Abstract
There is urgent need to develop novel treatment strategies to reduce antimicrobial resistance. Collateral sensitivity (CS), where resistance to one antimicrobial increases susceptibility to other drugs, might enable selection against resistance during treatment. However, the success of this approach would depend on the conservation of CS networks across genetically diverse bacterial strains. Here, we examine CS conservation across diverse Escherichia coli strains isolated from urinary tract infections. We determine collateral susceptibilities of mutants resistant to relevant antimicrobials against 16 antibiotics. Multivariate statistical analyses show that resistance mechanisms, in particular efflux-related mutations, as well as the relative fitness of resistant strains, are principal contributors to collateral responses. Moreover, collateral responses shift the mutant selection window, suggesting that CS-informed therapies may affect evolutionary trajectories of antimicrobial resistance. Our data allow optimism for CS-informed therapy and further suggest that rapid detection of resistance mechanisms is important to accurately predict collateral responses.
Resistance to one antibiotic can in some cases increase susceptibility to other antibiotics. Here, Podnecky et al. study these collateral responses in E. coli clinical isolates and show that efflux-related resistance mechanisms and relative fitness of the strains are principal contributors to this phenomenon.
Introduction
The evolution and increasing prevalence of antimicrobial resistance is driven by the consumption and misuse of antimicrobials in human medicine, agriculture, and the environment1–3. Historically, the threat of antimicrobial resistance was overcome by using novel antimicrobials with unique drug targets. However, the discovery rate of new antimicrobial agents has dwindled4–6 and severely lags behind the rate of resistance evolution7. While concerted scientific, corporate, and political focus is needed to recover antimicrobial pipelines8–10, there is an urgent need for alternative strategies that prolong the efficacy of existing antimicrobials and prevent or slow the emergence, spread, and persistence of antimicrobial resistance. Current global efforts to improve antimicrobial stewardship largely focus on reducing overall antimicrobial consumption and increasing awareness of resistance development9,11–13. While these efforts will affect the evolution and spread of resistance, mounting evidence suggests that these changes alone will not lead to large-scale reductions in the occurrence of antimicrobial resistance14–18.
Several recent studies have examined novel treatment strategies using multiple antimicrobials that could reduce the rate of resistance emergence and even reverse pre-existing resistance. These approaches, collectively termed selection inversion strategies, refer to cases where resistance becomes costly in the presence of other antimicrobial agents19. Among the most promising of these strategies are those based on a phenomenon first reported in 1952, termed collateral sensitivity (CS), where resistance to one antimicrobial simultaneously increases the susceptibility to another20. CS and its inverse, cross-resistance (CR), have been demonstrated for several bacterial species and across different classes of antimicrobials21–27. These results have formed the basis of proposed CS-informed antimicrobial strategies that combine drug pairs22,28 or alter temporal administration, e.g. drug cycling21,29. CS-informed strategies would force bacteria to evolve resistance along a predictable trajectory, resulting in CS; this predictability could be exploited to ultimately reverse resistance and prevent the fixation of resistance and multi-drug resistance development at the population level of bacterial communities.
Initial in vitro experiments support using CS-based strategies to re-sensitize resistant strains21 and reduce rates of resistance development29; however, the broader application of this principle depends on predictable bacterial responses during antimicrobial therapy. This predictability must be general for a given drug class and should not vary across strains of the same species. To date, most studies of CS and CR have focused on describing collateral networks21–23 using resistant mutants derived from single laboratory-adapted strains and limited numbers of clinical isolates. Two studies on Pseudomonas aeruginosa have investigated CS in collections of clinical isolates30,31. However, these studies lack either baseline controls30 or sufficient genetic diversity among tested strains31. As valuable as earlier work has been, the responses of single strains (laboratory or clinical) may not be representative of CS and CR responses in other strains.
To address this limitation, here we focus on understanding collateral networks in clinical urinary tract isolates of Escherichia coli with selected resistance to drugs widely used for the treatment of urinary tract infections: ciprofloxacin, trimethoprim, nitrofurantoin, and mecillinam. We investigate collateral networks to 16 antimicrobials from diverse drug classes in 10 genetically diverse clinical strains (corresponding to 49 laboratory-generated mutants) to assess the factors contributing to collateral responses (both CS and CR). This approach allows us to identify variation in the sign and magnitude of collateral responses and identify mechanisms of CS and CR that are preserved in various genetic backgrounds. Using multivariate statistical modeling, we show that resistance mutations, particularly those affecting efflux pumps, and the relative fitness of resistant isolates are more important determinants of collateral networks than genetic background. Our results support the idea that collateral responses may be predictable.
Results
Collateral responses vary between and across resistance groups
We examined collateral responses to antimicrobial resistance in a panel of 10 genetically diverse (Supplementary Fig. 1a–b) E. coli strains isolated from urinary tract infections. For each of these pan-susceptible strains (Supplementary Fig. 1c)32, a single resistant mutant was generated to each of four individual antimicrobials used to treat urinary tract infections: ciprofloxacin, trimethoprim, nitrofurantoin, and mecillinam. Here we define resistance group as the collection of mutants from the 10 different genetic backgrounds that were selected for resistance to the same antimicrobial. Mutants resistant to mecillinam required only a single selection step, while multiple selection steps were required to select for resistance above clinical breakpoints for the remaining antimicrobials. In total, 40 resistant mutants were generated with resistance levels above clinical breakpoints, as determined by antimicrobial susceptibility testing using both gradient strip diffusion (Supplementary Table 1) and inhibitory concentration 90% (IC90)21 testing (Table 1). The two methods are correlated, but IC90 measurements allow for more robust detection of small relative differences in susceptibility33,34. Changes in the IC90 of resistant mutants from each respective wild-type strain (Supplementary Fig. 2) were compared for 16 antimicrobials (Table 2). Overall, collateral responses were observed in 39% (233/590) of possible instances (Supplementary Table 2); of these 49% (115/233) were associated with only a 1.5-fold change in IC90. Such small changes would not be observed by typical two-fold antimicrobial susceptibility testing methods frequently used in clinical laboratories.
Table 1.
Description of Escherichia coli strains used in the study and average IC90 changes following antimicrobial selection
| Strain | STa | Origin | CIPb | MECb | NITb | TMPb | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| WTc | cCIPR | WTc | cMECR | WTc | cNITR | WTc | cTMPR | |||
| K56-2 | 73 | Greece | 0.014 | 16 | 0.146 | >30 | 8 | >64 | 0.225 | >28 |
| K56-12 | 104 | Portugal | 0.016 | 1.67 | 0.273 | 28 | 7.33 | >64 | 0.563 | >32 |
| K56-16d | 127 | Portugal | 0.009 | 3 | 0.167 | 18.7 | 4 | >64 | 0.25 | >30 |
| K56-41 | 73 | Greece | 0.016 | 2.33 | 0.104 | 13.3 | 6 | >64 | 0.25 | 6.67 |
| K56-44d | 12 | Greece | 0.013 | 1.67 | 0.141 | 16 | 6.67 | >64 | 0.375 | 6 |
| K56-50 | 100 | Greece | 0.012 | 3 | 0.141 | 10.7 | 12 | >64 | 0.172 | 18 |
| K56-68 | 95 | Sweden | 0.014 | 4 | 0.141 | 30 | 6.67 | >64 | 0.208 | 18.7 |
| K56-70 | 537 | Sweden | 0.007 | 2.67 | 0.083 | >32 | 4.67 | >64 | 0.25 | 14.7 |
| K56-75e | 69 | UK | 0.008 | 1.17 | 0.063 | 13 | 6 | >64 | 0.167 | 5.33 |
| K56-78 | 1235 | UK | 0.015 | 6 | 0.141 | 16 | 8 | >64 | 0.5 | 7.33 |
aMulti-locus sequence type (ST)
bThe average IC90 values (µg mL−1) of three or more biological replicates for wild type (WT) and resistant (R) mutants to ciprofloxacin (CIP), mecillinam (MEC), nitrofurantoin (NIT), and trimethoprim (TMP). Individual results above detection limits (MEC = 32 µg mL−1, NIT = 64 µg mL−1, TMP = 32 µg mL−1) were analyzed as those values, yielding final results with uncertainty (>average). EUCAST Clinical Breakpoints v 7.1 for Enterobacteriaceae63 were: >0.5 µg mL−1 CIP, >8 µg mL−1 MEC, >64 µg mL−1 NIT, and >4 µg mL−1 TMP
cThe strain number names the WT, and designations CIPR, MECR, NITR, and TMPR describe which drug the isolates were selected with, and resistance achieved
d, eStrains containing the Col156 or Col(MP18) replicon, respectively
Table 2.
