Abstract
The ectopic HLA-G expression in malignancies has been extensively explored and clinical significance of the molecule was widely acknowledged. Besides previously well-documented seven isoforms (HLA-G1~-G7), other novel isoforms of HLA-G have been reported but their clinical relavenace remians evaluated. In this study, lesion HLA-G expression in 379 case-matched serial section primary colorectal cancers (CRC) were evaluated with mAb 4H84 (recognizing an epitope in HLA-G α1 domain), and mAb 5A6G7 (recognizing an epitope encoded by intron 4), respectively. Data showed that HLA-G positive staining with mAbs 4H84 and 5A6G7 was 70.7 and 60.4%, respectively. When percentage of HLA-G expression detected with mAb 4H84 subtracted that with mAb 5A6G7, the difference (ΔHLA-G) with negative (ΔHLA-Gneg), comparable (ΔHLA-Gcom) and positive (ΔHLA-Gpos) were observed in 64 (16.9%), 159 (42.0%), and 156 (41.2%) cases, respectively. Noteworthy, unexpected immunostaining was observed in 44 (11.6%) lesions that no staining was detected with mAb 4H84 but positive with mAb 5A6G7 (4H84neg5A6G7pos). This staining pattern was unpredictable because all seven known HLA-G isoforms containing the α 1 domain could be recognized by the mAb 4H84. Moreover, patients with ΔHLA-Gneg had obviously better survival than those with ΔHLA-Gcom and ΔHLA-Gpos (p = 0.017), and ΔHLA-G could be an independent prognostic factor for CRC patients (p = 0.008). Our findings provides the first report that potential unidentified HLA-G isoforms is of distinct clinical significance in CRC patients.
Keywords: HLA-G, isoform, antibodies, colorectal cancer, prognosis
Introduction
The cancer promoting function of HLA-G, such as rendering comprehensive suppressive roles to various types of immune component cells and enhancing the proliferation and accumulation of immune regulatory cells have been extensively investigated (1). Different degree of aberrant ectopic expression of HLA-G in cancers has been frequently found in most of the cancers studied so far, and the conception that HLA-G expression plays critical basically and clinical parts in cancer biology or therapy was established (2).
Besides application of the clinical tumor lesions, the direct roles of HLA-G participating in cancer progression have been demonstrated in previous studies with murine models. Data obtained from xenotumor or syngeneic immunocompetent murine tumor model revealed that both HLA-G1 or HLA-G5 expression promoted tumor development by impairment of innate and adaptive immune responses but favoring the immune suppressive cells such as CD11b+Gr1+ myeloid-derived suppressor cells (MDSC) expansion (3, 4). Fortunately, these could be reversed with HLA-G and/or receptor blockade by certain antibodies. With an immunodeficient murine ovarian cancer xenotumor model, we found that HLA-G1 expressing ovarian cancer cells had stronger invasion and metastasis potential compared that with the HLA-G-negative parental cells (5). We also presented the underlying mechanisms of HLA-G in tumor progression not only related to the inhibition of NK lysis, but to the specific induction of matrix metalloproteinase-15 (MMP-15) expression (6). This finding was solidified by later studies that HLA-G5 could induce the expression and activity of MMP-2 and MMP-9 in trophoblastic cells, while knock-down the HLA-G experssion resulted in dramatically decreased expression of MMPs was also observed (7, 8).
It's well documented thus far, with the alternative splicing of the primary transcript of HLA-G, seven isoforms including four-membrane bound (HLA-G1, -G2,-G3, and -G4) and three soluble (HLA-G5, -G6, and -G7) molecules were validated. Each unique HLA-G isoform contains one to three extracellular globular domains encoded by exon 2 (α 1), exon 3 (α2) and exon 4 (α3) respectively, and presence or absence of residues encoded by intronic sequences (IMGT/HLA database). However, these seven isoforms all contain the extracellular α 1 domain (9, 10). Among these isoforms, HLA-G1 and soluble HLA-G5 molecule have been studied more extensively due to the commercially available antibodies. However, it's reasonable that other novel unrecognized HLA-G isoforms remains unveiled. Indeed, based on immunohistochemical and deep sequencing, an important study by Tronik-Le Roux et al. (11) recently revealed that unrecognized novel HLA-G isoforms such as isoforms without α1 domain were presented in renal cancer samples. The findings raises the questions whether these novel HLA-G proteins are of clinical significance.
In our study, mAbs 4H84 and 5A6G7 were used to analyze the HLA-G expression by immunohistochemistry in primary colorectal cancer (CRC) samples. The percentage of HLA-G expression in tumor cells by mAbs 4H84 and 5A6G7 were compared, and the difference of the HLA-G expression (ΔHLA-G) was obtained by the percentage of HLA-G expression detected with HLA-G mAb 4H84 subtracted that with mAb 5A6G7. ΔHLA-G with negative (ΔHLA-Gneg), comparable (ΔHLA-Gcom) and positive (ΔHLA-Gpos) were observed. Our findings revealed that patients with ΔHLA-Gneg had much better survival than patients with ΔHLA-Gcom and ΔHLA-Gpos, and the ΔHLA-G could be an independent prognostic factor for CRC patients.
Materials and methods
CRC patients
A cohort of 379 consecutive primary CRC lesions were provided by Tissue Bank of Taizhou Hospital of Zhejiang Province (National human genetic resources platform of China YCZYPT[2017]02). Patients were diagnosed and treated at the department of Gastrointestinal Surgery in Taizhou Hospital of Zhejiang Province from November 2004 to September 2012.
The clinical stage including stage I (n = 86), II (n = 114), III (n = 169), and IV (n = 11) respectively, were classified according to the UICC and the AJCC 7th TNM staging system (12). Details of the clinical history of the patients was recorded and the last follow-up was performed at April,15th, 2014. During the period, 38 patients was lost follow-up in the cohort. Overall survival was calculated from the date of surgical operation to the event (patient death, n = 113) or censored (last follow-up, n = 228) with the median follow-up of 45.0 months. 113 cancer-related deaths was observed in 10 stage I, 26 stage II, 71 stage III and 6 stage IV CRC patients. Informed written consent was provided by all participated patients and the study protocol was approved by the institutional ethic review board, Taizhou Hospital of Zhejiang Province.
