Abstract
Lung cancer is one of the most prevalent malignancies worldwide; it has been ranked the most lethal type of cancer. Non-small cell lung carcinoma (NSCLC) comprises >80% of all types of lung cancer. Although certain achievements have been made in the treatment of NSCLC, including the targeted gene drug epidermal growth factor receptor-tyrosine kinase inhibitor (EGFR-TKI), the five-year survival rate of patients has not significantly increased. A previous study demonstrated that B7-H5, a novel co-stimulatory molecule in the B7 molecule family, was negatively correlated with EGFR in pancreatic cancer. Thus, in the present study, we aimed to investigate whether EGFR participates in immune evasion, probably through regulation of B7-H5 expression. NCI-H1299 NSCLCL cells were divided into control, mock, small interfering-EGFR and EGFR-TKI groups. The cell viability and apoptosis rate were analysed by a Cell Counting Kit-8 assay and flow cytometry. The transforming growth factor (TGF)-β and interleukin (IL)-10 content was measured using an ELISA. The expression levels of EGFR, B7-H5, Survivin, apoptosis regulator Bax, apoptosis regulator Bcl-2 (Bcl-2), TGF-β, vascular endothelial growth factor (VEGF), IL-10 and cyclooxygenase (COX)-2 were assessed via quantitative PCR and western blotting. The activation of the tyrosine-protein kinase JAK2 (JAK2)/signal transducer and activator of transcription 3 (STAT3) signalling pathway was detected using western blotting. The results demonstrated a notable negative correlation between EGFR and B7-H5 expression levels in cancer tissues and cell lines. Inhibition of EGFR expression via gene silencing and EGFR inhibition markedly decreased cell viability and increased the apoptosis of NCI-H1299 cells, by upregulating survivin and Bcl-2 expression. The protein expression levels of TGF-β, VEGF, IL-10 and COX-2 were additionally decreased, with weak activation of the JAK2/STAT3 signalling pathway. EGFR may be involved in immune evasion, possibly through regulation of B7-H5 expression in NSCLC.
Keywords: non-small cell lung carcinoma, epidermal growth factor receptor, B7-H5, transforming growth factor-β, vascular endothelial growth factor, interleukin-10, cyclooxygenase-2, tyrosine-protein kinase JAK2, signal transducer and activator of transcription 3
Introduction
Lung cancer is one of the most common malignant tumours worldwide, with an increasing incidence each year; its morbidity has increased >10-fold in male and female patients in the last five decades (1). Non-small cell lung cancer (NSCLC) represents >80% of lung cancer cases. A total of 40–50% of patients with NSCLC are at an advanced stage and are treated with radiotherapy and chemotherapy, although these therapies lack sensitivity (2).
With the discovery of a series of tumour-targeting genes, a number of targeted gene drugs against NSCLC, including epidermal growth factor receptor-tyrosine kinase inhibitor (EGFR-TKI), are being developed. In recent years, EGFR, a transmembrane receptor glycoprotein, has been an important target of gene therapy against NSCLC (2,3). In tumours, including NSCLC, thyroid cancer and colorectal cancer, EGFR signalling pathways are frequently abnormally activated or highly expressed, which indicates the correlation between tumour progression and EGFR-associated gene expression. The abnormal expression of any protein in the downstream transduction pathways of EGFR promotes the erroneous transduction of proliferation signals to the cell nucleus, thus inducing unusual hyperplasia of cells as a consequence (4–6). EGFR, which was reported to be abnormally activated and overexpressed, is the initial point of these pathways, and therefore becomes an important effector target for drug therapy against NSCLC (7). Competitively combining with the ATP binding site of tyrosine kinase domain to ATP, EGFR-TKI is able to inhibit the phosphorylation and activation of EGFR tyrosine kinase, and delay the EGFR signal transduction system, to restrain cell proliferation and accelerate apoptosis, thus achieving the purpose of tumour inhibition (7,8). For over a decade, EGFR-TKIs, including gefitinib and erlotinib, have been clinically identified to have a specific curative effect on patients with an EGFR gene mutation (9,10). However, like other chemotherapeutics, the problem of drug resistance has gradually emerged with the promotion of clinical applications (11). Recently, immunotherapy has become the most revolutionary treatment in patients with NSCLC (12). Wang et al (13) demonstrated that inactivity of the EGFR/mitogen-activated protein kinase pathway was associated with the reversal of PD-L1-mediated immune evasion in NSCLC in an in vivo study. Therefore, research into effective targets of EGFR and the internal mechanisms involved in the immune evasion of NSCLC is of significance.
The B7 family is the principal co-stimulatory molecule family in T-lymphocyte activation, and includes B7-1, B7-2, B7-H1, B7-H2, H7-H3 and B7-4 (14–16). Inamura et al (17) reported a significant association between high B7-H3 expression with wild-type EGFR and smoking in patients, indicating the potential effectiveness of an anti-B7-H3 therapy for EGFR wild-type or smoking-associated lung cancer. In 2013, Zhu et al (18) identified a novel co-stimulatory pathway regulating human T-cell responses, the B7-H5/CD28 homologue (CD28H) pathway. A recent study in pancreatic cancer indicated the loss of B7-H5, one of the co-stimulatory molecules in the B7 molecule family, which may contribute to immune evasion (19). To the best of our knowledge, there has been no direct research into the association between and mechanism of B7-H5 and EGFR. The present study aimed to determine the possible association between EGFR and B7-H5 in the immune evasion of NSCLC, and attempted to investigate the associated pathway.
Materials and methods
Tissues and cells
A total of 42 patients with NSCLC at The First People's Hospital of Huzhou (Huzhou, China) were included in the present study. All cancer tissues specimens were obtained from surgical tumour resections, and their adjacent normal lung tissue specimens were obtained simultaneously as the negative control. The normal and cancer tissues represented matched pairs from each patient. Basic clinical and pathological data for these patients was collected with their written informed consent. The study was approved by the ethics committee of The First People's Hospital of Huzhou. The cell lines BEAS-2B, A549, NCI-H1299, NCI-H1755 and 95D were all obtained from Shanghai Yansheng Industrial Co. Ltd. (Shanghai, China). Cells were maintained in Dulbecco's modified Eagle medium (DMEM; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing with 10% fetal bovine serum, and 1% streptomycin/penicillin (Gibco; Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2.
Grouping
NCI-H1299 cells were divided into four groups: Control group, mock group, silencing (si)EGFR group and EGFR-TKI group. Cells in the mock group were infected with a blank vector. Cells in the siEGFR group were infected with the recombinant plasmids of siEGFR. In the EGFR-TKI group, cells were treated with gefitinib as a positive control (Cardinal Health, Inc., Dublin, OH, USA).
Cell infection and treatment
Recombinant plasmids of siEGFR and siB7-H5 were purchased from Shanghai Quanyang Biotechnology Co., Ltd. (Shanghai, China) and the sequences were as follows: siEGFR antisense 3′-UUCCGCAUUCCUCGUCUAUUU-5′ and sense 5′-GGCGUAAGGAGCAGAUAAAUU-3′; siB7-H5 antisense 3′-UUCGUCGCACAAUUCACAAAU-5′ and sense 5′-GCAGCGUGUUAAGUGUUUAUU-3′; and a negative siRNA control antisense 3′-TTAAGAGGCUUGCACAGUGCA-5′ and sense 5′-UUCUCCGAACGUGUCACGUTT-3′. Cells in the logarithmic growth phase were seeded into a 6-well plate at a density of 2×106 to culture for 24 h. Recombinant plasmids were transfected into cells, according to the manufacturer's protocol of the Invitrogen Lipofectamine® LTX (Thermo Fisher Scientific, Inc., Waltham, MA, USA). A total of 2 µg pIRES2-ZsGreen1-vector or pIRES2-ZsGreen1-ARHGAP18 (Sangon Biotech Co., Ltd., Shanghai, China), 5 µl Lipofectamine® LTX (Thermo Fisher Scientific, Inc.) and 250 µl Opti-Minimum Essential Medium (Shanghai Haoran Biological Technology Co., Ltd., Shanghai, China) was prepared, mixed and incubated at room temperature for 25 min. Subsequently, 500 µl mixture was added into the 6-well plate with RPMI 1640 medium (Thermo Fisher Scientific, Inc.). Finally, following a 48-h culture period, transfected cells were harvested for the following experiments.