List of antimicrobials used in this study
| Antimicrobiala | Abbreviation | Drug class | Drug target(s) | Solvent |
|---|---|---|---|---|
| Amoxicillin | AMX | β-lactam (Penicillin) | Cell wall synthesis | Phosphate bufferb |
| Azithromycin | AZT | Macrolide | Protein synthesis (50S) | ≥95% Ethanol |
| Ceftazidime | CAZ | β-lactam (Cephalosporin) | Cell wall synthesis | Water + 10% (w w-1) Na2CO3 |
| Chloramphenicol | CHL | Amphenicol | Protein synthesis (50S) | ≥95% Ethanol |
| Ciprofloxacin | CIP | Fluoroquinolone | DNA replication, cell division | 0.1 N HCl |
| Colistin | COL | Polymyxin | Cell wall & cell membrane | Water |
| Ertapenem | ETP | β-lactam (Carbapenem) | Cell wall synthesis | Water |
| Fosfomycin | FOS | Phosphonic | Cell wall synthesis (MurA) | Water |
| Gentamicin | GEN | Aminoglycoside | Protein synthesis (30S) | Water |
| Mecillinam | MEC | β-lactam (Penicillin) | Cell wall synthesis (PBP2) | Water |
| Nitrofurantoin | NIT | Nitrofuran | Multiplec | Dimethyl sulfoxide |
| Trimethoprim | TMP | Antifolate | Folate synthesis (FolA) | Dimethyl sulfoxide |
| Sulfamethoxazole | SMX | Antifolate | Folate synthesis (FolP) | Dimethyl sulfoxide |
| TMP + SMX (1:19) | SXT | Antifolate | Folate synthesis (FolA + FolP) | Dimethyl sulfoxide |
| Temocillin | TEM | β-lactam (Penicillin) | Cell wall synthesis | Water |
| Tetracycline | TET | Tetracycline | Protein synthesis (30S) | Water |
| Tigecycline | TGC | Tetracycline | Protein synthesis (30S) | Water |
aWhen available, final antimicrobial concentration was determined using manufacturer-provided or calculated drug potencies, otherwise potency was assumed to be 100%. Aliquots were stored at −20 or −80 °C in single-use vials. All antimicrobials and chemical solvents were obtained from Sigma-Aldrich (St. Louis, MO, USA) with the exception of ciprofloxacin (Biochemika, now Sigma-Aldrich) and temocillin (Negaban®)
b0.1 mol L−1, pH 6.0 phosphate buffer supplemented with 6.5% (v v−1) 1 M NaOH (sodium hydroxide)
cNitrofurantoin is thought to target macromolecules including DNA and ribosomal proteins, affecting multiple cellular processes, including protein, DNA, RNA, and cell wall synthesis
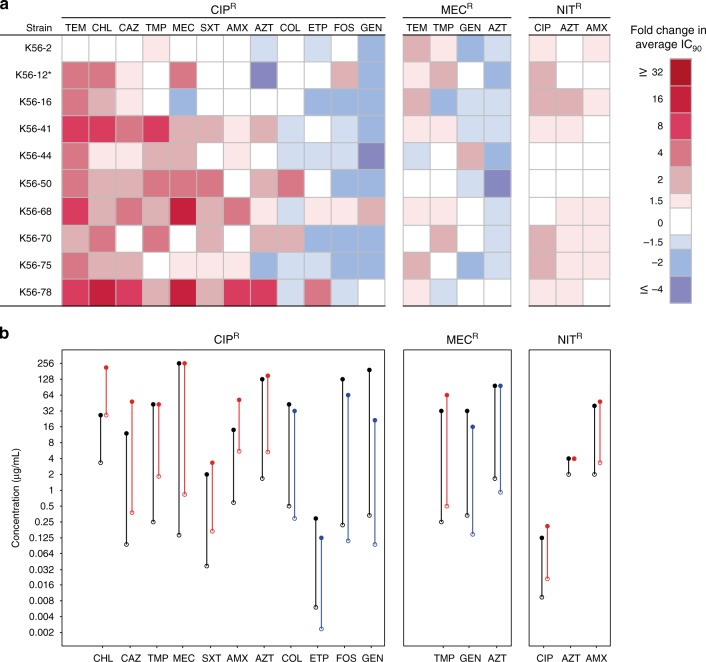
Overall CR was more frequent than CS, 141 versus 92 instances (Supplementary Table 2), and collateral networks varied considerably between resistance groups. We observed 19 cases of conserved collateral responses (Fig. 1a), where CR or CS to a specific antimicrobial was found in ≥50% of the mutants within a resistance group, defined as CR50 or CS50, respectively. For each CR50 and CS50 observation, IC90 results were further assessed by generating dose–response curves of representative strain:drug combinations (Supplementary Fig. 3). Inhibition of growth was shown to vary across antimicrobial concentrations between resistant mutants and respective wild-type strains, confirming the changes in antimicrobial susceptibility determined by the IC90 assays.
Fig. 1.
Conserved collateral responses in antimicrobial resistant mutants. a Relative change in antimicrobial susceptibility was determined by comparing average IC90 values of resistant mutants to the respective wild-type strain. Collateral responses that were found in ≥50% of the strains are displayed, excluding CR observed in all trimethoprim-resistant mutants to trimethoprim-sulfamethoxazole (see Supplementary Fig. 2). Antimicrobials are ordered by most frequent CR (red; left) to most frequent CS (blue; right) for each group. *The slow growing K56-12 CIPR was incubated an additional 24 h for IC90 determination. b The average IC90 (open circles) and average mutation prevention concentration (MPC; filled circles) were determined and compared between resistant mutants (colored) with collateral responses, either CS (blue) or CR (red), and their respective wild-type strain (black) in strain:drug combinations representing conserved collateral responses, excluding temocillin. The mutant selection window (vertical lines) was defined as the range between IC90 (lower bound) and MPC (upper bound). K56-16 NITR had equivalent IC90 and MPC values for azithromycin, thus no mutation selection window was reported. Generally, changes in MPC values reflected observed IC90 changes, shifting the mutation selection window upwards or downwards accordingly. In 8/10 tested combinations an increase in IC90 value (CR) from wild-type to resistant mutant correlated with at least a small increased MPC, with the remaining combinations showing no change in MPC value. Similarly, decreased IC90 values (CS) correlated with decreased MPCs (5/7)
During the selection of resistant mutants, we often observed colonies of varying size for all resistance groups, suggesting changes to bacterial fitness. To test this, we measured the growth rates of mutants relative to the respective wild-type strains (Supplementary Fig. 4). In general, mutants resistant to ciprofloxacin and mecillinam displayed severely reduced growth rates, suggesting high costs of resistance. Relative growth rates varied between 0.34–0.75 with a mean of 0.53 for ciprofloxacin-resistant mutants and between 0.49–0.79 with a mean of 0.64 for mecillinam-resistant mutants. Mutants resistant to nitrofurantoin and trimethoprim displayed lower fitness effects, and several resistant mutants harbored apparent cost-free resistance mutations (Supplementary Fig. 4). Only two of ten nitrofurantoin-resistant mutants and four of ten trimethoprim-resistant mutants displayed an apparent cost of resistance. Relative growth rates varied between 0.93–1.05 and 0.68–1.07 with averages of 0.99–0.94 for nitrofurantoin- resistant and trimethoprim-resistant mutants, respectively.
Ciprofloxacin resistance linked to conserved collateral responses
Nearly half (108/233, 46%) of the observed collateral responses were in ciprofloxacin-resistant mutants, while the remaining 125 were distributed between the other three resistance groups (Supplementary Table 2). Within the ciprofloxacin-resistant group, the majority of collateral responses were CR (70/108, 65%). Additionally, CS responses in ciprofloxacin-resistant mutants were the most conserved in our dataset, with CS to gentamicin occurring in 8 of 10 strains and CS to fosfomycin in 7 of 10 strains (Fig. 1a). Gentamicin and other aminoglycosides are important for the treatment of a wide range of infections35, while fosfomycin is primarily used for treatment of uncomplicated urinary tract infections36,37. The ciprofloxacin-resistant mutants were also unique in the magnitude of observed changes, with cases of CR close to 30-fold and CS as high as six-fold changes in IC90 (Supplementary Fig. 2).
Characterization of antimicrobial-resistant mutants
We hypothesized that CS and CR variation in and between resistance groups could be attributed to different mutations causing resistance in each strain. Using whole genome sequencing, we identified a total of 149 mutations in the resistant mutants (Supplementary Data 1–4). Of these, 88 mutations affect previously described or putative antimicrobial resistance-associated genes, gene-regions, or pathways (Supplementary Data 1–4). The remaining mutations were found in other cellular processes not known to affect antimicrobial susceptibility (e.g. metabolic pathways and virulence factors), such as mutation to the FimE regulator of FimA that was frequently observed in mecillinam-resistant mutants (Supplementary Data 2). Aside from FimE, we did not observe mutations in regions unrelated to resistance across mutants of the same resistance group (parallel evolution), suggesting that such mutations had limited, if any, effect on collateral responses in this study.
