Abstract
Esophageal cancer ranks fourth in cancer-associated mortality in China and the incidence of esophageal adenocarcinoma has risen dramatically over the past two decades. MicroRNA (miRNA/miR) serves a pivotal role in human cancer cell growth, invasion and migration. MiR-675-3p is highly expressed in esophageal squamous cell cancer (ESCC) tissues, and may have an influence on ESCC cell migration and invasion. ESCC tumor tissue samples from 35 patients were profiled. MiR-675-3p expression was confirmed by reverse transcription-quantitative polymerase chain reaction. Manipulation of miR-675-3p via knockdown was carried out with subsequent evaluation of effects on cell proliferation, invasion, migration, and use of western blotting and ELISA assays. MiR-675-3p was overexpressed in ESCC tissues compared with normal tissues, and had higher expression levels in ESCC cells compared with the healthy esophageal epithelial cell line. The results revealed a predominant upregulation of cell migration and invasion ability. MiR-675-3p inhibitor inhibited ESCC cell proliferation, migration and invasion ability. It was also demonstrated that downregulation of miR-675-3p decreased the levels of matrix metalloproteinase (MMP) 2 and 9 and increased the level of E-cadherin. In addition, the effects of miR-675-3p inhibitor on ESCC cell lines were eliminated by con-transfection with miR-675-3p inhibitor and miR-675-3p mimic. In conclusion, the results indicated that miR-675-3p may be involved in the progression of ESCC through regulating ESCC cell migration and invasion capacity via modulating epithelial mesenchymal transition markers (MMP2, MMP 9 and E-cadherin).
Keywords: microRNA-675-3p, esophageal squamous cell cancer, migration, invasion
Introduction
Cancer is a leading global cause of mortality that accounts for almost 13% of mortality worldwide (1). According to Globocan (globocan.iarc.fr/Default.aspx), it is estimated that by 2020, there will be between 15 and 17 million newly diagnosed cases of cancer every year, 60% of which will be in developing countries (1). Asia accounts for 60% of the world population and half the global burden of cancer. With current resources, one-third of cancers could be preventable, and another two-third of newly diagnosed cancer patients could experience increased survival or early-stage detection (1). Prevention as well as early detection of cancers leads to both better health outcomes and considerable savings in treatment costs (2). Despite this, progress in global cancer control has been slow, largely due to the weak and fragmented nature of both the global and national responses to treatment.
Esophageal cancer (EC) is one of the most common diseases in gastroenterology, and additionally one of the most fatal (3). EC is the fourth most common gastrointestinal cancer in the United States of America with a <20% 5-year survival (4). The incidence of esophageal adenocarcinoma has risen dramatically over the past two decades (3). EC also ranks fourth among cancer-associated mortalities in China (5). Surgical resection is the main treatment method for EC, with a postoperative 5-year survival rate of only 34–36% (6). Conventional treatment strategies including neoadjuvant radiochemotherapy, preoperative neoadjuvant chemotherapy, and three-field lymph node dissection are helpful for improving the 5-year survival rate of EC patients. However, the outcomes are still unsatisfactory, and local recurrence and distant metastasis results in patient mortality (7). Therefore, biomarkers can be used to identify the recurrence or metastasis of EC in order to facilitate timely diagnosis and treatment strategies, thus improving the prognosis of EC patients.
MicroRNA (miRNA/miR) is a class of widely distributed non-coding RNA of 20–22 nucleotides, usually serving a role in regulation of post-transcriptional gene expression (8). The biogenesis of miRNA involves a complex protein system, including members of the Argonaute family, Polymerase II-dependent transcription and the RNase III Drosha and Dicer 13 (9). MiRNA is generally transcribed as a primary transcript (pre-miRNA) through Polymerase II and is processed to a 70-nucleotide stem-loop by the Microprocessor (Drosha/Pasha). The resulting pre-miRNA is transported into the cytoplasm via Exportin-5. Dicer proceeds to cleave the pre-miRNA to produce a mature miRNA: miRNA duplex. TAR RNA binding protein/Loquacious is used to bind the miRNA to Argonaute and RNA-induced silencing complex is formed, then, the miRNA strand is degraded (8). MiRNA has been demonstrated to be involved in multiple biological processes, such as cell differentiation, proliferation, oncogenesis, angiogenesis, tumor invasion and tumor metastasis. It has been demonstrated that miRNA serves a pivotal role in human cancer cell growth, invasion and migration (10). Furthermore, miRNA expression is associated with a variety of cancers, and miRNA may function as ‘oncogenes’ or ‘tumor suppressor genes’. Approximately 50% of miRNA is located on the genome at the fragile sites associated with the tumor, indicating that miRNA serves a crucial role in the development and progression of tumors.
MiR-675 is derived from long non-coding RNA H19, an miRNA embedded within H19's first exon, and H19 can generate two mature microRNAs, miRNA-675-3p and miRNA-675-5p in a Drosha and Dicer splicing dependent manner. MiRNA-675 is expressed exclusively in the placenta from the gestational time point when placental growth normally ceases, but placentas that lack H19 continue to grow. The main physiological role of H19 is inhibiting the growth of the placenta prior to birth, by regulating processing of miR-675 (11,12). The inverse association between miRNA-675 and placenta growth indicates that miRNA-675 can act as a growth restrictor during embryonic development (11). MiRNA-675 represses the expression of tumor suppressor retinoblastoma in a classical way and promotes the proliferation of colon cancer cells (13). In addition, miRNA-675-5p is expressed in EC and its over-expression in some esophageal squamous cancer cell lines result in their proliferation and migration. In the present study, the expression of miR-675-3p was investigated in a healthy esophageal epithelial cell line (HECC), and its function on esophageal squamous cell cancer (ESCC) proliferation and migration was evaluated.