Cell Counting Kit-8 (CCK-8) assay
Cell viability in each group was detected with a CCK-8 Κit (Beyotime Institute of Biotechnology, Haimen, China). Cells at a density of 3×104 cells/ml were grouped and seeded into 96-well plates at 100 µl per well, and incubated at 37°C in a 5% CO2 incubator for 4 h. Following the addition of 10 µl CCK reagent to each well, cells were placed into the 5% CO2 incubator at 37°C for 1–4 h. The optical density (OD) value of each group was observed at 450 nm using a spectrophotometer (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany).
Flow cytometry (FCM)
Cells in the logarithmic phase were collected and seeded into 6-well plates at a density of 1×105 cells per well. Cells were digested with EDTA-free trypsin (Beyotime Institute of Biotechnology), stained with Annexin V-fluorescein isothiocyanate and propidium iodide (Shanghai BestBio Science Co., Ltd., Shanghai, China), and incubated in the dark at 4°C for 15 min. The cell cycle distribution and apoptosis rate of each group was detected using an EPICS XL-MCL flow cytometer (Beckman Coulter, Inc., Brea, CA, USA) with an excitation wavelength of 488 nm and an emission wavelength of 530 nm. Data was analysed using FCS Express version 3.0 (De Novo Software, Glendale, CA, USA).
ELISA analysis
The transforming growth factor (TGF)-β and interleukin (IL)-10 content in cells were measured using a TGF-β1 ELISA kit (Abcam, Cambridge, UK; cat. no. ab100647) and an IL-10 ELISA kit (Abcam; cat. no. ab100549). Detection was performed according to the manufacturer's protocol. OD values were read at 420 nm using a spectrophotometer (Sigma-Aldrich; Merck KGaA).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Cells were seeded into 6-well plates at a density of 2×106 cells/well. Total RNA was extracted using TRIzol (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The concentration of the extracted RNA was assessed using a UV spectrophotometer (Thermo Fisher Scientific, Inc.). cDNA was synthesized using reverse transcriptase, a poly (A) polymerase and tagged oligo (dT) primers in a 20 µl reaction volume for RT-qPCR (miScript; Qiagen, Inc., Valencia, CA, USA) according to the manufacturer's protocol. SYBR Green PCR Master Mix (Takara Bio, Inc., Otsu, Japan) was used to quantitate the expression levels of target genes. The amplification assay was performed on an Applied Biosystems PCR system (ABI 7500; Thermo Fisher Scientific, Inc.). PCR was performed by activating the DNA polymerase at 95°C for 5 min, followed by 40 cycles of two-step PCR (95°C for 10 sec and 60°C for 30 sec) and a final extension at 75°C for 10 min then holding at 4°C. GAPDH was applied as the internal control to monitor the efficiency of qPCR. All primers in the study were designed by Sangon Biotech Co., Ltd. The specific primer sequences for each gene are listed as follows: Forward 5′GGTGAGTGGCTTGTCTGGAA3′ and reverse 5′CCTTACGCCCTTCACTGTGT3′ for EGFR (product, 152 bp); forward 5′GAACTGGATGCCCTTGTGGT3′ and reverse 5′TGCTGTCTGTGCCTTCATGT3′ for B7-H5 (product, 123 bp); forward 5′CCAGGCGGTGATGTGGATCT3′ and reverse 5′GTGCTGGTAGCCATGAGGGT3′ for Survivin (product, 139 bp); forward 5′GCCAGCAAACTGGTGCTCAA3′ and reverse 5′CCAACCACCCTGGTCTTGGA3′ for apoptosis regulator Bax (Bax; product, 126 bp); forward 5′CACTGGCCAGGGTCAGAGTT3′ and reverse 5′TGGCCATAGACCCTGTCAGC3′ for apoptosis regulator Bcl-2 (Bcl-2; product, 85 bp); forward 5′GGGCTACCATGCCAACTTCT3′ and reverse 5′GACACAGAGATCCGCAGTCC3′ for TGF-β (product, 384 bp); forward 5′GGCTTGGGGCTTCCTAACTG3′ and reverse 5′GGGAATCCCTCCGAGACACT3′ for IL-10 (product, 99 bp); forward 5′GATCCCCAGGGCTCAAACAT3′ and reverse 5′GAAAAGGCGCAGTTTACGCT3′ for cyclooxygenase-2 (COX-2; product, 126 bp) and forward 5′AATGGGCAGCCGTTAGGAAA3′ and reverse 5′GCGCCCAATACGACCAAATC3′ for GAPDH (product, 168 bp). Each reaction was run in triplicate. The 2−ΔΔCq method was used for quantification (20).
Western blotting
Cells were seeded in 6-well plates at a density of 2×106 cells/well, and grouped. Cells were harvested and washed twice with PBS, and lysed in ice-cold radioimmunoprecipitation assay buffer (Whiga Technology Co., Ltd., Guangzhou, China) with a freshly mixed 0.01% protease inhibitor (phenylmethylsulfonyl fluoride; BeijingBioLab Science and Technology Co., Ltd., Beijing, China), followed by incubation for 30 min on ice. Cell lysates were centrifuged at 10,000 × g for 5 min at 4°C, and the supernatants containing 20–30 µg protein were collected, and protein concentration was determined using a BCA protein assay kit (Thermo Fisher Scientific, Inc.). Samples were run on a 10% SDS-PAGE gel, and electrophoretically transferred to a nitrocellulose membrane (Merck KGaA). To block the nonspecific proteins, 5% fat-free milk was incubated with the membrane for 2 h at room temperature. Membranes were incubated with the following primary specific antibodies at 4°C for 6 h and then at room temperature for 4 h: Anti-EGFR antibody (1:1,000; Abcam; cat. no. ab52894), anti-B7-H5 antibody (1:1,000; Abcam; cat. no. ab201565), anti-Survivin antibody (1:5,000; Abcam; cat. no. ab76424), anti-Bax antibody (1:1,000; Abcam; cat. no. ab32503), anti-Bcl-2 antibody (1:1,000; Abcam; cat. no. ab692), anti-TGF-β antibody (1:1,000; Abcam; cat. no. ab92486), anti-vascular endothelial growth factor (VEGF) receptor 2 antibody (1:1,000; Abcam; cat. no. ab10972), anti-IL-10 antibody (1:800; Abcam; cat. no. ab34843), anti-COX-2 antibody (1:1,000; Abcam; cat. no. ab15191), anti-tyrosine-protein kinase JAK2 (JAK2; phosphorylated Y1007) antibody (1:1,000; Abcam; cat. no. ab195055), anti-JAK2 antibody (1:1,000; Abcam; cat. no. ab39636), anti-signal transducer and activator of transcription 3 (STAT3; phosphorylated Y705) antibody (1:2,000; Abcam; cat. no. ab76315), anti-STAT3 antibody (1:1,000; Abcam; cat. no. ab68153), and anti-GAPDH antibody (1:2,000; Abcam; cat. no. ab8245). The horseradish peroxidase secondary antibodies were goat anti-mouse immunoglobulin (Ig)G heavy and light chains (H&L; 1:2,000; Abcam; cat. no. ab6789), goat anti-rabbit IgG H&L (1:2,000; Abcam; cat. no. ab6721), and donkey anti-goat IgG H&L (1:2,000; Abcam; cat. no. ab6885). The membranes were then incubated at room temperature for 1 h. Blots were visualized via enhanced chemiluminescence (ECL; Thermo Fisher Scientific, Inc.). An ECL system (Amersham; GE Healthcare, Chicago, IL, USA) was used to detect the bands. The density of the blots was read with the Quantity One software version 2.4 (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Statistical analysis
Data is expressed as the mean ± standard deviation. Prism GraphPad version 6.0 software (GraphPad Software, Inc., La Jolla, CA, USA) was used to analyze the data. Differences among groups were evaluated via one-way analysis of variance followed by Tukey's multiple comparisons test. The χ2 test was used for the analysis of the significance of EGFR and clinicopathological characteristics. P<0.05 was considered to indicate a statistically significant difference.
