Abstract
Gastric cancer has become a serious disease in the past decade. It has the second highest mortality rate among the four most common cancer types, leading to ~700,000 mortalities annually. Previous studies have attempted to elucidate the underlying biological mechanisms of gastric cancer. The present study aimed to obtain useful biomarkers and to improve the understanding of gastric cancer mechanisms at the genetic level. The present study used bioinformatics analysis to identify 1,829 differentially expressed genes (DEGs) which were obtained from the GSE54129 dataset. Using protein-protein interaction information from the Search Tool for the Retrieval of Interacting Genes database, disease modules were constructed for gastric cancer using Cytoscape software. In the Gene Ontology analysis of biology processes, upregulated genes were significantly enriched in ‘extracellular matrix organization’, ‘cell adhesion’ and ‘inflammatory response’, whereas downregulated DEGs were significantly enriched in ‘xenobiotic metabolic process’, ‘oxidation-reduction process’ and ‘steroid metabolic process’. During Kyoto Encyclopedia of Genes and Genomes analysis, upregulated DEGs were significantly enriched in ‘extracellular matrix-receptor interaction’, ‘focal adhesion’ and ‘PI3K-Akt signaling pathway’, whereas the downregulated DEGs were significantly enriched in ‘chemical carcinogenesis’, ‘metabolism of xenobiotics by cytochrome P450’ and ‘peroxisome’. The present study additionally identified 10 hub genes from the DEGs: Tumor protein p53 (TP53), C-X-C motif chemokine ligand 8 (CXCL8), tetraspanin 4 (TSPAN4), lysophosphatidic acid receptor 2 (LPAR2), adenylate cyclase 3 (ADCY3), phosphoinositide-3-kinase regulatory subunit 1 (PIK3R1), neuromedin U (NMU), C-X-C motif chemokine ligand (CXCL12), fos proto-oncogene, AP-1 transcription factor subunit (FOS) and sphingosine-1-phosphate receptor 1 (S1PR1), which have high degrees with other DEGs. The survival analysis revealed that the high expression of ADCY3, LPAR2, S1PR1, TP53 and TSPAN4 was associated with a lower survival rate, whereas high expression of CXCL8, FOS, NMU and PIK3R1 was associated with a higher survival rate. No significant association was identified between CXCL12 and survival rate. Additionally, TSPAN1 and TSPAN8 appeared in the top 100 DEGs. Finally, it was observed that 4 hub genes were highly expressed in gastric cancer tissue compared with para-carcinoma tissue in the 12 patients; the increased TSPAN4 was significant (>5-fold). Tetraspanin family genes may be novel biomarkers of gastric cancer. The findings of the present study may improve the understanding of the molecular mechanisms underlying the development of gastric cancer.
Keywords: gastric cancer, pathway, biomarker, enrichment, different expression
Introduction
Gastric cancer has become an important health risk, as it is the fourth most common cancer after breast, lung and ovarian cancer and the second leading cause of cancer mortality worldwide (1). It is estimated that ~934,000 novel cases are diagnosed and that there are ~700,000 mortalities annually (2). In China, there are >220,000 mortalities due to gastric cancer annually, which is approximately one-half of the global gastric cancer mortalities (3).
Gastric cancer has been subdivided into two primary subtypes based on histological appearance, including the well-differentiated intestinal type and the poorly differentiated diffuse type (4). Gastric cancer has a poor prognosis, as the 5-year survival for patients with gastric cancer in China is only 30% (5). However, the majority of gastric cancer cases are diagnosed in the middle to late stage (6). Therefore, the development of methods for timely diagnosis and effective symptomatic treatment of gastric cancer is urgent. Therefore, the present study investigated the potential biomarkers of gastric cancer using bioinformatics tools and aimed to provide guidance on the diagnosis and treatment of gastric cancer.
Bioinformatics tools, techniques and resources are critical to biomarker discovery, assessment, validation, qualification, standardization and market acceptance into clinical practice. Major scientific efforts and economic investment over the past 20 years have resulted in thousands of novel candidate biomarkers for diseases; however, relatively few biomarkers have been used in clinical practice (7).
The present study used a high-throughput bioinformatics platform to analyze microarray data and identify differentially expressed genes (DEGs) for further investigation. The current study used the microarray dataset GSE54129, which was downloaded from Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo/), containing 111 human gastric cancer tissues and 21 non-cancerous gastric tissues. The present study used R software to obtain the expression profiles and DEGs. Gene enrichment analysis and network analysis were also used to provide interpretation of high-throughput results. To confirm whether the hub genes associated with poor prognosis were involved in the pathogenesis of gastric cancer, gastric cancer and para-carcinoma tissues from 12 patients were subjected to reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis. The present study identified potential biomarkers for further research.
Materials and methods
Patients and ethics statement
The use of human gastric cancer specimens and the database was approved by the institutional review board of the Hospital of Qingdao University (Qingdao, China). Human tissues used in this study were obtained from patients with gastric cancer (recruited between September 2014 and September 2016; mean age, 62.5 years; range, 54 to 72 years; male, 58.3%, female 41.7%) following resection at the Hospital of Qingdao University. All patients received standard adjuvant radiotherapy following surgery. Written informed consent was obtained from all patients.
RNA isolation and RT-qPCR analysis
Total RNA was isolated from cells using TRIzol reagent (cat. no. 15596026; Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). A total of 4 µg RNA was reverse transcribed into cDNA using the RevertAid First Strand cDNA Synthesis kit (cat. no. 1622; Fermentas; Thermo Fisher Scientific, Inc.). qPCR was performed using the Power SYBR Green PCR Master Mix (cat. no. 4367659; Applied Biosystems; Thermo Fisher Scientific, Inc.) on a 7500 system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The following primers were used to perform qPCR: Adenylate cyclase 3 (ADCY3), 5′-TCTCCGAGCCCGAATACT-3′ (forward) and 5′-GTTCCGGACCGAGATTTCAT-3′ (reverse); lysophosphatidic acid receptor 2 (LPAR2), 5′-TGACTGGAGGCCCAGAT-3′ (forward) and 5′-GCTCTTTGCCACTGTTG-3′ (reverse); sphingosine-1-phosphate receptor 1 (S1PR1), 5′-GCTCTCCGAACGCAACTT-3′ (forward) and 5′-CGATGAGTGATCCAGGC-3′ (reverse); tetraspanin 4 (TSPAN4), 5′-CGTCAAGTACCTCATGTTC-3′ (forward) and 5′-ACGGGAAGGAAGAGGAC-3′ (reverse); and GAPDH (reference gene) 5′-TCTCTCCGTCCTCGGAT-3′ (forward) and 5′-CGTAGTTGTGCTGATG-3′ (reverse). The thermocycling conditions were as follows: Pre-denaturation at 94°C for 5 min; 40 cycles of denaturation at 94°C for 30 sec, annealing at 60°C for 20 sec and extension at 72°C for 20 sec; and a final extension at 72°C for 10 min. All RT-qPCR reactions yielded products with a single dissociation peak. Results were analyzed using the 2−ΔΔCq method (8).
5-bromo-2-deoxyuridine (BrdU) assay
To perform the BrdU assay, cells were grown in 96-well plates (1×104 cells/well) (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany; cat. no. CLS3595-50EA). After 18 h, BrdU (10 µg/ml; Sigma-Aldrich; Merck KGaA) was added to each well. A total of 3 h subsequently, the culture media were discarded, and BrdU incorporation was measured using a Cell Proliferation ELISA kit (cat. no. 11647229001; Roche Diagnostics, Basel, Switzerland), according to the manufacturer's protocol. Absorbance was normalized to the cell number.