For each of the 40 resistant mutants at least one putative resistance mechanism was identified, including mutations to previously described antimicrobial drug targets and promoters of drug targets, drug-modifying (activating) enzymes, regulators of efflux pumps, RNA polymerases and mutations to other metabolic and biochemical processes that may contribute to resistance (Table 3). Briefly, all but one ciprofloxacin-resistant mutant contained mutations in both gyrA and efflux regulatory genes and/or gene-regions likely affecting efflux expression (acrAB and/or mdtK), while one strain had only drug target mutations and displayed the well-described GyrA (S83L) and ParC (G78D) mutation combination (Supplementary Data 1). Both efflux and drug target mutations are frequently found in surveys of clinical isolates38–41. Nitrofurantoin-resistant mutants had mutations in one or both nitro-reductases (nfsA, nfsB) and the majority of strains had additional mutations in mprA, which encodes an efflux regulator of EmrAB-TolC pump expression (Supplementary Data 3). Mutants resistant to trimethoprim contained mutations either in folA and/or its promoter or genetic amplification of a large region containing folA (Supplementary Data 4). The mecillinam-resistant mutants are unique in that they evolved as single step mutants, where a single mutation could confer clinical resistance to mecillinam. Resistance development for the remaining three drugs required several steps, as multiple mutations were required for resistance above clinical breakpoints. In total, 12 different mutations in genes and/or cellular processes previously linked to mecillinam resistance were identified in this resistance group (Supplementary Data 2)42.
Table 3.
The number of antimicrobial resistant mutants with resistance-associated mutations
| Resistance mechanism | CIPR | MECR | NITR | TMPR | |
|---|---|---|---|---|---|
| Drug target | Modification | 10a | 6 | ||
| Overproduction | 6 | ||||
| Drug activation | Nitroreductase disruption | 10 | |||
| Drug uptake | Porin mutation | 1 | |||
| Efflux | AcrAB-TolC | 7 | 1 | ||
| MdtK | 9 | 1 | |||
| MdfA | 1 | ||||
| EmrAB-TolC | 7 | ||||
| ABC transport | 1 | ||||
| ppGpp synthesis (stringent response activation) | Stringent response | 4 | |||
| tRNA synthesis | 4 | ||||
| tRNA processing | 1 | ||||
| Cellular metabolism | 3 |
aAll mutants resistant to ciprofloxacin contained one mutation in the gyrA gene, except the K56-2 CIPR mutant that contained two mutations in gyrA and a mutation in parC
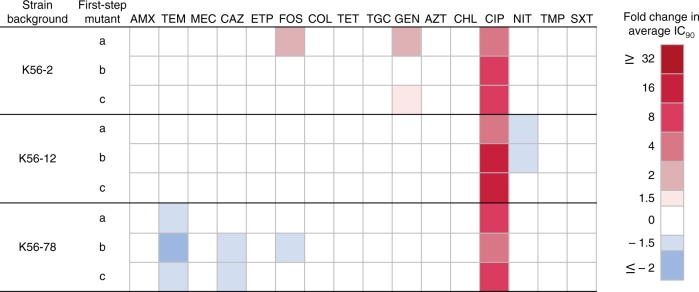
The ciprofloxacin-resistant group displayed a clear trend where conserved CR responses were strongly linked to mutations in efflux regulatory regions suggesting that gyrA drug target mutations had a limited effect on CS and CR. Trimethoprim-resistant mutants also had few collateral responses, likely due to the specific mechanism of resistance affecting a single unique drug target (i.e. overexpression/alteration of FolA). To further investigate the effects of drug target mutations, we assessed the collateral responses of mutants generated following a single selection-step with ciprofloxacin. These first-step mutants contained single, non-synonymous mutations to gyrA and no other mutations (e.g. in efflux pumps) linked to ciprofloxacin resistance (Supplementary Data 1). The IC90 of these strains was uniformly lower than in ciprofloxacin-resistant strains containing multiple resistance mutations. Few collateral responses were observed in these first-step mutants (Fig. 2), and none were conserved across different strain backgrounds. These results suggest that most collateral responses observed in the ciprofloxacin-resistant mutants are due to the observed efflux mutations.
Fig. 2.
Collateral effects in gyrA mutants with decreased susceptibility to ciprofloxacin. Relative changes in antimicrobial susceptibilities, CS (blue) and CR (red), were determined by comparing average IC90 values of nine first-step mutants to their respective wild-type strain. Antimicrobials are ordered by antimicrobial class, as in Supplementary Fig. 2
Efflux and fitness are main contributors to collateral responses
Multivariate statistical approaches were used to investigate the extent to which genetic (strain) background, resistance group, the putative mechanism of resistance (in particular efflux-related mutations), growth rate, and the fitness cost of resistance explain the variation in collateral responses. All factors were investigated individually (Supplementary Fig. 5a–e). Throughout the remaining analyses we focus mainly on efflux-related mutations, rather than resistance group, to explicitly address putative mechanisms of resistance, and relative fitness rather than growth rate.
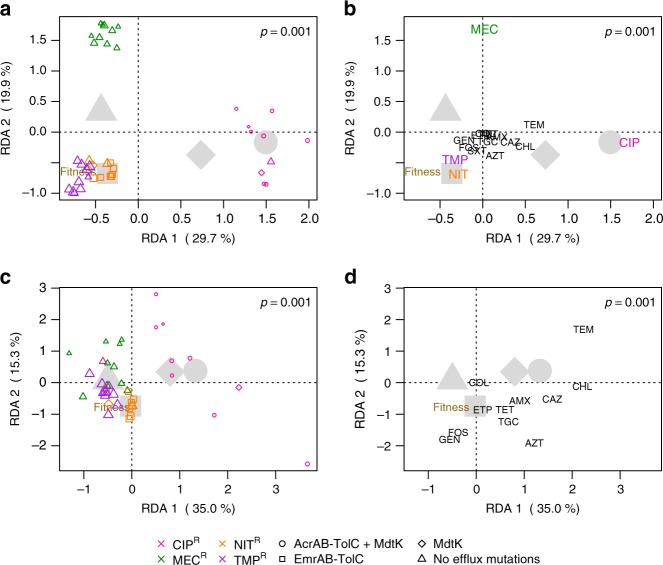
We estimated several models with individual, or a combination of, factors to assess their effect size and significance given some level of collinearity between fitness and efflux-type (Fig. 3, Supplementary Fig. 5a–r). A model including strain background, relative fitness, and efflux-related mutations as factors explained 62.5% of the total variation in IC90 values (Fig. 3a, b, Supplementary Table 3). In this three-factor model there was clear separation of the mutants by resistance group (Fig. 3a). The ciprofloxacin-resistant mutants showed strong CR towards temocillin, chloramphenicol, ceftazidime, and amoxicillin, separating this resistance group from the others along the first ordination axis (Fig. 3a, b). Along the second ordination axis, mecillinam-resistant isolates were distinct, had CR to temocillin, and were more likely to have CS towards drugs, such as azithromycin and chloramphenicol (Fig. 3a, b). Both efflux-type and relative fitness were significant predictors when tested alone and in combination (Supplementary Table 3). The model (Fig. 3a, b) also revealed that strain background had a non-significant (p = 0.993) contribution (Supplementary Table 3). Even when modeled alone (Supplementary Fig. 5a), strain background only accounted for 6.5% of the variation and was non-significant (Supplementary Table 3).
Fig. 3.