Immunohistochemistry and staining evaluation
Immunohistochemistry analysis for HLA-G expression was performed for each case-matched serial section primary CRC lesions on 4-μm paraffin-embedded sections according to our previous study with the two anti-HLA-G murine monoclonal antibodies (mAb) 4H84 (1:500, Exbio, Prague, Czech Republic) and 5A6G7 (1:500, Exbio, Prague, Czech Republic), respectively (13, 14). mAb 4H84 (IgG1) was immunized with amino acids 61–83 in the HLA-G alpha 1 domain, which recognizes an epitope located in the α 1 domain in denatured heavy chain to all α 1 domain containing HLA-G isoforms such as HLA-G1~HLA-G7. mAb 5A6G7 (IgG1) was immunized with C-terminal amino acid sequence (22-mer) of HLA-G5 and -G6 proteins coupled to ovalbumin, which recognizes HLA-G isoforms such as HLA-G5 and -G6 encoded by the retained intron 4 (IMGT/HLA database), but not cross react with the HLA-G1 isoform.
The percentage of HLA-G expression was evaluated by two reviewers who were blind to the patient clinicopathological information. The percentage of HLA-G positive tumor cells was based on the presence of HLA-G staining while irrespective the staining intensity. HLA-G positive CRC cells >5% in a section was considered as positive (15).
Difference of the percentage of HLA-G positve tumor cells (ΔHLA-G) in the case-matched CRC samples was calculated by the percentage of HLA-G expression detected with mAb 4H84 subtracted that with mAb 5A6G7. According to the value of ΔHLA-G, three groups were obtained as follows: ΔHLA-Gneg (ΔHLA-G> −5.0%), ΔHLA-Gcom (−5.0%≤ΔHLA-G≤5.0%), and ΔHLA-Gpos (ΔHLA-G>5.0%).
Statistical analysis
Pearson chi-square test was used for categorical data analysis. Survival probability analysis were performed with Kaplan-Meier method and log-rank test. Cox proportional hazards model was used for multivariate analysis. P < 0.05 (two-tailed) was considered as statistically significant. All statistical analysis was performed with the SPSS 13.0 software (SPSS, Inc., Chicago, IL, USA).
Results
Expression of HLA-G probed with mAbs 4H84 and 5A6G7 in CRC
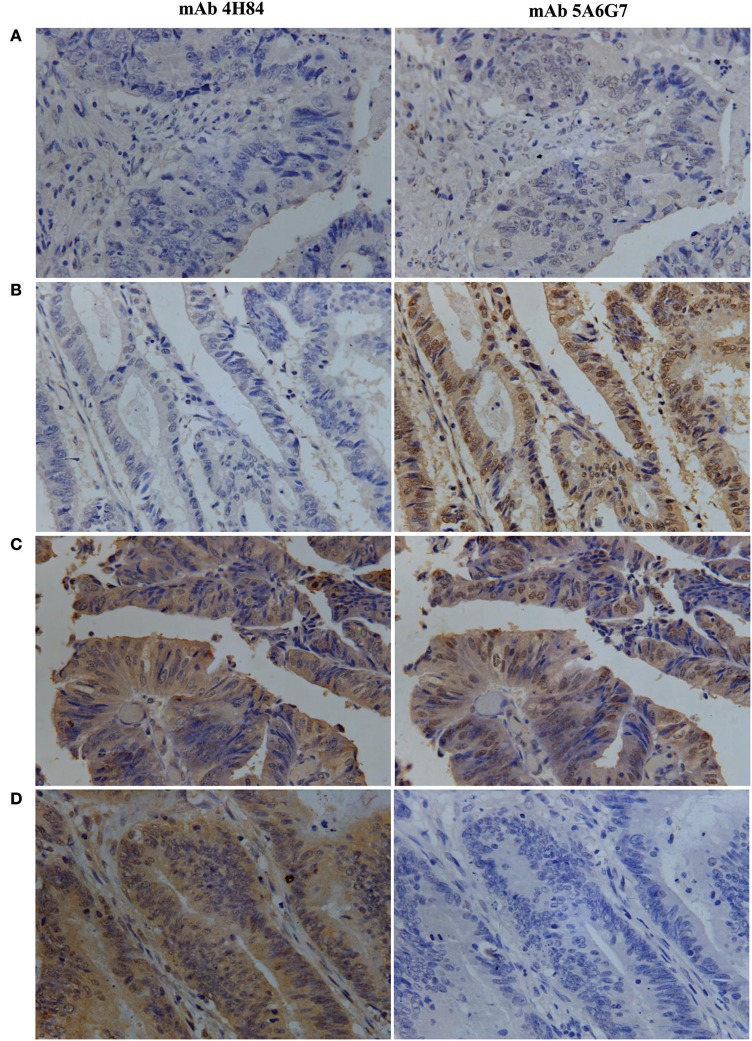
The IHC immunostaining pattern with mAbs 4H84 and 5A6G7 was shown in Figure 1. In these case-matched serial section primary CRC lesions, positive HLA-G expression was observed in 70.7% (268/379) with mAb 4H84 and in 60.4% (219/379) with mAb 5A6G7 in CRC patients. The percentage of the HLA-G expression detected with both antibodies ranges from negative to 99%. Neither the HLA-G expression status detected with mAb 4H84, nor with mAb 5A6G7 was statistical significantly related to the clinicopathological parameters such as gender, age, TNM status and AJCC clinical disease stage of CRC patients (Table 1).
Figure 1.
Representative immunohistochemistry analysis of HLA-G expression patterns in primary CRC serial section lesions with mAbs 4H84 and 5A6G7. (A) mAbs 4H84neg5A6G7neg; (B) mAbs 4H84neg 5A6G7pos; (C) mAbs 4H84pos5A6G7pos; and (D) mAbs 4H84pos5A6G7neg. (400×).
Table 1.
HLA-G expression detected with mAbs 4H84 and 5A6G7 related to the clinical parameters in CRC patients.