Transfection
Transfection with plasmids and short hairpin (sh)RNA experiments were performed according to protocols provided by Invitrogen (Thermo Fisher Scientific, Inc.). DNA oligonucleotides carrying shRNA were constructed into the pLKO.1 plasmid (Addgene, Inc., Cambridge, MA, USA). Packaging plasmid psPAX2 (4 µg/ml) and 4 µg envelope plasmid pMD2.G (both from Addgene, Inc.) were co-transfected into 293 cells (American Type Culture Collection, Manassas, VA, USA) achieving 80% density with 4 µg recombinant plasmids using Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.). The 293 cells were grown in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), streptomycin (100 ug/ml), and penicillin (100 U/ml) and incubated at 37°C with 5% CO2 in a humidified atmosphere. The supernatant containing lentivirus particles was collected after 48 h. Target cells were infected 3 times with medium containing lentivirus (108 TU/ml) for 6 h, and recovered in normal growth medium for 24 h before next infection. The purchased shRNA target oligos were as follows: TSPAN4 shRNA#1, 5′-CTGAGCACCGCCTGGTCTCTT-3′; TSPAN4 shRNA#2, 5′-GCCTTTGTCATGGCCATCGGC-3′; TSPAN4 shRNA#3, 5′-GGTGGCCACGTGCTGGCTGCG-3′; and control shRNA (non-targeting), 5′-AACTGGACTTCCAGAAGAACA-3′.
Microarray data
The microarray dataset, GSE54129, was downloaded from the GEO database. It was based on the Affymetrix GPL570 platform (Affymetrix Human Genome U133 Plus 2.0 Array). This expression profile contained 132 samples; 111 human gastric cancer samples and 21 non-cancerous gastric samples were analyzed by high-density oligonucleotide microarray.
DEG identification
The expression profiles GSE54129 contained 132 CEL files, and in order to obtain the expression data the present study used the Affy package version 1.52.0 in R (version 3.3.1) (9) and the ‘rma’ method to perform the background correction, normalization and expression calculations. Subsequently, the limma package version 3.0.1 (10) was used obtain the DEGs, with the Benjamini-Hochberg adjustment method to identify DEGs with log2|fold-change (FC)| >1 and adjusted P-value cut-off <0.05 as statistically significant candidate genes. When the DEGs were obtained, the gplot package version 3.3.2 (11) was used to draw a heatmap that revealed the regulation of the top 100 DEGs (50 upregulated and 50 downregulated), and a volcano plot for the distribution of the DEGs in this dataset.
Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) analysis
The KEGG database (www.kegg.jp) may provide functional meanings to genes and genomes at the molecular and higher levels (12). GO (www.geneontology.org) is a community-based bioinformatics resource that supplies information about gene product functions using ontologies to represent biological knowledge. The GO database may be used to identify the molecular function, cellular component and biological process of input gene sets (13). The present study placed all DEGs in the online analysis tool Database for Annotation, Visualization and Integrated Discovery (DAVID) to obtain the relevant biological annotation. DAVID integrates a variety of online gene annotation tools, which facilitate the comprehensive analysis of gene function (14).
Disease module search
After the DEGs were obtained from the gene expression profiles, the interactions between DEGs were determined using the Search Tool for the Retrieval of Interacting Genes (STRING) database. The STRING database contains information on >5,200,000 proteins from >2,000 organisms. It provides assessment and integration of protein-protein interactions, including direct (physical) and indirect (functional) associations (15). All DEGs were mapped to STRING and only interactions with a high interaction score >0.9 were selected. The DEG network was constructed using Cytoscape software (version 3.2.1; http://www.cytoscape.org/), disease modules were extracted from the whole DEG network using the MCODE application in Cytoscape, with the following criteria: MCODE score >10 and each node >3 degrees.
Survival analysis
Kaplan Meier plots (KMplots) are capable of assessing the effect of 54,675 genes on survival using 10,293 cancer samples. These include 5,143 breast, 1,648 ovarian, 2,437 lung and 1,065 gastric cancer patients with a mean follow-up period of 69, 40, 49, and 33 months, respectively. The primary purpose of the tool is a meta-analysis based biomarker assessment (16). The top 10 hub genes of the disease module were placed in the KMplot database to examine the association between alterations in their gene expression levels and the 5-year survival rate of a patient.
Tumor xenografts
All animal experiments were approved by Qingdao University (Qingdao, China) and performed in accordance with the Institutional Animal Care and Use Committee guidelines. All experiments were performed with female FOX CHASE severe combined immunodeficient (SCID) mice (purchased from Model Animal Research Center of Nanjing University). SCID mice were used for experiments when 6 weeks old. The weight of the mice was 18–20 g and there were 5 mice in both the control and the test group. The mice were housed at 22±2°C with a relative humidity of ~50–80%, a working illumination of 50~300 lx, 12-h light/dark cycle, ventilation frequency of ~16 −20 times/h with an airflow speed, <0.18 m/sec and an ammonia concentration, <20 ppm. NCI-N87 or SGC-7901 (both American Type Culture Collection, Manassas, VA, USA) cells with stably downregulated TSPAN4 or control cells were subcutaneously injected into mice. When the tumor diameter in the right flank of mice reached 5 mm, mice were sacrificed using an injection of pentobarbital and tumor tissue was collected.
Statistical analysis
Statistical analyses were calculated in GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA) using the test appropriate for each comparison. Data are presented as the mean ± standard deviation for continuous data. One-way analysis of variance or multivariate analysis of variance was used and Dunnett's test was used for pairwise comparisons of multiple treatment groups. All experimental groups were compared with the control groups. P<0.05 was considered to indicate a statistically significant difference.
Results
DEG identification
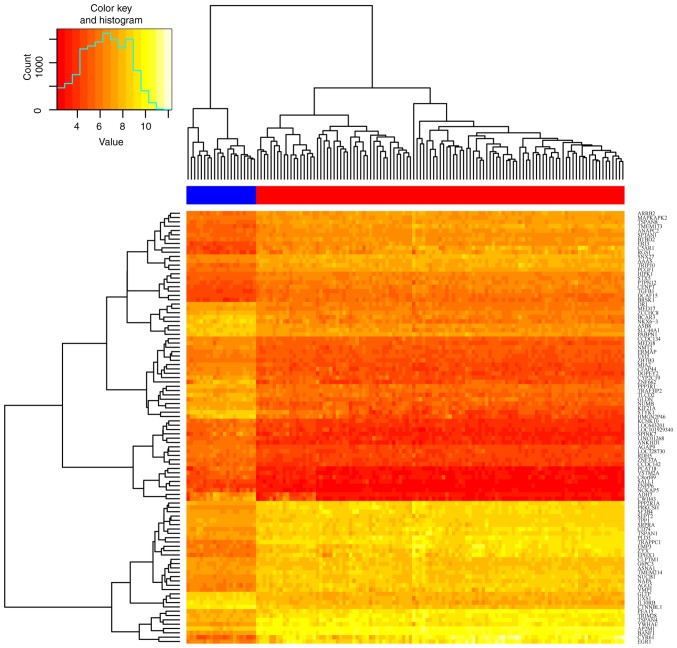
The dataset GSE54129 used in the present study contains 132 samples: 21 normal tissues samples and 111 tumor tissue samples. Each sample set was read and analyzed using the Affy and limma packages in R software. Following preprocessing, results for the whole dataset were obtained using the ‘rma’ method in the Affy package, with P<0.05 and fold control (log2|FC|) >1 as thresholds to screen out satisfactory DEGs from the expression matrix. A total of 1,829 DEGs were identified, in which there were 838 upregulated genes and 991 downregulated genes. To obtain a brief overview of these DEGs, the top 100 DEGs (50 up-regulated & 50 down-regulated) were captured in a heatmap using the gplot package (Fig. 1), and a volcano plot to illustrate the distributions of all DEGs (Fig. 2).