Results of multivariate statistical modeling. Graphical representations of two redundancy analyses (RDA, triplot) results relating various parameters to the observed changes in IC90 between resistant mutants and respective wild-type strains for (a, b) 16 antimicrobials tested and (c, d) a subset of these antimicrobials, excluding ciprofloxacin, mecillinam, nitrofurantoin, trimethoprim, and trimethoprim-sulfamethoxazole. Each RDA is broken down into two plots; (a, c) where weighted averages of resistant mutants are plotted as colored symbols (color indicates resistance group, shape the assigned efflux group, and symbol size proportional to relative fitness, see Supplementary Fig. 4). In (b, d) antimicrobial drug names indicate the tip of vectors that pass through the origin in the direction of increasing IC90 fold change or CR (direction of steepest ascent). Vectors can be used to interpret the change in IC90 for the antimicrobials shown. For both statistical models, the first and second RDA axes shown display the majority of explained variation in IC90 changes. Large gray symbols show centroids (average effect) for all resistant mutants within a given efflux group (shape). The vector tip of relative fitness (brown) is also shown. a The majority of explained variation is driven by primary resistances, where ciprofloxacin (pink)-resistant and mecillinam (green)-resistant mutants cluster distinctly from the other resistance groups, which showed higher relative fitness. b Resistant mutants possessing MdtK mutations alone (diamond) or together with AcrAB-TolC mutations (circle) are likely to show CR to chloramphenicol, ceftazidime, temocillin, and azithromycin, but sensitivity to gentamicin, fosfomycin, and trimethoprim. Whereas those without efflux mutations (triangle) are more likely to display low-level CS or no change to most antimicrobials tested. The analysis of the subset RDA (c, d) shows patterns consistent with the full model, but with less clustering of mutants by resistance group (c). The combination of AcrAB-TolC and MdtK efflux mutations displayed the greatest fitness costs, while mutants lacking efflux-related mutations were the most fit (d). RDA significance was assessed by permutation tests (1000 permutations), where p ≤ 0.05 was considered significant. For more comprehensive multivariate models see Supplementary Fig. 5–6
We initially hypothesized that genetic background would significantly affect collateral responses. Our initial analysis suggests that it does not. Arguably, the inclusion of IC90 data from the drugs to which primary resistance was selected could confound the analysis, despite our efforts to minimize these effects using log-transformed data. We used the same approaches to assess a subset of collateral responses, excluding data for all of the 40 resistant mutants to five antimicrobials containing the drugs used for selection (ciprofloxacin, mecillinam, nitrofurantoin, trimethoprim) and trimethoprim-sulfamethoxazole. Within the subset model, patterns consistent with the full model were observed, but with a lower degree of clustering by resistance group (Fig. 3c). For example, K56-2 CIPR is now co-localized with the mecillinam-resistant isolates, indicating that this isolate is distinct from other ciprofloxacin-resistant mutants (Fig. 3c), which still showed strong tendencies of CR to temocillin, chloramphenicol, ceftazidime, and amoxicillin (Fig. 3c, d). Despite these changes, efflux-type and fitness were still significant predictors of collateral networks, and strain background remained non-significant (Supplementary Table 3) when modeled alone (Supplementary Fig. 5f) and in two-factor combinations (Supplementary Fig. 5n–o), but had a limited, significant contribution (p = 0.040), determined by permutation tests, in the three-factor model (Fig. 3c, d, Supplementary Table 3). However, mutations in efflux-related genes and gene regulators were the strongest predictor of collateral responses tested, explaining over 33% of the variation in the subset. Fitness alone also had significant predictive value, but to a lesser extent (17% variation explained). It is important to note that we observed a correlation between efflux mutations and relative fitness that is likely explained by reduced fitness resulting from the cost of overexpression of efflux pump(s)39.
To investigate the influence of resistance mechanism on IC90 variation at a higher resolution, we modeled each resistance group separately relating the putative resistance mechanism (beyond efflux-type) and fitness separately and in combination (Supplementary Fig. 6a–o). However, potentially due to a lower number of samples within each resistance group that were separated into more detailed classifications of resistance mechanism, these factors had varying degrees of contribution. For mutants resistant to ciprofloxacin (Supplementary Fig. 6a) and trimethoprim (Supplementary Fig. 6j), resistance mechanism was non-significant, but it was a significant factor for those resistant to mecillinam (Supplementary Fig. 6d) and nitrofurantoin (Supplementary Fig. 6g). Fitness was a significant factor only for the mecillinam resistance group (Supplementary Fig. 6e) and similarly, models containing both resistance mechanism and fitness were non-significant for all resistance groups, with the exception of the mecillinam-resistant mutants (Supplementary Fig. 6f).
In the first-step (GyrA) ciprofloxacin mutants, strain background was a significant factor for collateral responses (Supplementary Fig. 6m). However, this was not the case when the original ciprofloxacin-resistant mutants from the same strain backgrounds were added to the analysis (Supplementary Fig. 6n), suggesting again that other factors are more important than strain background. Overall, in comparison to the ciprofloxacin-resistant mutants, collateral responses of first-step mutants were far less frequent and more closely resembled those of the GyrA/ParC mutation-containing K56-2 CIPR mutant. A final redundancy analysis was performed on all ciprofloxacin-resistant and first-step mutants (Supplementary Fig. 6o), and showed a significant effect of resistance mechanism, supporting that mechanism, efflux in particular, is a major driver of collateral responses.
Collateral responses shift the mutation selection window
The mutant selection window can be defined as the concentration space between the lowest antimicrobial concentration that selects for and enriches resistant mutants43 and the concentration that prevents the emergence of first-step resistant mutants, the mutation prevention concentration (MPC)44,45. In theory, if drug concentrations remain above the MPC during treatment, antimicrobial resistance is less likely to evolve44,45. It was recently demonstrated in E. coli MG1655 that changes in MPC correlated with collateral responses in resistant mutants21. We determined the MPC for 17 strain:drug combinations that exemplified conserved collateral responses (Fig. 1b). The MPC for each resistant mutant and its respective wild-type were compared. In 12/17 (70.6%) the change in MPC was consistent with the sign of collateral responses as determined by IC90. This demonstrates that even small CS/CR changes can affect the mutant selection window, correspondingly shifting it down or up. In 4/17 (23.5%) the MPC displayed no change between the wild-type and mutant. This was observed when testing the MPC for mecillinam, trimethoprim, and azithromycin, though we speculate that increasing the precision of the MPC assay (as was done with IC90 testing) might negate these discrepancies. Changes in MPC results with azithromycin were inconsistent with the change in IC90 for a ciprofloxacin-resistant mutant and the mutants resistant to mecillinam and nitrofurantoin, which displayed a decreased MPC instead of an expected increase or no change, respectively.
Discussion
Here, we identify conserved collateral responses in antimicrobial susceptibility across genetically diverse clinical E. coli strains following antimicrobial resistance development. Our findings are relevant beyond urinary-tract infections because uropathogenic E. coli are shown to also stably colonize the bladder and gut46 and to cause bloodstream infections47. Our data show that CS and CR are pervasive in clinical E. coli strains, consistent with earlier results based on laboratory-adapted strains of various species21–23,25,30,48 and a limited number of clinical isolates21,30. Resistance to ciprofloxacin resulted in a greater number of collateral responses than resistance to mecillinam, nitrofurantoin, or trimethoprim. This is likely due to mutations to known regulators of the AcrAB-TolC and MdtK efflux pumps. Both have broad substrate specificities to diverse antimicrobials including fluoroquinolones, β-lactams, tetracycline, chloramphenicol, trimethoprim-sulfamethoxazole, and some macrolides for the AcrAB-TolC efflux pump49,50, and fluoroquinolones, chloramphenicol, trimethoprim, and some β-lactams for the MdtK pump39,51. Interestingly, both overexpression of MdtK51 and RpoB39 mutations (that were linked to MdtK expression) have been shown to reduce susceptibility to fosfomycin, as was observed in the ciprofloxacin-resistant mutants in this study (Fig. 1a). Overall, CR was much more prevalent than CS, and the magnitude of collateral responses were most often small, consistent with other reports21–23. We observed that collateral responses varied substantially by resistance group, but variation was also observed within resistance groups.
Using CS50 and CR50 thresholds to identify conserved responses, we found that conserved CR was more than twice as common as conserved CS. Whereas many of the conserved collateral responses identified in this study support the findings in previous work using single laboratory-adapted strains, we observed several clinically relevant differences. For example, our finding of conserved CS in ciprofloxacin-resistant mutants to gentamicin was previously reported in E. coli K1222 but not in E. coli MG165521. In mutants resistant to ciprofloxacin we also observed conserved CR towards chloramphenicol, as reported in ref. 21, but not in ref. 23. We identified conserved CR of nitrofurantoin-resistant mutants to amoxicillin, and this was not reported in MG165521. These observations underscore the importance of exploring collateral networks in multiple mutants of different clinical strain backgrounds and with different resistance mechanisms to assess their potential clinical application.
Visual inspection of the data revealed a few clinically relevant examples of CS phenotypes that appeared independent of putative mechanism of resistance. We show that E. coli strains resistant to ciprofloxacin display CS towards gentamicin, fosfomycin, ertapenem, and colistin, and these phenotypes were conserved across multiple mechanisms of resistance. These results parallel those of a recent study on P. aeruginosa clinical isolates from cystic fibrosis patients, where resistance to ciprofloxacin was associated with CS to gentamicin, fosfomycin, and colistin31. Taken together these data support the presence of general, conserved collateral networks that may both affect the population dynamics of antimicrobial resistance during treatment and counter-select for resistance, as recently indicated31.
