Abstract
Endometriosis affects 6–10% of women of reproductive age. Though a significant amount of research has explored the pathogenesis of endometriosis, little is clear. Elucidating the mechanisms is urgently required for improving the therapeutic efficiency of endometriosis treatment. Long non-coding RNAs (lncRNAs) have recently acquired extensive attention as regulatory components in a variety of biological processes and diseases. However, the functions of many lncRNAs in endometriosis are poorly understood. Therefore, the exploration of the dysregulated genes in endometriosis, particularly lncRNAs, is of importance. In the present study, datasets for endometriosis, including GSE7305, GSE7846, GSE29981 and E-MTAB-694, were downloaded from Gene Expression Omnibus and ArrayExpress. Then, the limma and Affy packages were used to analyze the CEL file. The RankProd method was used to conduct meta-analysis. Long intergenic non-protein coding RNA 1279 (LINC01279) was significantly upregulated in the three datasets, and was the most upregulated lncRNA as determined by the RankProd method. Gene set enrichment and Gene Ontology analyses were conducted, which revealed that LINC01279 is likely to function as a cell cycle mediator in endometriosis. Finally, it was identified that LINC01279 is strongly associated with certain previously identified key factors in the development of endometriosis, including cyclin-dependent kinase 14 and C-X-C motif chemokine ligand 12. Thus, it was demonstrated that LINC01279 may be associated with the pathogenesis of endometriosis. This may potentially represent a target in the therapy of endometriosis.
Keywords: endometriosis, long non-coding RNA, long intergenic non-protein coding RNA 1279, gene expression omnibus, ArrayExpress, bioinformatics
Introduction
Endometriosis, the presence of endometrial tissue outside of the uterus, affects 6–10% of women of reproductive age (1,2). Among patients with endometriosis, 40–50% of them experience problems with fertility, ~50% experience severe chronic pelvic pain, and some may have dyspareunia and irregular uterine bleeding. Endometriosis significantly affects life quality and work productivity (3–5). Though a significant amount of research has explored the pathogenesis of endometriosis, little is clear. Illuminating the mechanisms is urgent for improving the therapeutic efficiency of endometriosis treatment.
In recent years, the rapid advance of high-throughput sequencing-based gene expression profiling has facilitated the identification of more and more long non-coding RNAs (lncRNAs). Defined as non-coding RNAs over 200 nt in length, lncRNAs were initially considered to represent transcriptional noise, but have since received extensive attention as novel regulatory components in a variety of biological processes and diseases, including cancer, heart disease and celiac disease (6–8). In addition, studies have reported that a number of lncRNAs mediate biological processes in endometriosis. H19 regulated stromal cell growth via the IGF signaling pathway in the endometrium of patients with endometriosis (9). In addition, Lee et al (10) demonstrated an association between genetic polymorphisms of CDKN2B-AS and WNT4, and Korean patients with endometriosis. Nakaoka et al (11) identified an allelic imbalance in the regulation of ANRIL mediated by chromatin interaction at the 9p21 endometriosis risk locus. Although the ENCODE project and GENCODE annotation have identified thousands of lncRNAs, the effect of many lncRNAs to endometriosis is not yet understood (12,13).
In recent decades, microarrays have been widely used to identify candidate biomarkers and therapeutic targets through investigating the alteration of gene expression at a genome-wide level. Several studies on endometriosis have been performed to investigate the dysregulated genes and essential pathways involved in its pathogenesis. For example, Houshdaran et al (14) reported the aberrant endometrial DNA methylome and associated gene expression in patients with endometriosis; Wang et al (15) analyzed the serum microRNA profile by Solexa sequencing in patients with endometriosis; Wang et al (16) reported the differential expression of microRNAs in ectopic endometrial tissue during the implantation window for endometriosis patients with related infertility. Unfortunately, only a small number of significantly dysregulated genes between the normal endometrium and endometriosis have been identified. This motivated us to explore the dysregulated genes in endometriosis, specifically lncRNAs.
Materials and methods
Reannotation of Affymetrix human genome U133 plus 2.0
Reannotation of the probe IDs for Affymetrix Human Genome U133 Plus 2.0 (Affymetrix; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was required to obtain data regarding lncRNA expression. Initially, we downloaded the HG-U133 Plus 2 annotation file from Affymetrix (CVS format; edition no. 36; www.affymetrix.com/estore/index.jsp; Affymetrix; Thermo Fisher Scientific, Inc.), which included 54,674 probe IDs for coding RNAs, microRNAs, lncRNAs and other non-coding RNAs. The criteria for assigning lncRNA probe IDs were as follows: i) The Refseq ID started with NR_ or XR_, indicative of non-coding RNAs; ii) the Ensemble gene IDs' annotations were antisense, processed transcripts, sense overlapping, non-sense mediated decay, sense intronic or lincRNA; and iii) pseudogenes, rRNAs, microRNAs, and other small RNAs including tRNAs, snRNAs and snoRNAs were excluded. Finally, 3,809 probe IDs were reannotated as lncRNAs in Affymetrix Human Genome U133 Plus 2.0 (Affymetrix; Thermo Fisher Scientific, Inc.) representing 2,964 different lncRNAs (17).