Figure 1.
Heatmap of the top 100 differentially expressed genes. The blue bar contains 21 control samples, and the red bar contains 111 tumor samples. Red, downregulated; yellow, upregulated.
Figure 2.
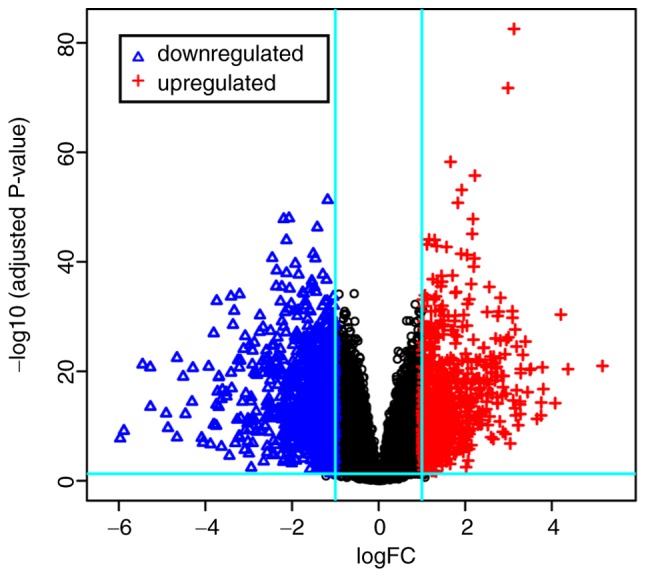
Volcano plot of the distribution of differentially expressed genes the dataset. A total of 838 genes were upregulated and 991 were downregulated. Red, upregulated; blue, downregulated. FC, fold-change.
KEGG pathway analysis
KEGG pathway analysis and GO enrichment results were obtained by uploading all DEGs to the online tool DAVID. The results of the KEGG analysis (Table I) illustrated the most significantly enriched pathways of DEGs in gastric cancer (top five upregulated and top five downregulated). The upregulated DEGs were significantly enriched in ‘ECM-receptor interaction’, ‘focal adhesion’, ‘PI3K-Akt signaling pathway’, ‘staphylococcus aureus infection’ and ‘protein digestion and absorption’, while the downregulated DEGs were significantly enriched in ‘chemical carcinogenesis’, ‘metabolism of xeno-biotics by cytochrome P450’, ‘peroxisome’, ‘fatty acid degradation’ and ‘gastric acid secretion’.
Table I.
Kyoto Encyclopedia of Genes and Genomes pathway analysis of differentially expressed genes associated with gastric cancer.
| A, Upregulated | ||||
|---|---|---|---|---|
| Pathway ID | Name | Count | P-value | Genes |
| hsa04512 | ECM-receptor interaction | 26 | 2.17×10−12 | TNC, COL3A1, COMP, COL6A3, THBS2, COL6A2, COL6A1, LAMC1, THBS1, COL11A1, SPP1, FN1, THBS4, COL4A2, COL4A1, HSPG2, ITGA1, ITGA4, COL5A3, ITGA7, COL5A2, COL5A1, LAMA4, ITGA5, COL1A2, COL1A1 |
| hsa04510 | Focal adhesion | 40 | 3.37×10–12 | TNC, COL3A1, MYL9, RAC2, COMP, ILK, COL6A3, ZYX, COL6A2, COL6A1, THBS4, THBS1, COL11A1, THBS2, PIK3R1, SPP1, FN1, IGF1, COL4A2, COL4A1, ITGA1, ACTN1, ITGA4, FLNC, COL5A3, PARVB, COL5A2, COL5A1, FLNA, KDR, VEGFC, LAMA4, FYN, ITGA5, ITGA7, COL1A2, PDGFRB, COL1A1, LAMC1, MYLK |
| hsa04151 | PI3K-Akt signaling pathway | 49 | 2.87×10−12 | HSP90AB1, OSMR, FGF10, TLR4, CSF3R, PIK3AP1, MYC, ANGPT2, COL11A1, PPP2R1A, TP53, VEGFC, COL1A2, PDGFRB, GNB4, COL1A1, LAMC1, MCL1, TNC, COL3A1, BCL2L1, IL7R, G6PC3, COMP, COL6A3, SPP1, COL6A2, COL6A1, IL2RG, THBS1, THBS2, PIK3R1, THBS4, FN1, CSF1R, IL6, COL4A2, IL2RA, COL4A1, ITGA1, IGF1, ITGA4, COL5A3, YWHAE, KDR, COL5A2, COL5A1, ITGA5, ITGA7 |
| hsa05150 | Staphylococcus aureus infection | 17 | 1.35×10-8 | ICAM1, SELP, C3AR1, C5AR1, C3, FPR3, C1R, ITGB2, C1S, HLA-DMB, C1QA, C1QB, FPR1, C1QC, HLA-DQA1, HLA-DPA1, FCGR3B |
| hsa04974 | Protein digestion and absorption | 21 | 3.16×10-8 | COL18A1, COL4A2, COL4A1, COL21A1, COL3A1, ELN, COL15A1, ATP1A2, COL5A3, COL5A2, COL5A1, SLC1A5, COL14A1, COL6A3, COL1A2, COL6A2, COL12A1, COL6A1, COL1A1, COL11A1, COL10A1 |
| B, Downregulated | ||||
| Pathway ID | Name | Count | P-value | Genes |
| hsa05204 | Chemical carcinogenesis | 18 | 2.86×10-7 | CYP3A5, CYP2C19, SULT2A1, CYP2C9, CYP2C18, NAT1, ADH1C, ADH1A, ADH7, CYP3A7-CYP3A51P, MGST2, ALDH3A1, CBR1, SULT1A1, SULT1A2, GSTP1, UGT2A3, UGT2B15 |
| hsa00980 | Metabolism of xeno-biotics by cytochrome P450 | 16 | 2.72×10-6 | CYP3A5, SULT2A1, CYP2C9, ADH1C, ADH1A, ADH7, ALDH3A1, AKR1C2, CBR1, AKR1C4, AKR7A3, UGT2A3, UGT2B15, AKR1C1, GSTP1, MGST2 |
| hsa04146 | Peroxisome | 16 | 1.2×10-5 | XDH, ACOX1, NUDT12, EHHADH, EPHX2, PRDX5, PHYH, PEX7, ACOX3, PEX11A, FAR1, PXMP2, IDH1, ACSL3, SCP2, SLC27A2 |
| hsa00071 | Fatty acid degradation | 11 | 4.57×10-5 | ACOX1, CPT2, EHHADH, ADH1C, ALDH2, ADH1A, ADH7, HADH, ACSL3, ALDH3A2, ACOX3 |
| hsa04971 | Gastric acid secretion | 11 | 3.2×10-3 | KCNJ16, KCNJ15, PLCB3, PLCB4, ATP4A, ATP4B, KCNE2, GAST, CA2, SST, KCNK10 |
GO enrichment analysis
The results of the GO analysis (Table II) demonstrated that with respect to biological processes (BP), upregulated DEGs were significantly enriched in ‘extracellular matrix organization’, ‘cell adhesion’ and ‘inflammatory response’, while downregulated DEGs were significantly enriched in ‘xenobiotic metabolic process’, ‘oxidation-reduction process’ and ‘steroid metabolic process’. With respect to molecular function (MF), upregulated DEGs were significantly enriched in ‘heparin binding’, ‘extracellular matrix structural constituent’ and ‘integrin binding’, while downregulated DEGs were significantly enriched in ‘oxidoreductase activity’, ‘NAD+ binding’ and ‘aldo-keto reductase (NADP) activity’. With respect to cellular component, upregulated DEGs were significantly enriched in ‘extracellular matrix’, ‘extracellular region’ and ‘proteinaceous extracellular matrix’, while downregulated DEGs were significantly enriched in ‘extracellular exosome’, ‘peroxisomal matrix’ and ‘apical plasma membrane’.