Results
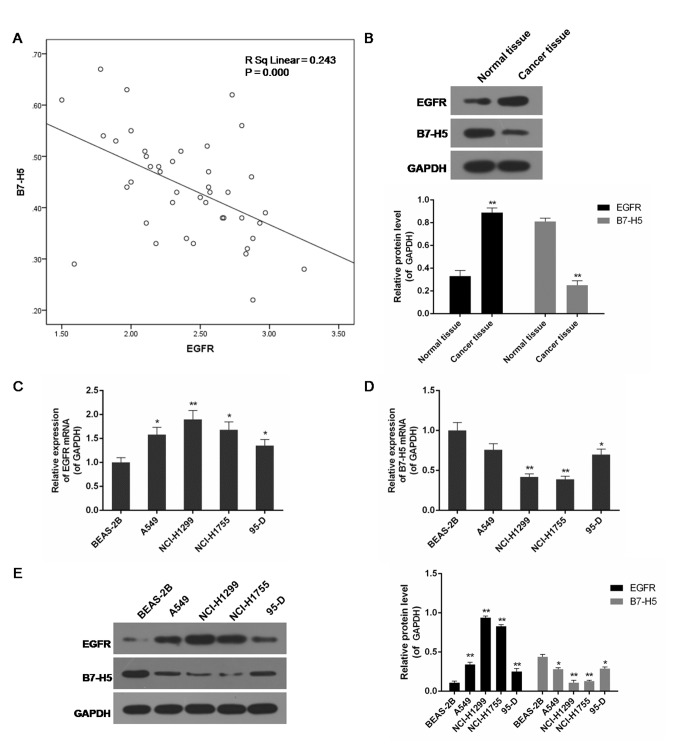
Negative correlation between EGFR and B7-H5 expression in lung cancer tissues and cell lines
The present study included a total of 42 patients from our hospital. The expression of EGFR and B7-H5 in surgically-resected NSCLC specimens and adjacent normal cancer tissues was assessed. Compared with normal tissues, the level of EGFR in cancer specimens was increased, while the expression of B7-H5 was decreased, and the correlation analysis indicated a negative association between them (P<0.01; Fig. 1A and B). EGFR mRNA and B7-H5 mRNA were observed to be negatively correlated. Examination of the correlation between EGFR expression and clinical pathological features demonstrated a positive association between increased EGFR expression levels and tumour-node-metastasis staging (Table I). A negative association between EGFR and B7-H5 mRNA and protein expression levels was observed in lung cell lines. In the lung cancer cell lines A549, NCI-H1299, NCI-H1755 and 95-D, particularly NCI-H1299 and NCI-H1755, EGFR expression was significantly increased, whereas B7-H5 was significantly inhibited compared with the normal lung cell line BEAS-2B (P<0.05 or P<0.01; Fig. 1C-E).
Figure 1.
Negative correlation between EGFR and B7-H5 expression in 42 lung cancer tissues and their adjacent normal tissues, and five cell lines, including BEAS-2B, A549, NCI-H1299, NCI-H1755 and 95-D. (A) A negative correlation between EGFR and B7-H5 expression was detected in 42 lung cancer tissues and their adjacent normal tissues. (B) Increased EGFR protein expression and decreased B7-H5 protein expression was detected in a representative lung cancer tissue sample compared with its adjacent normal tissue. *P<0.05 and **P<0.01 vs. respective normal tissue. (C) EGFR mRNA was highly expressed in lung cancer cell lines, particularly NCI-H1299. (D) B7-H5 mRNA was weakly expressed in lung cancer cell lines, particularly NCI-H1299 and NCI-H1755. (E) Increased EGFR protein expression and decreased B7-H5 protein expression was detected in lung cancer cell lines, particularly NCI-H1299. Data are presented as the mean ± standard deviation; n=3. *P<0.05 and **P<0.01 vs. respective BEAS-2B cell group. EGFR, epidermal growth factor receptor.
Table I.
Association between EGFR and clinical data of patients with non-small cell lung cancer.
| Cancer staging | Sex, male/female | Age, <59/≥59 years | EGFR expression, lower/higher |
|---|---|---|---|
| TNM | |||
| I | 5/2 | 4/3 | 5/2 |
| II | 8/3 | 5/6 | 4/7 |
| III | 15/9 | 9/15 | 5/19 |
| P-values | 0.802 | 0.639 | 0.043a |
P<0.05, χ2 test. TNM, tumour-node-metastasis; EGFR, epidermal growth factor receptor.
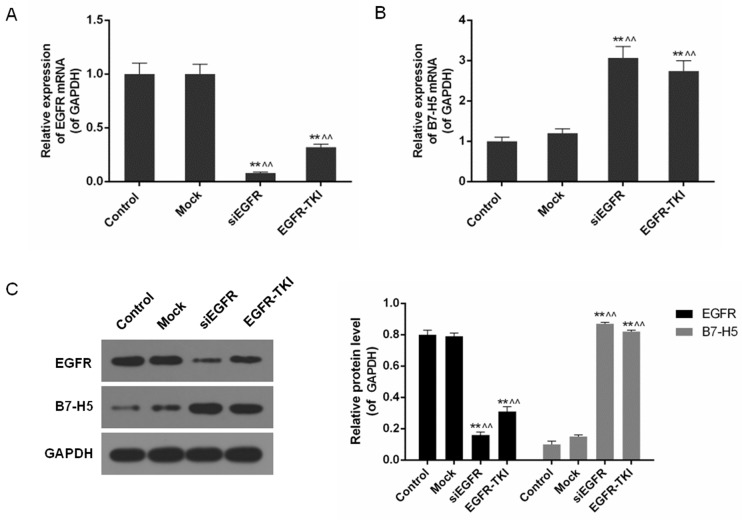
Low expression of EGFR increases B7-H5 levels in siEGFR- or EGFR-TKI-treated NCI-H1299 cells
It was detected that EGFR gene silencing and EGFR-TKI significantly inhibited EGFR mRNA and protein expression in NCI-H1299 cells, particularly in the siEGFR group (P<0.01; Fig. 2). In contrast to the low levels of EGFR expression, B7-H5 gene products were significantly increased (P<0.01; Fig. 2B and C). B7-H5 protein levels in the siEGFR and EGFR-TIKI groups were >4 times higher compared with the control and mock groups, where EGFR was highly expressed (P<0.01; Fig. 2C).
Figure 2.