Materials and methods
Ethics statement
Written informed consent was obtained from all participants, and the study protocol was approved by the ethics committee of JiangSu Cancer Hospital (Nanjing, China).
Clinical samples and cell lines
ESCC and paired normal esophageal tissues (>5 cm away from the tumor margin) were obtained from 35 patients (median age 63.7 years, range 48–76 years, female:male=1:6) who underwent esophagus resection between September 2016 and June 2017 at JiangSu Cancer Hospital. All patients had no history of previous malignancies, chemotherapy and radiotherapy. All the samples were immediately snap-frozen in liquid nitrogen and stored at −80°C for RNA extraction. All tumors and paired normal tissues were histologically confirmed by two pathologists. Human ESCC cell lines (Te-1 and Kyse-150) were obtained from Chinese Academy of Science cell bank (Shanghai, China), Human Healthy Esophageal Epithelial cell line (HECC) were obtained from iCell Bioscience Inc (Shanghai, China). Cells were cultured and maintained in RPMI 1640 (HyClone; GE Healthcare, Chicago, IL, USA) supplemented with 10% fetal bovine serum (FBS, Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in an incubator with 5% CO2 at 37°C. All culture materials were purchased from Gibco; Thermo Fisher Scientific, Inc.
Mir-675-3p overexpression and interference oligonucleotide transfection assay
Kyse-150 and Te-1 cells were seeded at a density of 5×104 cells/ml in six-well plates and incubated overnight at 37°C. According to the protocol, they were transiently transfected with the 20 µmol interference oligonucleotide which included negative control (NC; 5′-3′ CAGUACUUUUGUGUAGUACAA), mir-675-3p inhibitor (5′-3′ UGAGCGGUGAGGGCAAUACAG) or mir-675-3p mimic (sense 5′-3′ CUGUAUGCCCUCACCGCUCA; anti-sense 5′-3′ AGCGGUGAGGGCAUACAGUU) using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Following 6 h transfection at 37°C, the medium was replaced with RPMI1640 media containing 10% FBS. Three group were used in this experiment, which included negative control (NC), miR-675-3p inhibitor group and miR-675-3p inhibitor + miR-675-3p mimic. The group of NC and miR-675-3p inhibitor were only transfected with NC or miR-675-3p inhibitor, respectively, and the group of miR-675-3p inhibitor + miR-675-3p mimic, were transfected with miR-675-3p mimics 6 h following transfection with miR-675-3p inhibitor.
Total RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
At 24 h post-transfection, cells were treated with TRIzol (Takara Bio, Inc., Otsu, Japan) to extract total RNA, which was reverse transcribed to first strand cDNA using Primescript RT Reagent (Takara Bio, Inc.). The expression levels of miR-675-3p (sense 5′-CGGAGAGGGCCCACAGTG-3′; anti-sense was an universal primer which was obtained from GeneCopoeia, USA), MMP2 (sense 5′-GCTGACGGTAAGGACGGACTC-3′; anti-sense 5′-CGTTGCCATTGAACAAGAAGG-3′), MMP9 (sense 5′-TGTGCTACAGGGAGAGATAAGA-3′; anti-sense 5′-GTGGGTGGAGCAGAGTAAATAA-3′) and E-cadherin (sense 5′-CTCAGTTGGAACAGGGTGAATA-3′; anti-sense 5′-GTGCAGGACACTCAAATCAAAG-3′) were examined using RT-qPCR in a Step one plus system (Roche Molecular Diagnostics, Pleasanton, CA, USA) using 2−ΔΔCq analysis. In this system, all reactions were performed in SybrGreen (ChamQ™ SYBR qPCR Master Mix; Vazyme, Piscataway, NJ, USA), in a volume of 20 ul containing 2 ul cDNA, according to the manufacturer's protocol. All reactions were performed in triplicate. The thermal cycling conditions included a step of 5 min at 95°C followed by 40 cycles of 95°C for 10 sec and 60°C for 30 sec. U6 was used as internal control. Final amounts of target in cells were determined as follows: Target amount=2−ΔΔCq, where ΔΔCq=[Cq (target)-Cq (U6)]sample-[Cq (target)-Cq (U6)]internal standard. Final amounts of target in tissues were determined as target amount=2−ΔΔCq (14).
Cell proliferation assay
Cell proliferation was monitored by a colorimetric assay using the Cell Counting Kit-8 (CCK-8, Vazyme) according to the manufacturer's protocol. Briefly, cells were seeded on 6-well plates and transfected with NC, miR-675-3p inhibitor or miR-675-3p inhibitor + miR-675-3p mimics, after 12 h of transfection, the cells were transferred onto the 96-well plates (8×103 cells/well) and the cell proliferation assays were conducted and analyzed by a Microplate Reader (ELx808; BIO-TEK Instruments, Inc., Winooski, VT, USA) every 12 h for 3 days using CCK-8.
Scratch wound assay
Cells (3×105) were transfected with NC, miR-675-3p inhibitor or miR-675-3p inhibitor + miR-675-3p mimics and cultured in a cell culture dish (35×10 mm). Cell layers were scratched using a pipette tip to form wound gaps. The wound location in the cell culture dish was marked. Cells were imaged to record the wound width (0 h). A total of 12 h following this, images were taken again at the marked wound location to measure the cell migration ability.
Transwell invasion and migration ability assay
Cells (3×105) were transfected with NC, miR-675-3p inhibitor or miR-675-3p inhibitor + miR-675-3p mimics according to the protocol. Following incubation for 12 h, Kyse-150 cells or Te-1 cells were transferred to the upper Matrigel-coated or not Matrigel-coated invasion chambers (BD Biosciences, San Jose, CA, USA) in a serum-free RPMI 1640, and RPMI 1640 containing 10% FBS was added to the lower chambers. Following 24 h, non-migrated or non-invaded cells on the upper surface were removed, and the migrating or invading cells on the underside surface were fixed with 4% paraformaldehyde, stained with 0.1% crystal violet at room temperature for 15 min, and imaged. A total of three independent experiments were performed.