Table II.
GO analysis of differentially expressed genes associated with gastric cancer.
| A, Upregulated BP | |||
|---|---|---|---|
| GO term | Function | Count | P-value |
| GO:0030198 | Extracellular matrix organization | 60 | 8.71×10−33 |
| GO:0007155 | Cell adhesion | 84 | 2.77×10−28 |
| GO:0006954 | Inflammatory response | 71 | 4.15×10−23 |
| GO:0030574 | Collagen catabolic process | 27 | 7.35×10−19 |
| GO:0050900 | Leukocyte migration | 44 | 4.32×10−16 |
| B, Upregulated MF | |||
| GO term | Function | Count | P-value |
| GO:0008201 | Heparin binding | 40 | 5.50×10−19 |
| GO:0005201 | Extracellular matrix structural constituent | 27 | 1.27×10−18 |
| GO:0005178 | Integrin binding | 30 | 6.20×10−16 |
| GO:0005515 | Protein binding | 466 | 3.03×10-10 |
| GO:0050840 | Extracellular matrix binding | 12 | 4.26×10−9 |
| C, Upregulated CC | |||
| GO term | Function | Count | P-value |
| GO:0031012 | Extracellular matrix | 88 | 2.87×10−48 |
| GO:0005576 | Extracellular region | 186 | 1.45×10−36 |
| GO:0005578 | Proteinaceous extracellular matrix | 71 | 2.96×10−35 |
| GO:0005615 | Extracellular space | 161 | 5.22×10−33 |
| GO:0070062 | Extracellular exosome | 225 | 1.94×10−21 |
| D, Downregulated BP | |||
| GO term | Function | Count | P-value |
| GO:0006805 | Xenobiotic metabolic process | 24 | 5.27×10−13 |
| GO:0055114 | Oxidation-reduction process | 67 | 3.82×10−11 |
| GO:0008202 | Steroid metabolic process | 16 | 4.19×10−10 |
| GO:0030855 | Epithelial cell differentiation | 16 | 6.14×10−7 |
| GO:0007586 | Digestion | 15 | 9.10×10−7 |
| E, Downregulated MF | |||
| GO term | Function | Count | P-value |
| GO:0016491 | Oxidoreductase activity | 26 | 7.08×10−6 |
| GO:0070403 | NAD+ binding | 6 | 2.06×10−4 |
| GO:0004033 | Aldo-keto reductase (NADP) activity | 6 | 4.44×10−4 |
| GO:0004062 | Aryl sulfotransferase activity | 5 | 1.7×10−3 |
| GO:0004022 | Alcohol dehydrogenase (NAD) activity | 4 | 3.1×10−3 |
| F, Downregulated CC | |||
| GO term | Function | Count | P-value |
| GO:0070062 | Extracellular exosome | 198 | 2.96×10−9 |
| GO:0005782 | Peroxisomal matrix | 12 | 7.33×10−6 |
| GO:0016324 | Apical plasma membrane | 32 | 2.21×10−5 |
| GO:0005789 | Endoplasmic reticulum membrane | 69 | 2.47×10−5 |
| GO:0016021 | Integral component of membrane | 295 | 9.53×10−5 |
GO, gene ontology; BP, biological process; CC, cellular component; MF, molecular function.
Disease module search
To understand the interactions among the collected DEGs, we the STRING database was used to obtain the protein-protein interaction (PPI) information for each DEG. During this step, PPIs with the highest interaction scores (confidence >0.9) were selected. Using Cytoscape software to analyze the PPI information, the degree of each node was ranked and the top 10 hub nodes with higher degrees were obtained. These hub genes included tumor protein P53 (TP53), C-X-C motif chemokine ligand 8 (CXCL8), tetraspanin 4 (TSPAN4), lysophosphatidic acid receptor 2 (LPAR2), adenylate cyclase 3 (ADCY3), phosphoinositide-3-kinase regulatory subunit 1 (PIK3R1), neuromedin U (NMU), C-X-C motif chemokine ligand 12 (CXCL12), fos proto-oncogene, AP-1 transcription factor subunit (FOS) and sphingosine-1-phosphate receptor 1 (S1PR1). Among these genes, TP53 had the highest node degree (degree=53; Table III). In addition, it was noted that TSPAN4 appeared in the heat map of the top 100 DEGs, and a further two tetraspanin family genes, TSPAN1 and TSPAN8, also appeared in it. Tetraspanin family genes may be a highlight biomarker of gastric cancer.
Table III.
Top 10 hub genes ranked by degree.
| Gene symbol | Full name | Degree |
|---|---|---|
| TP53 | Tumor protein P53 | 53 |
| CXCL8 | C-X-C motif chemokine ligand 8 | 53 |
| TSPAN4 | Tetraspanin 4 | 51 |
| LPAR2 | Lysophosphatidic acid receptor 2 | 50 |
| ADCY3 | Adenylate cyclase 3 | 49 |
| PIK3R1 | Phosphoinositide-3-kinase regulatory subunit 1 | 48 |
| NMU | Neuromedin U | 44 |
| CXCL12 | C-X-C motif chemokine ligand 12 | 44 |
| FOS | Fos proto-oncogene, AP-1 transcription factor subunit | 44 |
| S1PR1 | Sphingosine-1-phosphate receptor 1 | 41 |
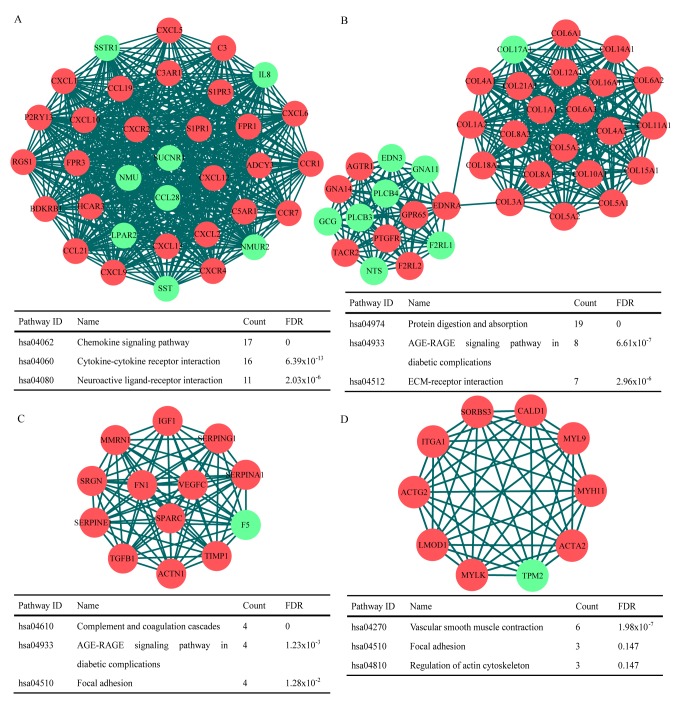
Furthermore, the interact information of 1,829 DEGs and 2,901 edges was used to construct the top 4 MCODE score disease modules of gastric cancer using the MCODE application in Cytoscape software. KEGG pathway enrichment results were obtained for each gene contained within the modules (Fig. 3). The results demonstrated that the genes contained in the modules were significantly enriched in ‘chemokine signaling pathway’, ‘protein digestion and absorption’, ‘complement and coagulation cascades’ and ‘AGE-RAGE signaling pathway in diabetic complications’.