Expression level of EGFR and B7-H5 in the control, mock, siEGFR and EGFR-TKI groups. (A) The expression of EGFR mRNA was significantly downregulated in EGFR-inhibited NCI-H1299 cells. (B) The expression of B7-H5 mRNA was significantly upregulated in EGFR-inhibited NCI-H1299 cells. (C) Inhibition of EGFR expression decreased the EGFR protein expression level and increased the B7-H5 protein expression level. Data are presented as the mean ± standard deviation; n=3. **P<0.01 vs. control group; ^^P<0.01 vs. mock group. EGFR, epidermal growth factor receptor; si, small interfering; TKI, tyrosine kinase inhibitor.
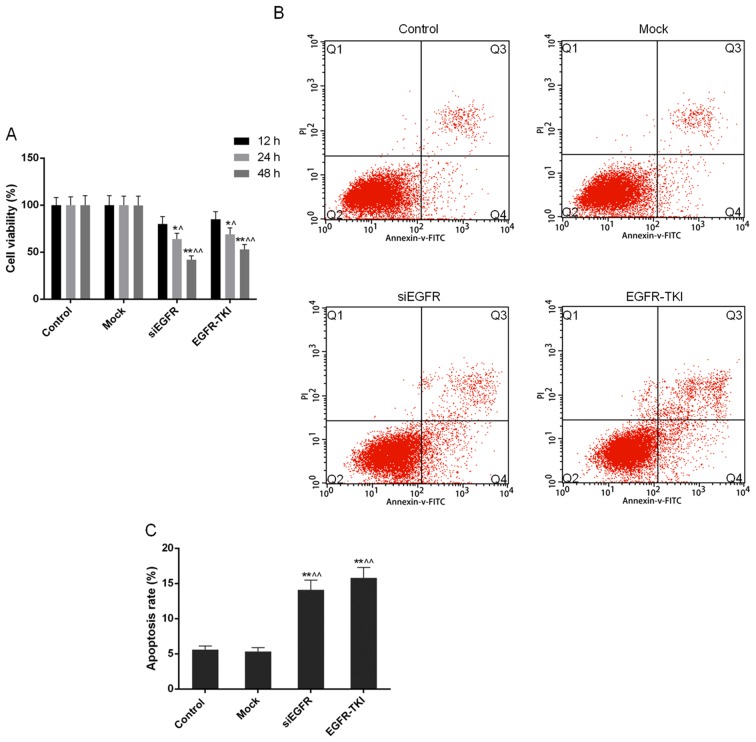
Cell viability is reduced when EGFR expression is inhibited in NCI-H1299 cells
Compared with the control and mock groups, it was observed through the CCK-8 assay that the inhibition of EGFR expression in siEGFR- and EGFR-TKI-treated cells was able to markedly decrease the cell viability and proliferation of NCI-H1299 cells; this effect occurred in a time-dependent manner. Cell viability in the siEGFR and EGFR-TKI groups, following incubation for 24 and 48 h, was significantly different compared with that in the control and mock groups (P<0.05 or P<0.01; Fig. 3A).
Figure 3.
Cell viability and apoptosis in the control, mock, siEGFR and EGFR-TKI groups. (A) Cell viability was markedly reduced by inhibiting EGFR expression, in a time-dependent manner. (B) Inhibition of EGFR expression increased the apoptosis of NCI-H1299 cells. (C) The apoptosis rate of NCI-H1299 cells was increased by inhibiting EGFR expression. Data are presented as the mean ± standard deviation; n=3. *P<0.05 and **P<0.01 vs. control group; ^P<0.05 and ^^P<0.01 vs. mock group. EGFR, epidermal growth factor receptor; si, small interfering; TKI, tyrosine kinase inhibitor; FITC, fluorescein isothiocyanate; PI, propidium iodide.
Apoptosis rate of NCI-H1299 cells increases when EGFR expression is inhibited
In the present study, the FCM results demonstrated that the apoptosis rate of NCI-H1299 cells was significantly increased to 14.09±1.38% in the siEGFR group and to 15.79±1.50% in the EGFR-TKI group, from ~5% in the control and mock groups (P<0.01; Fig. 3B and C).
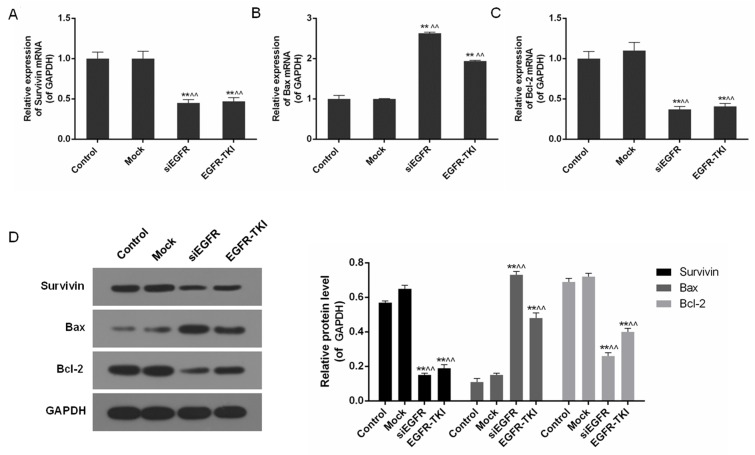
Inhibition of EGFR expression in NCI-H1299 cells downregulates Survivin and Bcl-2
In addition to the inhibition of EGFR, mRNA and protein levels of apoptosis-associated genes including Survivin, Bax and Bcl-2, were differentially-expressed in the siEGFR and EGFR-TKI groups, compared with the control and mock groups. In EGFR-inhibited NCI-H1299 cells, expression of the anti-apoptotic genes Survivin and Bcl-2 was significantly downregulated, whereas expression of the pro-apoptotic gene Bax was significantly increased, particularly in the siEGFR group (P<0.01; Fig. 4).
Figure 4.
Expression level of Survivin, Bax and Bcl-2 in the control, mock, siEGFR and EGFR-TKI groups. (A) The expression of Survivin mRNA was significantly downregulated in EGFR-inhibited NCI-H1299 cells. (B) The expression of Bax mRNA was significantly upregulated in EGFR-inhibited NCI-H1299 cells. (C) The expression of Bcl-2 mRNA was significantly downregulated in EGFR-inhibited NCI-H1299 cells. (D) Inhibition of EGFR expression decreased the Survivin and Bcl-2 protein expression level, and increased the Bax protein expression level. Data are presented as the mean ± standard deviation; n=3. **P<0.01 vs. respective control group; ^^P<0.01 vs. respective mock group. EGFR, epidermal growth factor receptor; si, small interfering; TKI, tyrosine kinase inhibitor; Bax, apoptosis regulator Bax; Bcl-2, apoptosis regulator Bcl-2.
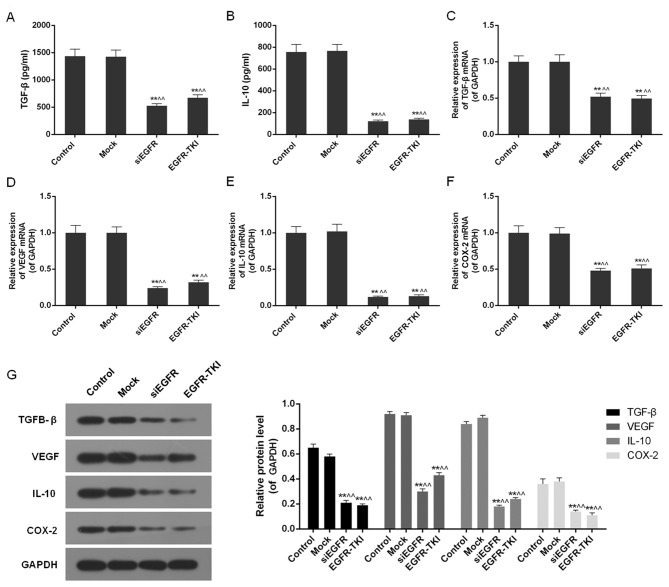
Inhibition of EGFR expression in NCI-H1299 cells downregulates TGF-β, VEGF, IL-10 and COX-2
Inhibition of EGFR expression in the siEGFR and EGFR-TKI groups was observed to dramatically reduce the expression of immune cell cytokines, including TGF-β, VEGF, IL-10 and COX-2. The ELISA detected a significant decrease in TGF-β and IL-10 in EGFR-inhibited cells (P<0.01; Fig. 5A and B). It was additionally observed via qPCR and western blotting that the mRNA and protein expression of TGF-β, VEGF, IL-10 and COX-2 was significantly downregulated compared with the control and mock groups (P<0.01; Fig. 5C-G).
