adenosine diphosphate-ribose) polymerase, cleaved-caspase-3, B-cell lymphoma-2 (Bcl-2) and Bcl-2-associated X protein. PYGB silencing increased the ROS content in PC3 cells, and affected nuclear factor (NF)-κB/nuclear factor-erythroid 2-related factor 2 (Nrf2) signaling pathways in PC3 cells. In conclusion, PYGB silencing suppressed the growth and promoted the apoptosis of prostate cancer cells by affecting the NF-κB/Nrf2 signaling pathway. The present study provided evidence that may lead to the development of a potential therapeutic strategy for prostate cancer.
Keywords: brain-type glycogen phosphorylase, prostate cancer, nuclear factor-κB, nuclear factor-erythroid 2-related factor 2
Introduction
Prostate cancer is a common malignant tumor that occurs in the prostate epithelium and is the second leading cause of cancer-associated mortalities among older men worldwide (1). Prostate cancer begins with intraepithelial neoplasia, and locally invasive prostate cancer gradually progresses to castration resistant prostate cancer, which represents the leading cause of prostate cancer-associated mortalities at present (2). This is a complex process involving alterations in the extracellular matrix microenvironment to support the invasion and increase of cell motility (3). However, the exact underlying mechanism of prostate cancer progression is unclear and determining the molecular mechanism is crucial for clinical diagnosis and therapy.
Nuclear factor (NF)-κB represents an important member of the transcription factor family, which widely exists in eukaryotes to participate in multiple physiological and pathological processes (4). It has been demonstrated that NF-κB may adjust cell apoptosis in a number of different ways, involving regulation of the expression of apoptosis-associated genes and interacting with other signaling pathways (5). A previous study also reported that in epidermal cells, the activation of NF-κB suppressed cell proliferation (6). Nuclear factor-erythroid 2-related factor 2 (Nrf2) serves an important role in the anti-oxidative signaling pathway in organisms (7). Nrf2 can be activated by the external stimuli from free radicals and is then transferred to the nucleus to combine with the antioxidant response element (8,9). It has been reported previously that the activity of antioxidases from Nrf2 knockdown mice was significantly reduced, while the expression of correlative cytokines was markedly enhanced, involving interleukin (IL)-6, IL-1β, tumor necrosis factor-α, inducible nitric oxide synthase and cyclo-oxygenase 2 (10,11). In addition, a large number of investigations have suggested that interactions exist between NF-κB and Nrf2. For example, NF-κB may suppress the activity of Nrf2 by selectively preventing the combination of Nrf2 and cyclic adenosine 3′,5′-monophosphate response element binding protein (12,13). However, to the best of our knowledge, the roles and mechanisms of the NF-κB/Nrf2 signaling pathway in prostate cancer have not been fully elucidated.
Brain-type glycogen phosphorylase (PYGB), as a glycogen phosphorylase, metabolizes glycogen. It is generally considered to provide energy for organisms in the emergency state (14–16). PYGB has been investigated previously in tumors of the gastrointestinal system and in lung carcinomas (17–20). In addition, it has been demonstrated that PYGB may inhibit the production of reactive oxygen species (ROS) and suppress the apoptosis of Escherichia coli (E. coli) cells (21). To the best of our knowledge, the effect of PYGB on prostate cancer tissues and cells has not yet been reported.
The present study analyzed the association between PYGB expression and prostate cancer tissues together with clinicopathological data. In addition, the present study explored the effect of PYGB silencing and the NF-κB/Nrf2 signaling pathway in the growth and apoptosis of prostate cancer cells.
Materials and methods
Tissue samples and clinical data collection
Patients that had been pathologically diagnosed with prostate cancer were included in the present study. Patients were excluded from the experiment if androgen or radiation therapy had been received. A total of 50 male patients (mean age, 70.5) undergoing resection therapy at the East Hospital, Tongji University School of Medicine (Shanghai, China) from June 2010 to August 2015 were included in this study. All patients recruited to the present study provided written informed consent for the utilization of their tissue samples for clinical research. The project protocol was approved by the Institutional Review Board of East Hospital, Tongji University School of Medicine. Clinicopathological data, involving patient age and sex, histological grade and tumor, nodes, metastasis (TNM) stage, was obtained from the patient database. Matched adjacent normal prostate tissues were also collected as negative controls. Following resection, all of the tissue samples were snap-frozen in liquid nitrogen and stored at −80°C immediately for the subsequent experiments.
Cell culture, genes and plasmids
Human prostate cancer cell lines, including LNCap, PC3 and DU145, and the normal prostate cell line PrEC were obtained from the Type Culture Collection of The Chinese Academy of Sciences (Shanghai, China). Cells were maintained in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in a 5% CO2 atmosphere at 37°C. PYGB small interfering (si)-RNA (5′-AGGCCTGGGAAATCACGAAG-3′) or negative siRNA control (5′-GGAACGGAGGCGAAGAGATAT-3′) was cloned into the pcDNA3.1(+) empty vector (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). si-PYGB or siRNA-NC was sub-cloned into the pSilencer 2.1-U6 vector (Thermo Fisher Scientific) for gene interference. The expression of PYGB was detected 20 h post-transfection.