Figure 3.
(A-D) Top four disease modules constructed from the differentially expressed genes, and the enrichment results for each module. Red nodes, upregulated genes; green nodes, downregulated genes; FDR, false discovery rate.
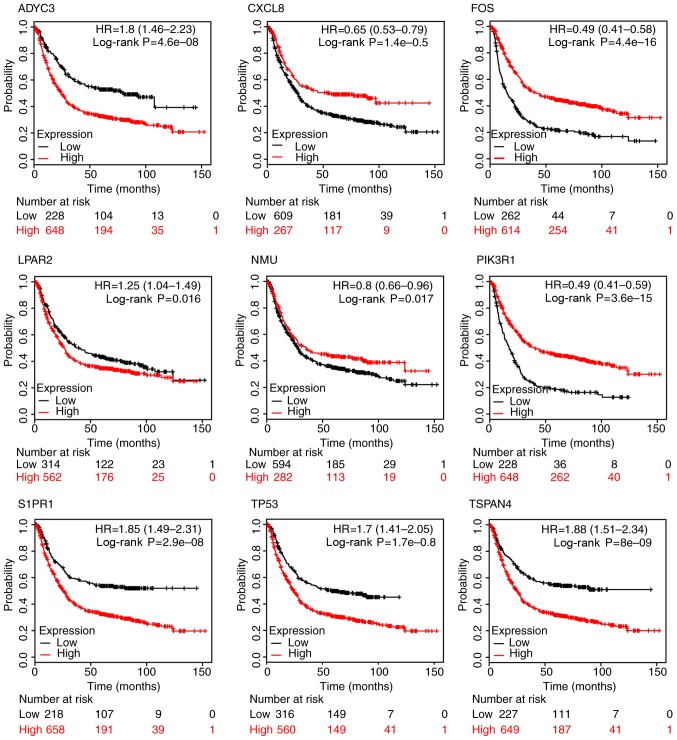
Survival analysis
It was necessary to validate the 10 hub genes screened from the DEGs. KMplot analysis was used to elucidate the influence on survival rate of the differential expression of hub genes. Among the 10 hub genes, increased expression of four genes was associated with a decreased survival rate; they were ADCY3, LPAR2, S1PR1 and TSPAN4. The increased expression of five genes was associated with an increased survival rate; they were TP53, CXCL8, FOS, NMU and PIK3R1 (Fig. 4). Quantitative alterations in the expression of CXCL12 were not significantly associated with survival rate.
Figure 4.
Survival analysis of hub genes which have a significant association with survival rate. ADCY3, adenylate cyclase 3; CXCL8, C-X-C motif chemokine ligand 8; FOS, fos proto-oncogene, AP-1 transcription factor subunit; LPAR2, lysophosphatidic acid receptor 2; NMU, neuromedin U; PIK3R1, phosphoinositide-3-kinase regulatory subunit 1; S1PR1, sphingosine-1-phosphate receptor 1; TP53, tumor protein p53; TSPAN4, tetraspanin 4.
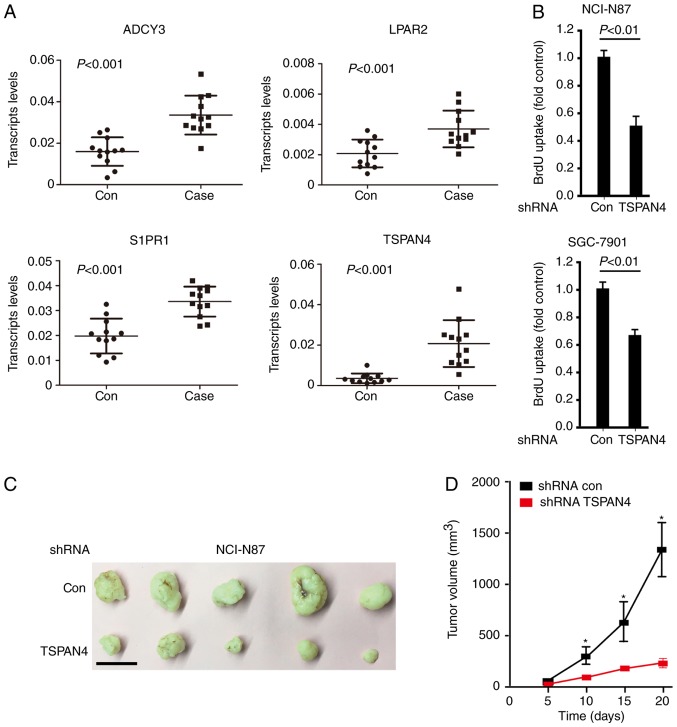
Tumor xenografts
To examine whether the four hub genes associated with poor prognosis were involved in pathogenesis of gastric cancer, gastric cancer and para-carcinoma tissues from 12 patients were subjected to qPCR analysis. It was observed that four genes were highly expressed in gastric cancer tissue compared with para-carcinoma tissue in the 12 patients (Fig. 5A). However, the increased TSPAN4 was most significant (>5-fold). To investigate whether TSPAN4 was associated with the proliferation of gastric cancer cells, a BrdU assay was performed. The results indicated that the downregulation of TSPAN4 markedly decreased the proliferation of NCI-N87 and SGC-7901 cells (Fig. 5B). To examine the role of TSPAN4 in tumorigenesis in vivo, TSPAN4 was stably downregulated in NCI-N87 cells using a lentivirus carrying TSPAN4 shRNA, and NCI-N87 cells with stably downregulated TSPAN4 or control cells were subcutaneously injected into athymic nude mice. A total of 20 days subsequently, all the control cells formed visible xenograft tumors; conversely, mice injected with NCI-N87 cells with stably downregulated TSPAN4 exhibited significantly delayed xenograft tumor growth, with a decreased mean tumor weight compared with the control group (P<0.01; Fig. 5C and D). Similar results were obtained when using SGC-7901 cells (data not shown), validating TSPAN4as a potential therapeutic target in gastric cancer caused by TSPAN4 overexpression.
Figure 5.
TSPAN4 is involved in the pathogenesis of gastric cancer. (A) Total RNA was prepared and subjected to reverse transcription-quantitative polymerase chain reaction analysis. GAPDH was used as a control. The results were analyzed using the 2−Δ∆Cq method. Data are presented as the mean ± standard deviation of three independent replicates, P-values as indicated. (B) Cellular proliferation of control and TSPAN4 shRNA gastric cancer cells was assessed using the BrdU assay. Data are presented as the mean ± standard error of the mean of five experiments. (C) NCI-N87 cells treated with lentivirus-delivered TSPAN4 knockdown were subcutaneously implanted into female athymic nude mice (n=5 per experimental condition). Tumor images were captured on day 20. Scale bar, 1 cm. (D) Tumor growth curves. Data are presented as the mean ± standard error of the mean. *P<0.01 vs. shRNA con. Con, control; TSPAN4, tetraspanin 4; ADCY3, adenylate cyclase 3; LPAR2, lysophosphatidic acid receptor 2; S1PR1, sphingosine-1-phosphate receptor 1; shRNA, short hairpin RNA.