Figure 5.
TGF-β, VEGF, IL-10 and COX-2 content in the control, mock, siEGFR and EGFR-TKI groups. (A) The TGF-β content was decreased in EGFR-inhibited NCI-H1299 cells. (B) The IL-10 content was decreased in EGFR-inhibited NCI-H1299 cells. (C) The expression of TGF-β mRNA was downregulated in EGFR-inhibited NCI-H1299 cells. (D) The expression of VEGF mRNA was downregulated in EGFR-inhibited NCI-H1299 cells. (E) The expression of IL-10 mRNA was downregulated in EGFR-inhibited NCI-H1299 cells. (F) The expression of COX-2 mRNA was downregulated in EGFR-inhibited NCI-H1299 cells. (G) Inhibition of EGFR expression decreased the protein expression levels of TGF-β, VEGF, IL-10 and COX-2. Data are presented as the mean ± standard deviation; n=3. **P<0.01 vs. control group; ^^P<0.01 vs. mock group. EGFR, epidermal growth factor receptor; si, small interfering; TKI, tyrosine kinase inhibitor; TGF-β, transforming growth factor-β; IL-10, interleukin-10; VEGF, vascular endothelial growth factor; COX-2, cyclooxygenase-2.
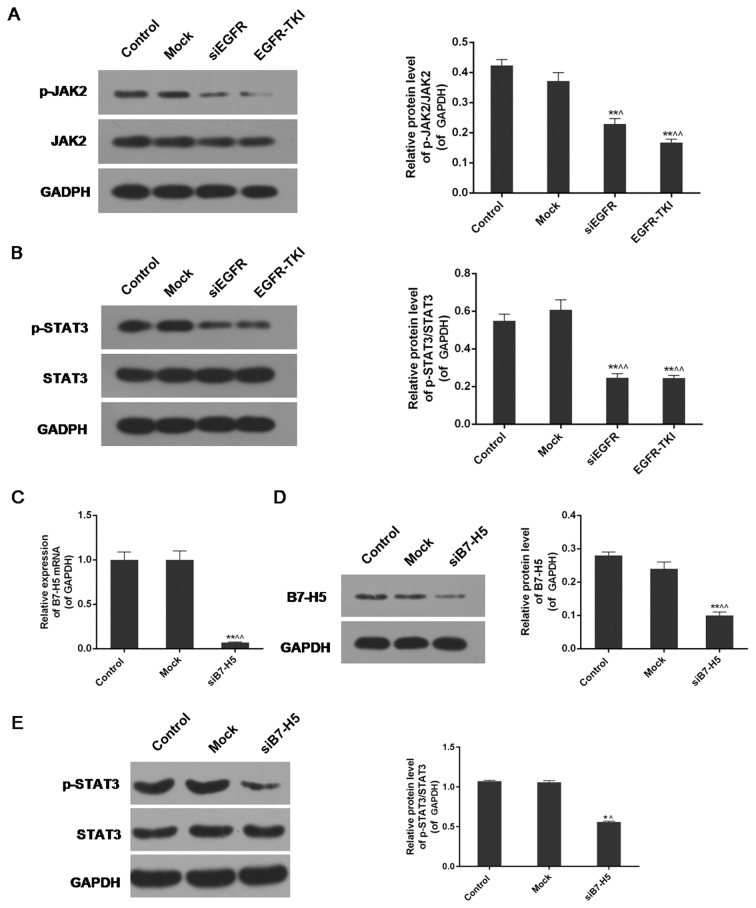
Inhibition of EGFR expression in NCI-H1299 cells inhibits the phosphorylation of JAK2 and STAT3
The expression levels of phosphorylated (p-)JAK2, JAK2, p-STAT3 and STAT3 were analysed via western blotting. The results indicated a significant decrease in the protein expression levels of p-JAK2 and p-STAT3 in the siEGFR and EGFR-TKI groups compared with the control and mock groups. For JAK2 and STAT3, there were no notable differences in their expression among the four groups. The expression levels of p-JAK2/JAK2 and p-STAT3/STAT3 were decreased in EGFR-inhibited cells compared with normal NCI-H1299 cells (P<0.01; Fig. 6A and B).
Figure 6.
Protein expression levels of p-JAK2, JAK2, p-STAT3 and STAT3 in EGFR-inhibited and B7-H5-inhibited cells. (A) The phosphorylation of JAK2 was inhibited in EGFR-inhibited NCI-H1299 cells. (B) The phosphorylation of STAT3 was inhibited in EGFR-inhibited NCI-H1299 cells. (C) The expression of B7-H5 mRNA was significantly downregulated in B7-H5 gene-silenced NCI-H1299 cells. (D) The protein expression level of B7-H5 was decreased in B7-H5-inhibited NCI-H1299 cells. (E) The phosphorylation of STAT3 was inhibited in B7-H5-inhibited NCI-H1299 cells. Data are presented as mean ± standard deviation; n=3. *P<0.05 and **P<0.01 vs. control group; ^P<0.05 and ^^P<0.01 vs. mock group. EGFR, epidermal growth factor receptor; si, small interfering; TKI, tyrosine kinase inhibitor; JAK2, tyrosine-protein kinase JAK2; STAT3, signal transducer and activator of transcription 3; p, phosphorylated.
B7-H5 silencing partially enhances the STAT3 signalling pathway under EGFR-TKI conditions
B7-H5 gene silencing significantly downregulated the expression of B7-H5 in NCI-H1299 cells, at the mRNA and protein levels (P<0.01; (Fig. 6C and D). The results of the western blotting revealed that weak expression of B7-H5 under treatment with EGFR-TKI resulted in reduced phosphorylation of STAT3. The p-STAT3/STAT3 ratio of protein expression was decreased from 1.07±0.01 and 1.06±0.02 in the control and mock groups, respectively, to 0.56±0.01 in the siB7-H5 group (P<0.01; Fig. 6E). The decrease was less marked compared with the EGFR-TKI group in Fig. 6B.
Discussion
In the present study, a negative correlation between EGFR and B7-H5 expression was demonstrated in lung cancer tissues, in addition to a number of lung cancer cell lines, including A549, NCI-H1299, NCI-H1755 and 95-D. In lung cancer, B7-H5 was weakly expressed, whereas EGFR expression was abundant; by contrast, high expression of B7-H5 and low expression of EGFR was detected in normal cells. In the present study, the inhibition of EGFR expression by EGFR gene silencing and EGFR-TKI decreased the viability of NCI-H1299 cells in a time-dependent manner; furthermore, the apoptosis rate was increased ~2-fold compared with normal NCI-H1299 cells, thus implying the inhibiting influence of EGFR downregulation on tumour cells.