Cell viability analysis
Cell Counting Kkit-8 (CCK-8; Beyotime Institute of Biotechnology, Haimen, China) assay was performed to determine cell viability. A total of 6×104 cells/ml PC3 cells in the logarithmic phase were seeded into the wells of 96-well plates and maintained in an incubator (37°C and 5% CO2) for 12 h. Following this, cells were transfected with siRNA negative vector (1 µg; mock group) or PYGB siRNA vector (1 µg; si-PYGB group) using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) for 20 h. The untreated cells served as a control. Cells were then maintained in the incubator (37°C, 5% CO2) for 12, 24 and 48 h. Subsequently, 10 µl CCK reagent was supplemented into each well. Cells were then maintained in 5% CO2 at 37°C for 3 h. A microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used to read the absorbance at a wavelength of 450 nm. Cell viability was evaluated by calculating the percentage of cell survival compared with the control.
Apoptosis assay
Flow cytometry (FCM) was used to assess the cell apoptosis. Following washing with PBS, PC3 cells were trypsinized using 0.25% Trypsin (Beyotime Institute of Biotechnology). An Annexin V-FITC/PI Apoptosis Detection kit was purchased from Invitrogen (Thermo Fisher Scientific, Inc.). Cells were centrifuged at 224 × g for 1 min at 4°C, and the supernatant was discarded. Subsequently, cells were suspended in Annexin-V binding buffer at a density of 1×106 cells/ml. Annexin V-fluorescein isothiocyanate and propidium iodide were then maintained with cells at room temperature in the dark for 15 min. Cell apoptosis was evaluated by using a FACSCalibur flow cytometer with BD CellQuest Pro 3.3 (BD Biosciences, Franklin Lakes, CA, USA).
Western blot analysis
Tissues were rapidly grinded in liquid nitrogen. Cells were lysed with NP40 lysis buffer (Beyotime) and protein was isolated using a Tissue or Cell Total Protein Extraction kit (Solarbio, China). Protein concentration was measured by BCA assay kit (Thermo, USA). Equal amount of proteins (25 µg/lane) were separated by 12% SDS-PAGE and then transferred onto a polyvinylidene difluoride membrane (EMD Millipore, Billerica, MA, USA). The membranes were blocked with 5% skimmed milk at room temperature for 1 h. The blotting was performed using specific primary antibodies at 4°C overnight, including anti-PYGB (1:2,000; Abcam, Cambridge, UK; cat. no. ab154969; rabbit anti-human), anti-poly (adenosine diphosphate-ribose) polymerase (PARP; 1:2,000; Abcam; cat. no. ab32138; rabbit anti-human), anti-cleaved-PARP (1:5,000; Abcam; cat. no. ab32064; rabbit anti-human), anti-cleaved-caspase-3 (1:1,000; Abcam; cat. no. ab2302; rabbit anti-human), anti-NF-κB (1:500; Abcam; cat. no. ab31412; rabbit anti-human), anti-Nrf2 (1:1,000; Abcam; cat. no. ab89443; mouse anti-human), anti-B-cell lymphoma-2 (Bcl-2; 1:1,000; Abcam; cat. no. ab32124; rabbit anti-human), anti-Bcl-2-associated X protein (Bax; 1:1,000; Abcam; cat. no. ab32503; rabbit anti-human) and anti-actin (1:500; Abcam; cat. no. ab205; mouse anti-human). A horseradish peroxidase-conjugated secondary antibody (1:1,000; cat. no. bs-0293M; BIOSS, Beijing, China) was incubated with the blots at room temperature for 1 h. Enhanced chemiluminescent reagents (EMD Millipore) using an ECL system (GE Healthcare, Chicago, IL, USA) was used for visualization. The density of the blots was determined with Quantity One 4.6.2 software (Bio-Rad Laboratories, Inc.).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues or cultured cell lines (PrEC, LNCap, PC3 and DU145) with TRIzol reagent (Thermo Fisher Scientific, Inc.). RNA was reverse transcribed to cDNA using a Transcriptor High Fidelity cDNA Synthesis kit (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) according to the manufacturer's protocol. The temperature protocol consisted of 25°C for 5 min, 37°C for 60 min and 85°C for 5 min. RT-qPCR analysis was performed using SYBR Green qPCR Master mix (MedChemExpress, Princeton, NJ, USA) on an ABI 7500 thermocycler (Applied Biosystems; Thermo Fisher Scientific, Inc.). The qPCR thermocycling conditions were as follows: 5 min pretreatment at 95°C, followed by 35 cycles of 95°C for 15 sec and 60°C for 30 sec, a final extension at 72°C for 10 min and then held at 4°C. Actin was used as the control of the input RNA level. The primers used in this analysis were designed by Invitrogen (Thermo Fisher Scientific, Inc.) and were as follows: PYGB, forward, 5′-AGGCCTGGGAAATCACGAAG-3′ and reverse, 5′-CCATGTTGATCCGCTTGCAG-3′ (product: 237 bp); NF-κB, forward, 5′-GCGAGAGGAGCACAGATACC-3′ and reverse, 5′-AGGGGTTGTTGTTGGTCTGG-3′ (product: 279 bp); Nrf2, forward, 5′-TGAGGTTTCTTCGGCTACGTT-3′ and reverse, 5′-AGCTCCTCCCAAACTTGCTC-3′ (product: 174 bp); Bax, forward, 5′-CAGCTCTGAGCAGATCATGAAGACA-3′ and reverse, 5′-GCCCATCTTCTTCCAGATGGTGAGC-3′ (product: 235 bp); Bcl-2, forward, 5′-ACTTGTGGCCCAGATAGGCACCCAG-3′ and reverse, 5′-CGACTTCGCCGAGATGTCCAGCCAG-3′ (product: 214 bp); and Actin, forward, 5′-CACAATGTGCGACGAAGACG-3′ and reverse, 5′-ATGATGCCGTGCTCGATAGG-3′ (product: 237 bp). Gene expression was quantified according to the 2−ΔΔCq method (22).