ELISA
Kyse-150 cells (3×105) or Te-1 cells (3×105) were cultured in six-well plates and following 24 h transfection with NC, miR-675-3p inhibitor or miR-675-3p inhibitor + miR-675-3p mimics, the related proteins in the cell culture were extracted by centrifuging at 300 × g for 10 min at 4°C following the manufacturer's protocol. The protein concentrations of matrix metalloproteinase (MMP)2, MMP9 and E-cadherin were measured by ELISA assay (Hangzhou Multisciences, Biotech Co., Ltd, Zhejiang, China, catalog number: EK1M022-96T, EK1M092-96T, EK12352-96T, respectively) following the manufacturer's protocol. All samples were measured in duplicate.
Western blot analysis
Protein levels of MMP2, MMP9 and E-cadherin were detected by western blotting 24 h following treatment. In brief, the total cell proteins were extracted from cells using lysis buffer (Cell Signaling Technology Inc., Danvers, MA, USA). BCA assay (Thermo Fisher Scientific, Inc.) was carried out to measure the protein concentrations. Equal amounts of protein samples (25 µg) were separated by 12% SDS-PAGE and then transferred onto polyvinylidene difluoride membranes. Following this, the membranes were blocked with 5% skim milk at room temperature for 2 h. Subsequently, incubation occurred with the primary antibodies against MMP2 (cat. no. 13132), MMP9 (cat. no. 2270) or E-cadherin (cat. no. 3195; all 1:1,000; all Cell Signaling Technology Inc.) overnight at 4°C. The membrane was then incubated with secondary antibodies (HRP-conjugated goat Anti-Rabbit, cat. no. Ab203-01, Vazyme, Piscataway, NJ, USA) at room temperature for 2 h. GAPDH (1:5,000; cat. no. 8884, Cell Signaling Technology Inc.) served as the internal control. Finally, to visualize the protein bands, an enhanced chemiluminescence detection system (Super Signal West Dura Extended Duration Substrate; Thermo Fisher Scientific, Inc.) was used.
Statistical analysis
Data were analyzed by GraphPad Prism5 (GraphPad Software, Inc., La Jolla, CA, USA) and were expressed as the mean ± standard deviation. Comparisons between groups were made using Student's t-test or one-way analysis of variance followed by Tukey's test. P<0.05 was considered to indicate a statistically significant difference.
Results
MiR-675-3p expression in ESCC
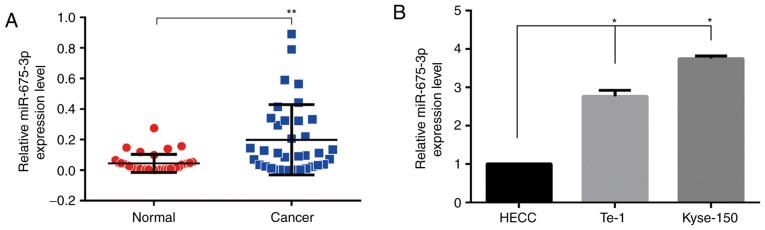
To explore the expression of miR-675-3p in ESCC tissues, RT-qPCR was used. As demonstrated in Fig. 1A, compared with the normal tissues, cancer tissues expressed higher levels of miR-675-3p, suggesting that the expression level of miR-675-3p was upregulated in ESCC tissues. To further investigate the functional effects of miR-675-3p in ESCC cells, RT-qPCR was also performed to investigate miR-675-3p expression level in human ESCC cell lines (Kyse-150 and Te-1) and the HECC. As demonstrated in Fig. 1B, compared with HECC, both ESCC cell lines expressed higher levels of miR-675-3p.
Figure 1.
Expression levels of miR-675-3p by reverse transcription-quantitative polymerase chain reaction. (A) ESCC patients' normal tissues and cancer tissues and (B) HECC and ESCC cells. *P<0.05 vs. HECC; **P<0.01 vs. normal. HECC, healthy esophageal epithelial cell line; miR, microRNA; ESCC, esophageal squamous cell cancer.
Knockdown of miR-675-3p inhibits ESCC cell growth in vitro
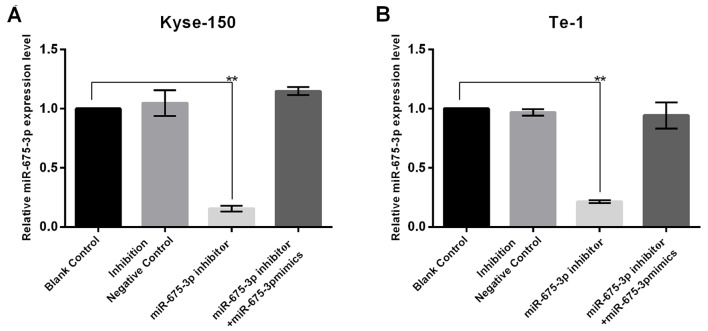
In order to investigate the influence of miR-675-3p on ESCC cell growth, the present study firstly transfected NC, miR-675-3p inhibitor, or miR-675-3p inhibitor+miR-675-3p mimics into cells, and the transfection efficiency was detected using RT-qPCR. The results indicated that miR-675-3p inhibitor significantly decreased miR-675-3p expression in KYSE150 and Te-1 cells and this reduction was reversed by miR-675-3p mimics (Fig. 2). In order to investigate the influence of miR-675-3p on ESCC cell growth, the CCK-8 assay was conducted. The results demonstrated that the proliferation and viability of Kyse-150 and Te-1 cells transfected with miR-675-3p inhibitor was significantly impaired compared with the negative control (NC), indicating an inhibitory effect. However, co-transfection with miR-675-3p inhibitor and miR-675-3p mimics eliminated this effect on Kyse-150 and Te-1 cells, and the proliferation and viability approached the level of the negative control (Fig. 3).