The anti-tumour action mechanism of the body primarily involves cell-mediated immunity and humoral immunity. Cell-mediated immunity is the principal mechanism of anti-tumour action; however, during tumorigenesis, tumour cells are able to evade monitoring and attack the immune system to continue mitosis and growth (in a variety of ways). Effective activation and functional mediation of T lymphocytes requires the synergistic effect of dual signal control. The first signal is the complex formed by an antigen peptide and the major histocompatibility complex, which is identified by a T cell receptor, and the second signal is the receptor combined with a B7-family antigen-presenting cell or other co-stimulatory molecules. In the absence of the second signal, although the first signal exists, the T lymphocyte may not effectively proliferate to modulate the immune response, resulting in tumour immune evasion (21,22). A recent study in pancreatic cancer indicated that the loss of B7-H5 may contribute to immune evasion (19). Zhu et al (18) reported that B7-H5 was constitutively expressed in macrophages and may be induced on dendritic cells, and additionally demonstrated that the B7-H5/CD28H interaction co-stimulated human T cell growth and cytokine production. It has been confirmed that alterations in tumour immunity result from alterations in gene activity and gene products in tumour cells. Tumorigenesis and metastasis are closely associated with the immunity of the human body and the internal environment of tumour cells (23,24). Recent studies have indicated that there may exist cells that have different functions against normal human tissues surrounding the microenvironment of tumour cells (25–27). Without an effective anti-tumour immune response, these cells may induce tumorigenesis and metastasis via inhibition of the immune response of local tumours. Immune cells in the tumour microenvironment are immunological effector cells that serve an important role in tumour recognition and immune defence to attack tumour cells (28–30). However, it was demonstrated that tumour cells are able to secrete a number of inhibitors, modify antigens on the cell surface, or negatively regulate gene expression so as to escape immune cell recognition and destruction. This may eventually promote the formation, development and metastasis of tumour cells (31). In the present study, a marked downregulation of the expression of the anti-apoptotic proteins Survivin and Bcl-2, and an upregulation of the pro-apoptotic protein Bax, was detected in EGFR-inhibited groups. The decreasing levels of Bcl-2/Bax promoted apoptosis, and resulted in increased apoptosis rates in the siEGFR and EGFR-TKI groups compared with normal cells.
A number of lines of evidence have indicated that numerous cellular factors, including TGF-β, VEGF, IL-10 and COX-2, are involved in immunosuppression (32–34). As a multifunctional cellular factor, TGF-β has an inhibitory effect on the immune system, and the immunosuppression activated by TGF-β in the microenvironment of tumours is considered to be one of the most important elements for cancer cells to evade immune surveillance. TGF-β inhibits the multiplication of T lymphocytes by suppressing the secretion of IL-2, which is one of the key factors in stimulating the proliferation and differentiation of T lymphocytes (35). IL-10 is primarily secreted by Th2 cells, mononuclear P macrophages, B lymphocytes, and keratinocytes under normal conditions; however, an unusual increase in its expression occurs during tumorigenesis. In the body of a cancer patient, the Th2 cell subset, the primary source of IL-10, increases abnormally. Furthermore, T regulatory cells, another source of IL-10, secrete high levels of IL-10 and low levels of IL-2; additionally, tumours themselves may generate IL-10, leading to a rise in the expression of IL-10 (36). Previous studies demonstrated that VEGF has a strong inhibitory effect on the differentiation of hematopoietic stem cells (CD34+) to dendritic cells, so as to further influence the proliferation of specific cytotoxic T lymphocytes, thus protecting tumours from the monitoring of the immune system (37,38). The membrane-conjugated protein COX-2 is able to induce apoptosis in immune cells by activating caspase-3, inhibit the antitumor activity of T lymphocytes and NK cells by inducing the production of prostaglandin E2, inhibit the generation of tumour necrosis factor and restrain antibody synthesis by B lymphocytes, consequently reducing the ability of the body to combat tumours while promoting tumour angiopoeisis (39). In the present study, inhibition of EGFR expression and high expression of B7-H5 significantly decreased expression of the four previously mentioned cellular factors, including TGF-β, VEGF, IL-10 and COX-2 in NCI-H1299 cells. Weak expression of these four factors suppresses tumorigenesis and development, and decreases the capability of a cell to escape immune surveillance.
STAT is a regulatory factor of signal transduction. STAT3 is the key signalling cascade compound in the STAT family, which is involved in the response of various cytokines, including the JAK/STAT signalling pathway. STAT3 is phosphorylated by JAK at a tyrosine residue, and subsequently transferred to the cell nucleus to activate its target gene. The JAK/STAT signalling pathway is associated with cell proliferation, differentiation and apoptosis; however, continuous activation of the pathway leads to abnormal cell proliferation and malignant transformation (40,41). p-STAT3, the activated form of STAT3, may act on specific DNA binding sites in the cell nucleus to directly or indirectly upregulate the expression of anti-apoptotic genes, including Bcl-2, Bcl-2-like protein 1 and cyclin D1/D2 to regulate the proliferation and apoptosis of cells (42,43). To date, high expression of activated p-STAT3 has been detected in a number of types of cancer, including gastric cancer, colon cancer, prostate cancer and breast cancer (44–46). Abnormal activation of the JAK/STAT signalling pathway may induce growth disorders, and promote malignant cell transformation and tumorigenesis (47). Compared with normal tissues and cells, the activation of STAT components, particularly STAT3, exists in the majority of human cancer tissues and cells to mediate cell proliferation, apoptosis, differentiation and other physiological activities (46). The present study demonstrated that in NCI-H1299 cells, the JAK2/STAT3 signalling pathway is activated, and inhibiting the expression of EGFR markedly reduced the phosphorylation of JAK2 and STAT3 to restrain its activation. The results indicated the important association between EGFR expression levels and activation of the JAK2/STAT3 signalling pathway. Due to the negative correlation between EGFR and B7-H5, the expression of B7-H5 was markedly increased while that of EGFR was inhibited, highlighting the possible role of B7-H5 in EGFR-regulated tumour immune escape. However, when EGFR and B7-H5 were inhibited, STAT3 activation remained suppressed, which may have resulted from the fact that the effects of EGFR on regulating the JAK2/STAT3 pathway are attributed to complex factors and mechanisms, and B7-H5 is only one of the influential elements. Apart from the significant association between EGFR and certain members of the B7 family, including B7-H5 and B7-H3, Zhang et al (48) reported that the expression of programmed death-ligand 1 may be regulated by EGFR activation in esophageal squamous cell carcinoma, and revealed the important role of EGFR-mediated immune escape. Furthermore, in triple-negative breast cancer cells, Lim et al (49) recently illustrated that EGFR signaling promoted tumor progression and immune evasion by elevating the level of aerobic glycolysis. The complete mechanism involved in EGFR-targeted therapy for immune escape requires further study.
The results of the present study demonstrated a notable negative correlation between EGFR and B7-H5 expression levels, in lung cancer tissues and cancer cell lines. Inhibiting the expression of EGFR by gene silencing and EGFR inhibition significantly reduced the cell viability and increased the apoptosis of NCI-H1299 cells, by upregulating the expression of Survivin and Bcl-2. The protein expression levels of TGF-β, VEGF, IL-10 and COX-2 were decreased by weak activation of the JAK2/STAT3 signalling pathway. EGFR may be involved in immune evasion, possibly through regulation of B7-H5 expression in NSCLC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Zhejiang Provincial Nature Fund (grant no. 2017C33178).