We assumed a priori that genetic background, resistance group, resistance mechanism, and the fitness cost of resistance could potentially affect the generality, sign, and magnitude of collateral networks in clinical E. coli strains. Despite the fact that some collateral responses are conserved across different strains and mechanisms of resistance, our multivariate statistical approaches show overall that mechanism of resistance is the key predictor of CS and CR variability. This is primarily the case for efflux-related mutations. However, mechanism of resistance also significantly contributed to the observed CS and CR variation in the mecillinam mutants where no efflux mutations were found. The presented data are consistent with earlier reports based on multiple resistant mutants derived from single strains with different resistance mechanisms towards specific antimicrobials22,23,52. Our finding that genetic background did not significantly contribute to collateral responses is an important addition to these earlier studies. Finally, we found that the fitness cost of resistance also contributed significantly to the observed variation in CS and CR, despite some collinearity between efflux-related mutations and reduced fitness. Taken together, our data and previous reports indicate that applied use of collateral networks in future treatment strategies may be dependent on rapid identification of specific resistance mechanisms. Moreover, clinical application of CS as a selection inversion strategy warrants further investigations to ideally explore CS in isogenic backgrounds, representing several diverse strains, with permutations of all known antimicrobial resistance-associated traits. Such extensive studies would likely provide valuable information on the mechanisms of CS. Other confounding factors such as mobile genetic elements with heterogeneous resistance determinants should also be investigated as they would likely influence and reduce the predictability of collateral networks.
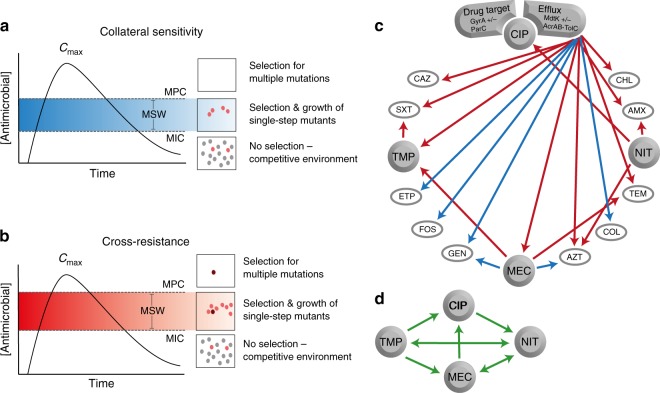
Selection inversion, as described by ref. 21, depends on the cycling of drug pairs that display reciprocal CS. We did not observe reciprocal CS between any of the four drugs studied here that are widely used for treatment of urinary tract infections. However, we asked if modest reductions and increases in antimicrobial susceptibilities would affect the mutant selection window44 for the most prevalent CS and CR phenotypes. We subjected conserved CS and CR phenotypes to MPC assays and revealed that even a small 1.5-fold change in IC90 could equally alter the MPC, resulting in a shift of the mutant selection window (Fig. 1b). These results suggest that antimicrobial treatment strategies informed by collateral networks could affect the evolutionary trajectories of antimicrobial resistance. Sequential treatment using drug pairs that display CR would, following resistance development, shift the mutant selection window towards higher antimicrobial concentrations, as was previously observed53, and increase the likelihood for resistance development to subsequent treatment options (Fig. 4a). Conversely, sequential treatment based on drug pairs that display CS can shift the mutant selection window down and reduce the window of opportunity for high-level resistance development (Fig. 4b). This result suggests that the initial choice of antimicrobial may set the stage for later resistance development.
Fig. 4.
Presentations of the potential effects and implications of CS and CR. a Sequential drug administration informed by CS (blue) could potentially narrow or shift the mutant selection window (MSW) downwards in concentration space whereas (b) CR (red) results in a widened or shifted upwards mutant selection window for secondary antimicrobials. This would affect the probability of acquiring second-step mutations leading to high-level resistance. Consequently, CS-informed secondary therapies could reduce selection and thus propagation of first-step mutants resulting in a reduced opportunity for second-step mutations to occur. Dots represent bacteria resistant to a primary antibiotic (gray), spontaneous mutants with reduced susceptibility to a secondary drug (pink), or those with high-level resistance to the secondary drug (dark red). Note that these are hypothetical schematics and in many cases the maximum concentration achieved (Cmax) may be below the MPC. c Arrows indicate conserved collateral responses, where CS (blue) and CR (red) are depicted. The collateral responses in this study are mainly predicted by efflux-related mutations in the ciprofloxacin-resistant mutants. These data suggest potential secondary treatment options that may reduce the rate of resistance evolution (a, b) following initial treatment failure. d Green arrows indicate putative temporal administration of four antimicrobials used for the treatment of urinary-tract infections, as informed by the collateral networks in (c)
Based on our in vitro findings, trimethoprim and nitrofurantoin are attractive from a clinical perspective, as resistance to these resulted in few collateral responses, preserving the innate sensitivity to available secondary antimicrobials (Fig. 4c, d). However, mecillinam could be even more attractive, as CS largely dominates the observed collateral responses in resistant mutants. Additionally, isolates resistant to mecillinam, especially those evolved in vivo, are associated with high fitness costs42. In contrast, exposure to ciprofloxacin was more likely to cause dramatic collateral responses that depend on the mechanism of resistance and could potentially negatively impact future therapeutic options (Fig. 4c). These observations align with antimicrobial treatment recommendations in Norway, where mecillinam, nitrofurantoin, and trimethoprim are recommended for first-line therapy of uncomplicated urinary-tract infections, and ciprofloxacin is reserved for otherwise complicated infections54. Similarly, in the United States nitrofurantoin, trimethoprim-sulfamethoxazole, and mecillinam are recommended before fluoroquinolones, such as ciprofloxacin, ofloxacin, and levofloxacin55.
Our conclusions are not without limitations. First, we acknowledge that including more clinical isolates from different infection foci, more diverse genetic backgrounds including different bacterial species, as well as other selective agents, could change the outcome of our statistical analyses. This would allow increased sensitivity for the assessment of the different factors controlling collateral responses. A more targeted approach to assess the impact of specific resistance mechanisms on CS and CR across genetically diverse clinical strains is lacking in the field. Our analyses suggest that the fitness cost of resistance explains some variability in the collateral networks reported here. We used relative growth rates as a proxy for relative fitness, and our data are consistent with reports demonstrating that growth rates affect susceptibilities to several antimicrobials56,57. It is unclear if collateral networks will be perturbed by compensatory evolution, which eliminates the fitness costs of primary resistance58–60. Finally, this and previous studies focus on antimicrobial resistance development to a single drug, and there is a complete lack of data on how multidrug resistance, including resistance genes on mobile genetic elements, will affect collateral networks. We are currently investigating these and other questions that will aid in our understanding of collateral networks and their potential therapeutic application.
Methods
Bacterial strains
We used 10 clinical, urinary-tract infection isolates of E. coli from the ECO-SENS collections61,62 originating from countries across Europe between 2000 and 2008 (Table 1). The isolates were chosen to represent pan-susceptible strains with diverse genetic backgrounds and were reported plasmid-free32. Subsequent analysis based on whole-genome sequencing discovered two changes to previously reported sequence types and the presence of plasmid replicons in three strains (Supplementary Table 4). E. coli ATCC 25922 was used for reference and quality control purposes. For general growth, bacterial strains were grown in either Miller Difco Luria–Bertani (LB) broth (Becton, Dickinson and Co., Sparks, MD, USA) or on LB agar; LB broth with select agar (Sigma-Aldrich) at 15 g L−1 and incubated at 37 °C.
Selection of antimicrobial-resistant mutants
Single antimicrobial-resistant mutants were selected at drug concentrations above the European Committee on Antimicrobial Susceptibility Testing (EUCAST) clinical breakpoints63 for ciprofloxacin, nitrofurantoin, trimethoprim, and mecillinam (Supplementary Table 1). Briefly, 100 µL of 10× concentrated overnight culture was spread on Mueller Hinton II agar (MHA-SA; Sigma-Aldrich) plates containing ciprofloxacin (Biochemika), nitrofurantoin (Sigma-Aldrich), or trimethoprim (Sigma-Aldrich) with two-fold increasing concentrations of the antimicrobial. After 24–48 h, a mixture of growth from the highest concentration plate with multiple colonies was used to start a new overnight culture at the same antimicrobial concentration. This was repeated until there was growth above the clinical breakpoint63. Mecillinam-resistant mutants and first-step ciprofloxacin mutants (gyrA mutation-containing) were selected as single-step mutants on LB or MHA-SA agar, respectively. Mutants were confirmed as E. coli using matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) analysis with MALDI BioTyper software (Bruker, MA, USA).
Antimicrobial susceptibility testing
Mutants were initially screened for resistance above EUCAST breakpoints63 with gradient diffusion strips following manufacturers guidelines (Liofilchem, Italy), on Mueller Hinton II agar (MHA-BD; Becton, Dickinson and Company). Plates with insufficient growth were incubated for an additional 24 h.