Figure 2.
Transfection efficiency of miR-675-3p. The expression level of miR-675-3p in inhibitor group of these cells were dropped by 80% compared with the negative control and the results confirmed that the transfection had the desired effect on miR-675-3p in (A) Kyse-150 and (B) Te-1 cells. **P<0.01 vs. Blank Control. miR, microRNA.
Figure 3.
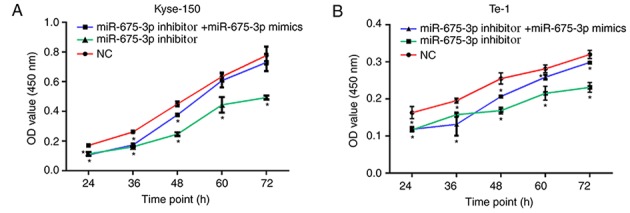
Cell Counting Kit-8 assays were performed to determine the proliferation of Kyse-150 and Te-1 cells. OD values reflecting the cell proliferation of (A) Kyse-150 and (B) Te-1 cells.
Knockdown of miR-675-3p inhibits ESCC cell migration and invasion
The effect of miR-675-3p on the migration of ESCC cells was first checked by a wound healing assay. The results of the in vitro wound healing assay demonstrated that compared with the negative control cells, the migration of Kyse-150 and Te-1 cells was significantly attenuated by miR-675-3p inhibitor. When the levels of miR-675-3p were increased by the miR-675-3p mimics, the migration of Kyse-150 and Te-1 cells recovered to the level of negative control group (Fig. 4).
Figure 4.
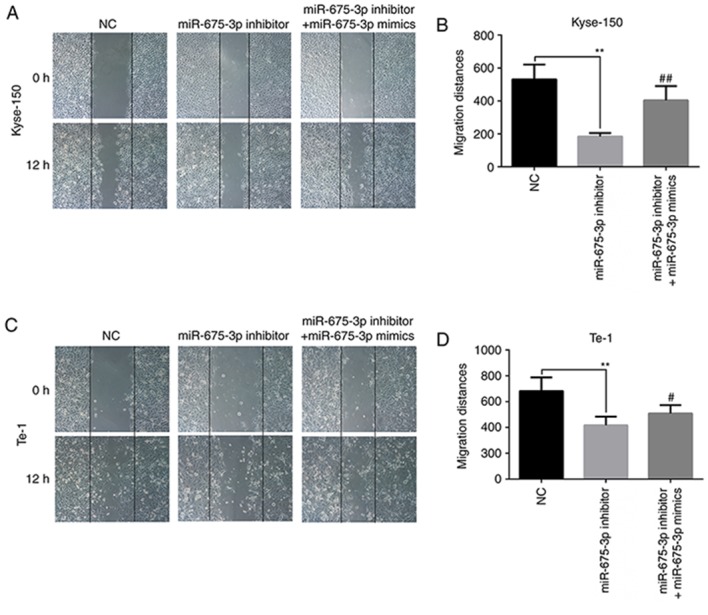
Effect of miR-675-3p on esophageal squamous cell cancer lines in wound healing assay. Representative images of the wound healing assay in (A) Kyle-150 and (B) its quantification. Representative images of the wound healing assay in (C) Te-1 cells and (D) its quantification. Magnification, ×100. **P<0.01 vs. NC; #P<0.05, ##P<0.01 vs. miR-675-3p inhibitor. miR, microRNA; NC, normal control.
To further verify the effects of miR-675-3p on cell migration and invasion ability in ESCC, a transwell assay was performed. An in vitro migration assay revealed that the migration ability of Kyse-150 and Te-1 cells transfected with miR-675-3p inhibitor were suppressed compared with the negative control, but co-transfection with miR-675-3p inhibitor and miR-675-3p mimics eliminated this effect on Kyse-150 and Te-1 cells (Fig. 5). Similarly, as presented in Fig. 6, compared with the negative control, downregulation of miR-675-3p effectively repressed the invasion capacity of Kyse-150 and Te-1 cells, however co-transfection with miR-675-3p inhibitor and miR-675-3p mimics resulted in migration and invasion capacities that approached the level of the negative control group (Figs. 5 and 6, respectively). These data may indicate the oncogenic role of miR-675-3p via the effects on the migration and invasion of ESCC.
Figure 5.
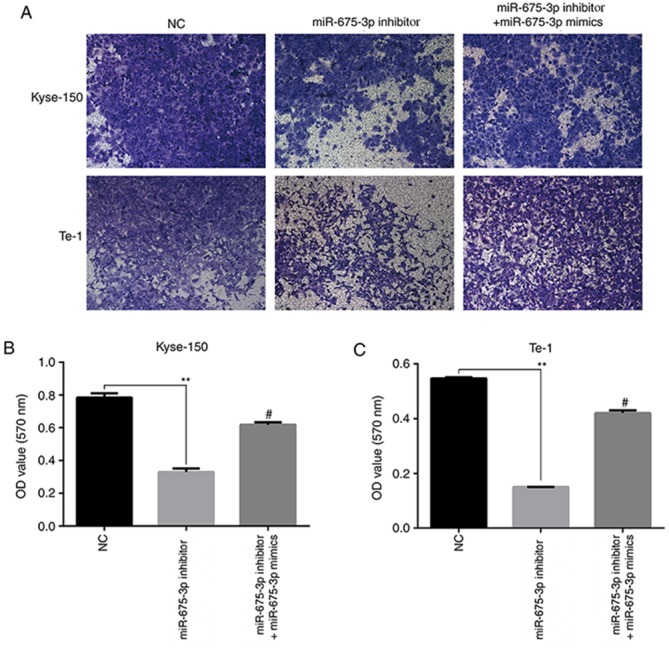
Transwell no matrigel-coated assay performed to determine the migration ability of the esophageal squamous cell cancer lines. (A) Representative images and quantification for (B) Kyse-150 and (C) Te-1. Magnification, ×100. **P<0.01 vs. NC; #P<0.05 vs. miR-675-3p inhibitor. miR, microRNA; NC, normal control; OD, optical density.