Discussion
The incidence and mortality of gastric cancer has made it the 4th most common cancer with the 2nd highest mortality rate (17). Due to the complex etiology of gastric cancer, identifying biomarkers and biological pathways has become valuable for the diagnosis and treatment of the disease. The dataset GSE54129 was used in the present study and for a comprehensive bioinformatics analysis, and 838 upregulated and 991 downregulated DEGs were identified.
In the KEGG analysis of the DEGs, the upregulated genes were significantly mapped in ‘ECM-receptor interaction’, ‘focal adhesion’, ‘PI3K-Akt signaling pathway’, ‘staphylococcus aureus infection’ and ‘protein digestion and absorption’. In previous studies, ‘ECM-receptor interaction’ and ‘focal adhesion’ were identified as representing some of the main differences between normal and tumor tissues of gastric cancer at a molecular level (18,19). Guo et al (20), demonstrated that focal adhesion kinase (FAK) was negatively correlated with olfactomedin 4 with respect to lymph node metastasis in gastric cancer tissues. In addition, FAK was positively correlated with the expression of caveolin 1, which is known as both a tumor promoter and suppressor in different cancer types (21). Furthermore, hepatocyte growth factor is able to upregulate heparanase expression and induce tumor cell migration via the phosphatidylinositol 3-kinase-RAC-α serine/threonine-protein kinase signaling pathway (22). In the present study, the downregulated DEGs were significantly enriched in ‘chemical carcinogenesis’, ‘metabolism of xeno-biotics by cytochrome P450’, ‘peroxisome’, ‘fatty acid degradation’ and ‘gastric acid secretion’. These results also coincided with the results of previous studies (23–27).
The results of the GO analysis demonstrated that in BP, the downregulated DEGs were significantly enriched in ‘oxidation-reduction process’ and ‘steroid metabolic process’ and previous structure-activity relationship analysis has demonstrated that the 7-methyl-substituted and 15-methyl-substituted steroid analogs lead to a marked increase in potency against the human gastric cancer cell line MGC-803 (28).
During the disease module construction based on the PPI network from STRING, ten hub genes from DEGs were screened out: TP53, CXCL8, TSPAN4, LPAR2, ADCY3, PIK3R1, NMU, CXCL12, FOS and S1PR1. From the results of the KM analysis, it was identified that the increased expression of five genes was negatively correlated with the 5-year survival rate. They were TP53, ADCY3, LPAR2, S1PR1 and TSPAN4. TP53 had the top degree among all DEGs. TP53 encodes a tumor suppressor protein containing transcriptional activation, DNA binding and oligomerization domains. These proteins induce cell cycle arrest, apoptosis, senescence, DNA repair or alterations in metabolism (29–32). Previous research has proved the association between TP53 and gastric cancer. Ando et al (33) studied 182 clinical samples of gastric cancer and observed that TP53-positive tumors were invaded more deeply and had more lymph node and liver metastases, and that some genes (PICT1, RPL11) participate in the cancer progression via TP53 (34). TP53 mutations occur late in gastric carcinogenesis, contributing to the final transition to cancer (35). LPAR2, a lysophosphatidic acid (LPA) receptor, contributes to Ca2+ mobilization, a critical cellular response to LPA in cells, through its association with Gi and Gq proteins. Experimental results have observed that LPAR2 is highly expressed in SGC-7901 cells, a human gastric cancer cell line, and the LPAR2/Gq/11/p38 pathway regulates LPA-induced gastric cancer cell line migration (36). ADCY3 is a membrane-associated enzyme and catalyzes the formation of the secondary messenger cyclic adenosine monophosphate (cAMP); a study reported that the expression of ADCY3 was regulated through an epigenetic mechanism, thus ADCY3 overexpression may exert its tumor-promoting effects via the cAMP/protein kinase A/cAMP response-element binding protein pathway (37). S1PR1, which is highly expressed in endothelial cells, has also been suggested to be a therapeutic target for gastric cancer in another study (38). S1PR1 was demonstrated to have a markedly increased expression level in benign tissues compared with malignant human tissues by Wang et al (39). TSPAN4 serves a role in the regulation of cell development, activation, growth and motility. TSPAN1 and TSPAN8 were additionally screened out from the top 100 DEGs, which are members of the transmembrane 4 superfamily. A study by Chen et al (40) suggested that the overexpression of TSPAN1 is positively correlated with clinical stage and negatively correlated with survival rate (at 3 and 5 years). The overexpression of TSPAN1 was reported to be negatively correlated with carcinoma differentiation, and TSPAN1 is positively correlated with the infiltration and lymph node status of a tumor (40). Furthermore, Lu et al (41) reported that the miR-573/TSPAN1 axis is important in the control of gastric carcinogenesis. In addition, TSPAN8 was suggested to be associated with tumor progression and is an independent prognostic factor in patients with gastric cancer; it has been reported to promote gastric cancer cell growth and metastasis, at least partially through the activation of the extracellular signal-regulated kinase/mitogen-activate protein kinase pathway (42,43).
The other genes, PIK3R1, NMU, FOS, CXCL8 and CXCL12, have a significant positive correlation with the 5-year survival rate. PIK3R1 is involved in the regulation of cell function, including proliferation and survival. PIK3R1 serves a critical role in the formation of a number of types of malignant tumors. The results reported by Fu et al (44) further demonstrated that downregulated PIK3R1 exerts inhibitory effects on the proliferation and invasion of SGC7901 and U251 cells. NMU has been regarded to have promoting effect on the generation of a biologically active neuropeptide which serves a role in pain, stress immune-mediated inflammatory diseases and feeding regulation; the gene has been reported to exhibit early alterations associated with cancer (45). NMU was observed to be the second most upregulated gene in HT29 colon cancer cells in a recent study (46). The FOS gene family consists of 4 members: FOS, FosB proto-oncogene, AP-1 transcription factor subunit, FOS like 1, AP-1 transcription factor subunit, and FOS like 2, AP-1 transcription factor subunit; they encode leucine zipper proteins that are able to dimerize with proteins of the JUN family, thereby forming the transcription factor complex AP-1. Research has demonstrated that AP-1 and the AP-1 binding sites of the matrix metalloproteinase (MMP)9 promoter (−670/MMP9) are activated by IL-1β-induced p38 activation, increased the migration of gastric adenocarcinoma cells (47).
CXCL8 and CXCL12 appeared in the top 10 genes. CXCL8 functions as a chemoattractant and is also a potent angiogenic factor. CXCL8 is able to modulate host immunity, neovascularization, and the growth and invasive behavior of tumors. Research has demonstrated that CXC chemokines have the ability to stimulate the formation of new blood vessels, facilitating tumor progression (48); this may explain why the high expression of CXCL8 was positively correlated with survival rate in the KM analysis performed in the present study. It is noteworthy that although CXCL12 did not have a significant association with survival rate in the KM analysis, high CXCL12 expression levels were significantly associated with larger tumor size, increased tumor depth, lymphatic invasion and poor prognosis in gastric cancer in the study of Izumi et al (49).
In conclusion, by combining bioinformatics methods and tools, the present study provided a comprehensive analysis of DEGs from gastric cancer. A number of pathways and biomarkers in the progression of gastric cancer were identified, which may provide guidance for further molecular biological study. Furthermore, via tumor xenografts, it was observed that downregulating TSPAN4 expression was able to inhibit tumorigenesis, indicating that the gene may have a delaying effect on the progression of gastric cancer. As a complex disease, the biological mechanisms of gastric cancer remain to be completely elucidated. Further research is required, including biological experiments and bioinformatics analysis.