Statistical analysis
The results of the present study are presented as the mean ± standard deviation of at least three independent experiments. All experimental data was analyzed by a Student's t test, or one-way analysis of variance followed by Tukey's post-hoc test. Kaplan-Meier survival analysis was performed and P-values were calculated with log-rank test. The differences between the categorial variables were determined with the Chi-square test. P<0.05 was considered to indicate a statistically significant difference. GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA, USA) was used to perform the data analysis.
Results
PYGB is upregulated in prostate cancer tissues and is associated with disease progression
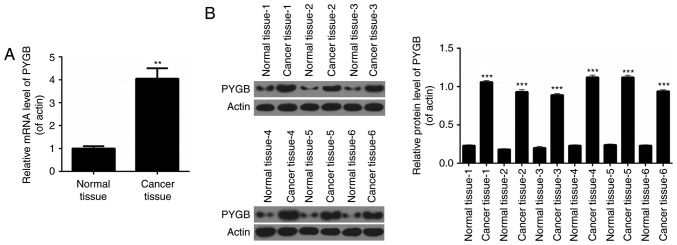
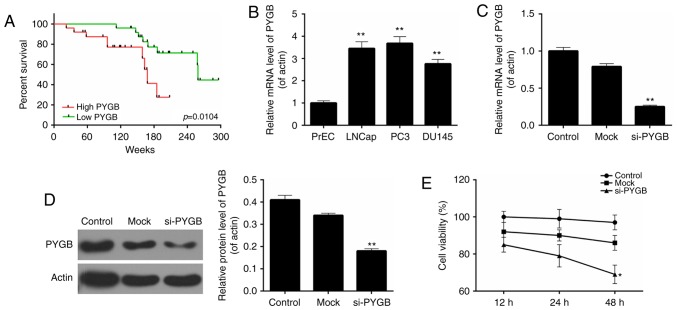
RT-qPCR analysis was performed to determine the expression levels of PYGB in pairs of human prostate cancer tissues and matched adjacent normal prostate tissues from 50 prostate cancer patients. It was revealed that the expression levels of PYGB in prostate cancer tissues were significantly higher than those observed in pair-matched adjacent non-tumorous tissues (Fig. 1A; P<0.01). Western blot analysis was performed to evaluate PYGB protein expression in 6 randomly selected pairs of human prostate cancer tissues and matched adjacent normal prostate tissues from prostate cancer patients. The results indicated that compared with pair-matched adjacent non-tumorous tissues, PYGB expression was significantly enhanced in prostate cancer tissues (Fig. 1B; P<0.001). These results implied that PYGB expression was upregulated in prostate cancer tissues. Clinicopathological analysis revealed that PYGB was associated with the degree of histological differentiation (P<0.05), TNM stage (P<0.01) and metastasis (P<0.01) of prostate cancer tissues (Table I). However, there was no significant association between PYGB and other clinicopathological characteristics, involving age and gender (Table I; P>0.05). In addition, the present study also assessed the association between PYGB expression in prostate cancer and patient survival to understand the prognostic significance of PYGB upregulation in prostate cancer. It was demonstrated that high expression levels of PYGB were significantly associated with a poor 5-year survival rate in prostate cancer patients (Fig. 2A; P<0.05). Low PYGB expression was associated with a higher 5-year survival rate in prostate cancer patients. Collectively, these results indicated that high PYGB expression may be associated with prostate cancer progression.
Figure 1.
PYGB is upregulated in prostate cancer tissues. (A) Reverse transcription-quantitative polymerase chain reaction was performed to determine the expression levels of PYGB in prostate cancer tissues and matched adjacent normal prostate tissues from 50 prostate cancer patients. (B) Western blot assay was performed to measure the expression levels of PYGB in prostate cancer tissues and matched adjacent normal prostate tissues from prostate cancer patients. **P<0.01 and ***P<0.001 vs. normal tissue. PYGB, brain-type glycogen phosphorylase.
Table I.
Associations between brain-type glycogen phosphorylase expression levels and the clinical data of prostate cancer patients.
| Factor | Number of patients | Low PYGB expression, n (%) | High PYGB expression, n (%) | P-value |
|---|---|---|---|---|
| Sex | ||||
| Male | 50 | 21 (100.0) | 29 (100.0) | |
| Age (years) | 0.054 | |||
| <55 | 18 | 4 (20.0) | 14 (46.7) | |
| ≥55 | 32 | 16 (80.0) | 16 (53.3) | |
| Histological grade | 0.012a | |||
| Well | 5 | 2 (13.3) | 3 (8.6) | |
| Moderately | 35 | 10 (66.7) | 25 (71.4) | |
| Poorly | 10 | 3 (20.0) | 7 (20.0) | |
| TNM stage | 0.008b | |||
| I/II | 23 | 5 (20.8) | 18 (75.0) | |
| III/IV | 27 | 19 (79.2) | 6 (25.0) | |
| Metastasis | <0.001 | |||
| No | 38 | 9 (47.4) | 29 (93.5) | |
| Yes | 12 | 10 (52.6) | 2 (6.5) |
P<0.05
P<0.01. Chi-square test. PYGB, brain-type glycogen phosphorylase; TNM, tumor, nodes, metastasis.
Figure 2.
