Abstract
Handling and consumption of chicken meat are risk factors for human campylobacteriosis. This study was performed to describe the Campylobacter population in broiler carcasses and environmental samples throughout the slaughter process. Moreover, the genetic diversity and antimicrobial resistance of the Campylobacter strains were evaluated. Cloacal swabs, samples from carcasses at different stages, and environmental samples were collected thrice from the different flocks at the same abattoir located in Central Jiangsu, China. Campylobacter isolated from the three batches (n = 348) were identified as Campylobacter jejuni (n = 117) and Campylobacter coli (n = 151) by multiplex PCR. Characterization by multilocus sequence typing revealed a specific genotype of Campylobacter for each batch. Antimicrobial sensitivity to 18 antibiotics were analyzed for all selected strains according to the agar diffusion method recommended by the Clinical and Laboratory Standards Institute. Antibiotic susceptibility tests indicated that the majority of the tested isolates were resistant to quinolones (>89.7%). Less resistance to macrolide (59.8%), gentamicin (42.7%), amikacin (36.8%) was observed. Results showed that 94.0% of the tested strains demonstrated multidrug resistance.
Keywords: Campylobacter, broiler, slaughter process, prevalence, MLST, antibiotic susceptibility
Introduction
Campylobacter is a leading cause of bacterial foodborne infections in developed countries. This infection has surpassed Salmonella several years ago and caused a significant economic burden (EFSA and ECDC, 2016). Although new species of Campylobacter have recently been discovered, human campylobacteriosis are dominated by two main species, Campylobacter jejuni and Campylobacter coli (Tresse et al., 2017). Campylobacter infection causes watery diarrhea, abortion, human acute enteritis, and several complications, such as Guillain-Barré syndrome and Reiter’s syndrome, in severe cases. Handling and consumption of poultry are the major sources for human infection (Boysen et al., 2014). Reducing the prevalence or number of Campylobacter in broilers at the primary stage could be an effective way to protect public health from Campylobacter infections (European Food Safety Authority [EFSA], 2010). However, despite the many biosecurity interventions at the farm, Campylobacter has not been well controlled in broiler flocks after the rearing period (Newell et al., 2011).
During slaughter, many opportunities may facilitate cross-contamination and spread of bacteria despite good hygiene (Althaus et al., 2017). Poultry meat from Campylobacter-negative flocks may be contaminated by previously slaughtered Campylobacter-positive flocks (Miwa et al., 2003; Allen et al., 2007). Campylobacter can be spread to the poultry meat in the slaughter line, especially after evisceration or from dirty surfaces (Corry and Atabay, 2001; Melero et al., 2012). The prevalence of Campylobacter within positive flocks at slaughter are high (approximately 80%) (Colles et al., 2010; European Food Safety Authority [EFSA], 2010). However, in China, few studies have been conducted on the contamination of broiler carcasses throughout the production chain. During slaughter, Campylobacter could also be recovered in the processing equipment and environmental samples (Berrang et al., 2000; Cason et al., 2007; Ellerbroek et al., 2010). Studies have shown that some Campylobacter strains recovered from the slaughterhouse environment can contaminate carcasses when several batches of poultry are slaughtered (Peyrat et al., 2008; Melero et al., 2012). Previous studies only assessed the slaughtering performance to identify operations that increase or decrease the contamination of carcasses (Habib et al., 2012; Seliwiorstow et al., 2015). Moreover, the possibility of Campylobacter in plants as a continuous source of contamination is still ambiguous (García-Sánchez et al., 2017).
Campylobacter outbreaks are sporadic and caused by cross-contamination, and these characteristics hamper the determination of the sources of contamination. Molecular methods play an important role in the epidemiological study of tracing sources and routing of pathogen transmission. Pulsed-field gel electrophoresis and multilocus sequence typing (MLST) have been employed successfully for the epidemiological study of Campylobacter from different sources and outbreaks (Dingle et al., 2005; Michaud et al., 2005; Thakur et al., 2009). The resistance of Campylobacter to antibiotics has also been a persistent issue generally related to the indiscriminate use of antibiotics for therapy or as a growth promoter (Chen et al., 2010; Rozynek et al., 2013). Determining the drug resistance of Campylobacter strains is important to control and prevent human infection. In China, molecular subtyping and antimicrobial susceptibilities of Campylobacter strains from different sources have been conducted (Zhang et al., 2014; Zeng et al., 2016). However, limited data are available on the comprehensive molecular characterization and antibiotic susceptibility of Campylobacter from broilers during slaughter.
This study was performed to determine the prevalence of Campylobacter in the slaughter and poultry processing environment. Moreover, the genotype characteristics and antibiotic susceptibility of these strains were assessed.
Materials and Methods
Sample Collection
Samples were collected from a slaughterhouse located in Central Jiangsu, China. This slaughterhouse processes 20,000 broilers per day. We selected three batches (birds from one flock slaughtered at the same day) in different farms from April 2017 to November 2017. During each visit, cloacal samples were individually collected using sterile swabs before slaughter. Then, samples from the broiler carcasses were collected after plucking, evisceration, washing, and chilling. Water from the cleaning pool and swabs from the operating table and workers’ gloves were also collected as environmental samples. Furthermore, sample collection was performed consecutively during 1 h of the slaughter process. All cloacal swabs were placed in Cary-Blair medium. Swab samples taken at different points were collected using cotton swabs moistened with sterile saline and stored in aseptic bags. Water samples were placed in sterile plastic containers. All samples were transported to the laboratory under cool conditions within 3 h and analyzed the same day. The number of samples collected at different points are shown in Table 1.
Table 1.