In addition, mRNA probe IDs were identified by excluding non-coding RNAs (lncRNAs, microRNAs, pseudogenes, rRNAs, tRNAs, snRNAs and snoRNAs). Probe IDs encoding more than one mRNA and those without RefSeq Protein IDs were excluded. Finally, 38,429 probe IDs remained, representing 17,510 mRNAs.
Individual microarray data analysis
To identify co-expressed mRNAs that were differentially expressed lncRNAs and lncRNA in endometriosis, the raw CEL files for GSE7305 (10 normal samples and 10 endometriosis samples) (18), GSE7846 (5 normal samples and 5 endometriosis samples) (19) and GSE29981 (20 endometriosis samples) were downloaded from GEO (www.ncbi.nlm.nih.gov/gds/), and E-MTAB-694 (17 normal samples and 18 endometriosis samples) (20) from ArrayExpress (www.ebi.ac.uk/arrayexpress). The limma and Affy packages were used to read and process the raw CEL files. The datasets were normalized using the robust multichip averaging (RMA) method in the Affy package to return log2-transformed expression intensities (21). The normalized datasets were then subjected to analysis in limma to identify the differentially expressed genes. Genes significantly dysregulated in endometriosis compared with normal tissue were defined by the thresholds of P<0.05, and log2 fold change (FC)>1 (upregulated) or <-1 (downregulated). The pheatmap package was used to produce a heap map of the differentially expressed genes, with a green, black and red color scale from low to high expression. Finally, the differentially expressed genes were overlapped among the GSE7305, GSE7846 and E-MTAB-694, and the results were represented with a Venn diagram produced with the Venny online tool (bioinfogp.cnb.csic.es/tools/venny/index.html).
Meta-analysis of multiple microarray datasets
Meta-analysis was performed for the GSE7305, GSE7846 and E-MTAB-694 which contain normal samples and endometriosis using the RankProd package (22) to identify the upregulated and downregulated genes between normal endometrial tissue and endometriosis. Initially, the raw CEL files were normalized using RMA in the Affy package to return log2-transformed intensities (21). The normalized datasets were then merged using the inSilicoMerging package, and batch effects were corrected using COMBAT (23). To identify the most differentially expressed probe sets, the RP advance function within the RankProd package was used (22). False discovery rates (pfp) for differential expression were determined using 1,000 permutations. Upregulated or downregulated probe sets were identified based on false discovery rate (pfP<0.01) and FC value (FC>1, upregulated or FC<1, downregulated). Probes that mapped to multiple genes were discarded, to avoid the misinterpretation of the results and to increase specificity.
GO analysis and gene set enrichment
In order to investigation the function of the overlapping lncRNAs, guilt-by-association (GBA) analysis was used, in which the function of a poorly characterized lncRNA gene can be inferred on the basis of the functions of co-expressed protein-coding genes (PCGs) (24). Initially, the top 500 positively Pearson's correlated protein coding genes were subjected to GO analysis to identify the functional of the poorly characterized genes. In addition, gene set enrichment analysis (GSEA) was conducted to further investigate the functions of the lncRNAs. In GSEA, the median LINC01279 expression value was used to divide samples into high and low groups according to the associated LINC01279 expression level. As a metric for ranking genes in GSEA, the difference between the mean expression between samples with low and high LINC01279 expression was used; other parameters were used at their default values. Besides, the motifs c2.cp.kegg.v6.1.symbols.gmt and c5.bp.v6.1.symbols.gmt were enrolled to our analysis for GSEA. Finally, the Pearson correlations between LINC01279 and the candidate genes were computed by the Cor package.