Acknowledgements
The authors would like to thank Yichen Yang from the Bioinfomatics Laboratory of Tianjin Medical University (Tianjin, China) for guidance on software.
Funding
The present study was supported by National Natural Science Foundation of China (grant nos. 81472338 and 81602068).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
WWQ and LS drafted the manuscript and took part in planning the execution of the high-throughput analysis experiments. NL took part in the development of the analysis code. JL collected the expression data and screened the different expressed genes. SZ produced the qPCR analysis. WSQ designed this experiment and revised the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The use of human gastric cancer specimens and the database was approved by the institutional review board of the Hospital of Qingdao University (Qingdao, China). Written informed consent was obtained from all patients. All animal experiments were approved by Qingdao University and performed in accordance with the Institutional Animal Care and Use Committee guidelines.
Patient consent for publication
Written informed consent was obtained from all patients.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Crew KD, Neugut AI. Epidemiology of gastric cancer. World J Gastroenterol. 2006;12:354–362. doi: 10.3748/wjg.v12.i3.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization (WHO), corp-author GLOBOCAN 2012: Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2012. WHO; Geneva: 2012. [Google Scholar]
- 4.Liu P, Wang X, Hu CH, Hu TH. Bioinformatics analysis with graph-based clustering to detect gastric cancer-related pathways. Genet Mol Res. 2012;11:3497–3504. doi: 10.4238/2012.September.26.5. [DOI] [PubMed] [Google Scholar]
- 5.Tian J, Wang XD, Chen ZC. Survival of patients with stomach cancer in Changle city of China. World J Gastroenterol. 2004;10:1543–1546. doi: 10.3748/wjg.v10.i11.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wagner AD, Grothe W, Haerting J, Kleber G, Grothey A, Fleig WE. Chemotherapy in advanced gastric cancer: A systematic review and meta-analysis based on aggregate data. J Clin Oncol. 2006;24:2903–2909. doi: 10.1200/JCO.2005.05.0245. [DOI] [PubMed] [Google Scholar]
- 7.Bejjani BA, Shaffer LG. Clinical utility of contemporary molecular cytogenetics. Annu Rev Genomics Hum Genet. 2008;9:71–86. doi: 10.1146/annurev.genom.9.081307.164207. [DOI] [PubMed] [Google Scholar]
- 8.Arocho A, Chen B, Ladanyi M, Pan Q. Validation of the 2-DeltaDeltaCt calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL P210 transcripts. Diagn Mol Pathol. 2006;15:56–61. doi: 10.1097/00019606-200603000-00009. [DOI] [PubMed] [Google Scholar]
- 9.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 10.Phipson B, Lee S, Majewski IJ, Alexander WS, Smyth GK. Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann Appl Stat: 2016;10:946–963. doi: 10.1214/16-AOAS920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warnes GR, Bolker B, Bonebakker L, Gentleman R, Andy Liaw WH, Lumley T, Maechler M, Magnusson A, Moeller S, Schwartz M, Venables B. https://cran.r-project.org/web/packages/gplots/index.html. [Mar 30;2016 ];gplots: Various R Programming Tools for Plotting Data. [Google Scholar]
- 12.Kanehisa M. The KEGG database. Novartis Found Symp. 2002;247:91–101. 244–252. doi: 10.1002/0470857897.ch8. discussion 101–103, 119–128. [DOI] [PubMed] [Google Scholar]
- 13.Martucci D, Masseroli M, Pinciroli F. Gene ontology application to genomic functional annotation, statistical analysis and knowledge mining. Stud Health Technol Inform. 2004;102:108–131. [PubMed] [Google Scholar]
- 14.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 15.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database Issue):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szász AM, Lánczky A, Nagy Á, Förster S, Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A, Győrffy B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferro A, Peleteiro B, Malvezzi M, Bosetti C, Bertuccio P, Levi F, Negri E, La Vecchia C, Lunet N. Worldwide trends in gastric cancer mortality (1980–2011), with predictions to 2015, and incidence by subtype. Eur J Cancer. 2014;50:1330–1344. doi: 10.1016/j.ejca.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 18.Yang S, Shin J, Park KH, Jeung HC, Rha SY, Noh SH, Yang WI, Chung HC. Molecular basis of the differences between normal and tumor tissues of gastric cancer. Biochim Biophys Acta. 2007;1772:1033–1040. doi: 10.1016/j.bbadis.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Z, Song Y, Piao D, Liu T, Zhao L. Identification of genes and long non-coding RNAs associated with the pathogenesis of gastric cancer. Oncol Rep. 2015;34:1301–1310. doi: 10.3892/or.2015.4129. [DOI] [PubMed] [Google Scholar]
- 20.Guo LL, He ZC, Yang CQ, Qiao PT, Yin GL. Epigenetic silencing of olfactomedin-4 enhances gastric cancer cell invasion via activation of focal adhesion kinase signaling. BMB Rep. 2015;48:630–635. doi: 10.5483/BMBRep.2015.48.11.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nam KH, Lee BL, Park JH, Kim J, Han N, Lee HE, Kim MA, Lee HS, Kim WH. Caveolin 1 expression correlates with poor prognosis and focal adhesion kinase expression in gastric cancer. Pathobiology. 2013;80:87–94. doi: 10.1159/000341685. [DOI] [PubMed] [Google Scholar]
- 22.Hao NB, Tang B, Wang GZ, Xie R, Hu CJ, Wang SM, Wu YY, Liu E, Xie X, Yang SM. Hepatocyte growth factor (HGF) upregulates heparanase expression via the PI3K/Akt/NF-κB signaling pathway for gastric cancer metastasis. Cancer Lett. 2015;361:57–66. doi: 10.1016/j.canlet.2015.02.043. [DOI] [PubMed] [Google Scholar]
- 23.Cho SJ, Kook MC, Lee JH, Shin JY, Park J, Bae YK, Choi IJ, Ryu KW, Kim YW. Peroxisome proliferator-activated receptor γ upregulates galectin-9 and predicts prognosis in intestinal-type gastric cancer. Int J Cancer. 2015;136:810–820. doi: 10.1002/ijc.29056. [DOI] [PubMed] [Google Scholar]
- 24.Choi JM, Park WS, Song KY, Lee HJ, Jung BH. Development of simultaneous analysis of tryptophan metabolites in serum and gastric juice-an investigation towards establishing a biomarker test for gastric cancer diagnosis. Biomed Chromatogr. 2016;30:1963–1974. doi: 10.1002/bmc.3773. [DOI] [PubMed] [Google Scholar]
- 25.Leung WK, Wu KC, Wong CY, Cheng AS, Ching AK, Chan AW, Chong WW, Go MY, Yu J, To KF, et al. Transgenic cyclooxygenase-2 expression and high salt enhanced susceptibility to chemical-induced gastric cancer development in mice. Carcinogenesis. 2008;29:1648–1654. doi: 10.1093/carcin/bgn156. [DOI] [PubMed] [Google Scholar]