Availability of data and materials
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
Authors' contributions
ZD and GZ conceived and designed the study. WX analyzed and interpreted the patient data regarding the use of cancer tissues and cell lines. LZ performed the cell experiments. This report has been approved by all authors.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of The First People's Hospital of Huzhou.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Nand M, Maiti P, Pant R, Kumari M, Chandra S, Pande V. Virtual screening of natural compounds as inhibitors of EGFR 696–1022 T790M associated with non-small cell lung cancer. Bioinformation. 2016;12:311–317. doi: 10.6026/97320630012311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Remon J, Besse B, Soria JC. Successes and failures: What did we learn from recent first-line treatment immunotherapy trials in non-small cell lung cancer? BMC Med. 2017;15:55. doi: 10.1186/s12916-017-0857-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hynes NE, Lane HA. ERBB receptors and cancer: The complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–354. doi: 10.1038/nrc1667. [DOI] [PubMed] [Google Scholar]
- 5.Hirsch FR, Varella-Garcia M, Bunn PA, Jr, Di Maria MV, Veve R, Bremmes RM, Baron AE, Zeng C, Franklin WA. Epidermal growth factor receptor in non-small-cell lung carcinomas: Correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol. 2003;21:3798–3807. doi: 10.1200/JCO.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki S, Dobashi Y, Sakurai H, Nishikawa K, Hanawa M, Ooi A. Protein overexpression and gene amplification of epidermal growth factor receptor in nonsmall cell lung carcinomas. An immunohistochemical and fluorescence in situ hybridization study. Cancer. 2005;103:1265–1273. doi: 10.1002/cncr.20909. [DOI] [PubMed] [Google Scholar]
- 7.Woodburn JR. The epidermal growth factor receptor and its inhibition in cancer therapy. Pharmacol Ther. 1999;82:241–250. doi: 10.1016/S0163-7258(98)00045-X. [DOI] [PubMed] [Google Scholar]
- 8.Wakeling AE, Guy SP, Woodburn JR, Ashton SE, Curry BJ, Barker AJ, Gibson KH. ZD1839 (Iressa): An orally active inhibitor of epidermal growth factor signaling with potential for cancer therapy. Cancer Res. 2002;62:5749–5754. [PubMed] [Google Scholar]
- 9.Inoue A, Suzuki T, Fukuhara T, Maemondo M, Kimura Y, Morikawa N, Watanabe H, Saijo Y, Nukiwa T. Prospective phase II study of gefitinib for chemotherapy-naive patients with advanced non-small-cell lung cancer with epidermal growth factor receptor gene mutations. J Clin Oncol. 2006;24:3340–3346. doi: 10.1200/JCO.2005.05.4692. [DOI] [PubMed] [Google Scholar]
- 10.Sonobe M, Manabe T, Wada H, Tanaka F. Mutations in the epidermal growth factor receptor gene are linked to smoking-independent, lung adenocarcinoma. Br J Cancer. 2005;93:355–363. doi: 10.1038/sj.bjc.6602707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa DB, Schumer ST, Tenen DG, Kobayashi S. Differential responses to erlotinib in epidermal growth factor receptor (EGFR)-mutated lung cancers with acquired resistance to gefitinib carrying the L747S or T790M secondary mutations. J Clin Oncol. 2008;26:1184–1186. doi: 10.1200/JCO.2007.14.9039. [DOI] [PubMed] [Google Scholar]
- 12.Rolfo C, Caglevic C, Santarpia M, Araujo A, Giovannetti E, Gallardo CD, Pauwels P, Mahave M. Immunotherapy in NSCLC: A Promising and Revolutionary Weapon. Adv Exp Med Biol. 2017;995:97–125. doi: 10.1007/978-3-319-53156-4_5. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Jia Y, Zhao S, Zhang X, Wang X, Han X, Wang Y, Ma M, Shi J, Liu L. BIN1 reverses PD-L1-mediated immune escape by inactivating the c-MYC and EGFR/MAPK signaling pathways in non-small cell lung cancer. Oncogene. 2017;36:6235–6243. doi: 10.1038/onc.2017.217. [DOI] [PubMed] [Google Scholar]
- 14.Lenschow DJ, Walunas TL, Bluestone JA. CD28/B7 system of T cell costimulation. Annu Rev Immunol. 1996;14:233–258. doi: 10.1146/annurev.immunol.14.1.233. [DOI] [PubMed] [Google Scholar]
- 15.Dong H, Chen L. B7-H1 pathway and its role in the evasion of tumor immunity. J Mol Med (Berl) 2003;81:281–287. doi: 10.1007/s00109-003-0430-2. [DOI] [PubMed] [Google Scholar]
- 16.Kryczek I, Wei S, Zhu G, Myers L, Mottram P, Cheng P, Chen L, Coukos G, Zou W. Relationship between B7-H4, regulatory T cells, and patient outcome in human ovarian carcinoma. Cancer Res. 2007;67:8900–8905. doi: 10.1158/0008-5472.CAN-07-1866. [DOI] [PubMed] [Google Scholar]
- 17.Inamura K, Yokouchi Y, Kobayashi M, Sakakibara R, Ninomiya H, Subat S, Nagano H, Nomura K, Okumura S, Shibutani T, Ishikawa Y. Tumor B7-H3 (CD276) expression and smoking history in relation to lung adenocarcinoma prognosis. Lung Cancer. 2017;103:44–51. doi: 10.1016/j.lungcan.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 18.Zhu Y, Yao S, Iliopoulou BP, Han X, Augustine MM, Xu H, Phennicie RT, Flies SJ, Broadwater M, Ruff Wetal. B7-H5 costimulates human T cells via CD28H. Nat Commun. 2013;4:2043. doi: 10.1038/ncomms3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Byers JT, Paniccia A, Kaplan J, Koenig M, Kahn N, Wilson L, Chen L, Schulick RD, Edil BH, Zhu Y. Expression of the Novel Costimulatory Molecule B7-H5 in Pancreatic Cancer. Ann Surg Oncol. 2015;22(Suppl 3):S1574–S1579. doi: 10.1245/s10434-014-4293-2. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Chen L. Co-inhibitory molecules of the B7-CD28 family in the control of T-cell immunity. Nat Rev Immunol. 2004;4:336–347. doi: 10.1038/nri1349. [DOI] [PubMed] [Google Scholar]
- 22.Woodland DL, Kohlmeier JE. Migration, maintenance and recall of memory T cells in peripheral tissues. Nat Rev Immunol. 2009;9:153–161. doi: 10.1038/nri2496. [DOI] [PubMed] [Google Scholar]
- 23.Katayama A, Ogino T, Bandoh N, Nonaka S, Harabuchi Y. Expression of CXCR4 and its down-regulation by IFN-gamma in head and neck squamous cell carcinoma. Clin Cancer Res. 2005;11:2937–2946. doi: 10.1158/1078-0432.CCR-04-1470. [DOI] [PubMed] [Google Scholar]
- 24.Muller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 25.Ghebeh H, Mohammed S, Al-Omair A, Qattan A, Lehe C, Al-Qudaihi G, Elkum N, Alshabanah M, Bin Amer S, Tulbah A, et al. The B7-H1 (PD-L1) T lymphocyte-inhibitory molecule is expressed in breast cancer patients with infiltrating ductal carcinoma: Correlation with important high-risk prognostic factors. Neoplasia. 2006;8:190–198. doi: 10.1593/neo.05733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamanishi J, Mandai M, Iwasaki M, Okazaki T, Tanaka Y, Yamaguchi K, Higuchi T, Yagi H, Takakura K, Minato N, et al. Programmed cell death 1 ligand 1 and tumor-infiltrating CD8+ T lymphocytes are prognostic factors of human ovarian cancer; Proc Natl Acad Sci USA; 2007; pp. 3360–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun J, Xu K, Wu C, Wang Y, Hu Y, Zhu Y, Chen Y, Shi Q, Yu G, Zhang X. PD-L1 expression analysis in gastric carcinoma tissue and blocking of tumor-associated PD-L1 signaling by two functional monoclonal antibodies. Tissue Antigens. 2007;69:19–27. doi: 10.1111/j.1399-0039.2006.00701.x. [DOI] [PubMed] [Google Scholar]