Results
LINC01279 and MSC-AS1 were upregulated in endometriosis
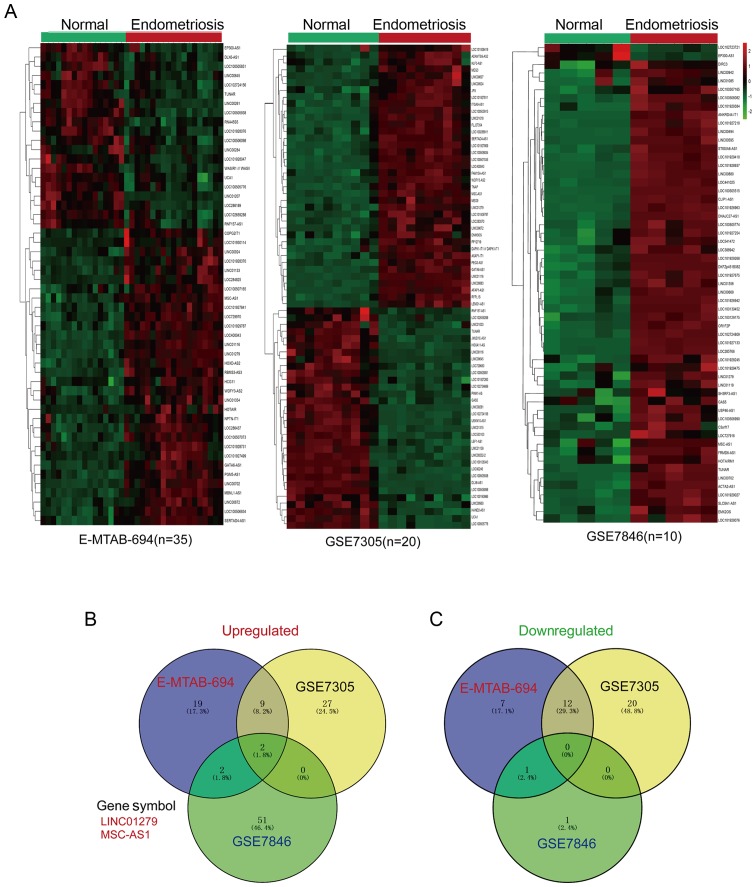
To identify the potentially dysregulated lncRNAs in endometriosis, we performed an integrative analysis of microarray expression profiles from GEO datasets and ArrayExpress datasets created with the Affymetrix Human Genome U133 Plus 2.0 platform (Affymetrix; Thermo Fisher Scientific, Inc.). To obtain the dysregulated lncRNAs, the GSE7305, GSE7846 and E-MTAB-694 were enrolled in our study. As GSE29981 do not include normal samples and we do not enrolled this profile for dys-regulated lncRNAs analysis. First, the upregulated and downregulated lncRNAs were identified with the limma package and presented as a heatmap (Fig. 1A). As depicted in Fig. 1A, based on the cutoffs of P<0.05 and log2 FC>1, there were 32 upregulated lncRNAs in E-MTAB-694, 38 in GSE7305 and 55 in GSE7846. In addition, there were 20 downregulated lncRNAs in E-MTAB-694, 32 in GSE7305 and 2 in GSE7846. The overlapping upregulated and downregulated lncRNAs were represented with a Venn diagram; as shown in Fig. 1B and C, LINC01279 and MSC-AS1 were consistently upregulated in all three datasets, while there were no downregulated genes common to all three datasets. As no previous studies have reported the functions of these two lncRNAs to the best of our knowledge, the exploration of the functions of LINC01279 and MSC-AS1 in endometriosis may be of importance.
Figure 1.
Differentially expressed lncRNAs in endometriosis. (A) Heatmaps of differently expressed lncRNAs in GSE7305 (n=20), GSE7846 (n=10) and E-MTAB-694 (n=35). (B) Overlapping upregulated lncRNAs in GSE7305, GSE7846 and E-MTAB-694. (C) Overlapping downregulated lncRNAs in GSE7305, GSE7846 and E-MTAB-694. lncRNAs, long non-coding RNAs.
LINC01279 was the most upregulated lncRNA in endometriosis, as determined with the RankProd package
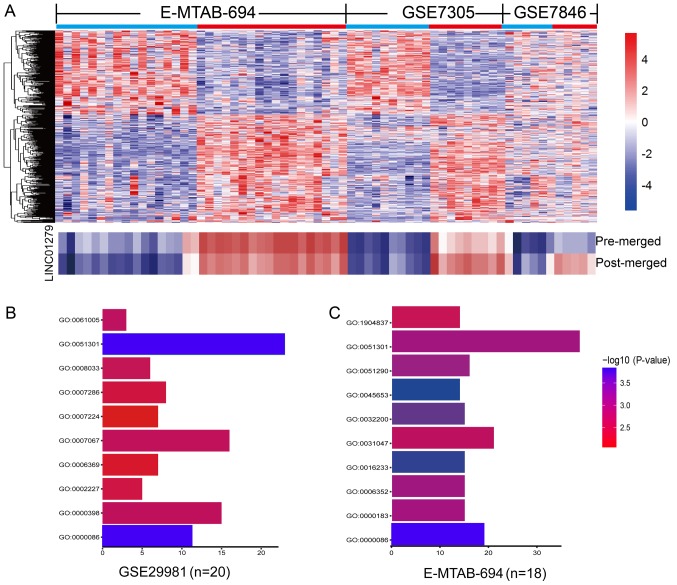
As described above, LINC01279 and MSC-AS1 were upregulated in three independent datasets. In order to identify which was the most upregulated in endometriosis, multiple microarray datasets were analyzed by the RankProd package (Fig. 2A). As shown in Table I, LINC01279 was the most upregulated lncRNA as identified in RankProd. These results suggested that LINC01279 may play an important role in the development of endometriosis.
Figure 2.