Contamination ratio of Campylobacter during slaughter process in three batches.
| Sample site | No. of Campylobacter positive samples /total no. of samples (%) |
|||||
|---|---|---|---|---|---|---|
| Batch 1 (Farm 1) |
Batch 2 (Farm 2) |
Batch 3 (Farm 3) |
||||
| C. jejuni | C.coli | C. jejuni | C. coli | C. jejuni | C. coli | |
| Cloacal swabs | 19/30 (63.3%) | 0 | 0 | 20/20 (100.0%) | 0 | 3/22 (13.6%) |
| After plucking | 22/30 (73.3%) | 0 | 0 | 18/21 (85.7%) | 4/15 (26.7%) | 10/15 (66.7%) |
| After evisceration | 24/30 (80.0%) | 0 | 0 | 17/22 (77.3%) | 0 | 12/15 (80.0%) |
| After washing | 23/30 (76.7%) | 0 | 0 | 20/21 (95.2%) | 3/20 (15.0%) | 11/20 (55.0%) |
| After chilling | 7/8 (87.5%) | 0 | 1/10 (10.0%) | 9/10 (90.0%) | 0 | 10/10 (100.0%) |
| Operating table | 4/5 (80.0%) | 0 | 0 | 5/5 (100.0%) | 1/10 (10.0%) | 9/10 (90.0%) |
| Workers’ gloves | 3/5 (60.0%) | 0 | 1/5 (20.0%) | 4/5 (80.0%) | 2/5 (40.0%) | 2/5 (40.0%) |
| Water | 3/3 (100.0%) | 0 | 0 | 1/3 (33.3%) | 0 | 0/3/0 |
| Total | 105/141 (74.5%) | 96/107 (89.7%) | 67/100 (67.0%) | |||
Campylobacter Isolation and Identification
All cloacal swab samples were placed in 1 mL of PBS (phosphate buffer saline) for full immersion. Then, the swabs were removed from the solution. After 10 times dilution, 100 μL of each solution was placed on modified charcoal-cefoperazone-deoxycholate agar (mCCDA) (Oxoid CM0739) with cefoperazone (LKT, C1630), amphotericin B (Wako, 011-1363), and rifampicin (Wako, 185-01003). All plates were incubated at 42°C for 36–42 h in a microaerophilic environment (5% O2, 10% CO2, and 85% N2).
Cotton swabs from carcasses and environment were enriched in 10 mL of Bolton Broth (BB, CM0983, Oxoid; supplemented with BB Selective Supplement, SR0183E, Oxoid) based on previous reports (Adzitey et al., 2012; Tissier et al., 2012). Water from the cleaning pool were filtered through filters with a 0.22 μm pore diameter. Then the membranes were introduced into 10 mL of BB. After 24 h of incubation at 42°C in glass jars under a microaerobic atmosphere, 10 μL of the resulting solutions were streaked onto mCCDA plates, and the plates were incubated at 42°C for 48 h.
After incubation, the suspected colonies were picked and subcultured in Mueller–Hinton (MH) agar (Difco, MD) with blood and incubated for 24–48 h at 42°C under microaerobic conditions. Multiplex PCR tests were used to confirm and identify whether the strains were Campylobacter jejuni or Campylobacter coli according to a previous study (Huang et al., 2009).
Antibiotic Susceptibility Testing
Susceptibility of Campylobacter to 18 antibiotics (Oxoid) from six classes of antibiotics was determined using the following antimicrobial impregnated disks (Oxoid, England, United Kingdom): β-lactams (ampicillin, AMP, 10 μg; amoxicillin, AML, 30 μg; cefotaxime, CEX, 10 μg; and ceftriaxone, CRO, 30 μg); aminoglycosides (streptomycin, S, 10 μg; gentamicin, GEN, 10 μg; kanamycin, K, 30 μg; amikacin, AMK, 30 μg; and tobramycin, TOB, 10 μg); quinolones (norfloxacin, NOR, 10 μg; ciprofloxacin, CIP, 5 μg; ofloxacin, OFX, 5 μg; nalidixic acid, NA, 30 μg; and enrofloxacin, ENR, 5 μg); macrolide (erythromycin, E, 15 μg; and azithromycin, AZM, 15 μg); tetracycline (TE, 30 μg); and clindamycin (DA, 2 μg). This process was carried out according to the Kirby-Bauer disk diffusion method (Bauer et al., 1966) and as recommended by the Clinical Laboratory Standards Institute (Clinical and Laboratory Standards Institute, 2012). Some isolates were revived from the glycerol stocks using BB with 10% lysed sheep blood. Then, these isolates were incubated for 36 h at 42°C under microaerobic conditions. Revived cultures were streaked using sterile cotton swabs on MH (Mueller Hinton) agar plates (Oxoid) supplemented with 10% sheep blood for another 36 h of incubation. The bacteria were scrapped to PBS, and the turbidity of the suspension was adjusted to 0.5 McFarland standard (Clinical and Laboratory Standards Institute, 2012). The suspension was stacked onto MH agar plates for 36 h of incubation under microaerophilic conditions. Escherichia coli ATCC 25922 and C. jejuni NCTC 11168 were used as reference strains.
The diameters of the inhibition zones around the antibiotic disk were measured. The breakpoints used to categorize isolates as susceptible, intermediate, and resistant–were based on the Clinical and Laboratory Standards Institute recommendations (Clinical and Laboratory Standards Institute, 2012). Isolates resistant to ≥3 unrelated antibiotic classes were classified as isolates with multidrug resistance (MDR).
MAR (Multiple Antimicrobial Resistance) was used to quantify the multi-resistance of Campylobacter isolates. MAR index = a/b. In this formula, “a” indicated the number of antibiotics to which the isolate was resistant and “b” indicated the total number of antibiotics to which the isolate was tested (Krumperman, 1983).
Multilocus Sequence Typing for Campylobacter
DNA was extracted from some representative strains using a commercial DNA Kit (Tiangen Biotech Inc., Beijing, China). MLST was conducted using primer sequences obtained from http://pubmlst.org/ Campylobacter as previously described (Dingle et al., 2001). The nucleotide sequences of the amplicons were determined by GenScript, Inc. (Nanjing, China). Allele numbers and STs (sequence types) were assigned using the Campylobacter PubMLST database.
Data Analysis
The difference in the prevalence levels across the batches in cloacal swabs and various sample points obtained after plucking, evisceration, washing and chilling were analyzed using a nonparametric test (Chi-square test) using SPSS software (version 17.0). P < 0.05 were considered statistically significant.
Sequence analysis and ST determinations of clonal complexes were performed using the PubMLST database1 for ST designation. Consensus tree was constructed using UPGMA cluster analysis based on the seven housekeeping gene sequences.