PYGB is associated with the disease progression of prostate cancer and PYGB silencing suppresses the cell viability of PC3 cells. (A) The overall survival rates of prostate cancer patients based on the expression levels of PYGB measured by Kaplan-Meier survival analysis, and the P-values were calculated by log-rank test. (B) RT-qPCR was performed to determine the expression levels of PYGB in several prostate cancer cell lines, including LNCap, PC3 and DU145, together with the normal prostate cell line PrEC. (C) RT-qPCR and (D) western blot analysis was performed to measure the expression levels of PYGB in PC3 cells transfected with the siRNA negative control (mock group) and PYGB siRNA (si-PYGB group); the control group was untreated. (E) Cell counting kit-8 assay was performed to evaluate the cell viability of PC3 cells. *P<0.05, **P<0.01 vs. mock group. No significant differences were detected between the control and mock groups. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; PYGB, brain-type glycogen phosphorylase; si-/siRNA, small interfering RNA.
PYGB silencing suppresses cell viability of PC3 cells
Based on the above results, the present study evaluated the cell viability of three different prostate cancer cell lines, including LNCap, PC3 and DU145, together with a normal prostate cell line PrEC. The results revealed that, when compared with the normal prostate cell line, the PYGB expression was significantly higher in the three prostate cancer cell lines (Fig. 2B). The highest PYGB expression was detected in the PC3 cell line. Therefore, the PC3 cell line was chosen as the research target of prostate cancer cells in the subsequent studies. The knockdown efficiency was ~75% in PC3 cells following cells being transfected with si-PYGB (Fig. 2C). According to the western blotting results, it was revealed that following transfection with si-PYGB, the expression level of the PYGB protein was significantly reduced (Fig. 2D; P<0.01). Thus, the CCK-8 assay was performed to determine the cell viability of PC3 cells for 12, 24 and 48 h following siRNA transfection to further evaluate the potential effects of PYGB silencing. The results showed that, when compared with the non-transfected control (control) and non-targeting transfected control (mock), the cell viability of PC3 cells treated with si-PYGB for 48 h was significantly reduced (Fig. 2E; P<0.05). However, no significant difference was revealed between the control and mock groups at each time point (Fig. 2E). These results suggested that PYGB silencing suppressed cell viability of PC3 cells.
PYGB silencing promotes the apoptosis of PC3 cells
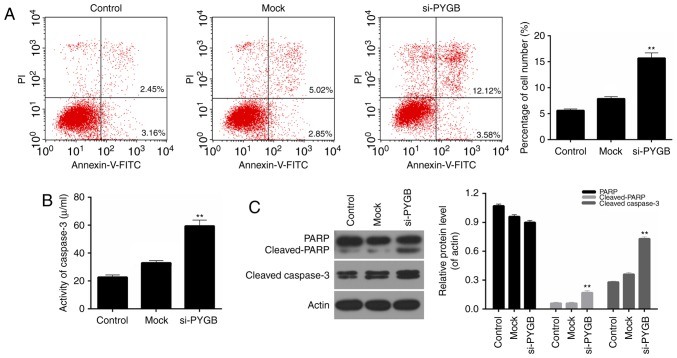
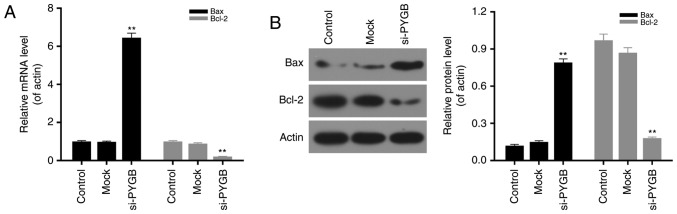
In addition, the cell apoptosis of PC3 cells was assessed by FCM in the present study. As the FCM results revealed (Fig. 3A), the percentage of apoptotic PC3 cells in the si-PYGB group was 15.70%, which was markedly higher than the control and mock groups (5.61 and 7.87%, respectively; P<0.05). These results suggested that PYGB silencing significantly strengthened the apoptotic capacity of PC3 cells. In addition, the activity of caspase-3 in PC3 cells was evaluated. It was noted that the activity of caspase-3 in PC3 treated with si-PYGB was significantly enhanced (Fig. 3B; P<0.01). The present study also investigated the expression levels of apoptotic-associated proteins in PC3 cells. According to the RT-qPCR and western blotting data, it was demonstrated that PARP expression in the three groups was not significantly different, whereas an increase in cleaved-PARP expression in PC3 cells was observed in the si-PYGB group (Fig. 3C; P<0.05). It was also revealed that there were high expression levels of cleaved-caspase-3 in PC3 cells treated with si-PYGB (Fig. 3C; P<0.05). In addition, the present study measured the expression levels of Bax and Bcl-2 in PC3 cells transfected with si-PYGB to further determine the associated apoptotic mechanisms. The results revealed that PTGB silencing significantly upregulated the mRNA and protein expression levels of Bax, and reduced Bcl-2 expression levels in PC3 cells (Fig. 4; P<0.05). Based on these results, it was concluded that PYGB silencing promoted the apoptosis of PC3 cells via regulation of cleaved-PARP, cleaved-caspase-3, Bax and Bcl-2 expression.
Figure 3.
PYGB silencing promotes the apoptosis of PC3 cells. PC3 cells were transfected with siRNA negative control (mock group) and PYGB siRNA (si-PYGB group). The cells in control group received no treatment. (A) Flow cytometric analysis was performed to determine the levels of cell apoptosis in PC3 cells. (B) Colorimetry was performed to measure the activity of caspase-3 in PC3 cells. (C) Western blotting was performed to measure the expression levels of PARP, cleaved-PARP and cleaved-caspase-3 in PC3 cells transfected with the siRNA negative control (mock group) and PYGB siRNA (si-PYGB group); the control group was untreated. **P<0.01 vs. mock group. No significant differences were detected between the control and mock groups. PYGB, brain-type glycogen phosphorylase; PARP, poly (adenosine diphosphate-ribose) polymerase; siRNA, small interfering RNA; PI, propidium iodide; FITC, fluorescein isothiocyanate.