To maximize the precision of our susceptibility estimates and in accord with related studies on CS, collateral changes were determined by IC90 testing21 with some modifications. IC90 values were determined following 18 h incubation at 700 rpm (3 mm stroke) in Mueller Hinton Broth (MHB, Becton, Dickinson and Co.). Slow-growing strains where positive growth controls did not reach OD600 nm of 0.3 after 18 h (i.e. K56-12 CIPR) were interpreted after 42 h incubation. Standard two-fold concentrations and the median values between them were used as a 1.5-fold testing scale. IC90 values were the lowest concentration tested that resulted in ≥90% inhibition of growth. Percent inhibition was calculated compared to the positive control (untreated sample) with background removed21. IC90 results were determined in at least three biological replicates on separate days. The final result reflects the average of a minimum of three replicates that met quality control standards, including the result of ATCC 25922 on each plate, positive growth control OD600 nm > 0.3, negative growth control OD600 nm < 0.05, and accepting no more than one skip (defined as a break in the inhibition pattern). When one skip was observed, the IC90 value was consistently interpreted as the lowest concentration tested that resulted in ≥90% inhibition of growth. Fold change in IC90 was calculated as the ratio between the resistant mutant and its respective wild-type. The IC90 testing varied for two antimicrobials (according to EUCAST recommendations), where fosfomycin was tested in MHB supplemented with 25 µg mL−1 glucose-6-phosphate (Sigma-Aldrich) and tigecycline was tested in fresh MHB media that was prepared daily.
Dose–response curves were generated with average OD600 values (background subtracted) for concentrations tested during IC90 testing. Averages were plotted for mutants and respective wild-type strains.
MPC testing
We determined the MPC for 17 of 20 conserved collateral responses. Temocillin was excluded due to lack of supply and trimethoprim-sulfamethoxazole was excluded for trimethoprim-resistant isolates due to fundamental CR between trimethoprim and trimethoprim-sulfamethoxazole. MPC determination was based on previous work by Marcusson et al.64. Briefly, 10 mL overnight culture was centrifuged and the pellet re-suspended in 1 mL MHB, estimated to contain ≥1010 CFU (actual values were 1.4 × 1010–7 × 1010 CFU). The inoculum was split and spread onto four large (14 cm diameter) MHA-SA agar plates for each antimicrobial concentration tested in a two-fold dilution series. The MPC was the lowest concentration with no visible growth after 48 h. Where growth/no growth was difficult to interpret, suspected growth was re-streaked on plates at the same antimicrobial concentration. Azithromycin and ertapenem were regularly inconsistent, making re-streaking essential. Resistant mutants and wild-types were tested in parallel, and results represent the average of at least two biological replicates.
Growth rate measurements
To obtain growth curves of wild-types and resistant mutants, single colonies were used to inoculate at least three biological replicates of MHB starter cultures (2 mL) that were incubated at 37 °C for 24 h shaking at 500 rpm. Each culture was diluted 1:100 in MHB (resulting in ~2 × 107 cell mL−1) and 250 µL was added in triplicate to a 96-well microtiter plate. The plate was incubated overnight at 37 °C in a Versamax plate reader (Molecular Devices Corporation, California, USA) with shaking for 9.2 min between reads. OD600 measurements were taken every 10 min and growth rates were estimated using the GrowthRates v.2.1 software65. To a varying extent, the ciprofloxacin-resistant mutants of K56-12, K56-16, K56-44, and K56-68, as well as, K56-44 MECR, K56-68 MECR, and K56-70 TMPR displayed noise in the growth curves due to clumping or non-homogeneous growth, and GrowthRates was unable to fit a line with R-value above the 0.98 cut-off value. Additional experiments and visual inspection of the log-transformed OD600 values were used to solve this issue. If GrowthRates failed to analyze the curves, a line was fitted within the log phase through at least five consecutive points that displayed log-linear growth. Growth rate (r) was calculated based on the slope65. Relative growth rates were calculated as r(resistant mutant) r(wild-type)−1.
Whole genome sequencing
Genomic DNA was isolated using the GenElute Bacterial Genomic DNA kit (Sigma-Aldrich) following guidelines for Gram-positive DNA extraction. Purity and quantification was determined with Nanodrop (Thermo Scientific) and Qubit High Sensitivity DNA assay (Life Technologies), respectively. For library preparation of wild-types and resistant mutants, 1 µg of DNA was sheared on a Covaris S2 to ≈400 bp using the recommended settings (intensity: 4, duty cycle: 10%, cycles per burst: 200, treatment time: 55 s). Libraries were then prepared and indexed using the DNA Ultra II Library Preparation Kit (New England Biolabs, E7645). For library preparation of DNA from first-step ciprofloxacin mutants, 1 ng of DNA was used with the Nextera XT DNA prep kit (Illumina, San Diego), according to the producer’s instructions. All libraries were quantified by Qubit High Sensitivity DNA assay and distributions and quality assessed by Bioanalyser DNA 1000 Chip (Agilent, 5067-1504) before normalizing and pooling. Illumina sequencing, NextSeq 550, and MiSeq, was used for the first-step ciprofloxacin mutants and the wild-types and resistant mutants, respectively, using paired-end reads. For NextSeq 550 a mid-output flowcell with 300 cycles was used. V2 chemistry was used for the MiSeq sequencing.
Wild-type genomes were assembled, as follows. Illumina adapters on wild-type reads were removed with Trimmomatic version 0.3666 using standard settings, then assembled with SPAdes67. Contigs less than 500 bp in length or less than 2.0 coverage were removed. Wild-type genome assemblies were inspected and compared to the E. coli MG1655 genome (GenBank U00096.2) using QUAST68. Final assembled genomes of wild-type strains were annotated using the automated Prokaryotic Genome Annotation Pipeline (https://www.ncbi.nlm.nih.gov/genome/annotation_prok/).
Wild-type and mutant sequences were compared to identify putative resistance mechanisms. First, wild-type genomes were annotated with Rapid Annotation using Subsystem Technology server (RAST, version 2.0) for E. coli69. SeqMan NGen (DNASTAR, Madison, WI) was used to align raw mutant reads to the corresponding, annotated wild-type genomes, using standard settings. Reported SNPs had ≥10× coverage depth and ≥90% variant base calls. SNPs present in the wild-type assembly or in at least two mutants resistant to different antimicrobials from the same strain background were excluded. Reported SNPs, indels, and rearrangements were manually inspected and gene annotations confirmed using Gene Construction Kit (Textco Biosoftware Inc., Raleigh, NC) and NCBI BLAST searches, respectively.
Multilocus sequence typing (MLST), as well as, plasmid replicon and acquired antimicrobial resistance gene content were determined for the wild-type genomes using MLST version 1.8, PlasmidFinder version 1.3, and ResFinder version 3.070, respectively (http://genomicepidemiology.org/). The MLST of each wild-type strain was confirmed compared to the original reported sequence type32 for all but two strains, K56-41 and K56-70. These strains were originally described as ST420 and ST550, but were ST73 and ST537 in our analysis, respectively (Supplementary Table 4). The wild-type strains were previously described as plasmid free, but we identified two small plasmid replicons, Col156 in K56-16 and K56-44 and Col(MP18) in K56-75 (Supplementary Table 4). Using ResFinder, we detected only one acquired genetic element, sul2 (linked to sulfonamide resistance) in K56-44, and two point mutations, PmrB V161G in K56-50 and K56-70 and ParE D475E in K56-78, that are linked to colistin and quinolone (ciprofloxacin) resistance, respectively (Supplementary Table 4). Though all of the wild-type strains were phenotypically pan-susceptible (Supplementary Fig. 1c), these resistance determinants could affect antimicrobial susceptibilities differentially in the presence of other mutations71,72.
To assess genetic diversity, a phylogenic tree was generated based on the genome sequences of wild-type strains. Assembled wild-type genomes were annotated with PROKKA73, the core gene-encoding regions were extracted and compared using ROARY74, and a maximum-likelihood tree with 100 bootstraps was generated using RAxML75. Genetic distances were calculated in R76 using previously described methods77.
Multivariate statistical analyses
The fold changes of mean IC90 values relative to the parental wild-type strain (collateral responses) were log transformed. Statistical analyses were performed on the complete data set, as well as a subset of the data excluding five antimicrobials (ciprofloxacin, mecillinam, nitrofurantoin, trimethoprim, and trimethoprim-sulfamethoxazole). To estimate and test the effects of strain background, resistance group, resistance mechanism, growth rate, and relative fitness we relied on multivariate modeling (redundancy analysis) to address the co-variation in IC90 across antimicrobials. A redundancy analysis is a constrained version of a principle component analysis that additionally allows for hypothesis testing. Linear constraint scores were plotted for each mutant. Response variables were overlaid with independent scaling to illustrate the direction of steepest ascent (increasing CR) from the origin for each antimicrobial. Data were inspected to check whether the assumptions underlying redundancy analysis were met. Significance testing of multivariate models and their factors (Supplementary Table 3) was done via permutation tests (1000 permutations), an approach robust to deviations from multivariate normality and variance homogeneity; p < 0.05 was considered significant. Analyses were done in R76 using the Vegan work package78.