Results
Prevalence of Campylobacter in the Slaughter Process
The isolation rates of Campylobacter and the samples collected from each point are listed in Table 1. All strains isolated in batch 1 were C. jejuni (105/105). However, in batches 2 and 3, C. coli was the predominant isolated strain (94/96 and 57/67, respectively). The contamination rate of Campylobacter on the carcasses at every point during slaughter was relatively high even after chilling (87.5–100%). As shown in Table 2, compared with batches 1 and 2, Campylobacter infection rate in the cloacal swabs in batch 3 was significantly lower (P < 0.05). However, significant correlations of infection rates were not observed after evisceration among different batches.
Table 2.
The statistical contrast across the three batches (P-value).
| Sample site | Batch (Farm) (No. of Campylobacter positive samples/total no. of samples) | P-value | |
|---|---|---|---|
| Cloacal swabs | 1 (19/30) | 2 (20/20) | 0.317 |
| 1 (19/30) | 3 (3/22) | 0.000∗ | |
| 2 (20/20) | 3 (3/22) | 0.000∗ | |
| After plucking | 1 (22/30) | 2 (18/21) | 0.295 |
| 1 (22/30) | 3 (14/15) | 0.118 | |
| 2 (18/21) | 3 (14/15) | 0.480 | |
| After evisceration | 1 (24/30) | 2 (17/22) | 0.814 |
| 1 (24/30) | 3 (12/15) | 1.000 | |
| 2 (17/22) | 3 (12/15) | 0.845 | |
| After washing | 1 (23/30) | 2 (20/21) | 0.076 |
| 1 (23/30) | 3 (14/20) | 0.602 | |
| 2 (20/21) | 3 (14/20) | 0.034∗ | |
| After chilling | 1 (7/8) | 2 (10/10) | 0.264 |
| 1 (7/8) | 3 (10/10) | 0.264 | |
| 2 (10/10) | 3 (10/10) | N | |
“N” means no statistics are computed; “∗” resembles P < 0.05.
At the batch level, we compared the contamination rates of Campylobacter at four sampling points (after plucking, evisceration, washing, and chilling) and found no significant difference except the point of after washing between batches 2 and 3 (P = 0.034) (Table 2). A total of 268 isolates were obtained from 348 samples (77.0%), including 117 C. jejuni and 151 C. coli isolates.
Campylobacter isolates were recovered from all environmental samples. The cotton swab samples from the operating table and workers’ gloves in the 3 batches showed relatively high contamination levels. However, Campylobacter species were not isolated from the cooled water in batch 3.
Antimicrobial Resistance
We selected 39 isolates (C. jejuni) from batch 1, 38 isolates (2 C. jejuni and 36 C. coli) from batch 2, and 40 isolates (1 C. jejuni and 39 C. coli) from batch 3 for antimicrobial resistance testing. A total of 18 antimicrobials classified under six antimicrobial groups were employed to test the selected Campylobacter isolates. Except for seven samples from batch 2, all isolates were resistant to at least one or more antimicrobials (Table 4). The majority of the tested isolates were resistant to quinolones (≥89.7%). Less resistance to macrolide (59.8%), gentamicin (42.7%), and amikacin (36.8%) was observed. The resistance to other antibiotics in this study was greater or equal to 65.0%. Resistance to macrolide was not detected from batch 1, but 81.6 and 100% from batches 2 and 3, respectively, were resistant to this antimicrobial (Table 3).
Table 4.
Resistance spectra of 117 Campylobacter to various antibiotic combinations.
| MAR index | No. of Campylobacter isolates |
Antibiotic | ||
|---|---|---|---|---|
| Batch 1 | Batch 2 | Batch 3 | ||
| 0 | 0 | 7 | 0 | – |
| 0.72 | 0 | 1 | 0 | AMP, AML, CEX, CRO, S, K, NOR, CIP, NA, E, AZM, TE, DA |
| 0.78 | 0 | 2 | 0 | AMP, AML, CEX, CRO, S, K, NOR, CIP, OFX, NA, E, AZM, TE, DA |
| 0.78 | 0 | 2 | 0 | AMP, AML, CEX, S, K, NOR, CIP, OFX, NA, ENR, E, AZM, TE, DA |
| 0.78 | 0 | 0 | 33 | CEX, CRO, S, K, TOB, NOR, CIP, OFX, NA, ENR, E, AZM, TE, DA |
| 0.83 | 0 | 5 | 0 | AMP, AML, CEX, S, K, TOB, NOR, CIP, OFX, NA, ENR, E, AZM, TE, DA |
| 0.83 | 0 | 2 | 0 | AMP, AML, CEX, CRO, S, K, TOB, NOR, CIP, NA, ENR, E, AZM, TE, DA |
| 0.83 | 0 | 4 | 0 | AMP, AML, CEX, CRO, S, K, NOR, CIP, OFX, NA, ENR, E, AZM, TE, DA |
| 0.83 | 0 | 0 | 2 | AMP, AML, CEX, S, GEN, K, TOB, NOR, CIP, OFX, NA, ENR, E, AZM, TE, |
| 0.83 | 0 | 1 | 0 | AMP, AML, CEX, S, GEN, K, TOB, NOR, CIP, OFX, NA, E, AZM, TE, DA |
| 0.89 | 0 | 9 | 0 | AMP, AML, CEX, CRO, S, K, TOB, NOR, CIP, OFX, NA, ENR, E, AZM, TE, DA |
| 0.89 | 0 | 1 | 0 | AMP, AML, CEX, GEN, K, AMK, TOB, NOR, CIP, OFX, NA, ENR, E, AZM, TE, DA |
| 0.89 | 0 | 1 | 0 | CEX, CRO, S, GEN, K, AMK, TOB, NOR, CIP, OFX, NA, ENR, E, AZM, TE, DA |
| 0.89 | 0 | 0 | 2 | AMP, AML, CEX, CRO, S, GEN, K, TOB, NOR, CIP, OFX, NA, ENR, E, AZM, TE |
| 0.89 | 39 | 0 | 0 | AMP, AML, CEX, CRO, S, GEN, K, AMK, TOB, NOR, CIP, OFX, NA, ENR, TE, DA |
| 0.94 | 0 | 3 | 1 | AMP, AML, CEX, CRO, S, GEN, K, TOB, NOR, CIP, OFX, NA, ENR, E, AZM, TE, DA |
| 1.00 | 0 | 0 | 2 | AMP, AML, CEX, CRO, S, GEN, K, AMK, TOB, NOR, CIP, OFX, NA, ENR, E, AZM, TE, DA |
Table 3.