Figure 4.
PYGB silencing modulates the expression levels of Bax and Bcl-2 in PC3 cells. PC3 cells were transfected with the siRNA negative control (mock group) and PYGB siRNA (si-PYGB group). The cells in control group received no treatment. (A) Reverse transcription-quantitative polymerase chain reaction and (B) western blotting were carried out to detect the mRNA and protein expression levels of Bax and Bcl-2, respectively, in PC3 cells transfected with the siRNA negative control (mock group) and PYGB siRNA (si-PYGB group); the control group was untreated. **P<0.01 vs. mock group. No significant differences were detected between the control and mock groups. PYGB, brain-type glycogen phosphorylase; Bcl-2, B-cell lymphoma-2; Bax, Bcl-2-associated X protein; si-/siRNA, small interfering RNA.
PYGB silencing increases the ROS content in PC3 cells
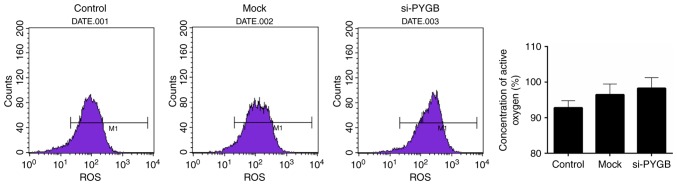
The present study also assessed the ROS content in PC3 cells. FCM results revealed that the ROS content appeared enhanced in the si-PYGB group when compared with the control group (Fig. 5); however, this was not significant.
Figure 5.
PYGB silencing enhances the ROS content in PC3 cells transfected with the siRNA negative control (mock group) and PYGB siRNA (si-PYGB group); the control group was untreated. Flow cytometric analysis was carried out to evaluate the ROS content in PC3 cells. ROS, reactive oxygen species; PYGB, brain-type glycogen phosphorylase; si-/siRNA, small interfering RNA.
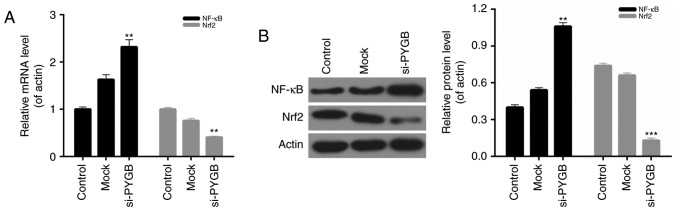
PYGB silencing affects the NF-κB/Nrf2 signaling pathway in PC3 cells
The present study also evaluated the expression of NF-κB and Nrf2 in PC3 cells from each group. The RT-qPCR and western blotting results indicated that the mRNA and protein expression levels of NF-κB in PC3 cells transfected with si-PYGB were significantly higher than the control and mock groups. However, a decrease in Nrf2 expression was observed in the PC3 cells treated with si-PYGB (Fig. 6; P<0.01). Therefore, it was concluded that the NF-κB/Nrf2 signaling pathway in PC3 cells may be affected by PYGB silencing.
Figure 6.
PYGB silencing affects the NF-κB/Nrf2 signaling pathway in PC3 cells. (A) Reverse transcription-quantitative polymerase chain reaction was performed to measure the expression levels of NF-κB and Nrf2 in PC3 cells. (B) Western blotting was performed to measure the expression levels of NF-κB and Nrf2 in PC3 cells. **P<0.01, ***P<0.001 vs. mock group. No significant differences were detected between the control and mock groups. PYGB, brain-type glycogen phosphorylase; NF-κB, nuclear factor-κB; Nrf2, nuclear factor-erythroid 2-related factor 2; si-/siRNA, small interfering RNA.
Discussion
PYGB is a glycogen phosphorylase and is primarily localized in adult brain tissues and embryo liver tissues, whose function is to provide energy to organisms (15). Increasing evidence has suggested that PYGB is frequently expressed in cancer, including gastric carcinoma (23), lung cancer (24) and renal cell cancer (25). For example, it has been previously demonstrated that PYGB expression was significantly upregulated in hepatocellular carcinoma, whereas the expression levels of PYGB in normal liver cells were low or not expressed (26). However, the PYGB expression levels in prostate cancer tissues or cells are currently not clear. Thus, the present study assessed PYGB expression in human prostate cancer tissues and matched adjacent normal prostate tissues from patients with prostate cancer. The results suggested that the expression levels of PYGB in prostate cancer tissues were markedly enhanced, while low PYGB expression was observed in normal tissues. In addition, the present study also evaluated PYGB expression in a number of different prostate cancer cell lines, and the results revealed that PYGB expression was relevant to the malignant degree of prostate cancer cells. These consequences were consistent with the previous studies in regard to PYGB expression in other cancer cells (17–20), and indicated that PYGB may act as a pathological diagnostic index for malignant prostate neoplasia's. According to the clinicopathological analysis data, the present study confirmed that PYGB was associated with the degree of histological differentiation, TNM stage and metastasis of prostate cancer tissues. Furthermore, via survival curve analysis of the expression levels of PYGB and the survival time of prostate cancer patients, it was observed that high PYGB expression was associated with a poor 5-year survival rate and low PYGB expression was associated with an improved 5-year survival rate in the prostate cancer patients. These results also implied that PYGB may serve as a prognostic indicator of prostate cancer. Overall, the present study proposed that PYGB may become an important target in the detection and therapy of prostate cancer.