| Variables | Cases | HLA-G (mAb 4H84) | HLA-G(mAb 5A6G7) | ||||
|---|---|---|---|---|---|---|---|
| Neg. | Pos. (%) | p* | Neg. | Pos. (%) | p* | ||
| Gender | |||||||
| Male | 214 | 69 | 145 (67.8%) | 0.150 | 95 | 119 (55.6%) | 0.329 |
| Female | 165 | 42 | 123 (74.5%) | 65 | 100 (60.6%) | ||
| Age | |||||||
| ≤ median (66 ys) | 188 | 52 | 136 (72.3%) | 0.490 | 78 | 110 (58.5%) | 0.776 |
| >median | 191 | 59 | 132 (69.1%) | 82 | 109 (57.1%) | ||
| TNM stage | |||||||
| Tumor status | |||||||
| T1+2 | 108 | 26 | 82 (75.9%) | 0.016 | 39 | 69 (63.9%) | 0.110 |
| T3 | 256 | 79 | 177 (69.1%) | 117 | 139 (54.3%) | ||
| T4 | 15 | 6 | 9 (60.0%) | 4 | 11 (73.3%) | ||
| Nodal status | |||||||
| N0 | 201 | 57 | 144 (71.6%) | 0.870 | 82 | 119 (59.2%) | 0.421 |
| N1 | 109 | 34 | 75 (68.8%) | 44 | 65 (54.3%) | ||
| N2 | 69 | 20 | 49 (71.0%) | 34 | 35 (73.3%) | ||
| Metastasis status | |||||||
| M0 | 368 | 110 | 258 (70.1%) | 0.135 | 156 | 212 (57.6%) | 0.690 |
| M1 | 11 | 1 | 10 (90.9%) | 4 | 7 (63.6%) | ||
| Disease stage | |||||||
| I | 85 | 23 | 62 (72.9%) | 0.434 | 28 | 57 (67.1%) | 0.207 |
| II | 114 | 34 | 80 (70.2%) | 54 | 60 (52.6%) | ||
| III | 169 | 53 | 116 (68.6%) | 74 | 95 (56.2%) | ||
| IV | 11 | 1 | 10 (90.9%) | 4 | 7 (63.6%) | ||
Comparison of HLA-G expression status between or among each variable using the Pearson chi-square test. TNM, lymph-node-metastasis and stage according to the TNM classification.
Be noted, differential HLA-G expression was commonly observed in the case-matched CRC samples between the samples detected with mAbs 4H84 and 5A6G7. In this CRC cohort, ΔHLA-Gneg in 64 (16.9%), ΔHLA-Gcom in 159 (42.0%), and ΔHLA-Gpos in 156 (41.2%) CRC cases was observed. Among the ΔHLA-Gneg group, unexpected immunostaining was observed in 44 (11.6%) case-matched CRC lesions that no staining was detected with the mAbs 4H84 but with the 5A6G7 (mAbs 4H84neg5A6G7pos). The data were unpredictable beacuse all seven known α1 domain containing HLA-G isoforms could be recognized by the mAb 4H84. However, both ΔHLA-G and its subgroups mAbs 4H84neg5A6G7pos and 4H84 pos5A6G7 neg status was unrelated to the clinicopathological parameters of CRC patients (Table 2).
Table 2.
Difference (ΔHLA-G) with mAbs 4H84 and 5A6G7 related to the clinical parameters in CRC patients.
| Variables | Cases | ΔHLA-G* | HLA-G (mAb 4H84 vs. 5A6G7) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Neg. | Com | Pos | p** | Cases | 4H84neg 5A6G7pos | 4H84pos 5A6G7neg | p** | ||
| Total | 379 | 64 | 159 | 156 | 137 | 44 | 93 | ||
| Gender | |||||||||
| Male | 214 | 35 | 96 | 83 | 0.418 | 76 | 25 | 51 | 0.828 |
| Female | 165 | 29 | 63 | 73 | 61 | 19 | 42 | ||
| Age | |||||||||
| ≤ median (66 ys) | 188 | 32 | 79 | 77 | 0.996 | 68 | 21 | 47 | 0.759 |
| >median | 191 | 32 | 80 | 79 | 69 | 23 | 46 | ||
| TNM stage | |||||||||
| Tumor status | |||||||||
| T1+2 | 108 | 19 | 47 | 42 | 0.821 | 33 | 10 | 23 | 0.175 |
| T3 | 256 | 41 | 107 | 108 | 100 | 31 | 69 | ||
| T4 | 15 | 4 | 5 | 6 | 4 | 3 | 1 | ||
| Nodal status | |||||||||
| N0 | 201 | 33 | 88 | 80 | 0.634 | 65 | 20 | 45 | 0.356 |
| N1 | 109 | 22 | 40 | 47 | 46 | 18 | 28 | ||
| N2 | 69 | 9 | 31 | 29 | 26 | 6 | 20 | ||
| Metastasis | |||||||||
| M0 | 368 | 63 | 154 | 151 | 0.782 | 134 | 44 | 90 | 0.228 |
| M1 | 11 | 1 | 5 | 5 | 3 | 0 | 3 | ||
| Disease stage | |||||||||
| I | 85 | 14 | 41 | 30 | 0.885 | 23 | 9 | 14 | 0.417 |
| II | 114 | 19 | 47 | 48 | 42 | 11 | 31 | ||
| III | 169 | 30 | 66 | 73 | 69 | 24 | 45 | ||
| IV | 11 | 1 | 5 | 5 | 3 | 0 | 3 | ||
ΔHLA-G: the difference of the percentage of HLA-G expression detected with mAb 4H84 subtracted that with mAb 5A6G7. ΔHLA-Gneg:ΔHLA-G >−5.0%; ΔHLA-Gcom:−5.0%≤ΔHLA-G≤5.0%; ΔHLA-Gpos: ΔHLA-G > 5.0%.
Comparison of HLA-G expression status between or among each variable using the Pearson chi-square test. TNM, lymph-node-metastasis and stage according to the TNM classification.
The mAb 5A6G7 specifically recognize an epitope located in the C-terminal amino acid sequence of HLA-G isoforms such as HLA-G5 and -G6. However, being with the α 1 domain, HLA-G5 and -G6 could also be recognized by the mAb 4H84. Therefore, to our prevailing knowledge, absence of mAb 4H84 labeling generally considered as the lack of HLA-G antigen expression. Noteworthy, the unexpected immunostaining pattern mAbs 4H84neg5A6G7pos now sounds reasonable by the recent findings by the study Tronik-Le Roux et al. (11) that novel HLA-G isoforms lacks the α1 domain indeed exist in renal cancers.
Significance of the ΔHLA-G status to CRC patient survival
Herein, Log-rank Mantel-Cox analysis of ΔHLA-G status and clinical parameters in CRC patient survival was evaluated. Data showed that, in addition to CRC patient tumor status, nodal status, metastasis status and AJCC clinical disease stage, both ΔHLA-G and its subgroups mAbs 4H84neg5A6G7pos and 4H84 pos5A6G7 neg status were significantly related to survival (Table 3).
Table 3.