LINC01279 is significantly upregulated in endometriosis. (A) (top panel) The differentially expressed long non-coding RNAs in GSE7305 (n=20), GSE7846 (n=10) and E-MTAB-694 (n=35) as identified with the RankProd method; (bottom panel) the heatmap of LINC01279 pre- and post-merged by RankProd. (B and C) GO analysis was performed for the top 500 genes positively correlated with LINC01279 in (B) GSE29981 and (C) E-MTAB-694. The significantly enriched terms included GO:0000086 (G2/M transition of mitotic cell cycle) and GO:0051301 (cell division). GO, Gene Ontology; LINC01279, long intergenic non-protein coding RNA 1279.
Table I.
Top 20 upregulated long non-coding RNAs in endometriosis by Rankprod analysis.
| Probe ID | ENTREZ_GENE_ID | Symbol | FC_class1/class2 | pfp | P-value |
|---|---|---|---|---|---|
| 227061_at | 100506621 | LINC01279 | 10.7411 | 0 | <0.0001 |
| 226582_at | 400043 | LOC400043 | 5.3191 | 0 | <0.0001 |
| 229669_at | 440416 | LOC339260 | 4.1442 | 0 | <0.0001 |
| 229669_at | 440416 | CCDC144NL-AS1 | 4.1442 | 0 | <0.0001 |
| 228564_at | 375295 | LINC01116 | 4.0437 | 0 | <0.0001 |
| 228438_at | 100132891 | MSC-AS1 | 3.1726 | 0 | <0.0001 |
| 236304_at | 100652988 | LINC00702 | 2.6954 | 0 | <0.0001 |
| 232298_at | 401093 | MBNL1-AS1 | 2.4114 | 0 | <0.0001 |
| 230595_at | 572558 | PGM5-AS1 | 2.3164 | 0 | <0.0001 |
| 226210_s_at | 55384 | MEG3 | 2.1848 | 0 | <0.0001 |
| 235606_at | 344595 | DUBR | 2.0825 | 0 | <0.0001 |
| 235362_at | 729970 | LOC729970 | 2.0117 | 0 | <0.0001 |
| 236118_at | 100128893 | GATA6-AS1 | 1.9728 | 0 | <0.0001 |
| 235173_at | 401093 | MBNL1-AS1 | 1.9670 | 0 | <0.0001 |
| 226211_at | 55384 | MEG3 | 1.9429 | 0 | <0.0001 |
| 238081_at | 404201 | WDFY3-AS2 | 1.8997 | 0 | <0.0001 |
| 236704_at | 101929787 | LOC101929787 | 1.8943 | 0 | <0.0001 |
| 228601_at | 401022 | HAGLR | 1.8205 | 0 | <0.0001 |
| 238360_s_at | 100505576 | LINC00672 | 1.8155 | 0 | <0.0001 |
| 239823_at | 101927841 | LOC101927841 | 1.8093 | 0 | <0.0001 |
pfp, false discovery rates; LINC01279, long intergenic non-protein coding RNA 1279.
LINC01279 acted as a cell cycle mediator in endometriosis
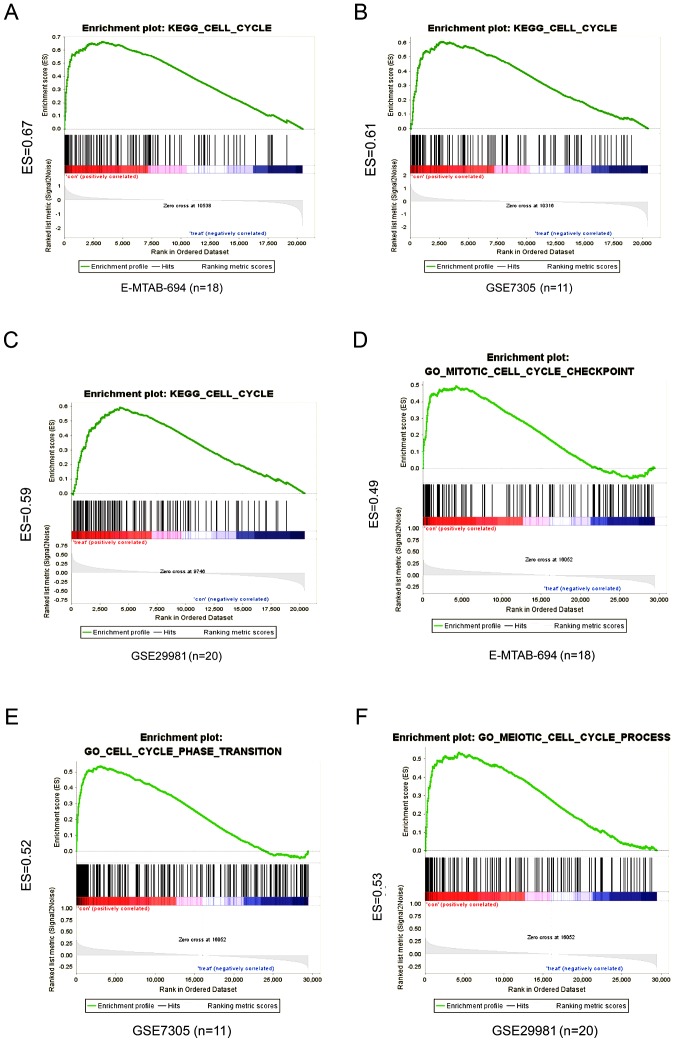
To explore the function of LINC01279, we conducted a GBA analysis in the E-MTAB-694 and GSE29981 microarray datasets, which contained normal endometrial tissue and endometriosis samples. Firstly, the Pearson correlations between LINC01279 and all protein coding genes were computed. Then we select the top 500 positively correlated genes were enrolled in GO analyses. The result showed that the functions of these genes were enriched in many biological processes; in particular, the overlapping genes from all three datasets were consistently enriched in cell cycle and cell division (Fig. 2B and C). Next, GSEA was conducted for E-MTAB-694, GSE29981 and GSE7305 between the LINC01279 low and high expression groups. Overlapping the results showed that both the high and low LINC01279 expression groups were enriched in the cell cycle pathway category, including E-MTAB-694 [enrichment score (ES)=0.67], GSE7305 (ES=0.61) and GSE29981 (ES=0.59) (Fig. 3A-C) with c2.cp.kegg.v6.1.symbols.gmt motif. With c5.bp.v6.1.symbols.gmt motif, the result were also enriched in cell cycle (Fig. 3D-F). These results indicated that the lncRNA LINC01279 may act as a cell cycle mediator in endometriosis.