Number and percentages of resistance of Campylobacter isolates from three batches.
| Antibiotic group | Antibiotic name | No. of resistant Campylobacter isolates (%) |
|||
|---|---|---|---|---|---|
| Batch 1 | Batch 2 | Batch 3 | Total | ||
| β-Lactams | AMP | 39/39 (100%) | 31/38 (81.6%) | 7/40 (17.5%) | 77/117 (65.8%) |
| AML | 39/39 (100%) | 31/38 (81.6%) | 7/40 (17.5%) | 77/117 (65.8%) | |
| CEX | 39/39 (100%) | 34/38 (89.5%) | 40/40 (100%) | 113/117 (96.6%) | |
| CRO | 39/39 (100%) | 20/38 (52.6%) | 40/40 (100%) | 99/117 (84.6%) | |
| Aminoglycosides | S | 39/39 (100%) | 31/38 (81.6%) | 40/40 (100%) | 109/117 (93.2%) |
| GEN | 39/39 (100%) | 5/38 (13.2%) | 6/40 (15.0%) | 50/117 (42.7%) | |
| K | 39/39 (100%) | 31/38 (81.6%) | 40/40 (100%) | 109/117 (93.2%) | |
| AMK | 39/39 (100%) | 2/38 (5.7%) | 2/40 (5.0%) | 42/117 (36.8%) | |
| TOB | 39/39 (100%) | 15/38 (39.5) | 40/40 (100%) | 94/117 (80.3%) | |
| Quinolones | NOR | 39/39 (100%) | 31/38 (81.6%) | 40/40 (100%) | 109/117 (93.2%) |
| CIP | 39/39 (100%) | 31/38 (81.6%) | 40/40 (100%) | 109/117 (93.2%) | |
| OFX | 39/39 (100%) | 31/38 (81.6%) | 40/40 (100%) | 109/117 (93.2%) | |
| NA | 39/39 (100%) | 31/38 (81.6%) | 40/40 (100%) | 109/117 (93.2%) | |
| ENR | 39/39 (100%) | 26/38 (68.4) | 40/40 (100%) | 105/117 (89.7%) | |
| Macrolide | E | 0 | 31/38 (81.6%) | 40/40 (100%) | 70/117 (59.8%) |
| AZM | 0 | 31/38 (81.6%) | 40/40 (100%) | 70/117 (59.8%) | |
| Tetracyclines | TE | 39/39 (100%) | 31/38 (81.6%) | 40/40 (100%) | 109/117 (93.2%) |
| Clindamycin | DA | 39/39 (100%) | 31/38 (81.6%) | 37/40 (92.5%) | 106/117 (90.6%) |
The MAR (Multiple Antimicrobial Resistance) indices of the tested isolates from the current study are indicated in Table 4. MDR (resistance to three or more antimicrobial families) was observed in the majority of the isolates. A total of 17 different antibiotic resistance patterns with MAR index ranging from 0 to 1.00 were observed. Multiple resistances were common with resistance from 14 to 16 to 18 antibiotics (MAR index 0.78–0.89). This characteristic was observed in most of the 117 Campylobacter strains. Strains from batch 1 showed resistance to 16 antibiotics (except AZM and E). The resistance spectra of the strains from batches 2 and 3 were more diverse than that of the strains from batch 1. A total of 94.0% of the tested strains demonstrated MDR.
MLST Analysis of Campylobacter Isolated From the Slaughter Process and the Environment
Isolates from cloacal swabs, environmental samples, and carcasses at different slaughter points were selected and subjected to MLST analysis. The STs, species, sources, and numbers of bacteria are summarized in Table 5. A total of eight STs, including one novel type, were observed from the 117 isolates. Three C. jejuni STs and 7 C. coli STs were identified. One clonal complex CC828 (55 isolates) was generated from these isolates, but 62 isolates could not be assigned to any of the defined CCs.
Table 5.
Distributions of STs for 117 Campylobacter isolates.
| Batch | species | ST | ST-CC | Source | Number |
|---|---|---|---|---|---|
| 1 | C. jejuni | 8089 | UA | Cloacal swab | 7 |
| 8089 | UA | After plucking | 7 | ||
| 8089 | UA | After evisceration | 8 | ||
| 8089 | UA | After washing | 4 | ||
| 8089 | UA | After chilling | 7 | ||
| 8089 | UA | Operating table | 2 | ||
| 8089 | UA | Workers’ gloves | 2 | ||
| 8089 | UA | Water | 2 | ||
| 2 | C. jejuni | 6186 | UA | After chilling | 1 |
| C. coli | 6186 | UA | Cloacal swab | 3 | |
| 5511 | 828 | Cloacal swab | 1 | ||
| 6186 | UA | After plucking | 6 | ||
| 6186 | UA | After evisceration | 3 | ||
| 5511 | 828 | After washing | 1 | ||
| 825 | 828 | After washing | 1 | ||
| 6186 | UA | After washing | 4 | ||
| NEW1 | – | After washing | 2 | ||
| 825 | 828 | After chilling | 2 | ||
| 860 | 828 | After chilling | 3 | ||
| 6186 | UA | After chilling | 3 | ||
| 872 | 828 | Operating table | 2 | ||
| 860 | 828 | Operating table | 1 | ||
| 872 | 828 | Workers’ gloves | 4 | ||
| 6186 | UA | Water | 1 | ||
| 3 | C. jejuni | 860 | 828 | Workers’ gloves | 2 |
| C. coli | 860 | 828 | Cloacal swab | 3 | |
| 860 | 828 | After plucking | 6 | ||
| 830 | 828 | After plucking | 1 | ||
| 860 | 828 | After evisceration | 6 | ||
| 860 | 828 | After washing | 5 | ||
| 6186 | 828 | After washing | 1 | ||
| 860 | 828 | After chilling | 7 | ||
| 830 | 828 | After chilling | 1 | ||
| 825 | 828 | After chilling | 1 | ||
| 860 | 828 | Operating table | 4 | ||
| 825 | 828 | Operating table | 2 | ||
| 860 | 828 | Workers’ gloves | 1 |
“ST-CC” means ST clonal complex; “UA” means unassigned.