Log-rank Mantel-Cox analysis of clinical parameters in survival in CRC patients.
| Variables | No. total | No. events | Mean survival | 95% CI | p-value | |
|---|---|---|---|---|---|---|
| Sex | Male | 192 | 65 | 72.0 | 66.2–77.7 | 0.927 |
| Female | 147 | 48 | 73.4 | 66.6–80.1 | ||
| Age | ≤ 66 ys | 164 | 55 | 73.4 | 67.1–79.6 | 0.755 |
| > 66 ys | 175 | 58 | 71.6 | 65.4–77.8 | ||
| Tumor status | T1+2 | 97 | 18 | 84.5 | 77.4–91.6 | 0.001 |
| T3 | 228 | 87 | 67.6 | 62.1–73.1 | ||
| T4 | 14 | 8 | 60.6 | 40.1–81.2 | ||
| Nodal status | N0 | 176 | 37 | 84.4 | 79.2–89.6 | <0.001 |
| N1 | 101 | 44 | 62.4 | 54.1–70.7 | ||
| N2 | 62 | 32 | 53.6 | 42.3–65.0 | ||
| Metastasis status | M0 | 328 | 107 | 73.4 | 68.8–77.8 | 0.089 |
| M1 | 11 | 6 | 51.9 | 25.2–78.7 | ||
| Clinical stage | I | 76 | 10 | 86.3 | 79.7–92.9 | <0.001 |
| II | 98 | 26 | 80.4 | 73.2–87.7 | ||
| III | 154 | 71 | 58.7 | 51.8–65.7 | ||
| IV | 11 | 6 | 51.9 | 25.1–78.7 | ||
| HLA-G (mAb 4H84) | Neg | 98 | 29 | 76.7 | 69.0–84.5 | 0.250 |
| Pos | 241 | 84 | 70.7 | 65.4–76.0 | ||
| HLA-G (mAb 5A6G7) | Neg | 142 | 54 | 69.5 | 62.9–76.2 | 0.268 |
| Pos | 197 | 59 | 74.9 | 69.0–80.8 | ||
| ΔHLA-G status* | ||||||
| ΔHLA-Gneg | 57 | 10 | 87.9 | 79.4–96.4 | 0.017 | |
| ΔHLA-Gcom | 143 | 48 | 71.0 | 64.0–77.9 | ||
| ΔHLA-Gpos | 139 | 55 | 67.2 | 60.3–74.1 | ||
| 4H84neg5A6G7pos | / | 40 | 8 | 85.5 | 74.8–96.2 | 0.046 |
| 4H84pos5A6G7neg | / | 84 | 33 | 68.1 | 59.2–77.0 | |
ΔHLA-G: the difference of the percentage of HLA-G expression detected with mAb 4H84 subtracted that with mAb 5A6G7. ΔHLA-Gneg:ΔHLA-G >−5.0%; ΔHLA-Gcom:−5.0%≤ΔHLA-G≤5.0%; ΔHLA-Gpos: ΔHLA-G > 5.0%.
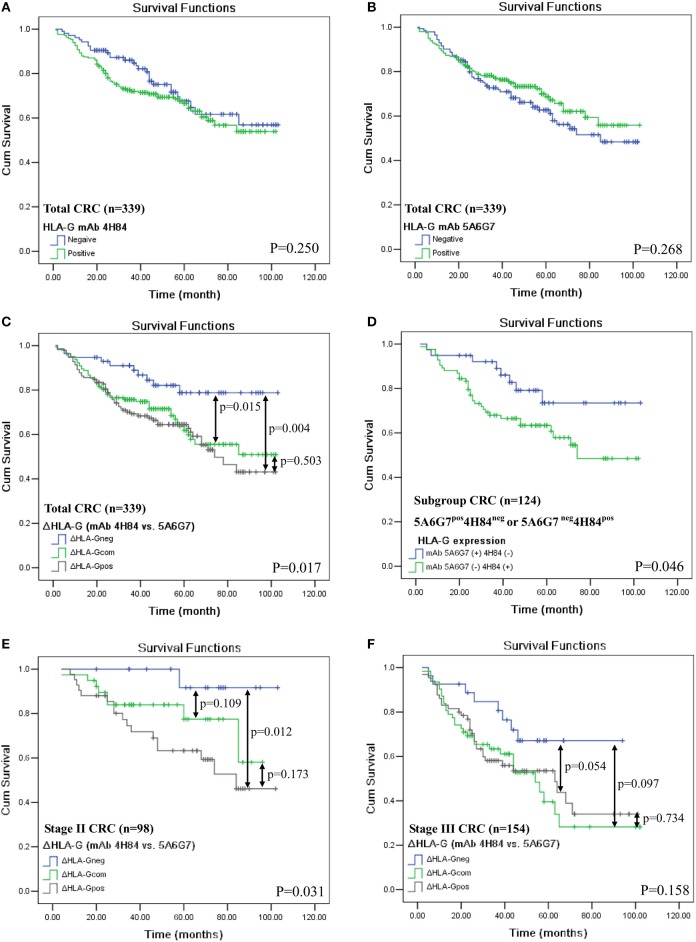
In addition, HLA-G expression status detected by either mAbs 4H84 (p = 0.250, Figure 2A) or 5A6G7 (p = 0.268, Figure 2B) were not associated with CRC patient survival. However, in the whole CRC cohort, ΔHLA-G status was dramatically relevant to the patient survival (p = 0.017, Figure 2C). The mean survival for ΔHLA-Gneg was 87.9 months (n = 57; 95% CI: 79.4–96.4), ΔHLA-Gcom was 71.0 months (n = 143; 95% CI: 64.0–77.9), and ΔHLA-Gpos was 67.2 months (n = 139; 95% CI: 60.3–74.1) respectively, where patients with ΔHLA-Gneg had a better outcome than that of patients with ΔHLA-Gcom and ΔHLA-Gpos. Interestingly, when compared the survival between the patients with mAbs 4H84neg5A6G7pos (n = 40) and mAbs 4H84 pos5A6G7neg (n = 84), we found that patients with mAbs 4H84neg5A6G7pos had a better survival than that of patients with mAbs 4H84pos5A6G7neg (p = 0.046, Figure 2D). The mean survival for mAbs 4H84neg5A6G7pos was 85.5 months (95% CI: 74.8–96.2), and mAbs 4H84pos5A6G7neg was 68.1 months (95% CI: 59.2–77.0), respectively.
Figure 2.
Kaplan-Meier survival analysis of HLA-G expression status with mAbs 4H84 or 5A6G7 in CRC patients. (A) Comparison between HLA-Gneg and HLA-Gpos with mAb 4H84 in the whole cohort of CRC patients (p = 0.250); (B) between HLA-Gneg and HLA-Gpos with mAb 5A6G7 in the whole cohort of CRC patients (p = 0.268); (C) among patient with ΔHLA-Gneg, ΔHLA-Gcom and ΔHLA-Gpos (p = 0.017) in the whole cohort of CRC patients. (D) between patients with HLA-G status of mAbs 5A6G7pos4H84neg and with mAbs 5A6G7neg4H84pos (p = 0.046); (E) among AJCC stage II CRC patient with ΔHLA-Gneg, ΔHLA-Gcom and ΔHLA-Gpos (p = 0.031); and (F) among AJCC stage III CRC patient with ΔHLA-Gneg, ΔHLA-Gcom and ΔHLA-Gpos (p = 0.158). Note: p-value in double-head arrow represents the comparison between subgroups of the CRC patients indicated.