Figure 3.
LINC01279 acts as a cell cycle mediator in endometriosis. (A-C) The GSEA with c2.cp.kegg.v6.1.symbols.gmt motif of the high and low LINC01279 expression groups identified an enrichment in the cell cycle pathway category for the (A) E-MTAB-694 (ES=0.67), (B) GSE7305 (ES=0.61) and (C) GSE29981 (ES=0.59). (D-F) The GSEA with c5.bp.v6.1.symbols.gmt motif of the high and low LINC01279 expression groups identified an enrichment in the cell cycle biological process category for the (D) E-MTAB-694 (ES=0.49), (E) GSE7305 (ES=0.53) and (F) GSE29981 (ES=0.52). LINC01279, long intergenic non-protein coding RNA 1279; GSEA, gene set enrichment analysis; ES, enrichment score.
Possible mechanism for LINC01279 in promoting endometriosis development
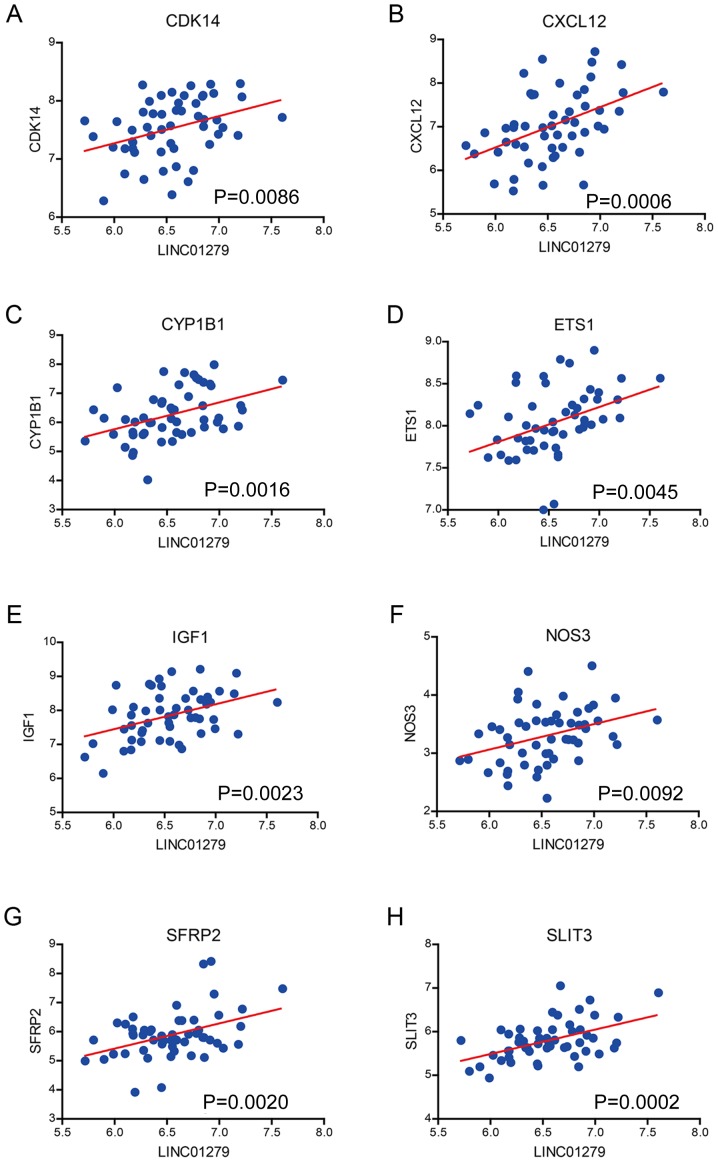
In order to identify the mechanism for the promotion of the development of endometriosis, we calculated the correlation between LINC01279 and all protein coding genes in a merged dataset, including the data from GSE7305, GSE7846, GSE29981 and E-MTAB-694, with the inSilicoMerging method. The result showed that previously identified regulators of endometriosis, such as cyclin-dependent kinase 14 (CDK14) (Fig. 4A), CSCL12 (Fig. 4B), cytochrome P450 family 1 subfamily B member 1 (CYP1B1) (Fig. 4C), E26 transformation specific (ETS1) (Fig. 4D), insulin-like growth factor 1 (IGF1) (Fig. 4E), nitric oxide synthase 3 (NOS3) (Fig. 4F), secreted frizzled related protein 2 (SFRP2) (Fig. 4G) and slit guidance ligand 3 (SLIT3) (Fig. 4H), were positively associated with LINC01279. This demonstrated that LINC01279 may regulate the expression of these genes to promote the development of endometriosis.