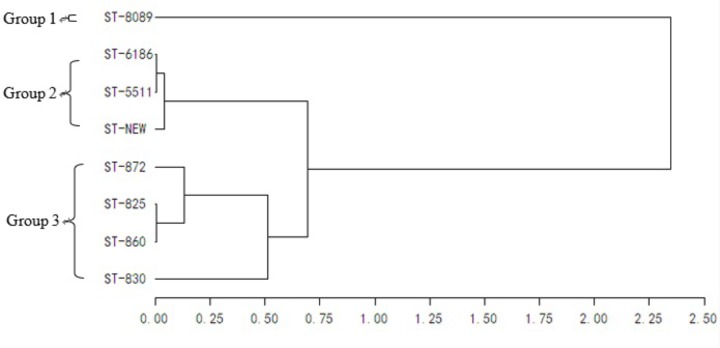
All 39 C. jejuni isolates from batch 1 were identified as ST8089. Compared with batch 1, batches 2 and 3 showed more diversity in STs. In batches 2 and 3, ST6186 (21/38, 55.3%) and ST860 (33/40, 82.5%) were the most frequently observed STs. This result was similar to the isolates from each cloacal sample. In addition, the STs from the environmental samples in each batch were highly consistent with those from the carcasses and cloacal swabs. In batch 2, strains after washing contained three traditional and one new ST types. All identified STs were further analyzed using UPGMA (Figure 1). Eight identified STs were classified into three clonal groups. All C. jejuni isolates from batch 1 belonged to group 1, and most of the Campylobacter isolates in batches 2 and 3 belonged to groups 2 and 3, respectively.
FIGURE 1.
Genetic relationships of the isolates based on the MLST. The consensus tree was developed using UPGMA cluster analysis.
Discussion
Campylobacter infection is considered one of the leading causes of bacterial gastroenteritis in developed and developing countries (Wei et al., 2014; Kaakoushm et al., 2015; Nohra et al., 2016). Several studies have associated the risk of human Campylobacter infection with highly contaminated broiler carcasses (Callicott et al., 2008; Nauta and Havelaar, 2008). Although China is one of the largest poultry producers worldwide, data on this pathogen are limited, especially during broiler production. In the present study, we showed the contamination rate of Campylobacter in samples collected along the slaughter line, that is, from the cloacal swabs in live birds to post chiller carcasses. The prevalence of Campylobacter after chilling was high (87.5–100%). This result was consistent with the finding of Seliwiorstow and collaborators, who compared the carcass contaminations before washing and after chilling in the slaughterhouse and found that the contamination rates were not reduced (Seliwiorstow et al., 2015). Accordingly, improved hygiene during slaughtering may reduce the number of Campylobacter in the carcasses, but the infection rates cannot be reduced because of cross-contamination. Despite the significantly low Campylobacter infection rate of cloacal swabs in batch 3 (P < 0.05), significant correlations of infection rates after evisceration among the different batches were not observed. These data explained that in the case of batch 3, which had lower positive cloacal numbers, the cross-contamination coming from the intestinal content of other flocks or its own flock still existed.
The investigation was conducted in 2012 in a commercial poultry production in Shanghai, China. A total of 23 C. jejuni (17.0%) and 33 C. coli (24.4%) were isolated from 135 broiler carcasses from the slaughterhouse (Ma et al., 2014). Wang and collaborators investigated the prevalence of Campylobacter from broiler chickens in slaughter houses in five Chinese provinces during 2008–2014, and isolated 977 C. jejuni (18.1%) and 1021 C. coli (19.0%) from 5,385 chickens (Wang et al., 2016). In southern Brazil, samples from the broiler slaughtering process were analyzed to directly count Campylobacter and results showed that 72 and 38% were C. jejuni and C. coli, respectively (Gonsalves et al., 2016). Similar results were demonstrated in the European Union, where 60.8% of broiler samples tested positive for C. jejuni, and 41.5% tested positive for C. coli (European Food Safety Authority [EFSA], 2010). In the present study, a total of 117 C. jejuni (33.6%) and 151 C. coli (43.4%) were isolated from 348 samples collected in the slaughterhouse. The isolation rates were higher than those obtained in previous years in China. More samples are needed for further study of the prevalence of Campylobacter species found in the different steps of the slaughter process.
Campylobacter isolates were recovered from all environmental samples. Campylobacter can form biofilms to survive outside the host and protect against chemical products, physical cleaning processes, and environmental stress, and these processes are proposed as a survival mechanism (Hanning et al., 2008). Thus, this mechanism may explain the presence of Campylobacter in the cool water despite the chemical treatment of the water. Defeathering and evisceration are considered as critical contamination steps in poultry processing (Sasaki et al., 2013; Gruntar et al., 2015). Samples from the operating table and workers’ gloves showed a high contamination level in this study. This result suggested an important cross-contamination rate between carcasses and processing equipment. Therefore, it is recommended that contaminated broiler flocks should be slaughtered at the end of the working day to reduce the cross-contamination among the flocks.
Antibiotic resistance is a persistent issue in veterinary medicine and human medical treatment because of the indiscriminate use of antibiotics in therapy or as a growth promoter. Although most Campylobacter infections are self-limiting and do not require any antibiotic treatment, antimicrobial treatment is necessary for some severe and prolonged cases. Fluoroquinolones and macrolides are usually administered to treat human campylobacteriosis in China (Zhang et al., 2014). Our results showed that more than 89.7% of the tested isolates were resistant to quinolones and 59.8% of the tested isolates were resistant to macrolide. This result was consistent with those of previous reports (Li et al., 2018). All tested isolates of C. jejuni from batch 1 were susceptible to macrolide (AZM and E), compare with 81.6 and 100% of the tested isolates of C. coli from batches 2 and 3, respectively, resistant to this antimicrobial. These data were in accordance with recent reports (Ruzauskas et al., 2011; Haruna et al., 2012; Fraqueza et al., 2014; Pergola et al., 2017), which indicated that C. jejuni was predominantly susceptible to erythromycin while C. coli was resistant. However, another possible reason for the results in the current study is that Campylobacter isolated from the same batch may have primarily the same antibiotic resistance profile.