To mitigate the heterogeneity of the samples on the prognistic value of ΔHLA-G, we futher analyzed the relevance of ΔHLA-G status to survival for patients with particular AJCC stages. The survival analysis was performed for patients with AJCC stage II and III, because limited evens were occurred in patietns with AJCC stage I (Nevent = 10) and stage IV (Nevent = 6). Data showed that, among patients with AJCC stage II, ΔHLA-G status was also obivously associated with the patient survival, the mean survival for ΔHLA-Gneg (n = 17), ΔHLA-Gcom (n = 39), and ΔHLA-Gpos (n = 42) was 99.3, 79.1, and 70.5 months, respectively (p = 0.031). For the subgroups of stage II patients, significant difference for survival was also observed between the patients with ΔHLA-Gneg vs. ΔHLA-Gpos (p = 0.012; Figure 2E). Among patients with AJCC stage III, though does not reach the statistical significance, the mean survival for ΔHLA-Gneg (n = 27, 72.7 months) was much longer than patients with, ΔHLA-Gcom (n = 62, 53.6 months) and ΔHLA-Gpos (n = 65, 56.7 months; p = 0.158, Figure 2F).
Value of ΔHLA-G status on prognosis for CRC patients
Finally, the prognostic value of the ΔHLA-G combined with CRC patient clinical parameters was analyzed (Table 4). By univariate analysis, primary tumor burden (T3+4 vs. T1+2, HR = 2.504, p < 0.001), regional lymph node (N1+2 vs. N0, HR = 3.064, p < 0.001), distant metastases (M1 vs. M0, HR = 2.009, p = 0.097), and clinical disease stage (III + IV vs. I + II, HR = 3.142, p < 0.001) was markedly associated with worse prognosis; However, ΔHLA-G neg (ΔHLA-G neg vs. ΔHLA-G com+pos, HR = 0.414, p = 0.008) was obviously associated with a better prognosis. Furthermore, by multivariate analysis, data showed that primary tumor (p = 0.030) and ΔHLA-G status (p = 0.008) remain statistic significantly, indicating that ΔHLA-G status could be an independent prognostic factor for CRC patients.
Table 4.
Cox proportional hazards model analysis of multi-variables in survival for CRC patients.
| Variables | Categories | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| Sex | Male vs. Female | 0.983 (0.667–1.427) | 0.927 | ||
| Age (years) | >66 vs. ≤ 66 | 1.060 (0.733–1.534) | 0.756 | ||
| Tumor status | T3+4 vs. T1+2 | 2.504 (1.512–4.147) | <0.001 | 1.793 (1.059–3.035) | 0.030 |
| Nodal status | N1+2 vs. N0 | 3.064 (2.062–4.554) | <0.001 | 0.919 (0.107–7.921) | 0.919 |
| Metastasis status | M1 vs.M0 | 2.009 (0.881–4.580) | 0.097 | 1.047 (0.419–2.612) | 0.922 |
| Clinical stage | III+IV vs. I+II | 3.142 (2.108–4.683) | <0.001 | 2.913 (0.322–26.34) | 0.341 |
| HLA-G (mAb 4H84) | Pos vs. Neg | 1.267 (0.830–1.933) | 0.272 | ||
| HLA-G (mAb 5A6G7) | Pos vs. Neg | 0.812 (0.561–1.175) | 0.270 | ||
| ΔHLA-G status* | ΔHLA-G neg vs. ΔHLA-G com+pos | 0.414 (0.216–0.792) | 0.008 | 0.416 (0.217–0.798) | 0.008 |
HR, hazard ratio; 95% CI = 95% confidence interval.
ΔHLA-G: the difference of the percentage of HLA-G expression detected with mAb 4H84 subtracted that with mAb 5A6G7. ΔHLA-Gneg:ΔHLA-G > −5.0%; ΔHLA-Gcom: −5.0%≤ΔHLA-G≤5.0%; ΔHLA-Gpos: ΔHLA-G > 5.0%.
Discussion
Since HLA-G was firstly found in cytotrophoblast in 1990 (16), immune regulation functions of HLA-G have been investigated extensively, and important roles of HLA-G in the maintenance for the fetal-maternal immunotolerance were established (17). However, with the HLA-G expression was observed in cancers for the first time in 1998, numerous studies have been carried out focusing on the basic biology and clinical significance of HLA-G in malignancies (2, 18). In clinical settings and pre-clinical murine models, HLA-G expression was strongly related to the capability of tumor cell invasiveness and metastasis, to advanced disease stage and poor survival in cancer patients (1, 13). Thus far, the importance of HLA-G in cancer promotion and progression has been widely acknowledged. Consequently, HLA-G as cancer immunotherapy target was proposed.
One of the distinct features of HLA-G is that, by gene alternative splicing, its primary transcript could generate at least seven isoforms (HLA-G1~HLA-G7) (9). However, it's reasonable more novel unrecognized HLA-G isoforms remains unveiled because different reading frame could be formed by the gene splicing. This is indeed in case of HLA-G as reported in a recent study (11). In that study, HLA-G expression in renal cancer lesions (RCC) was evaluated by immunohistochemistry (IHC) with different HLA-G antibodies including mAbs 4H84 and 5A6G7. Unexpectedly, in some RCC cases, IHC results showed that no immunostaining was detected with the mAb 4H84 but strongly with mAb 5A6G7 (mAbs 4H84neg5A6G7pos). The staining pattern seems controversy to our prevailing knowledge that absence of mAb 4H84 labeling generally accounts for the lack of HLA-G expression. This is based on the fact that all previously identified seven HLA-G isoforms contains α 1 domain and could be detected by the mAb 4H84.
In this context, we presented similar puzzled data of staining pattern (mAbs 4H84neg5A6G7pos) on the case-matched lung and ovarian cancers and lead to hot discussion by the participants during the 7th international conference on HLA-G (Paris, 2015. http://www.hlag2015.sitew.fr/#Program.C). Noteworthy, now the unpredictable and unexpected immunostaining pattern could be explained by the findings revealed by Tronik-Le Roux et al. (11), that unrecognized HLA-G isoforms heretofore such as α1 domain-absent isoforms were presented in CRC samples. The very instructive findings raises the questions whether these novel HLA-G proteins are of clinical significance because similar, distinct or opposing functions was observed among different isoforms.