Figure 6.
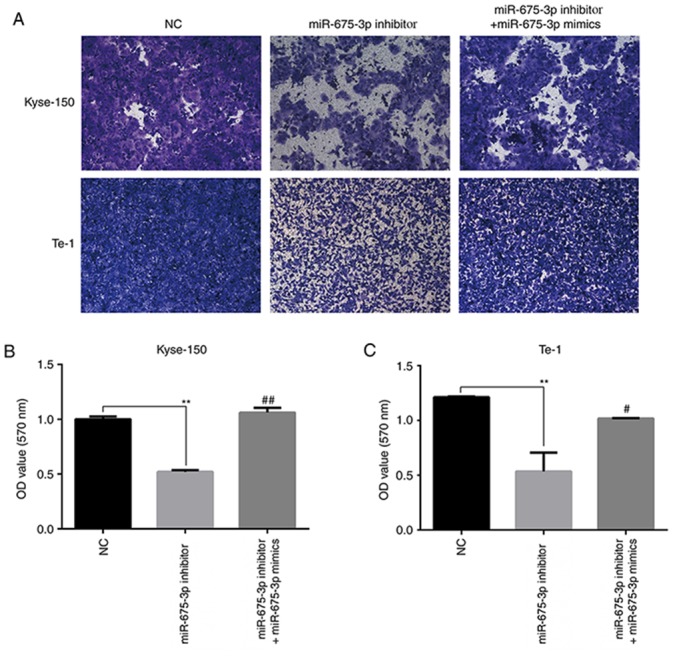
Transwell assay with matrigel-coated was performed to determine the invasion ability of the ESCC cells and 33% acetic acid was used to dissolve the crystal violet after photography. (A) Representative images and quantification for (B) Kyse-150 and (C) Te-1. Magnification, ×100. **P<0.01 vs. NC; #P<0.05, ##P<0.01 vs. miR-675-3p inhibitor. miR, microRNA; NC, normal control; OD, optical density.
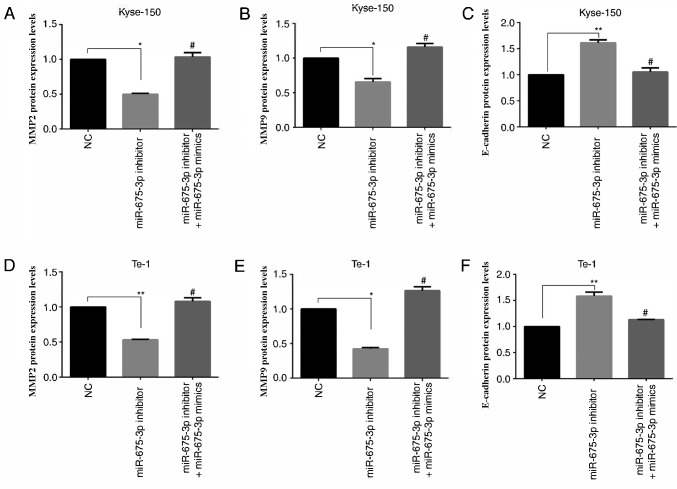
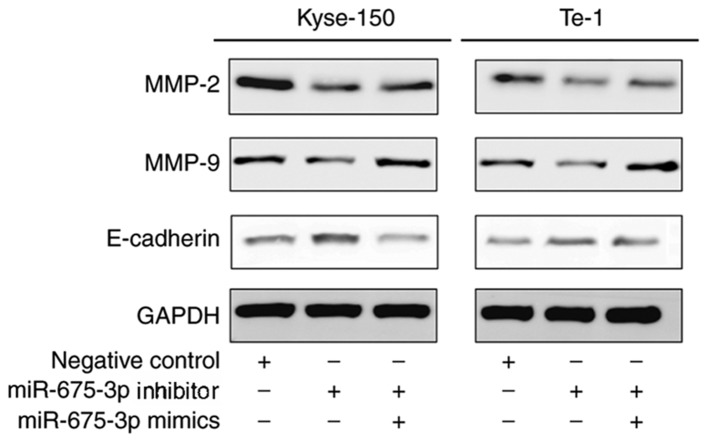
Effect of miR-675-3p on MMP2, MMP9 and E-cadherin expression in ESCC cell lines
MMP2 (15,16) and MMP9 (17,18) are involved in many events, such as cancer progression, and invasion, indicating that they may influence the invasion ability of cells. E-cadherin, is a calcium-dependent cell adhesion molecule. Loss of E-cadherin function or expression has been implicated in cancer progression and metastasis (19–21). E-cadherin downregulation decreases the strength of cellular adhesion within a tissue, resulting in an increase in cellular motility (22–24). This in turn may allow cancer cells to cross the basement membrane and invade surrounding tissues. Therefore, expression levels of MMP2, MMP9 and E-cadherin were analyzed by ELISA (Fig. 7) and western blot analysis (Fig. 8). Compared with the negative control, miR-675-3p inhibitor significantly decreased MMP2 and MMP9 expression whereas E-cadherin was enhanced. Co-transfection with miR-675-3p inhibitor and miR-675-3p mimics reversed these effects in Kyse-150 and Te-1 cells (Figs. 7 and 8).