MDR was observed in the majority of the tested isolates (94%) in this study. Higher frequency of MDR was also noted in C. coli isolates from different sources (99%) in China (Zhang et al., 2014). In the present study, 17 different antibiotic resistance patterns with MAR index ranging from 0 to 1.00 were observed. The majority of the MAR index calculated ranged from 0.78 to 0.89 among the 117 selected Campylobacter strains. Strains with a MAR index >0.2 have been identified from animals frequently treated with antimicrobials (Marian et al., 2012). In contrast, significantly lower resistance rates of ciprofloxacin were observed in Campylobacter from poultry meat in countries with strict antimicrobial controls (Miflin et al., 2007; Zhao et al., 2010). Hence, severe MDR, which may be a threat to public safety in China, should be given proper attention.
Campylobacter isolates from one farm showed primarily the same genotype and the same antibiotic resistance profile as previously reported (Pergola et al., 2017). Some studies have supported the hypothesis that the contamination of Campylobacter in broiler carcasses is mainly from the processed Campylobacter-positive birds within a batch (Rasschaert et al., 2006; Sasaki et al., 2014). In the present study, the composition of the MLST was relatively stable within a batch because of the predominance of certain MLST types. In each batch, the most frequently observed STs (ST8089, ST6186, and ST860) were similar to the STs of the isolates from each cloacal sample. Figure 1 shows the close genetic relationship with the dominant STs in each batch. These results may indicate that Campylobacter in slaughterhouses originated mainly from the farms. Thus, minimizing the Campylobacter colonization in the incoming broiler flock is important to reduce the public health risk.
Several isolates collected in this study shared identical genotypes (ST6186, ST825, ST830, ST860, and ST872) with those isolates from the feces of a diarrheal patient in China (Zhang et al., 2014). The consistency of STs in the environmental and carcass samples suggested that some environmental samples, such as those from the operating table and workers’ gloves, may reflect the potential source of contamination. This result further indicated the importance of good hygienic practices during the slaughter process.
Author Contributions
QZ, JZ, and XY performed the collection of samples. XZ and QZ did the Campylobacter detection and identification. MT and XZ performed the MLST and antibiotic susceptibility tests. QZ and MT did the data analysis. XZ prepared the manuscript. YG supervised and assisted in the manuscript preparation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Changzhou Lihua food company for their help with the sample collection.
Funding. This work was supported by the National Natural Science Foundation of China grant (31700005), Yangzhou Social Development Project (YZ2016058).
References
- Adzitey F., Rusul G., Huda N., Cogan T., Corry J. (2012). Prevalence, antibiotic resistance and RAPD typing of Campylobacter species isolated from ducks, their rearing and processing environments in Penang, Malaysia. Int. J. Food Microbiol. 154 197–205. 10.1016/j.ijfoodmicro.2012.01.006 [DOI] [PubMed] [Google Scholar]
- Allen V. M., Bull S. A., Corry J. E., Domingue G., Jørgensen F., Frost J. A., et al. (2007). Campylobacter spp. contamination of chicken carcasses during processing in relation to flock colonisation. Int. J. Food Microbiol. 113 54–61. 10.1016/j.ijfoodmicro.2006.07.011 [DOI] [PubMed] [Google Scholar]
- Althaus D., Zweifel C., Stephan R. (2017). Analysis of a poultry slaughter process: influence of process stages on the microbiological contamination of broiler carcasses. Ital. J. Food Saf. 6:7097. 10.4081/ijfs.2017.7097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer A. W., Kirby W. M., Sherris J. C., Turck M. (1966). Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45 493–496. 10.1093/ajcp/45.4_ts.493 [DOI] [PubMed] [Google Scholar]
- Berrang M. E., Buhr R. J., Cason J. A. (2000). Campylobacter recovery from external and internal organs of commercial broiler carcass prior to scalding. Poult. Sci. 79 286–290. 10.1093/ps/79.2.286 [DOI] [PubMed] [Google Scholar]
- Boysen L., Rosenquist H., Larsson J. T., Nielsen E. M., Sørensen G., Nordentoft S., et al. (2014). Source attribution of human campylobacteriosis in Denmark. Epidemiol. Infect. 142 1599–1608. 10.1017/S0950268813002719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callicott K. A., Harğardóttir H., Georgsson F., Reiersen J., Friğriksdóttir V., Gunnarsson E., et al. (2008). Broiler Campylobacter contamination and human campylobacteriosis in Iceland. Appl. Environ. Microbiol. 74 6483–6494. 10.1128/AEM.01129-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cason J. A., Hinton A., Jr., Northcutt J. K., Buhr R. J., Ingram K. D., Smith D. P., et al. (2007). Partitioning of external and internal bacteria carried by broiler chickens before processing. J. Food Prot. 70 2056–2062. 10.4315/0362-028X-70.9.2056 [DOI] [PubMed] [Google Scholar]
- Chen X., Naren G. W., Wu C. M., Wang Y., Dai L., Xia L. N., et al. (2010). Prevalence and antimicrobial resistance of Campylobacter isolates in broilers from China. Vet. Microbiol. 144 133–139. 10.1016/j.vetmic.2009.12.035 [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (2012). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement M11-S22. Wayne, PA: CLSI. [Google Scholar]
- Colles F. M., McCarthy N. D., Sheppard S. K., Layton R., Maiden M. C. (2010). Comparison of Campylobacter populations isolated from a free-range broiler flock before and after slaughter. Int. J. Food Microbiol. 137 259–264, 10.1016/j.ijfoodmicro.2009.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corry J. E., Atabay H. I. (2001). Poultry as a source of Campylobacter and related organisms. Symp. Ser. Soc. Appl. Microbiol. 30 96S–114S. 10.1046/j.1365-2672.2001.01358.x [DOI] [PubMed] [Google Scholar]