Substantial previous evidence has proved that HLA-G is a crucial tumor-driven immune escape factor involved in alterating the anti-tumor responses of immune contexture of solid tumors like CRC, and thus in determination of the fate of tumor development and patient clinical outcome (1). Through binding to the immune inhibitory receptors (particularly ILT2 and/or ILT4) expressed on various immune cells, HLA-G could directly or indirectly impair anti-tumor immune responses (2). Accumulating data revealed that HLA-G has comprehensive immune suppressive functions to T cells, B cells, NK cells and DCs, and to enhance the generation of immune regulatory cells such as MDSCs, DC-10 and M2-macrophages (19–23). Among HLA-G isofroms, the immune suppression mediated by HLA-G1 and HLA-G5 isoforms was well documented by different research groups, including ours. We found that HLA-G1 expressed in ovarian cancer cell lines NIH:OVCAR-3 and HO-8910 (6, 24), hepatocellular carcinoma cell line Hep G2 (25), lung adenocarcinoma cell line A549 (26) could dramatically decrease the NK cell cytolysis. We also found that by blocking the HLA-G with mAb 87G or its receptor ILT2 expressed on the effector NK-92 cell line could significantly reverse the NK cell lysis function (24, 26). Furthermore, HLA-G1 and HLA-G5 isoforms in NK cell cytolysis suppression is dependent on the level of both HLA-G1 and HLA-G5 expression. Compared to HLA-G1, HLA-G5 has a more potent inhibition effect on the NK cytolysis. Moreover, HLA-G1 and HLA-G5 isoforms have an additive effect on NK cytolysis suppression (27, 28). HLA-G1/ILT2 engagement on T cells could result in inhibition of CD4+ T cell alloproliferation and CD8+ and Vγ9Vδ2 T cell cytolysis and IFN-γ production (29–31). HLA-G5/ILT2 mediated signal was observed to supress B cell immunoglobulin production, and HLA-G5/ILT4 interaction impair phagocytosis and reactive oxygen species production of neutrophils (32, 33). Other functions such as impairment of the expression and function of chemokine receptors in T cells, NK cells and B cells mediated by HLA-G5/ILT2 signaling pathway was also reported (34).
As expected that, like other HLA antigens, subtle structural alterations in HLA-G molecules can have dramatic effects on innate and adaptive immune modulation, and consequently, related to different clinical significance (35, 36). In this scenario, recent studies revealed that the nature of the bound peptide could influence the substructure and stability of HLA-G, and a single amino acid exchange in the α2 domain in allelic HLA-G subtypes of HLA-G*01:01 and HLA-G*01:04 has obivously different capability in NK cell lysis inhibition (36, 37). HLA-G3 was found less efficient than HLA-G1 at inducing HLA-E surface expression (38). Moreover, unlike HLA-G1, HLA-G3 could not inhibit human monocyte/macrophage-mediated swine endothelial cell lysis or NK cells against K562 cells (39, 40). These findings indicated that distinct structure of HLA-G molecules or isoforms features with unique biological functions and could impact on its recognition by receptors of both innate and adaptive immune systems.
To evaluate whether different structural HLA-G molecaules have different clinical relavence, in this study, HLA-G expression in 379 case-matched primary CRC lesions were analyzed by IHC with mAb 4H84 and mAb 5A6G7. Different percentage of HLA-G expression was commonly observed between the case-matched serial sections when using the mAb 4H84 and mAb 5A6G7. Furthermore, clinical relevance of the difference of HLA-G expression (ΔHLA-G) between mAbs 4H84 and 5A6G7 was evaluated. Our data revealed that lower percentage of HLA-G expression detected with mAb 4H84 than mAb 5A6G7 (ΔHLA-Gneg) was found in 16.9% (64/379), comparable percentage detected with both antibodies (ΔHLA-Gcom) was found in 42.0% (159/379), higher percentage of HLA-G expression detected with mAb 4H84 than mAb 5A6G7 (ΔHLA-Gpos) was found in 41.2% (156/379) of CRC samples, respectively. Patients with ΔHLA-Gneg had significantly better survival than those with ΔHLA-Gcom and ΔHLA-Gpos CRC patients (p = 0.017). A better predictive value of ΔHLA-Gneg was also observed for patients with either AJCC stage II or III. Moreover, patients with staining pattern 4H84neg5A6G7pos had a better survival than that of patients with 4H84pos5A6G7neg (p = 0.046). Multivariate analysis by Cox proportional hazards model also showed that ΔHLA-G could be an independent prognostic factor for CRC patients (p = 0.008).
Different clinical significance of HLA-G expression in cancers detected with mAb 4H84 and mAb 5A6G7 was also reported in previous studies. HLA-G expression detected with mAb 4H84 was found strongly relative to clinical stage and poor prognosis in lung cancers, while no such significance was observed for HLA-G5/-G6 detected with mAb 5A6G7 (26, 41). Unlike detected with mAb 4H84, HLA-G expression detected with mAb 5A6G7 was also found to unrelated to clinical parameters such as FIGO stage and prognosis in ovarian cancers (14, 42). Thus, the immunostaining pattern for HLA-G expression with 4H84neg5A6G7pos strengthen the existence of novel α1 domain-absent HLA-G isoforms which could not be probed by the mAb 4H84. However, their specific function in physiological and pathological conditions still remains to be determined. To this end, being lack of currently available commercial antibodies for those novel HLA-G isoforms, a large campaign to develop new antibodies is extremely necessary and even inevitable.
Summary, our preliminary findings revealed that novel unidentified HLA-G isoforms recognized by mAbs 5A6G7 but not 4H84 might be existed in colorectal cancers. Moreover, HLA-G isoforms recognized by mAb 5A6G7 is of different clinical significant from that of the isoforms recognized by mAb 4H84. In the era of the precision medicine, the inter- and intra-tumor heterogeneity caused by complexity of the validated and even novel HLA-G isoforms, interpretation of the significance of HLA-G in cancers must be with extreme caution before their distinct roles are elucidated.
Author contributions
AL and W-HY: study design; XZ and AL: performed experiments; R-LZ, J-GZ, and W-JZ: material support and data acquisition; AL and W-HY: performed statistical analysis and drafted the manuscript. All authors read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by grants from National Natural Science Foundation of China (31370920, 81372247), Science and Technology Bureau of Zhejiang Province (2013C33112) and by Zhejiang Provincial program for the cultivation of high-level innovative health talents.