- 26.Kim HS, Kwack SJ, Lee BM. Alteration of cytochrome P-450 and glutathione S-transferase activity in normal and malignant human stomach. J Toxicol Environ Health A. 2005;68:1611–1620. doi: 10.1080/15287390500182867. [DOI] [PubMed] [Google Scholar]
- 27.Song H, Peng JS, Yao DS, Liu DL, Yang ZL, Du YP, Xiang J. Metabolic disorders of fatty acids and fatty acid amides associated with human gastric cancer morbidity. Chin Med J (Engl) 2012;125:757–763. [PubMed] [Google Scholar]
- 28.Li C, Qiu W, Yang Z, Luo J, Yang F, Liu M, Xie J, Tang J. Stereoselective synthesis of some methyl-substituted steroid hormones and their in vitro cytotoxic activity against human gastric cancer cell line MGC-803. Steroids. 2010;75:859–869. doi: 10.1016/j.steroids.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 29.Nicolai S, Rossi A, Di Daniele N, Melino G, Annicchiarico-Petruzzelli M, Raschellà G. DNA repair and aging: The impact of the p53 family. Aging (Albany NY) 2015;7:1050–1065. doi: 10.18632/aging.100858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rufini A, Tucci P, Celardo I, Melino G. Senescence and aging: The critical roles of p53. Oncogene. 2013;32:5129–5143. doi: 10.1038/onc.2012.640. [DOI] [PubMed] [Google Scholar]
- 31.Wawryk-Gawda E, Chylińska-Wrzos P, Lis-Sochocka M, Chłapek K, Bulak K, Jędrych M, Jodłowska-Jędrych B. P53 protein in proliferation, repair and apoptosis of cells. Protoplasma. 2014;251:525–533. doi: 10.1007/s00709-013-0548-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan L, Zhang Y, Xia J, Liu B, Zhang Q, Liu J, Luo L, Peng Z, Song Z, Zhu R. Resveratrol induces cell cycle arrest via a p53-independent pathway in A549 cells. Mol Med Rep. 2015;11:2459–2464. doi: 10.3892/mmr.2014.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ando K, Oki E, Zhao Y, Ikawa-Yoshida A, Kitao H, Saeki H, Kimura Y, Ida S, Morita M, Kusumoto T, Maehara Y. Mortalin is a prognostic factor of gastric cancer with normal p53 function. Gastric Cancer. 2014;17:255–262. doi: 10.1007/s10120-013-0279-1. [DOI] [PubMed] [Google Scholar]
- 34.Uchi R, Kogo R, Kawahara K, Sudo T, Yokobori T, Eguchi H, Sugimachi K, Maehama T, Mori M, Suzuki A, et al. PICT1 regulates TP53 via RPL11 and is involved in gastric cancer progression. Br J Cancer. 2013;109:2199–2206. doi: 10.1038/bjc.2013.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Busuttil RA, Zapparoli GV, Haupt S, Fennell C, Wong SQ, Pang JM, Takeno EA, Mitchell C, Di Costanzo N, Fox S, et al. Role of p53 in the progression of gastric cancer. Oncotarget. 2014;5:12016–12026. doi: 10.18632/oncotarget.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang D, Yang W, Zhang Q, Hu Y, Bao L, Damirin A. Migration of gastric cancer cells in response to lysophosphatidic acid is mediated by LPA receptor 2. Oncol Lett. 2013;5:1048–1052. doi: 10.3892/ol.2013.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hong SH, Goh SH, Lee SJ, Hwang JA, Lee J, Choi IJ, Seo H, Park JH, Suzuki H, Yamamoto E, et al. Upregulation of adenylate cyclase 3 (ADCY3) increases the tumorigenic potential of cells by activating the CREB pathway. Oncotarget. 2013;4:1791–1803. doi: 10.18632/oncotarget.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo Y, Zhang C, Tang F, Zhao J, Shen C, Wang C, Yu P, Wang M, Li Y, Di JI, et al. Bioinformatics identification of potentially involved microRNAs in Tibetan with gastric cancer based on microRNA profiling. Cancer Cell Int. 2015;15:115. doi: 10.1186/s12935-015-0266-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang C, Mao J, Redfield S, Mo Y, Lage JM, Zhou X. Systemic distribution, subcellular localization and differential expression of sphingosine-1-phosphate receptors in benign and malignant human tissues. Exp Mol Pathol. 2014;97:259–265. doi: 10.1016/j.yexmp.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen L, Li X, Wang GL, Wang Y, Zhu YY, Zhu J. Clinicopathological significance of overexpression of TSPAN1, K167 and CD34 in gastric carcinoma. Tumori. 2008;94:531–538. doi: 10.1177/030089160809400415. [DOI] [PubMed] [Google Scholar]
- 41.Lu Z, Luo T, Nie M, Pang T, Zhang X, Shen X, Ma L, Bi J, Wei G, Fang G, Xue X. TSPAN1 functions as an oncogene in gastric cancer and is downregulated by miR-573. FEBS Lett. 2015;589:1988–1994. doi: 10.1016/j.febslet.2015.05.044. [DOI] [PubMed] [Google Scholar]
- 42.Anami K, Oue N, Noguchi T, Sakamoto N, Sentani K, Hayashi T, Naito Y, Oo HZ, Yasui W. TSPAN8, identified by Escherichia coli ampicillin secretion trap, is associated with cell growth and invasion in gastric cancer. Gastric Cancer. 2016;19:370–380. doi: 10.1007/s10120-015-0478-z. [DOI] [PubMed] [Google Scholar]
- 43.Wei L, Li Y, Suo Z. TSPAN8 promotes gastric cancer growth and metastasis via ERK MAPK pathway. Int J Clin Exp Med. 2015;8:8599–8607. [PMC free article] [PubMed] [Google Scholar]
- 44.Fu Y, Zhang Q, Kang C, Zhang J, Zhang K, Pu P, Wang G, Wang T. Inhibitory effects of adenovirus mediated Akt1 and PIK3R1 shRNA on the growth of malignant tumor cells in vitro and in vivo. Cancer Biol Ther. 2009;8:1002–1009. doi: 10.4161/cbt.8.11.8285. [DOI] [PubMed] [Google Scholar]
- 45.Yamashita K, Upadhyay S, Osada M, Hoque MO, Xiao Y, Mori M, Sato F, Meltzer SJ, Sidransky D. Pharmacologic unmasking of epigenetically silenced tumor suppressor genes in esophageal squamous cell carcinoma. Cancer Cell. 2002;2:485–495. doi: 10.1016/S1535-6108(02)00215-5. [DOI] [PubMed] [Google Scholar]
- 46.Przygodzka P, Papiewska-Pajak I, Bogusz H, Kryczka J, Sobierajska K, Kowalska MA, Boncela J. Neuromedin U is upregulated by Snail at early stages of EMT in HT29 colon cancer cells. Biochim Biophys Acta. 2016;1860:2445–2453. doi: 10.1016/j.bbagen.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 47.Huang Q, Lan F, Wang X, Yu Y, Ouyang X, Zheng F, Han J, Lin Y, Xie Y, Xie F, et al. IL-1β-induced activation of p38 promotes metastasis in gastric adenocarcinoma via upregulation of AP-1/c-fos, MMP2 and MMP9. Mol Cancer. 2014;13:18. doi: 10.1186/1476-4598-13-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verbeke H, Geboes K, Van Damme J, Struyf S. The role of CXC chemokines in the transition of chronic inflammation to esophageal and gastric cancer. Biochim Biophys Acta. 2012;1825:117–129. doi: 10.1016/j.bbcan.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 49.Izumi D, Ishimoto T, Miyake K, Sugihara H, Eto K, Sawayama H, Yasuda T, Kiyozumi Y, Kaida T, Kurashige J, et al. CXCL12/CXCR4 activation by cancer-associated fibroblasts promotes integrin β1 clustering and invasiveness in gastric cancer. Int J Cancer. 2016;138:1207–1219. doi: 10.1002/ijc.29864. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.