Figure 4.
Correlation between the expression of LINC01279 and key factors in endometriosis. LINC01279 expression is positively correlated with (A) CDK14, (B) CXCL12, (C) CYP1B1, (D) EST1, (E) IGF1, (F) NOS3, (G) SFRP2 and (H) SLIT3. LINC01279, long intergenic non-protein coding RNA 1279; CDK14, cyclin-dependent kinase 14; CXCL12, C-X-C motif chemokine ligand 12; CYP1B1, cytochrome P450 family 1 subfamily B member 1; ETS1, E26 transformation specific; IGF1, insulin-like growth factor 1; NOS3, nitric oxide synthase 3; SFRP2, secreted frizzled related protein 2; SLIT3, slit guidance ligand 3.
Discussion
In recent years, with the development of microarray technology for the determination of SNPs, copy number, methylation status and mRNA expression, more and more large-scale genotyping analyses using microarrays have been performed to identify the genomic regions associated with endometriosis (25–28). Computational models have been widely used to compare the genotypes of healthy subjects and patients, thus identifying variants associated with a certain disease. lncRNAs, which may act as biological mediators in various diseases, have recently gained significant attention. This inspired us to compare the lncRNA expression profiles of normal endometrial tissue and endometriosis.
In our study, we found that LINC01279 and MSC-AS1 were consistently upregulated in three independent datasets. However, there was no overlap in downregulated lncRNAs in the same datasets. The RankProd method identified that LINC01279 was the most up-regulated lncRNA in endometriosis. To infer the function of this lncRNA in endometriosis, we conducted a GBA analysis, in which the co-expressed mRNAs were identified, and GO and GSEA analyses were performed to predict the function of LINC01279. This method may allow us to explore the functions of previously uncharacterized lncRNA.
As described above, from the GBA analysis, it was identified that LINC01279 may act as a cell cycle mediator. In order to further predict the possible mechanisms of LINC01279 in endometriosis, we conducted the Pearson correlation coefficient for all protein coding genes with LINC01279. Some previously identified key factors in the development of endometriosis were found to be strongly correlated with LINC01279, including the cell cycle regulating factor CDK14 (29). Endometriosis is conventionally viewed as a proliferative disorder characterized by the growth of endometrial cells in ectopic locations, and previous data have shown proliferation defects in the eutopic endometrium of women with endometriosis compared with women without endometriosis. For example, CSCL12 may promote the proliferation, migration and invasion of endometriosis (30). In addition, CYP1B1 (31), ETS1 (32), IGF1 (33), NOS3 (34), SFRP2 (35) and SLIT3 (36), which were previously identified to serve important roles in endometriosis, were positively correlated with LINC01279.
LINC01279 is 5,176 bp in length. In recent years, many studies have reported the mechanisms of how lncRNAs may regulate biological processes. For example, some lncRNAs may act as competing endogenous RNAs (ceRNAs) to ‘sponge’ microRNAs, thus affecting the expression of their target mRNAs. Yang et al (37) found that UCA1 could function as an endogenous sponge by directly binding miR-485-5p to regulate the expression of its target gene, matrix metallopeptidase 14, thus suppressing epithelial ovarian cancer metastasis. Sun et al (38) reported that HOXA11-AS acted as a ceRNA for miR-1297, thereby depressing EZH2 expression and imposing an additional level of post-transcriptional regulation in gastric cancer cells. Yu et al (39) reported that GAS5 functioned as a ceRNA for miR-222 to increase the level of p27 protein, thereby inhibiting the proliferation of hepatic stellate cells. Previous studies have also reported that some lncRNAs may scaffold the chromatin modification factors PRC2, LSD1 and DNMT1, or bind EZH2 to regulate cellular biological processes. For instance, the intronic lncRNA ANRASSF1 recruited PRC2 to the RASSF1A promoter, thus reducing the expression of RASSF1A and increasing cell proliferation. The lncRNA HOXA-AS2 has been reported to repress P21 and KLF2 expression transcription by binding with EZH2 and LSD1 in colorectal cancer (40). Yao et al (41) reported that the lncRNA ADAMTS9-AS2 was regulated by DNMT1, and inhibited migration in glioma cells. However, the mechanisms of how lncRNAs regulate biological processes are still unclear, and more research efforts are required.