References
- 1.Lin A, Yan WH. HLA-G expression in cancers: roles in immune evasion, metastasis and target for therapy. Mol Med. (2015) 21:782–91. 10.2119/molmed.2015.00083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carosella ED, Rouas-Freiss N, Tronik-Le Roux D, Moreau P, LeMaoult J. HLA-G: an immune checkpoint molecule. Adv Immunol. (2015) 127:33–144. 10.1016/bs.ai.2015.04.001 [DOI] [PubMed] [Google Scholar]
- 3.Agaugue S, Carosella ED, Rouas-Freiss N. Role of HLA-G in tumor escape through expansion of myeloid-derived suppressor cells and cytokinic balance in favor of Th2 versus Th1/Th17. Blood (2011) 117:7021–31. 10.1182/blood-2010-07-294389 [DOI] [PubMed] [Google Scholar]
- 4.Zhang W, Liang S, Wu J, Horuzsko A. Human inhibitory receptor immunoglobulin-like transcript 2 amplifies CD11b+Gr1+ myeloid-derived suppressor cells that promote long-term survival of allografts. Transplantation (2008) 86:1125–34. 10.1097/TP.0b013e318186fccd [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin A, Zhang X, Xu HH, Xu DP, Ruan YY, Yan WH. HLA-G expression is associated with metastasis and poor survival in the Balb/c nu/nu murine tumor model with ovarian cancer. Int J Cancer (2012) 131:150–7. 10.1002/ijc.26375 [DOI] [PubMed] [Google Scholar]
- 6.Lin A, Xu HH, Xu DP, Zhang X, Wang Q, Yan WH. Multiple steps of HLA-G in ovarian carcinoma metastasis: alter NK cytotoxicity and induce matrix metalloproteinase-15 (MMP-15) expression. Hum Immunol. (2013) 74:439–46. 10.1016/j.humimm.2012.11.021 [DOI] [PubMed] [Google Scholar]
- 7.Liu H, Liu X, Jin H, Yang F, Gu W, Li X. Proteomic analysis of knock-down HLA-G in invasion of human trophoblast cell line JEG-3. Int J Clin Exp Pathol. (2013) 6:2451–9. [PMC free article] [PubMed] [Google Scholar]
- 8.Guo Y, Lee CL, So KH, Gao J, Yeung WS, Yao Y, et al. Soluble human leukocyte antigen-g5 activates extracellular signal-regulated protein kinase signaling and stimulates trophoblast invasion. PLoS ONE (2013) 8:e76023. 10.1371/journal.pone.0076023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paul P, Cabestre FA, Ibrahim EC, Lefebvre S, Khalil-Daher I, Vazeux G, et al. Identification of HLA-G7 as a new splice variant of the HLA-G mRNA and expression of soluble HLA-G5, -G6, and -G7 transcripts in human transfected cells. Hum Immunol. (2000) 61:1138–49. 10.1016/S0198-8859(00)00197-X [DOI] [PubMed] [Google Scholar]
- 10.Yan WH, Fan LA. Residues Met76 and Gln79 in HLA-G alpha1 domain involve in KIR2DL4 recognition. Cell Res. (2005) 15:176–82. 10.1038/sj.cr.7290283 [DOI] [PubMed] [Google Scholar]
- 11.Tronik-Le Roux D, Renard J, Verine J, Renault V, Tubacher E, LeMaoult J, et al. Novel landscape of HLA-G isoforms expressed in clear cell renal cell carcinoma patients. Mol Oncol. (2017) 11:1561–78. 10.1002/1878-0261.12119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. (2010) 17:1471–4. 10.1245/s10434-010-0985-4 [DOI] [PubMed] [Google Scholar]
- 13.Zhang RL, Zhang X, Dong SS, Hu B, Han QY, Zhang JG, et al. Predictive value of different proportion of lesion HLA-G expression in colorectal cancer. Oncotarget (2017) 8:107441–51. 10.18632/oncotarget.22487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Han QY, Li JB, Ruan YY, Yan WH, Lin A. Lesion HLA-G5/-G6 isoforms expression in patients with ovarian cancer. Hum Immunol. (2016) 77:780–4. 10.1016/j.humimm.2015.12.003 [DOI] [PubMed] [Google Scholar]
- 15.Chew SF, Kanaan C, Tait BD. HLA expression and cancer−14th IHIWS immunohistochemistry quality control exercise exchange results. Tissue Antigens (2007) 69(Suppl. 1):248–51. 10.1111/j.1399-0039.2006.774_2.x [DOI] [PubMed] [Google Scholar]
- 16.Kovats S, Main EK, Librach C, Stubblebine M, Fisher SJ, DeMars R. A class I antigen, HLA-G, expressed in human trophoblasts. Science (1990) 248:220–3. 10.1126/science.2326636 [DOI] [PubMed] [Google Scholar]
- 17.Carosella ED, Moreau P, Le Maoult J, Le Discorde M, Dausset J, Rouas-Freiss N. HLA-G molecules: from maternal-fetal tolerance to tissue acceptance. Adv Immunol. (2003) 81:199–252. 10.1016/S0065-2776(03)81006-4 [DOI] [PubMed] [Google Scholar]
- 18.Paul P, Rouas-Freiss N, Khalil-Daher I, Moreau P, Riteau B, Le Gal FA, et al. HLA-G expression in melanoma: a way for tumor cells to escape from immunosurveillance. Proc Natl Acad Sci USA. (1998) 95:4510–5. 10.1073/pnas.95.8.4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bainbridge DR, Ellis SA, Sargent IL. HLA-G suppresses proliferation of CD4+ T-lymphocytes. J Reprod Immunol. (2000) 48:17–26. 10.1016/S0165-0378(00)00070-X [DOI] [PubMed] [Google Scholar]
- 20.Baudhuin J, Lesport E, Sousa S, Migraine J, Vigneron J, Lemaoult J, et al. HLA-G inhibition of NK-cell cytolytic function is uncoupled from tumor cell lipid raft reorganization. Eur J Immunol. (2012) 42:700–9. 10.1002/eji.201141930 [DOI] [PubMed] [Google Scholar]
- 21.Kostlin N, Ostermeir AL, Spring B, Schwarz J, Marme A, Walter CB, et al. HLA-G promotes myeloid-derived suppressor cell accumulation and suppressive activity during human pregnancy through engagement of the receptor ILT4. Eur J Immunol. (2017) 47:374–84. 10.1002/eji.201646564 [DOI] [PubMed] [Google Scholar]