In order to further investigate the role and underlying mechanism of PYGB in prostate cancer, the present study selected the PC3 cell line as the research target. The results of the present study demonstrated that the expression levels of PYGB in PC3 cells were higher than in the other prostate cancer cell lines and the normal prostate cell line (PrEC). In addition, an siRNA vector target of PYGB, si-PYGB, was constructed in the present study. To the best of our knowledge, the present study is the first to investigate the PYGB silencing effect on cell growth and apoptosis of prostate cancer cells. Following treatment with si-PYGB, the expression levels of PYGB in PC3 cells were reduced, which indicated that a good knockdown efficiency was achieved. The present study further evaluated the cell viability of PC3 cells transfected with si-PYGB, and the results demonstrated that PYGB silencing suppressed the cell viability of PC3 cells. In addition, the apoptotic capacity of PC3 cells treated with the empty vector and si-PYGB was assessed. The data revealed that when compared with other groups, PYGB silencing promoted the apoptosis of PC3 cells. Furthermore, based on the previous studies (27,28), the present study also measured the activity of caspase-3 and investigated the expression levels of several apoptotic-associated proteins in PC3 cells transfected with the empty vector and si-PYGB. The results indicated that PYGB silencing enhanced the activity of caspase-3 in PC3 cells. It was also demonstrated that PYGB silencing upregulated the expression levels of cleaved-PARP, cleaved-caspase-3 and Bax, and reduced the Bcl-2 expression levels in PC3 cells. Therefore, it was concluded that PYGB silencing promoted the apoptosis of PC3 cells by modulating the expression levels of cleaved-PARP, cleaved-caspase-3, Bax and Bcl-2. It has been previously demonstrated that PYGB inhibited the production of ROS, and suppressed the apoptosis and necrosis of E. coli cells (21). In the present study, it was confirmed that PYGB silencing promoted the apoptosis of PC3 cells; however, whether PYGB silencing induces ROS production required further investigation. Therefore, ROS content in PC3 cells transfected with the empty vector and si-PYGB was assessed. The results showed that PYGB silencing increased the production of ROS in PC3 cells, and these data also verified the cell apoptosis results. Thus, it was concluded that PYGB silencing increased ROS production in PC3 cells, which may further cause increased cell apoptosis of PC3 cells.
It has previously been reported that NF-κB is involved in the growth, invasion and apoptosis of human prostate cancer cells (29–32). Additionally, previous studies have also demonstrated the roles that Nrf2 served in prostate cancer (33–35). It was also demonstrated that NF-κB and Nrf2 possessed anti-inflammatory and anti-oxidative activities (36). Furthermore, the NF-κB and Nrf2 signaling pathways have been confirmed to contribute to the inhibition of colorectal carcinogenesis and prevent breast cancer (37,38). However, the roles and underlying mechanism of the NF-κB/Nrf2 signaling pathway in prostate cancer are not clear. Thus, the present study investigated the expression levels of NF-κB and Nrf2 in PC3 cells treated with si-PYGB. It was revealed that PYGB silencing significantly upregulated the expression levels of NF-κB in PC3 cells. Nrf2 expression in PC3 cells was reduced by PYGB silencing. Therefore, it was concluded that PYGB silencing affected the NF-κB/Nrf2 signaling pathway in PC3 cells.
Taken together, the results of the present study demonstrated that PYGB silencing suppressed the growth and promoted the apoptosis of prostate cancer cells by affecting the NF-κB/Nrf2 signaling pathway. This may provide a novel research focus for understanding the pathogenesis of prostate cancer, and may aid the diagnosis and therapy of prostate cancer.
However, further study is required to confirm the role of PYGB in prostate cancer. For example, the data of the present study would be supported by PYGB overexpression experiments, to further examine its effect on the growth and apoptosis of prostate cancer cells in vitro, or by exploring the effect of PYGB on prostate cancer progression in vivo. Furthermore, studies with larger sample sizes should be performed.
In conclusion, the present study highlighted that PYGB silencing suppressed the growth and promoted the apoptosis of prostate cancer cells by affecting the NF-κB/Nrf2 signaling pathway. The findings of the present study may influence the understanding of the underlying mechanisms of PYGB and prostate cancer cells. The potential effects of PYGB on the growth and apoptosis of prostate cancer cells suggested that PYGB may be an effective target for anti-tumor therapies.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
ZW and GH wrote the manuscript. ZW, GH, QL and WZ performed the experiments. ZW and JW designed the study. GH and QL performed the data analysis. ZW, GH and JW revised the manuscript. All authors reviewed the manuscript.
Ethics approval and consent to participate
All patients recruited to the present study provided written informed consent for the utilization of their tissue samples for clinical research. The project protocol was approved by the Institutional Review Board of East Hospital, Tongji University School of Medicine (Shanghai, China).