Electronic supplementary material
Description of Additional Supplementary Information
Acknowledgements
We thank Tony Brooks at UCL Genomics and Hagar Taman at UiT Genomics Support Centre Tromsø for genome sequencing, Ane L.G. Utnes for generation of nitrofurantoin-resistant mutants, Jessica N. Tran for generation of first-step ciprofloxacin mutants, Jessin Janice for phylogenetic analysis, Søren Overballe-Petersen for preliminary processing of wild-type genomes, and Maria Chiara Di Luca for analysis of the genomes for MLST and identification of plasmid replicons. Funding for this project was provided through the Northern Norway Regional Health Authority and UiT—The Arctic University of Norway (Project SFP1292-16), and JPI-EC-AMR (Project 271176/H10).
Author contributions
P.J.J. and Ø.S. conceived the project; N.L.P., E.G.A.F., J.K. and V.S. designed and performed experiments; R.P. designed and R.P. and N.L.P. performed the multivariate statistical modeling; all authors analyzed, interpreted and discussed the data; and N.L.P., E.G.A.F., A.P.R., D.E.R. and P.J.J. wrote the manuscript with contributions from the other authors.
Data availability
Whole-genome sequencing data are available at NCBI (BioProject PRJNA419689). All other relevant data are available within this article, the Supplementary Information, or from the corresponding author upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Nicole L. Podnecky, Email: nicole.podnecky@uit.no
Pål J. Johnsen, Email: paal.johnsen@uit.no
Electronic supplementary material
Supplementary Information accompanies this paper at 10.1038/s41467-018-06143-y.
References
- 1.Holmes AH, et al. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet. 2016;387:176–187. doi: 10.1016/S0140-6736(15)00473-0. [DOI] [PubMed] [Google Scholar]
- 2.Cantas L, et al. A brief multi-disciplinary review on antimicrobial resistance in medicine and its linkage to the global environmental microbiota. Front. Microbiol. 2013;4:96. doi: 10.3389/fmicb.2013.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson DI, Hughes D. Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 2014;12:465–478. doi: 10.1038/nrmicro3270. [DOI] [PubMed] [Google Scholar]
- 4.Silver LL. Challenges of antibacterial discovery. Clin. Microbiol. Rev. 2011;24:71–109. doi: 10.1128/CMR.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walsh C. Where will new antibiotics come from? Nat. Rev. Microbiol. 2003;1:65–70. doi: 10.1038/nrmicro727. [DOI] [PubMed] [Google Scholar]
- 6.Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science. 2009;325:1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies J. Where have all the antibiotics gone? Can. J. Infect. Dis. Med. Microbiol. 2006;17:287–290. doi: 10.1155/2006/707296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Infectious Diseases Society of America. The 10 x ‘20 initiative: pursuing a global commitment to develop 10 new antibacterial drugs by 2020. Clin. Infect. Dis. 2010;50:1081–1083. doi: 10.1086/652237. [DOI] [PubMed] [Google Scholar]
- 9.World Health Organization. WHO Global Strategy of Containment of Antimicrobial Resistance. WHO/CDS/CSR/DRS/2001.2 (World Health Organization: Geneva, Switzerland, 2001).
- 10.O’Neill J. Tackling Drug-resistant Infections Globally: Final Report and Recommendations. https://amr-review.org/Publications.html (2016).
- 11.Centers for Disease Control and Prevention (U.S.). The Core Elements of Hospital Antibiotic Stewardship Programs. CS273578-A (Centers for Disease Control and Prevention (U.S.): Atlanta, GA, 2014). [DOI] [PMC free article] [PubMed]
- 12.Harbarth, S. et al. Antimicrobial resistance: one world, one fight! Antimicrob Resist Infect Control4. 10.1186/s13756-015-0091-2 (2015).
- 13.High-Level Meeting on Antimicrobial Resistance [press release]. New York, NY, USA: OPGA/WHO/FAO/OIE, 21 Sept 2016. https://www.un.org/pga/71/2016/09/21/press-release-hl-meeting-on-antimicrobial-resistance.
- 14.Johnsen PJ, et al. Factors affecting the reversal of antimicrobial-drug resistance. Lancet Infect. Dis. 2009;9:357–364. doi: 10.1016/S1473-3099(09)70105-7. [DOI] [PubMed] [Google Scholar]
- 15.Johnsen PJ, et al. Retrospective evidence for a biological cost of vancomycin resistance determinants in the absence of glycopeptide selective pressures. J. Antimicrob. Chemother. 2011;66:608–610. doi: 10.1093/jac/dkq512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enne VI, Livermore DM, Stephens P, Hall LMC. Persistence of sulphonamide resistance in Escherichia coli in the UK despite national prescribing restriction. Lancet. 2001;357:1325–1328. doi: 10.1016/S0140-6736(00)04519-0. [DOI] [PubMed] [Google Scholar]
- 17.Sundqvist M, et al. Little evidence for reversibility of trimethoprim resistance after a drastic reduction in trimethoprim use. J. Antimicrob. Chemother. 2010;65:350–360. doi: 10.1093/jac/dkp387. [DOI] [PubMed] [Google Scholar]
- 18.Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat. Rev. Microbiol. 2010;8:260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 19.Baym M, Stone LK, Kishony R. Multidrug evolutionary strategies to reverse antibiotic resistance. Science. 2016;351:aad3292. doi: 10.1126/science.aad3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szybalski W, Bryson V. Genetic studies on microbial cross resistance to toxic agents. I. Cross resistance of Escherichia coli to fifteen antibiotics. J. Bacteriol. 1952;64:489–499. doi: 10.1128/jb.64.4.489-499.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imamovic L, Sommer MO. Use of collateral sensitivity networks to design drug cycling protocols that avoid resistance development. Sci. Transl. Med. 2013;5:204ra132. doi: 10.1126/scitranslmed.3006609. [DOI] [PubMed] [Google Scholar]
- 22.Lazar V, et al. Bacterial evolution of antibiotic hypersensitivity. Mol. Syst. Biol. 2013;9:700. doi: 10.1038/msb.2013.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazar V, et al. Genome-wide analysis captures the determinants of the antibiotic cross-resistance interaction network. Nat. Commun. 2014;5:4352. doi: 10.1038/ncomms5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanders CC, Sanders WEJ, Goering RV, Werner V. Selection of multiple antibiotic resistance by quinolones, β-lactams, and aminoglycosides with special reference to cross-resistance between unrelated drug classes. Antimicrob. Agents Chemother. 1984;26:797–801. doi: 10.1128/AAC.26.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzales PR, et al. Synergistic, collaterally sensitive beta-lactam combinations suppress resistance in MRSA. Nat. Chem. Biol. 2015;11:855–861. doi: 10.1038/nchembio.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macvanin M, Hughes D. Hyper-susceptibility of a fusidic acid-resistant mutant of Salmonella to different classes of antibiotics. FEMS Microbiol. Lett. 2005;247:215–220. doi: 10.1016/j.femsle.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 27.Barbosa C, et al. Alternative evolutionary paths to bacterial antibiotic resistance cause distinct collateral effects. Mol. Biol. Evol. 2017;34:2229–2244. doi: 10.1093/molbev/msx158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S, Lieberman TD, Kishony R. Alternating antibiotic treatments constrain evolutionary paths to multidrug resistance. Proc. Natl Acad. Sci. USA. 2014;111:14494–14499. doi: 10.1073/pnas.1409800111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuentes-Hernandez A, et al. Using a sequential regimen to eliminate bacteria at sublethal antibiotic dosages. PLoS Biol. 2015;13:e1002104. doi: 10.1371/journal.pbio.1002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jansen G, et al. Association between clinical antibiotic resistance and susceptibility of Pseudomonas in the cystic fibrosis lung. Evol. Med. Public Health. 2016;2016:182–194. doi: 10.1093/emph/eow016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imamovic L, et al. Drug-driven phenotypic convergence supports rational treatment strategies of chronic infections. Cell. 2018;172:121–134.e114. doi: 10.1016/j.cell.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bengtsson S, Naseer U, Sundsfjord A, Kahlmeter G, Sundqvist M. Sequence types and plasmid carriage of uropathogenic Escherichia coli devoid of phenotypically detectable resistance. J. Antimicrob. Chemother. 2012;67:69–73. doi: 10.1093/jac/dkr421. [DOI] [PubMed] [Google Scholar]
- 33.Munck C, Gumpert HK, Wallin AI, Wang HH, Sommer MO. Prediction of resistance development against drug combinations by collateral responses to component drugs. Sci. Transl. Med. 2014;6:262ra156. doi: 10.1126/scitranslmed.3009940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munck, C., Sommer, M. Antibiotic Resistance: Adaptive Evolution & Dissemination of Resistance Genes. http://orbit.dtu.dk/en/publications/antibiotic-resistance(9d3346b1-8519-4167-848d-5407bf2fcd6c).html (Department of Systems Biology, Technical University of Denmark, Denmark, 2014).