Figure 7.
(A and D) MMP-2, (B and E) MMP-9 and (C and F) E-cadherin protein expression levels as measured by ELISA. *P<0.05, **P<0.01 vs. NC; #P<0.05 vs. miR-675-3p inhibitor. miR, microRNA; MMP, matrix metalloproteinase; NC, normal control; OD, optical density.
Figure 8.
MMP-2, MMP-9 and E-cadherin protein expression levels in Kyse-150 and Te-1 cells which transfected with NC, miR-675-3p inhibitor and miR-675-3p mimics. MMP-2, MMP-9 and E-cadherin protein expression levels were measured by western blotting. GAPDH was used as a loading control. miR, microRNA; MMP, matrix metalloproteinase; NC, normal control.
Discussion
Although great progress regarding the role of miRNA in cancer pathogenesis has been made, the roles of miRNA in the carcinogenesis of ESCC are far from being adequately elucidated. In the present study, miR-675-3p was demonstrated to serve an important role in promoting the migration and invasion ability of ESCC cell lines through inhibiting or activating epithelial mesenchymal transition (EMT) marker expression levels. In the beginning of the present study, the expression of miR-675-3p was upregulated in ESCC tissues and cell lines compared with the normal control. Therefore, the high level of miR-675-3p may be associated with the progression and development of tumor and the migration and invasion ability of ESCC cells. Previous studies have highlighted the prospect that certain miRNAs may be used as biomarkers for prognosis assessment or tumor therapeutic targets in human cancer (25–27). MiRNAs are known for their dual role as oncogenes (28,29) or tumor suppressors (30–33) and they also have been implicated in the regulatory network of various cancer types.
MiR-675-5p has been demonstrated to affect tumor migration or invasion. He et al (34) suggested that miR-675-5p is downregulated in non-small cell lung cancer tissues compared with normal tissues, and miR-675-5p inhibition could promote the migration and invasion ability of non-small cell lung cancer cells. Zhou et al (35) demonstrated that downregulation of miR-675-5p inhibits the migration and invasion ability of the ESCC cells. In the present study, it was demonstrated that miR-675-3p was highly expressed in ESCC tumor tissues compared with the normal tissues. Therefore, it may be hypothesized that miR-675-3p as another variant of miR-675 may also have the biological activity which could affect the migration and invasion ability of ESCC cells. As demonstrated in the present results, miR-675-3p affected the migration and invasion abilities of ESCC cells. When miR-675-3p was downregulated in the ESCC cells, the migration and invasion ability of ESCC cells was downregulated, and when miR-675-3p levels were then upregulated, the migration and invasion ability of the ESCC cells recovered to levels similar to the control group. These results indicated that miR-675-3p could serve as a biomarker for tumor progression and development research, or for clinical diagnosis, but this needs to be further investigated.
MMP2, MMP9 (36–40) and E-cadherin, markers of EMT, are usually involved in cancer progression (24), cancer cell invasion events (41), and also may participate in regulating cell migration (42–44) and invasion ability (45). Therefore, to reveal whether miR-675-3p participates in the regulation of ESCC cell migration and invasion, miR-675-3p was downregulated in ESCC cells using an miR-675-3p inhibitor. It was demonstrated that inhibition of miR-675-3p in Kyse-150 and Te-1 cells significantly inhibited cell growth, as well as cellular migration and invasion capabilities. Furthermore, it was also demonstrated that miR-675-3p may serve an important role in influencing some proteins, such as MMP2, MMP9, and E-cadherin, which may affect the ability of migration and invasion in ESCC cells, however the underlying mechanism has not yet been investigated. Therefore, understanding the key role of miR-675-3p in ESCC may lead to the discovery of a novel biomarker, or identification of novel therapeutic targets for treating esophageal cancer, however this requires further investigation.
In conclusion, the present results are at the forefront of research for miR-675-3p, and further investigation is required for miR-675-3p potential targets and its function in tumorigenesis.
Acknowledgements
Not applicable.
Funding
The present study was supported by the key development plan for social development of Jiangsu Province (grant no. SBE2016750057) and Jiangsu Provincial Key R & D Program Social Development Clinical Frontier Technology Project (Application of Image-guided Precise Tumor Surgical Equipment in Esophageal Cancer Surgery; grant no. BE2016731).
Availability of data and materials
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
Authors' contributions
QX conceived, designed, drafted and performed the experiments. TC and YW performed some experiments and analyzed the data. WW participated in analyzing and interpreting the data. YX provided help in conceiving and designing the study and revising the manuscript. ZG contributed to the conception, design and guided assays, gave approval of the version to be published, and supervised the project. SC conceived the main goal for the study, designed the experiment, guided assays, provided financial support of the present study and the research to gave final approval of the version of the manuscript. All authors have approved the final manuscript.