Patient consent for publication
Informed consent was obtained from all participants for the publication of their data.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Chen W, Zheng R, Zeng H, Zhang S, He J. Annual report on status of cancer in China, 2011. Chin J Cancer Res. 2015;27:2–12. doi: 10.1186/s40880-015-0001-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cai C, Chen S, Ng P, Bubley GJ, Nelson PS, Mostaghel EA, Marck B, Matsumoto AM, Simon NI, Wang H, et al. Intratumoral de novo steroid synthesis activates androgen receptor in castration-resistant prostate cancer and is upregulated by treatment with CYP17A1 inhibitors. Cancer Res. 2011;71:6503–6513. doi: 10.1158/1538-7445.AM2011-LB-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steeg PS. Metastasis suppressors alter the signal transduction of cancer cells. Nat Rev Cancer. 2003;3:55–63. doi: 10.1038/nrc967. [DOI] [PubMed] [Google Scholar]
- 4.Baldwin AS., Jr The NF-kappa B and I kappa B proteins: New discoveries and insights. Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 5.Dutta J, Fan Y, Gupta N, Fan G, Gélinas C. Current insights into the regulation of programmed cell death by NF-kappaB. Oncogene. 2006;25:6800–6816. doi: 10.1038/sj.onc.1209938. [DOI] [PubMed] [Google Scholar]
- 6.Clarkson RW, Watson CJ. NF-kappaB and apoptosis in mammary epithelial cells. J Mammary Gland Biol Neoplasia. 1999;4:165–175. doi: 10.1023/A:1018725207969. [DOI] [PubMed] [Google Scholar]
- 7.Ma Q. Role of nrf2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol. 2013;53:401–426. doi: 10.1146/annurev-pharmtox-011112-140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baird L, Dinkova-Kostova AT. The cytoprotective role of the Keap1-Nrf2 pathway. Arch Toxicol. 2011;85:241–272. doi: 10.1007/s00204-011-0674-5. [DOI] [PubMed] [Google Scholar]
- 9.Li W, Kong AN. Molecular mechanisms of Nrf2-mediated antioxidant response. Mol Carcinog. 2009;48:91–104. doi: 10.1002/mc.20465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyanapalli SS, Paredes-Gonzalez X, Fuentes F, Zhang C, Guo Y, Pung D, Saw CL, Kong AN. Nrf2 knockout attenuates the anti-inflammatory effects of phenethyl isothiocyanate and curcumin. Chem Res Toxicol. 2014;27:2036–2043. doi: 10.1021/tx500234h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung KL, Lee JH, Khor TO, Wu TY, Li GX, Chan J, Yang CS, Kong AN. Nrf2 knockout enhances intestinal tumorigenesis in Apc(min/+) mice due to attenuation of anti-oxidative stress pathway while potentiates inflammation. Mol Carcinog. 2014;53:77–84. doi: 10.1002/mc.21950. [DOI] [PubMed] [Google Scholar]
- 12.Liu GH, Qu J, Shen X. NF-kappaB/p65 antagonizes Nrf2-ARE pathway by depriving CBP from Nrf2 and facilitating recruitment of HDAC3 to MafK. Biochim Biophys Acta. 2008;1783:713–727. doi: 10.1016/j.bbamcr.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Ziady AG, Sokolow A, Shank S, Corey D, Myers R, Plafker S, Kelley TJ. Interaction with CREB binding protein modulates the activities of Nrf2 and NF-κB in cystic fibrosis airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2012;302:L1221–L1231. doi: 10.1152/ajplung.00156.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newgard CB, Hwang PK, Fletterick RJ. The family of glycogen phosphorylases: Structure and function. Crit Rev Biochem Mol Biol. 1989;24:69–99. doi: 10.3109/10409238909082552. [DOI] [PubMed] [Google Scholar]
- 15.Rich SS, Goodarzi MO, Palmer ND, Langefeld CD, Ziegler J, Haffner SM, Bryer-Ash M, Norris JM, Taylor KD, Haritunians T, et al. A genome-wide association scan for acute insulin response to glucose in Hispanic-Americans: The insulin resistance atherosclerosis family study (IRAS FS) Diabetologia. 2009;52:1326–1333. doi: 10.1007/s00125-009-1373-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimada S, Maeno M, Akagi M, Hatayama I, Sato T, Sato K. Immunohistochemical detection of glycogen phosphorylase isoenzymes in rat and human tissues. Histochem J. 1986;18:334–338. doi: 10.1007/BF01675212. [DOI] [PubMed] [Google Scholar]
- 17.Barbosa AJ, Castro LP. BGP expression in gastric epithelium and early gastric cancer. Gastric Cancer. 2002;5:123–124. doi: 10.1007/s101200200021. [DOI] [PubMed] [Google Scholar]
- 18.Lee MK, Kim JH, Lee CH, Kim JM, Kang CD, Kim YD, Choi KU, Kim HW, Kim JY, Park DY, et al. Clinicopathological significance of BGP expression in non-small-cell lung carcinoma: Relationship with histological type, microvessel density and patients' survival. Pathology. 2006;38:555–560. doi: 10.1080/00313020601024029. [DOI] [PubMed] [Google Scholar]
- 19.Shiomori K, Shimada S, Marutsuka T, Hatayama I, Ogawa M. Genetic pathways of ‘de novo’ colorectal carcinomas with reference to fetal-type glycogen phosphorylase positive foci. Int J Oncol. 2003;22:65–74. [PubMed] [Google Scholar]