- 22.Amodio G, Comi M, Tomasoni D, Gianolini ME, Rizzo R, LeMaoult J, et al. HLA-G expression levels influence the tolerogenic activity of human DC-10. Haematologica (2015) 100:548–57. 10.3324/haematol.2014.113803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nunez SY, Ziblat A, Secchiari F, Torres NI, Sierra JM, Raffo Iraolagoitia XL, et al. Human M2 macrophages limit NK cell effector functions through secretion of TGF-beta and engagement of CD85j. J Immunol. (2018) 200:1008–15. 10.4049/jimmunol.1700737 [DOI] [PubMed] [Google Scholar]
- 24.Lin A, Yan WH, Xu HH, Gan MF, Cai JF, Zhu M, et al. HLA-G expression in human ovarian carcinoma counteracts NK cell function. Ann Oncol. (2007) 18:1804–9. 10.1093/annonc/mdm356 [DOI] [PubMed] [Google Scholar]
- 25.Lin A, Chen HX, Zhu CC, Zhang X, Xu HH, Zhang JG, et al. Aberrant human leucocyte antigen-G expression and its clinical relevance in hepatocellular carcinoma. J Cell Mol Med. 14:2162–71. 10.1111/j.1582-4934.2009.00917.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin A, Zhu CC, Chen HX, Chen BF, Zhang X, Zhang JG, et al. Clinical relevance and functional implications for human leucocyte antigen-g expression in non-small-cell lung cancer. J Cell Mol Med. 14:2318–29. 10.1111/j.1582-4934.2009.00858.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang WQ, Xu DP, Liu D, Li YY, Ruan YY, Lin A, et al. HLA-G1 and HLA-G5 isoforms have an additive effect on NK cytolysis. Hum Immunol. (2014) 75:182–9. 10.1016/j.humimm.2013.11.001 [DOI] [PubMed] [Google Scholar]
- 28.Chen BG, Xu DP, Lin A, Yan WH. NK cytolysis is dependent on the proportion of HLA-G expression. Hum Immunol. (2013) 74:286–9. 10.1016/j.humimm.2012.12.005 [DOI] [PubMed] [Google Scholar]
- 29.Lesport E, Baudhuin J, Sousa S, LeMaoult J, Zamborlini A, Rouas-Freiss N, et al. Inhibition of human gamma delta [corrected] T-cell antitumoral activity through HLA-G: implications for immunotherapy of cancer. Cell Mol Life Sci. (2011) 68:3385–99. 10.1007/s00018-011-0632-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Gal FA, Riteau B, Sedlik C, Khalil-Daher I, Menier C, Dausset J, et al. HLA-G-mediated inhibition of antigen-specific cytotoxic T lymphocytes. Int Immunol. (1999) 11:1351–6. 10.1093/intimm/11.8.1351 [DOI] [PubMed] [Google Scholar]
- 31.LeMaoult J, Krawice-Radanne I, Dausset J, Carosella ED. HLA-G1-expressing antigen-presenting cells induce immunosuppressive CD4+ T cells. Proc Natl Acad Sci USA. (2004) 101:7064–9. 10.1073/pnas.0401922101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baudhuin J, Migraine J, Faivre V, Loumagne L, Lukaszewicz AC, Payen D, et al. Exocytosis acts as a modulator of the ILT4-mediated inhibition of neutrophil functions. Proc Natl Acad Sci USA. (2013) 110:17957–62. 10.1073/pnas.1221535110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naji A, Menier C, Morandi F, Agaugue S, Maki G, Ferretti E, et al. Binding of HLA-G to ITIM-bearing Ig-like transcript 2 receptor suppresses B cell responses. J Immunol. (2014) 192:1536–46. 10.4049/jimmunol.1300438 [DOI] [PubMed] [Google Scholar]
- 34.Morandi F, Rouas-Freiss N, Pistoia V. The emerging role of soluble HLA-G in the control of chemotaxis. Cytokine Growth Factor Rev. (2014) 25:327–35. 10.1016/j.cytogfr.2014.04.004 [DOI] [PubMed] [Google Scholar]
- 35.Aptsiauri N, Ruiz-Cabello F, Garrido F. The transition from HLA-I positive to HLA-I negative primary tumors: the road to escape from T-cell responses. Curr Opin Immunol. (2018) 51:123–32. 10.1016/j.coi.2018.03.006 [DOI] [PubMed] [Google Scholar]
- 36.Walpole NG, Kjer-Nielsen L, Kostenko L, McCluskey J, Brooks AG, Rossjohn J, et al. The structure and stability of the monomorphic HLA-G are influenced by the nature of the bound peptide. J Mol Biol. (2010) 397:467–80. 10.1016/j.jmb.2010.01.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Celik AA, Simper GS, Huyton T, Blasczyk R, Bade-Doding C. HLA-G mediated immune regulation is impaired by a single amino acid exchange in the alpha 2 domain. Hum Immunol. (2018) 79:453–62. 10.1016/j.humimm.2018.03.010 [DOI] [PubMed] [Google Scholar]
- 38.Teklemariam T, Zhao L, Hantash BM. Full-length HLA-G1 and truncated HLA-G3 differentially increase HLA-E surface localization. Hum Immunol. (2012) 73:898–905. 10.1016/j.humimm.2012.06.007 [DOI] [PubMed] [Google Scholar]
- 39.Eguchi H, Maeda A, Lo PC, Matsuura R, Esquivel EL, Asada M, et al. HLA-G1, but not HLA-G3, Suppresses human Monocyte/Macrophage-mediated swine endothelial cell Lysis. Transpl Proc. (2016) 48:1285–7. 10.1016/j.transproceed.2015.10.070 [DOI] [PubMed] [Google Scholar]
- 40.Zhao L, Teklemariam T, Hantash BM. Mutated HLA-G3 localizes to the cell surface but does not inhibit cytotoxicity of natural killer cells. Cell Immunol. (2014) 287:23–6. 10.1016/j.cellimm.2013.11.005 [DOI] [PubMed] [Google Scholar]
- 41.Yan WH, Liu D, Lu HY, Li YY, Zhang X, Lin A. Significance of tumour cell HLA-G5/-G6 isoform expression in discrimination for adenocarcinoma from squamous cell carcinoma in lung cancer patients. J Cell Mol Med. (2015) 19:778–85. 10.1111/jcmm.12400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jung YW, Kim YT, Kim SW, Kim S, Kim JH, Cho NH, et al. Correlation of human leukocyte antigen-G (HLA-G) expression and disease progression in epithelial ovarian cancer. Reprod Sci. (2009) 16:1103–11. 10.1177/1933719109342131 [DOI] [PubMed] [Google Scholar]