- Dingle K. E., Colles F. M., Falush D., Maiden M. C. (2005). Sequence typing and comparison of population biology of Campylobacter coli and Campylobacter jejuni. J. Clin. Microbiol. 43 340–347. 10.1128/JCM.43.1.340-347.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingle K. E., Colles F. M., Wareing D. R., Ure R., Fox A. J., Bolton F. E., et al. (2001). Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39 14–23. 10.1128/JCM.39.1.14-23.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA and ECDC (2016). European Food Safety Authority, European Centre for Disease Prevention and Control: the European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2014. EFSA J. 13:4329 10.2903/j.efsa.2015.4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellerbroek L. I., Lienau J. A., Klein G. (2010). Campylobacter spp. in broiler flocks at farm level and the potential for cross-contamination during slaughter. Zoonoses Public Health 57 e81–e88. 10.1111/j.1863-2378.2009.01267 [DOI] [PubMed] [Google Scholar]
- European Food Safety Authority [EFSA] (2010). Analysis of the baseline survey on the prevalence of Campylobacter in broiler batches and of Campylobacter and Salmonella on broiler carcasses in the EU, 2008. EFSA J. 8 1503–1602. [Google Scholar]
- Fraqueza M. J., Martins A., Borges A. C., Fernandes M. H., Fernandes M. J., Vaz Y., et al. (2014). Antimicrobial resistance among Campylobacter spp. strains isolated from different poultry production systems at slaughterhouse level. Poult. Sci. 93 1578–1586. 10.3382/ps.2013-03729 [DOI] [PubMed] [Google Scholar]
- García-Sánchez L., Melero B., Jaime I., Hänninen M. L., Rossi M., Rovira J. (2017). Campylobacter jejuni survival in a poultry processing plant environment. Food Microbiol. 65 185–192. 10.1016/j.fm.2017.02.009 [DOI] [PubMed] [Google Scholar]
- Gonsalves C. C., Borsoi A., Perdoncini G., Rodrigues L. B., do Nascimento V. P. (2016). Campylobacter in broiler slaughter samples assessed by direct count on mCCDA and Campy-Cefex agar. Braz. J. Microbiol. 47 764–769. 10.1016/j.bjm [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruntar I., Biasizzo M., Kušar D., Pate M., Ocepek M. (2015). Campylobacter jejuni contamination of broiler carcasses: population dynamics and genetic profiles at slaughterhouse level. Food Microbiol. 50 97–101. 10.1016/j.fm.2015.03.007 [DOI] [PubMed] [Google Scholar]
- Habib I., Berkvens D., De Zutter L., Dierick K., Van Huffel X., Speybroeck N., et al. (2012). Campylobacter contamination in broiler carcasses and correlation with slaughterhouses operational hygiene inspection. Food Microbiol. 29 105–112. 10.1016/j.fm.2011.09.004 [DOI] [PubMed] [Google Scholar]
- Hanning I., Jarquin R., Slavik M. (2008). Campylobacter jejuni as a secondary colonizer of poultry biofilms. J. Appl. Microbiol. 105 1199–1208. 10.1111/j.1365-2672.2008.03853.x [DOI] [PubMed] [Google Scholar]
- Haruna M., Sasaki Y., Murakami M., Ikeda A., Kusukawa M., Tsujiyama Y., et al. (2012). Prevalence and antimicrobial susceptibility of Campylobacter in broiler flocks in Japan. Zoonoses Public Health 59 241–245. 10.1111/j.1863-2378.2011.01441.x [DOI] [PubMed] [Google Scholar]
- Huang J. L., Xu H. Y., Bao G. Y., Zhou X. H., Ji D. J., Zhang G., et al. (2009). Epidemiological surveillance of Campylobacter jejuni in chicken, dairy cattle and diarrhoea patients. Epidemiol. Infect. 137 1111–1120. 10.1017/S0950268809002039 [DOI] [PubMed] [Google Scholar]
- Kaakoushm N. O., Castaño-Rodríguez N., Mitchell H. M., Man S. M. (2015). Global epidemiology of Campylobacter infection. Clin. Microbiol. Rev. 28 687–720. 10.1128/CMR.00006-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumperman P. H. (1983). Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 46 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhang S., He M., Zhang Y., Fu Y., Liang H., et al. (2018). Prevalence and molecular characterization of Campylobacter spp. isolated from patients with diarrhea in Shunyi, Beijing. Front. Microbiol. 9:52. 10.3389/fmicb.2018.00052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Wang Y., Shen J., Zhang Q., Wu C. (2014). Tracking Campylobacter contamination along a broiler chicken production chain from the farm level to retail in China. Int. J. Food Microbiol. 181 77–84. 10.1016/j.ijfoodmicro.2014.04.023 [DOI] [PubMed] [Google Scholar]
- Marian M. N., Sharifah Aminah S. M., Zuraini M. I., Son R., Maimunah M., Lee H. Y., et al. (2012). MPN-PCR detection and antimicrobial resistance of Listeria monocytogenes isolated from raw and ready-to-eat foods in Malaysia. Food Control 28 309–314. 10.1016/j.foodcont.2012.05.030 [DOI] [Google Scholar]
- Melero B., Juntunen P., Hänninen M. L., Jaime I., Rovira J. (2012). Tracing Campylobacter jejuni strains along the poultry meat production chain from farm to retail by pulsed-field gel electrophoresis, and the antimicrobial resistance of isolates. Food Microbiol. 32 124–128. 10.1016/j.fm.2012.04.020 [DOI] [PubMed] [Google Scholar]
- Michaud S., Ménard S., Arbeit R. D. (2005). Role of real-time molecular typing in the surveillance of Campylobacter enteritis and comparison of pulsed-field gel electrophoresis profiles from chicken and human isolates. J. Clin. Microbiol. 43 1105–1111. 10.1128/JCM.43.3.1105-1111.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miflin J. K., Templeton J. M., Blackall P. J. (2007). Antibiotic resistance in Campylobacter jejuni and Campylobacter coli isolated from poultry in the south-east Queensland region. J. Antimicrob. Chemother. 59 775–778. 10.1093/jac/dkm024 [DOI] [PubMed] [Google Scholar]