- 28.Sun Y, Wang Y, Zhao J, Gu M, Giscombe R, Lefvert AK, Wang X. B7-H3 and B7-H4 expression in non-small-cell lung cancer. Lung Cancer. 2006;53:143–151. doi: 10.1016/j.lungcan.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 29.Ghebeh H, Barhoush E, Tulbah A, Elkum N, Al-Tweigeri T, Dermime S. FOXP3+ Tregs and B7-H1+/PD-1+ T lymphocytes co-infiltrate the tumor tissues of high-risk breast cancer patients: Implication for immunotherapy. BMC cancer. 2008;8:57. doi: 10.1186/1471-2407-8-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pettit SJ, Ali S, O'Flaherty E, Griffiths TR, Neal DE, Kirby JA. Bladder cancer immunogenicity: Expression of CD80 and CD86 is insufficient to allow primary CD4+ T cell activation in vitro. Clin Exp Immunol. 1999;116:48–56. doi: 10.1046/j.1365-2249.1999.00857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, Fitz LJ, Malenkovich N, Okazaki T, Byrne MC, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192:1027–1034. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim S, Buchlis G, Fridlender ZG, Sun J, Kapoor V, Cheng G, Haas A, Cheung HK, Zhang X, Corbley Metal. Systemic blockade of transforming growth factor-beta signaling augments the efficacy of immunogene therapy. Cancer Res. 2008;68:10247–10256. doi: 10.1158/0008-5472.CAN-08-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaspar NJ, Li L, Kapoun AM, Medicherla S, Reddy M, Li G, O'Young G, Quon D, Henson M, Damm D Letal. Inhibition of transforming growth factor beta signaling reduces pancreatic adenocarcinoma growth and invasiveness. Mol Pharmacol. 2007;72:152–161. doi: 10.1124/mol.106.029025. [DOI] [PubMed] [Google Scholar]
- 34.Erkanli S, Bolat F, Kayaselcuk F, Demirhan B, Kuscu E. COX-2 and survivin are overexpressed and positively correlated in endometrial carcinoma. Gynecol Oncol. 2007;104:320–325. doi: 10.1016/j.ygyno.2006.08.044. [DOI] [PubMed] [Google Scholar]
- 35.Li MO, Sanjabi S, Flavell RA. Transforming growth factor-beta controls development, homeostasis, and tolerance of T cells by regulatory T cell-dependent and -independent mechanisms. Immunity. 2006;25:455–471. doi: 10.1016/j.immuni.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 36.Hoechst B, Ormandy LA, Ballmaier M, Lehner F, Kruger C, Manns MP, Greten TF, Korangy F. A new population of myeloid-derived suppressor cells in hepatocellular carcinoma patients induces CD4(+)CD25(+)Foxp3(+) T cells. Gastroenterology. 2008;135:234–243. doi: 10.1053/j.gastro.2008.03.020. [DOI] [PubMed] [Google Scholar]
- 37.Kopfstein L, Veikkola T, Djonov VG, Baeriswyl V, Schomber T, Strittmatter K, Stacker SA, Achen MG, Alitalo K, Christofori G. Distinct roles of vascular endothelial growth factor-D in lymphangiogenesis and metastasis. Am J Pathol. 2007;170:1348–1361. doi: 10.2353/ajpath.2007.060835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kamstock D, Elmslie R, Thamm D, Dow S. Evaluation of a xenogeneic VEGF vaccine in dogs with soft tissue sarcoma. Cancer Immunol Immunother. 2007;56:1299–1309. doi: 10.1007/s00262-007-0282-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu XH, Kirschenbaum A, Yu K, Yao S, Levine AC. Cyclooxygenase-2 suppresses hypoxia-induced apoptosis via a combination of direct and indirect inhibition of p53 activity in a human prostate cancer cell line. J Biol Chem. 2005;280:3817–3823. doi: 10.1074/jbc.M406577200. [DOI] [PubMed] [Google Scholar]
- 40.Bahrenberg G, Behrmann I, Barthel A, Hekerman P, Heinrich PC, Joost HG, Becker W. Identification of the critical sequence elements in the cytoplasmic domain of leptin receptor isoforms required for Janus kinase/signal transducer and activator of transcription activation by receptor heterodimers. Mol Endocrinol. 2002;16:859–872. doi: 10.1210/mend.16.4.0800. [DOI] [PubMed] [Google Scholar]
- 41.Bjorbaek C, Uotani S, da Silva B, Flier JS. Divergent signaling capacities of the long and short isoforms of the leptin receptor. J Biol Chem. 1997;272:32686–32695. doi: 10.1074/jbc.272.51.32686. [DOI] [PubMed] [Google Scholar]
- 42.Kanda N, Seno H, Konda Y, Marusawa H, Kanai M, Nakajima T, Kawashima T, Nanakin A, Sawabu T, Uenoyama Yetal. STAT3 is constitutively activated and supports cell survival in association with survivin expression in gastric cancer cells. Oncogene. 2004;23:4921–4929. doi: 10.1038/sj.onc.1207606. [DOI] [PubMed] [Google Scholar]
- 43.Du XL, Yang H, Liu SG, Luo ML, Hao JJ, Zhang Y, Lin DC, Xu X, Cai Y, Zhan QM, Wang MR. Calreticulin promotes cell motility and enhances resistance to anoikis through STAT3-CTTN-Akt pathway in esophageal squamous cell carcinoma. Oncogene. 2009;28:3714–3722. doi: 10.1038/onc.2009.237. [DOI] [PubMed] [Google Scholar]
- 44.Zhang X, Zhang J, Wang L, Wei H, Tian Z. Therapeutic effects of STAT3 decoy oligodeoxynucleotide on human lung cancer in xenograft mice. BMC Cancer. 2007;7:149. doi: 10.1186/1471-2407-7-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang WT, Yang SF, Wu CC, Chen WT, Huang YC, Su YC, Chai CY. Expression of signal transducer and activator of transcription 3 and suppressor of cytokine signaling 3 in urothelial carcinoma. Kaohsiung J Med Sci. 2009;25:640–646. doi: 10.1016/S1607-551X(09)70569-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yakata Y, Nakayama T, Yoshizaki A, Kusaba T, Inoue K, Sekine I. Expression of p-STAT3 in human gastric carcinoma: Significant correlation in tumour invasion and prognosis. Int J Oncol. 2007;30:437–442. [PubMed] [Google Scholar]
- 47.Huang S. Regulation of metastases by signal transducer and activator of transcription 3 signaling pathway: Clinical implications. Clin Cancer Res. 2007;13:1362–1366. doi: 10.1158/1078-0432.CCR-06-2313. [DOI] [PubMed] [Google Scholar]
- 48.Zhang W, Pang Q, Yan C, Wang Q, Yang J, Yu S, Liu X, Yuan Z, Wang P, Xiao Z. Induction of PD-L1 expression by epidermal growth factor receptor-mediated signaling in esophageal squamous cell carcinoma. Onco Targets Ther. 2017;10:763–771. doi: 10.2147/OTT.S118982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lim SO, Li CW, Xia W, Lee HH, Chang SS, Shen J, Hsu JL, Raftery D, Djukovic D, Gu Hetal. EGFR signaling enhances aerobic glycolysis in triple-negative breast cancer cells to promote tumor growth and immune escape. Cancer Res. 2016;76:1284–1296. doi: 10.1158/0008-5472.CAN-15-2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.