Ethics approval and consent to participate
Written informed consent was obtained from all participants, and the study protocol was approved by the ethics committee of JiangSu Cancer Hospital (Nanjing, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.López-Gómez M, Malmierca E, de Górgolas M, Casado E. Cancer in developing countries: The next most preventable pandemic. The global problem of cancer. Crit Rev Oncol Hematol. 2013;88:117–122. doi: 10.1016/j.critrevonc.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 2.Sankaranarayanan R, Ramadas K, Qiao YL. Managing the changing burden of cancer in Asia. BMC Med. 2014;12:3. doi: 10.1186/1741-7015-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katzka DA. Recent advances in non-invasive esophageal tissue sampling. Curr Gastroenterol Rep. 2017;19:9. doi: 10.1007/s11894-017-0550-4. [DOI] [PubMed] [Google Scholar]
- 4.Peery AF, Crockett SD, Barritt AS, Dellon ES, Eluri S, Gangarosa LM, Jensen ET, Lund JL, Pasricha S, Runge T, et al. Burden of gastrointestinal, liver, and pancreatic diseases in the United States. Gastroenterology. 2015;149:1731–1741.e3. doi: 10.1053/j.gastro.2015.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 6.Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381:400–412. doi: 10.1016/S0140-6736(12)60643-6. [DOI] [PubMed] [Google Scholar]
- 7.Zhu ZJ, Hu Y, Zhao YF, Chen XZ, Chen LQ, Chen YT. Early recurrence and death after esophagectomy in patients with esophageal squamous cell carcinoma. Ann Thorac Surg. 2011;91:1502–1508. doi: 10.1016/j.athoracsur.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 8.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends in Mol Med. 2014;20:460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 9.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 10.Li C, Lei B, Huang S, Zheng M, Liu Z, Li Z, Deng Y. H19 derived microRNA-675 regulates cell proliferation and migration through CDK6 in glioma. Am J Transl Res. 2015;7:1747–1764. [PMC free article] [PubMed] [Google Scholar]
- 11.Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14:659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu M, Chen Q, Liu X, Sun Q, Zhao X, Deng R, Wang Y, Huang J, Xu M, Yan J, Yu J. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 2014;281:3766–3775. doi: 10.1111/febs.12902. [DOI] [PubMed] [Google Scholar]
- 13.Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung JJ, Kwok TT. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31:350–358. doi: 10.1093/carcin/bgp181. [DOI] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Jiang Z, Zhang H, Liu C, Yin J, Tong S, Lv J, Wei S, Wu S. β3GnT8 promotes colorectal cancer cells invasion via CD147/MMP2/Galectin3 axis. Front Physiol. 2018;9:588. doi: 10.3389/fphys.2018.00588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang DY, Sp N, Kim DH, Joung YH, Lee HG, Park YM, Yang YM. Salidroside inhibits migration, invasion and angiogenesis of MDA-MB 231 TNBC cells by regulating EGFR/Jak2/STAT3 signaling via MMP2. Int J Oncol. 2018;53:877–885. doi: 10.3892/ijo.2018.4430. [DOI] [PubMed] [Google Scholar]
- 17.Marshall DC, Lyman SK, McCauley S, Kovalenko M, Spangler R, Liu C, Lee M, O'Sullivan C, Barry-Hamilton V, Ghermazien H, et al. Selective allosteric inhibition of MMP9 is efficacious in preclinical models of ulcerative colitis and colorectal cancer. PLoS One. 2015;10:e0127063. doi: 10.1371/journal.pone.0127063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grauzam S, Brock AM, Holmes CO, Tiedeken JA, Boniface SG, Pierson BN, Patterson DG, Coaxum SD, Neskey DM, Rosenzweig SA. NEDD9 stimulated MMP9 secretion is required for invadopodia formation in oral squamous cell carcinoma. Oncotarget. 2018;9:25503–25516. doi: 10.18632/oncotarget.25347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan YL, Wang YM, Liu H, Qin GF, Tang AG, Duan Y. Aberrant expression of E-cadherin in lung tissues of patients with probable lung cancer. Asian Pac J Cancer Prev. 2012;13:5149–5153. doi: 10.7314/APJCP.2012.13.10.5149. [DOI] [PubMed] [Google Scholar]
- 20.Wong TS, Gao W, Chan JY. Interactions between E-cadherin and microRNA deregulation in head and neck cancers: the potential interplay. Biomed Res Int. 2014;2014:126038. doi: 10.1155/2014/126038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Repetto O, De Paoli P, De Re V, Canzonieri V, Cannizzaro R. Levels of soluble E-cadherin in breast, gastric, and colorectal cancers. Biomed Res Int. 2014;2014:408047. doi: 10.1155/2014/408047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Techasen A, Loilome W, Namwat N, Khuntikeo N, Puapairoj A, Jearanaikoon P, Saya H, Yongvanit P. Loss of E-cadherin promotes migration and invasion of cholangiocarcinoma cells and serves as a potential marker of metastasis. Tumour Biol. 2014;35:8645–8652. doi: 10.1007/s13277-014-2087-6. [DOI] [PubMed] [Google Scholar]
- 23.Wang CA, Drasin D, Pham C, Jedlicka P, Zaberezhnyy V, Guney M, Li H, Nemenoff R, Costello JC, Tan AC, Ford HL. Homeoprotein Six2 promotes breast cancer metastasis via transcriptional and epigenetic control of E-cadherin expression. Cancer Res. 2014;74:7357–7370. doi: 10.1158/0008-5472.CAN-14-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kreiseder B, Orel L, Bujnow C, Buschek S, Pflueger M, Schuett W, Hundsberger H, de Martin R, Wiesner C. α-Catulin downregulates E-cadherin and promotes melanoma progression and invasion. Int J Cancer. 2013;132:521–530. doi: 10.1002/ijc.27698. [DOI] [PubMed] [Google Scholar]