- 35.Kushner B, Allen PD, Crane BT. Frequency and demographics of gentamicin use. Otol. Neurotol. 2016;37:190–195. doi: 10.1097/MAO.0000000000000937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karageorgopoulos DE, Wang R, Yu XH, Falagas ME. Fosfomycin: evaluation of the published evidence on the emergence of antimicrobial resistance in Gram-negative pathogens. J. Antimicrob. Chemother. 2012;67:255–268. doi: 10.1093/jac/dkr466. [DOI] [PubMed] [Google Scholar]
- 37.Dijkmans, A. C. et al. Fosfomycin: pharmacological, clinical and future perspectives. Antibiotics (Basel)6. 10.3390/antibiotics6040024 (2017). [DOI] [PMC free article] [PubMed]
- 38.Wang H, Dzink-Fox JL, Chen M, Levy SB. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob. Agents Chemother. 2001;45:1515–1521. doi: 10.1128/AAC.45.5.1515-1521.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pietsch F, et al. Ciprofloxacin selects for RNA polymerase mutations with pleiotropic antibiotic resistance effects. J. Antimicrob. Chemother. 2016;72:75–84. doi: 10.1093/jac/dkw364. [DOI] [PubMed] [Google Scholar]
- 40.Kern WV, Oethinger M, Jellen-Ritter AS, Levy SB. Non-target gene mutations in the development of fluoroquinolone resistance in Escherichia coli. Antimicrob. Agents Chemother. 2000;44:814–820. doi: 10.1128/AAC.44.4.814-820.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huseby DL, et al. Mutation supply and relative fitness shape the genotypes of ciprofloxacin-resistant Escherichia coli. Mol. Biol. Evol. 2017;34:1029–1039. doi: 10.1093/molbev/msx052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thulin E, Sundqvist M, Andersson DI. Amdinocillin (Mecillinam) resistance mutations in clinical isolates and laboratory-selected mutants of Escherichia coli. Antimicrob. Agents Chemother. 2015;59:1718–1727. doi: 10.1128/AAC.04819-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gullberg E, et al. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 2011;7:e1002158. doi: 10.1371/journal.ppat.1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Drlica K, Zhao X. Mutant selection window hypothesis updated. Clin. Infect. Dis. 2007;44:681–688. doi: 10.1086/511642. [DOI] [PubMed] [Google Scholar]
- 45.Liang B, et al. Mutant prevention concentration-based pharmacokinetic/pharmacodynamic indices as dosing targets for suppressing the enrichment of levofloxacin-resistant subpopulations of Staphylococcus aureus. Antimicrob. Agents Chemother. 2011;55:2409–2412. doi: 10.1128/AAC.00975-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nielsen KL, et al. Adaptation of Escherichia coli traversing from the faecal environment to the urinary tract. Int. J. Med. Microbiol. 2016;306:595–603. doi: 10.1016/j.ijmm.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Subashchandrabose, S., Mobley, H. L. Virulence and fitness determinants of uropathogenic Escherichia coli. Microbiol. Spectr.3. 10.1128/microbiolspec.UTI-0015-2012 (2015). [DOI] [PMC free article] [PubMed]
- 48.Pal C, Papp B, Lazar V. Collateral sensitivity of antibiotic-resistant microbes. Trends Microbiol. 2015;23:401–407. doi: 10.1016/j.tim.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poole K. Efflux-mediated multiresistance in Gram-negative bacteria. Clin. Microbiol. Infect. 2004;10:12–26. doi: 10.1111/j.1469-0691.2004.00763.x. [DOI] [PubMed] [Google Scholar]
- 50.Elkins CA, Mullis LB. Substrate competition studies using whole-cell accumulation assays with the major tripartite multidrug efflux pumps of Escherichia coli. Antimicrob. Agents Chemother. 2007;51:923–929. doi: 10.1128/AAC.01048-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nishino K, Yamaguchi A. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 2001;183:5803–5812. doi: 10.1128/JB.183.20.5803-5812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiao, Y. J., Baym, M., Veres, A., Kishony, R. Population diversity jeopardizes the efficacy of antibiotic cycling. Preprint at htt//www.biorxiv.org/content/early/2016/10/20/082107 (2016).
- 53.Li X, Mariano N, Rahal JJ, Urban CM, Drlica K. Quinolone-resistant Haemophilus influenzae: determination of mutant selection window for ciprofloxacin, garenoxacin, levofloxacin, and moxifloxacin. Antimicrob. Agents Chemother. 2004;48:4460–4462. doi: 10.1128/AAC.48.11.4460-4462.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Norwegian Directorate of Health. Antibiotic use in Norwegian Primary Healthcare. National Guidelines, Version 1.3http://www.antibiotikaiallmennpraksis.no (2012).
- 55.Gupta K, et al. International clinical practice guidelines for the treatment of acute uncomplicated cystitis and pyelonephritis in women: a 2010 update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious Diseases. Clin. Infect. Dis. 2011;52:e103–e120. doi: 10.1093/cid/ciq257. [DOI] [PubMed] [Google Scholar]
- 56.Tuomanen E, Cozens R, Tosch W, Zak O, Tomasz A. The rate of killing of Escherichia coli by beta-lactam antibiotics is strictly proportional to the rate of bacterial growth. J. Gen. Microbiol. 1986;132:1297–1304. doi: 10.1099/00221287-132-5-1297. [DOI] [PubMed] [Google Scholar]
- 57.Greulich P, Scott M, Evans MR, Allen RJ. Growth-dependent bacterial susceptibility to ribosome-targeting antibiotics. Mol. Syst. Biol. 2015;11:796–796. doi: 10.15252/msb.20145949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schrag SJ, Perrot V, Levin BR. Adaptation to the fitness costs of antibiotic resistance in Escherichia coli. Proc. R. Soc. Lond. B. 1997;264:1287–1291. doi: 10.1098/rspb.1997.0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Björkman J, Nagaev I, Berg OG, Hughes D, Andersson DI. Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science. 2000;287:1479-–11482. doi: 10.1126/science.287.5457.1479. [DOI] [PubMed] [Google Scholar]
- 60.Starikova I, et al. A trade-off between the fitness cost of functional integrases and long-term stability of integrons. PLoS Pathog. 2012;8:e1003043. doi: 10.1371/journal.ppat.1003043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kahlmeter G. The ECO.SENS project: a prospective, multinational, multicentre epidemiological survey of the prevalence and antimicrobial susceptibility of urinary tract pathogens—interim report. J. Antimicrob. Chemother. 2000;46(Suppl. 1):15–22. doi: 10.1093/jac/46.suppl_1.15. [DOI] [PubMed] [Google Scholar]
- 62.Kahlmeter G, Poulsen HO. Antimicrobial susceptibility of Escherichia coli from community-acquired urinary tract infections in Europe: the ECO.SENS study revisited. Int. J. Antimicrob. Agents. 2012;39:45–51. doi: 10.1016/j.ijantimicag.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 63.The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 7.1. http://www.eucast.org (2017).
- 64.Marcusson LL, Olofsson SK, Komp Lindgren P, Cars O, Hughes D. Mutant prevention concentrations of ciprofloxacin for urinary tract infection isolates of Escherichia coli. J. Antimicrob. Chemother. 2005;55:938–943. doi: 10.1093/jac/dki136. [DOI] [PubMed] [Google Scholar]
- 65.Hall BG, Acar H, Nandipati A, Barlow M. Growth rates made easy. Mol. Biol. Evol. 2014;31:232–238. doi: 10.1093/molbev/mst187. [DOI] [PubMed] [Google Scholar]
- 66.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aziz RK, et al. The RAST server: rapid annotations using subsystems technology. BMC Genom. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zankari E, et al. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Komp Lindgren P, Karlsson A, Hughes D. Mutation rate and evolution of fluoroquinolone resistance in Escherichia coli isolates from patients with urinary tract infections. Antimicrob. Agents Chemother. 2003;47:3222–3232. doi: 10.1128/AAC.47.10.3222-3232.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moon DC, et al. Emergence of a new mutation and its accumulation in the topoisomerase IV gene confers high levels of resistance to fluoroquinolones in Escherichia coli isolates. Int. J. Antimicrob. Agents. 2010;35:76–79. doi: 10.1016/j.ijantimicag.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 73.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 74.Page AJ, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2016. [Google Scholar]
- 77.Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 78.Dixon P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003;14:927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Information
Data Availability Statement
Whole-genome sequencing data are available at NCBI (BioProject PRJNA419689). All other relevant data are available within this article, the Supplementary Information, or from the corresponding author upon request.