- 20.Tashima S, Shimada S, Yamaguchi K, Tsuruta J, Ogawa M. Expression of brain-type glycogen phosphorylase is a potentially novel early biomarker in the carcinogenesis of human colorectal carcinomas. Am J Gastroenterol. 2000;95:255–263. doi: 10.1111/j.1572-0241.2000.01692.x. [DOI] [PubMed] [Google Scholar]
- 21.Kim SY, Nishioka M, Hayashi S, Honda H, Kobayashi T, Taya M. The gene yggE functions in restoring physiological defects of Escherichia coli cultivated under oxidative stress conditions. Appl Environ Microbiol. 2005;71:2762–2765. doi: 10.1128/AEM.71.5.2762-2765.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Uno K, Shimada S, Tsuruta J, Matsuzaki H, Tashima S, Ogawa M. Nuclear localization of brain-type glycogen phosphorylase in some gastrointestinal carcinoma. Histochem J. 1998;30:553–559. doi: 10.1023/A:1003239302471. [DOI] [PubMed] [Google Scholar]
- 24.Lee CH, Son HI, Kim JH, Choi KU, Kim JY, Park DY, Sol MY. Clinicopathologic significance of brain-type glycogen phosphorylase expression in non-small-cell lung carcinomas. Lung Cancer. 2004:S41–S42. [Google Scholar]
- 25.Takashi M, Koshikawa T, Kurobe N, Kato K. Elevated concentrations of brain-type glycogen phosphorylase in renal cell carcinoma. Jpn J Cancer Res. 1989;80:975–980. doi: 10.1111/j.1349-7006.1989.tb01636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato K, Morris HP, Weinhouse S. Phosphorylase: A new isozyme in rat hepatic tumors and fetal liver. Science. 1972;178:879–881. doi: 10.1126/science.178.4063.879. [DOI] [PubMed] [Google Scholar]
- 27.Royle JS, Ross JA, Ansell I, Bollina P, Tulloch DN, Habib FK. Nitric oxide donating nonsteroidal anti-inflammatory drugs induce apoptosis in human prostate cancer cell systems and human prostatic stroma via caspase-3. J Urol. 2004;172:338–344. doi: 10.1097/01.ju.0000132367.02834.41. [DOI] [PubMed] [Google Scholar]
- 28.Tang DG, Li L, Zhu Z, Joshi B. Apoptosis in the absence of cytochrome c accumulation in the cytosol. Biochem Biophys Res Commun. 1998;242:380–384. doi: 10.1006/bbrc.1997.7969. [DOI] [PubMed] [Google Scholar]
- 29.Hafeez BB, Siddiqui IA, Asim M, Malik A, Afaq F, Adhami VM, Saleem M, Din M, Mukhtar H. A dietary anthocyanidin delphinidin induces apoptosis of human prostate cancer PC3 cells in vitro and in vivo: Involvement of nuclear factor-kappaB signaling. Cancer Res. 2008;68:8564–8572. doi: 10.1158/0008-5472.CAN-08-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lü L, Tang D, Wang L, Huang LQ, Jiang GS, Xiao XY, Zeng FQ. Gambogic acid inhibits TNF-α-induced invasion of human prostate cancer PC3 cells in vitro through PI3K/Akt and NF-κB signaling pathways. Acta Pharmacol Sin. 2012;33:531–541. doi: 10.1038/aps.2011.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raj GV, Sekula JA, Guo R, Madden JF, Daaka Y. Lysophosphatidic acid promotes survival of androgen-insensitive prostate cancer PC3 cells via activation of NF-kappaB. Prostate. 2004;61:105–113. doi: 10.1002/pros.20083. [DOI] [PubMed] [Google Scholar]
- 32.Shaikh IA, Brown I, Schofield AC, Wahle KW, Heys SD. Docosahexaenoic acid enhances the efficacy of docetaxel in prostate cancer cells by modulation of apoptosis: The role of genes associated with the NF-kappaB pathway. Prostate. 2008;68:1635–1646. doi: 10.1002/pros.20830. [DOI] [PubMed] [Google Scholar]
- 33.Jayakumar S, Kunwar A, Sandur SK, Pandey BN, Chaubey RC. Differential response of DU145 and PC3 prostate cancer cells to ionizing radiation: Role of reactive oxygen species, GSH and Nrf2 in radiosensitivity. Biochim Biophys Acta. 2014;1840:485–494. doi: 10.1016/j.bbagen.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 34.Nair S, Barve A, Khor TO, Shen GX, Lin W, Chan JY, Cai L, Kong AN. Regulation of Nrf2- and AP-1-mediated gene expression by epigallocatechin-3-gallate and sulforaphane in prostate of Nrf2-knockout or C57BL/6J mice and PC-3 AP-1 human prostate cancer cells. Acta Pharmacol Sin. 2010;31:1223–1240. doi: 10.1038/aps.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang C, Su ZY, Khor TO, Shu L, Kong AN. Sulforaphane enhances Nrf2 expression in prostate cancer TRAMP C1 cells through epigenetic regulation. Biochem Pharmacol. 2013;85:1398–1404. doi: 10.1016/j.bcp.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Surh YJ. NF-kappa B and Nrf2 as potential chemopreventive targets of some anti-inflammatory and antioxidative phytonutrients with anti-inflammatory and vantioxidative activities. Asia Pac J Clin Nutr. 2008;17(Suppl 1):S269–S272. [PubMed] [Google Scholar]
- 37.Mandal A, Bhatia D, Bishayee A. Anti-Inflammatory Mechanism Involved in Pomegranate-Mediated Prevention of Breast Cancer: The Role of NF-κB and Nrf2 Signaling Pathways. Nutrients. 2017;9(pii):E436. doi: 10.3390/nu9050436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yao J, Zhao L, Zhao Q, Zhao Y, Sun Y, Zhang Y, Miao H, You QD, Hu R, Guo QL. NF-κB and Nrf2 signaling pathways contribute to wogonin-mediated inhibition of inflammation-associated colorectal carcinogenesis. Cell Death Dis. 2014;5:e1283. doi: 10.1038/cddis.2014.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.