- 25.Shin VY, Chu KM. MiRNA as potential biomarkers and therapeutic targets for gastric cancer. World J Gastroenterol. 2014;20:10432–10439. doi: 10.3748/wjg.v20.i30.10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshizawa JM, Wong DT. Salivary microRNAs and oral cancer detection. Methods Mol Biol. 2013;936:313–324. doi: 10.1007/978-1-62703-083-0_24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rocci A, Hofmeister CC, Pichiorri F. The potential of miRNAs as biomarkers for multiple myeloma. Expert Rev Mol Diagn. 2014;14:947–959. doi: 10.1586/14737159.2014.946906. [DOI] [PubMed] [Google Scholar]
- 28.Pan Y, Liang H, Chen W, Zhang H, Wang N, Wang F, Zhang S, Liu Y, Zhao C, Yan X, et al. microRNA-200b and microRNA-200c promote colorectal cancer cell proliferation via targeting the reversion-inducing cysteine-rich protein with Kazal motifs. RNA Biol. 2015;12:276–289. doi: 10.1080/15476286.2015.1017208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamada N, Tsujimura N, Kumazaki M, Shinohara H, Taniguchi K, Nakagawa Y, Naoe T, Akao Y. Colorectal cancer cell-derived microvesicles containing microRNA-1246 promote angiogenesis by activating Smad 1/5/8 signaling elicited by PML down-regulation in endothelial cells. Biochim Biophys Acta. 2014;1839:1256–1272. doi: 10.1016/j.bbagrm.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 30.Jin M, Zhang T, Liu C, Badeaux MA, Liu B, Liu R, Jeter C, Chen X, Vlassov AV, Tang DG. miRNA-128 suppresses prostate cancer by inhibiting BMI-1 to inhibit tumor-initiating cells. Cancer Res. 2014;74:4183–4195. doi: 10.1158/0008-5472.CAN-14-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao R, Li C, Chai B. miRNA-144 suppresses proliferation and migration of colorectal cancer cells through GSPT1. Biomed Pharmacother. 2015;74:138–144. doi: 10.1016/j.biopha.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 32.Liu HT, Xing AY, Chen X, Ma RR, Wang YW, Shi DB, Zhang H, Li P, Chen HF, Li YH, Gao P. MicroRNA-27b, microRNA-101 and microRNA-128 inhibit angiogenesis by down-regulating vascular endothelial growth factor C expression in gastric cancers. Oncotarget. 2015;6:37458–37470. doi: 10.18632/oncotarget.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tao J, Zhi X, Zhang X, Fu M, Huang H, Fan Y, Guan W, Zou C. miR-27b-3p suppresses cell proliferation through targeting receptor tyrosine kinase like orphan receptor 1 in gastric cancer. J Exp Clin Cancer Res. 2015;34:139. doi: 10.1186/s13046-015-0253-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He D, Wang J, Zhang C, Shan B, Deng X, Li B, Zhou Y, Chen W, Hong J, Gao Y, et al. Down-regulation of miR-675-5p contributes to tumor progression and development by targeting pro-tumorigenic GPR55 in non-small cell lung cancer. Mol Cancer. 2015;14:73. doi: 10.1186/s12943-015-0342-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou YW, Zhang H, Duan CJ, Gao Y, Cheng YD, He D, Li R, Zhang CF. miR-675-5p enhances tumorigenesis and metastasis of esophageal squamous cell carcinoma by targeting REPS2. Oncotarget. 2016;7:30730–30747. doi: 10.18632/oncotarget.8950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Yang B, She Y, Ye Y. The lncRNA TP73-AS1 promotes ovarian cancer cell proliferation and metastasis via modulation of MMP2 and MMP9. J Cell Biochem. 2018 Jun 15; doi: 10.1002/jcb.27158. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 37.Li H, Zhang Y, Hai J, Wang J, Zhao B, Du L, Geng X. Knockdown of TRIM31 suppresses proliferation and invasion of gallbladder cancer cells by down-regulating MMP2/9 through the PI3K/Akt signaling pathway. Biomed Pharmacother. 2018;103:1272–1278. doi: 10.1016/j.biopha.2018.04.120. [DOI] [PubMed] [Google Scholar]
- 38.Kalhori V, Törnquist K. MMP2 and MMP9 participate in S1P-induced invasion of follicular ML-1 thyroid cancer cells. Mol Cell Endocrinol. 2015;404:113–122. doi: 10.1016/j.mce.2015.01.037. [DOI] [PubMed] [Google Scholar]
- 39.Jacob A, Jing J, Lee J, Schedin P, Gilbert SM, Peden AA, Junutula JR, Prekeris R. Rab40b regulates trafficking of MMP2 and MMP9 during invadopodia formation and invasion of breast cancer cells. J Cell Sci. 2013;126:4647–4658. doi: 10.1242/jcs.126573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J, Ping W, Zu Y, Sun W. Correlations of lysyl oxidase with MMP2/MMP9 expression and its prognostic value in non-small cell lung cancer. Int J Clin Exp Pathol. 2014;7:6040–6047. [PMC free article] [PubMed] [Google Scholar]
- 41.Qiu X, Cheng JC, Chang HM, Leung PC. COX2 and PGE2 mediate EGF-induced E-cadherin-independent human ovarian cancer cell invasion. Endocr Relat Cancer. 2014;21:533–543. doi: 10.1530/ERC-13-0450. [DOI] [PubMed] [Google Scholar]
- 42.Cai D, Chen SC, Prasad M, He L, Wang X, Choesmel-Cadamuro V, Sawyer JK, Danuser G, Montell DJ. Mechanical feedback through E-cadherin promotes direction sensing during collective cell migration. Cell. 2014;157:1146–1159. doi: 10.1016/j.cell.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen D, Wu Z, Luo LJ, Huang X, Qian WQ, Wang H, Li SH, Liu J. E-cadherin maintains the activity of neural stem cells and inhibits the migration. Int J Clin Exp Pathol. 2015;8:14247–14251. [PMC free article] [PubMed] [Google Scholar]
- 44.Brett A, Pandey S, Fraizer G. The Wilms' tumor gene (WT1) regulates E-cadherin expression and migration of prostate cancer cells. Mol Cancer. 2013;12:3. doi: 10.1186/1476-4598-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Canel M, Serrels A, Frame MC, Brunton VG. E-cadherin-integrin crosstalk in cancer invasion and metastasis. J Cell Sci. 2013;126:393–401. doi: 10.1242/jcs.100115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.