- Miwa N., Takegahara Y., Terai K., Kato H., Takeuchi T. (2003). Campylobacter jejuni contamination on broiler carcasses of C. jejuni-negative flocks during processing in a Japanese slaughterhouse. Int. J. Food Microbiol. 84 105–109. 10.1016/S0168-1605(02)00398-7 [DOI] [PubMed] [Google Scholar]
- Nauta M. J., Havelaar A. H. (2008). Risk-based standards for Campylobacter in the broiler meat chain. Food Control 19 372–381. 10.1016/j.foodcont.2007.04.016 [DOI] [Google Scholar]
- Newell D. G., Elvers K. T., Dopfer D., Hansson I., Jones P., James S., et al. (2011). Biosecurity-based interventions and strategies to reduce Campylobacter spp. on poultry farms. Appl. Environ. Microbiol. 77 8605–8614. 10.1128/AEM.01090-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohra A., Grinberg A., Midwinter A. C., Marshall J. C., Collins-Emerson J. M., French N. P. (2016). Molecular epidemiology of Campylobacter coli strains isolated from different sources in New Zealand between 2005 and 2014. Appl. Environ. Microbiol. 82 4363–4370. 10.1128/AEM.00934-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pergola S., Franciosini M. P., Comitini F., Ciani M., De Luca S., Bellucci S., et al. (2017). Genetic diversity and antimicrobial resistance profiles of Campylobacter coli and Campylobacter jejuni isolated from broiler chicken in farms and at time of slaughter in central Italy. J. Appl. Microbiol. 122 1348–1356. 10.1111/jam.13419 [DOI] [PubMed] [Google Scholar]
- Peyrat M. B., Soumet C., Maris P., Sanders P. (2008). Recovery of Campylobacter jejuni from surfaces of poultry slaughterhouses after cleaning and disinfection procedures: analysis of a potential source of carcass contamination. Int. J. Food Microbiol. 124 188–194. 10.1016/j.ijfoodmicro.2008.03.030 [DOI] [PubMed] [Google Scholar]
- Rasschaert G., Houf K., Van Hende J., De Zutter L. (2006). Campylobacter contamination during poultry slaughter in Belgium. J. Food Prot. 69 27–33. 10.4315/0362-028X-69.1.27 [DOI] [PubMed] [Google Scholar]
- Rozynek E., Maćkiw E., Kamińska W., Tomczuk K., Antos-Bielska M., Dzierżanowska-Fangrat K., et al. (2013). Emergence of macrolide-resistant Campylobacter strains in chicken meat in Poland and the resistance mechanisms involved. Foodborne Pathog. Dis. 10 655–660. 10.1089/fpd.2012.1333 [DOI] [PubMed] [Google Scholar]
- Ruzauskas M., Virgailis M., Siugzdiniene R., Zienius D., Mockeliunas R. (2011). Differences in antimicrobial resistance of Campylobacter jejuni isolated from broiler intestines and drumsticks in Lithuania. J. Food Saf. 31 379–385. 10.1111/j.1745-4565.2011.00310.x [DOI] [Google Scholar]
- Sasaki Y., Haruna M., Mori T., Kusukawa M., Murakami M., Tsujiyama Y., et al. (2014). Quantitative estimation of Campylobacter cross-contamination in carcasses and chicken products at an abattoir. Food Control 43 10–17. 10.1016/j.foodcont.2014.02.015 [DOI] [Google Scholar]
- Sasaki Y., Maruyama N., Zou B., Haruna M., Kusukawa M., Murakami M., et al. (2013). Campylobacter cross-contamination of chicken products at an abattoir. Zoonoses Public Health 60 134–140. 10.1111/j.1863-2378.2012.01509.x [DOI] [PubMed] [Google Scholar]
- Seliwiorstow T., Baré J., Van Damme I., Uyttendaele M., De Zutter L. (2015). Campylobacter carcass contamination throughout the slaughter process of Campylobacter-positive broiler batches. Int. J. Food Microbiol. 194 25–31. 10.1016/j.ijfoodmicro.2014.11.004 [DOI] [PubMed] [Google Scholar]
- Thakur S., White D. G., McDermott P. F., Zhao S., Kroft B., Gebreyes W., et al. (2009). Genotyping of Campylobacter coli isolated from humans and retail meats using multilocus sequence typing and pulsed-field gel electrophoresis. J. Appl. Microbiol. 106 1722–1733. 10.1111/j.1365-2672.2008.04142.x [DOI] [PubMed] [Google Scholar]
- Tissier A., Denis M., Hartemann P., Gassilloud B. (2012). Development of a rapid and sensitive method combining a cellulose ester microfilter and a real-time quantitative PCR assay to detect Campylobacter jejuni and Campylobacter coli in 20 liters of drinking water or low-turbidity waters. Appl. Environ. Microbiol. 78 839–845. 10.1128/AEM.06754-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresse O., Alvarez-Ordóñez A., Connerton I. F. (2017). Editorial: about the foodborne pathogen Campylobacter. Front. Microbiol. 8:1908. 10.3389/fmicb.2017.01908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Dong Y., Deng F., Liu D., Yao H., Zhang Q., et al. (2016). Species shift and multidrug resistance of Campylobacter from chicken and swine, China, 2008-14. J. Antimicrob. Chemother. 71 666–669. 10.1093/jac/dkv382 [DOI] [PubMed] [Google Scholar]
- Wei B., Cha S. Y., Kang M., Roh J. H., Seo H. S., Yoon R. H., et al. (2014). Antimicrobial susceptibility profiles and molecular typing of Campylobacter jejuni and Campylobacter coli from ducks in South Korea. Appl. Environ. Microbiol. 80 7604–7610. 10.1128/AEM.02469-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng D., Zhang X., Xue F., Wang Y., Jiang L., Jiang Y. (2016). Phenotypic characters and molecular epidemiology of Campylobacter jejuni in East China. J. Food Sci. 81 106–113. 10.1111/1750-3841.13146 [DOI] [PubMed] [Google Scholar]
- Zhang M., Liu X., Xu X., Gu Y., Tao X., Yang X., et al. (2014). Molecular subtyping and antimicrobial susceptibilities of Campylobacter coli isolates from diarrheal patients and food-producing animals in China. Foodborne Pathog. Dis. 11 610–619. 10.1089/fpd.2013.1721 [DOI] [PubMed] [Google Scholar]
- Zhao S., Young S. R., Tong E., Abbott J. W., Womack N., Friedman S. L., et al. (2010). Antimicrobial resistance of Campylobacter isolates from retail meat in the United States between 2002 and 2007. Appl. Environ. Microbiol. 76 7949–7956. 10.1128/AEM.01297-10 [DOI] [PMC free article] [PubMed] [Google Scholar]