To the best of our knowledge, this is the first study to date to report that the lncRNA LINC01279 acted as a cell cycle mediator in endometriosis, as determined by GBA analysis. Over recent decades, many CDK inhibitors have been used in treating cancer, such as the PARP inhibitor, olaparib, in ovarian cancer (42) and bortezomib in multiple myeloma (43). The results of this study indicate that LINC01279 may have potential as a new target for treating endometriosis. As described above, various mechanisms for the regulation of cellular functions by lncRNAs have been reported. Nevertheless, the exact mechanism of how LINC01279 affected the cell cycle in endometriosis is unclear and the in vivo and in vitro study should be done in further study.
Taken together, an lncRNA, LINC01279, was overexpressed in endometriosis. Furthermore, GO and GSEA analyses determined that LINC01279 potentially acted as a cell cycle mediator in endometriosis.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Maternal and Children Health Research Project of Jiangsu Province (grant no. F201626) and the Maternal and Children Health Care Association Project of Jiangsu Province (grant no. FYX201604).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
JL and QW were responsible for the analysis and interpretation of the data. RZ, CZ and JHL performed the statistical analysis. XH designed the study and drafted the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Colette S, Donnez J. Endometriosis. N Engl J Med. 2009;360:1911–1912. doi: 10.1056/NEJMc090328. [DOI] [PubMed] [Google Scholar]
- 2.Giudice LC. Clinical practice. Endometriosis. N Engl J Med. 2010;362:2389–2398. doi: 10.1056/NEJMcp1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldman MB, Cramer DW. The epidemiology of endometriosis. Prog Clin Biol Res. 1990;323:15–31. [PubMed] [Google Scholar]
- 4.Giudice LC, Kao LC. Endometriosis. Lancet. 2004;364:1789–1799. doi: 10.1016/S0140-6736(04)17403-5. [DOI] [PubMed] [Google Scholar]
- 5.Moradi M, Parker M, Sneddon A, Lopez V, Ellwood D. Impact of endometriosis on women's lives: A qualitative study. BMC Womens Health. 2014;14:123. doi: 10.1186/1472-6874-14-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atala A. Re: The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. J Urol. 2014;192:613. doi: 10.1016/j.juro.2014.05.031. [DOI] [PubMed] [Google Scholar]
- 7.Han P, Li W, Lin CH, Yang J, Shang C, Nuernberg ST, Jin KK, Xu W, Lin CY, Lin CJ, et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014;514:102–106. doi: 10.1038/nature13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castellanos-Rubio A, Fernandez-Jimenez N, Kratchmarov R, Luo X, Bhagat G, Green PH, Schneider R, Kiledjian M, Bilbao JR, Ghosh S. A long noncoding RNA associated with susceptibility to celiac disease. Science. 2016;352:91–95. doi: 10.1126/science.aad0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghazal S, Mckinnon B, Zhou J, Mueller M, Men Y, Yang L, Mueller M, Flannery C, Huang Y, Taylor HS. H19 lncRNA alters stromal cell growth via IGF signaling in the endometrium of women with endometriosis. EMBO Mol Med. 2015;8:996–1003. doi: 10.15252/emmm.201505245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee GH, Choi YM, Hong MA, Yoon SH, Kim JJ, Hwang K, Chae SJ. Association of CDKN2B-AS and WNT4 genetic polymorphisms in Korean patients with endometriosis. Fertil Steril. 2014;102:1393–1397. doi: 10.1016/j.fertnstert.2014.07.1237. [DOI] [PubMed] [Google Scholar]
- 11.Nakaoka H, Gurumurthy A, Hayano T, Ahmadloo S, Omer WH, Yoshihara K, Yamamoto A, Kurose K, Enomoto T, Akira S, et al. Allelic imbalance in regulation of ANRIL through chromatin interaction at 9p21 endometriosis risk locus. PLoS Genet. 2016;12:e1005893. doi: 10.1371/journal.pgen.1005893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.ENCODE Project Consortium, corp-author. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. GENCODE: The reference human genome annotation for the ENCODE project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Houshdaran S, Nezhat CR, Vo KC, Zelenko Z, Irwin JC, Giudice LC. Aberrant endometrial DNA methylome and associated gene expression in women with endometriosis. Biol Reprod. 2016;95:93. doi: 10.1095/biolreprod.116.140434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L, Huang W, Ren C, Zhao M, Jiang X, Fang X, Xia X. Analysis of serum microRNA profile by solexa sequencing in women with endometriosis. Reprod Sci. 2016;23:1359–1370. doi: 10.1177/1933719116641761. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Ma CH, Qiao J. Differential expression of microRNA in eutopic endometrium tissue during implantation window for patients with endometriosis related infertility. Zhonghua Fu Chan Ke Za Zhi. 2016;51:436–441. doi: 10.3760/cma.j.issn.0529-567X.2016.06.007. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 17.Zhang X, Sun S, Pu JK, Tsang AC, Lee D, Man VO, Lui WM, Wong ST, Leung GK. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol Dis. 2012;48:1–8. doi: 10.1016/j.nbd.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Hever A, Roth RB, Hevezi P, Marin ME, Acosta JA, Acosta H, Rojas J, Herrera R, Grigoriadis D, White E, et al. Human endometriosis is associated with plasma cells and overexpression of B lymphocyte stimulator; Proc Natl Acad Sci USA; 2007; pp. 12451–12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sha G, Wu D, Zhang L, Chen X, Lei M, Sun H, Lin S, Lang J. Differentially expressed genes in human endometrial endothelial cells derived from eutopic endometrium of patients with endometriosis compared with those from patients without endometriosis. Hum Reprod. 2007;22:3159–3169. doi: 10.1093/humrep/dem266. [DOI] [PubMed] [Google Scholar]
- 20.Sohler F, Sommer A, Wachter DL, Agaimy A, Fischer OM, Renner SP, Burghaus S, Fasching PA, Beckmann MW, Fuhrmann U, et al. Tissue remodeling and nonendometrium-like menstrual cycling are hallmarks of peritoneal endometriosis lesions. Reprod Sci. 2013;20:85–102. doi: 10.1177/1933719112451147. [DOI] [PubMed] [Google Scholar]
- 21.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taminau J, Meganck S, Lazar C, Steenhoff D, Coletta A, Molter C, Duque R, de Schaetzen V, Weiss Solís DY, Bersini H, Nowé A. Unlocking the potential of publicly available microarray data using inSilicoDb and inSilicoMerging R/Bioconductor packages. BMC Bioinformatics. 2012;13:335. doi: 10.1186/1471-2105-13-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: A simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 24.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Li Y, Yang Z, Liu K, Wang D. Genome-wide microarray analysis of long non-coding RNAs in eutopic secretory endometrium with endometriosis. Cell Physiol Biochem. 2015;37:2231–2245. doi: 10.1159/000438579. [DOI] [PubMed] [Google Scholar]
- 26.Fassbender A, Verbeeck N, Börnigen D, Kyama CM, Bokor A, Vodolazkaia A, Peeraer K, Tomassetti C, Meuleman C, Gevaert O, et al. Combined mRNA microarray and proteomic analysis of eutopic endometrium of women with and without endometriosis. Hum Reprod. 2012;27:2020–2029. doi: 10.1093/humrep/des127. [DOI] [PubMed] [Google Scholar]
- 27.Yang RQ, Teng H, Xu XH, Liu SY, Wang YH, Guo FJ, Liu XJ. Microarray analysis of microRNA deregulation and angiogenesis-related proteins in endometriosis. Genet Mol Res. 2016;15 doi: 10.4238/gmr.15027826. [DOI] [PubMed] [Google Scholar]
- 28.Ping S, Ma C, Liu P, Yang L, Yang X, Wu Q, Zhao X, Gong B. Molecular mechanisms underlying endometriosis pathogenesis revealed by bioinformatics analysis of microarray data. Arch Gynecol Obstet. 2016;293:797–804. doi: 10.1007/s00404-015-3875-y. [DOI] [PubMed] [Google Scholar]
- 29.Choi YJ, Anders L. Signaling through cyclin D-dependent kinases. Oncogene. 2014;33:1890–1903. doi: 10.1038/onc.2013.137. [DOI] [PubMed] [Google Scholar]
- 30.Leconte M, Chouzenoux S, Nicco C, Chéreau C, Arkwright S, Santulli P, Weill B, Chapron C, Dousset B, Batteux F. Role of the CXCL12-CXCR4 axis in the development of deep rectal endometriosis. J Reprod Immunol. 2014;103:45–52. doi: 10.1016/j.jri.2013.12.121. [DOI] [PubMed] [Google Scholar]
- 31.Piccinato CA, Neme RM, Torres N, Sanches LR, Cruz Derogis PB, Brudniewski HF, E Silva JC, Ferriani RA. Increased expression of CYP1A1 and CYP1B1 in ovarian/peritoneal endometriotic lesions. Reproduction. 2016;151:683–692. doi: 10.1530/REP-15-0581. [DOI] [PubMed] [Google Scholar]
- 32.Sakaguchi H, Fujimoto J, Aoki I, Toyoki H, Sato E, Tamaya T. Expression of E26 transformation specific (ETS-1) related to angiogenesis in ovarian endometriosis. Fertil Steril. 2004;82:507–510. doi: 10.1016/j.fertnstert.2004.02.103. [DOI] [PubMed] [Google Scholar]
- 33.Mu F, Hankinson SE, Schernhammer E, Pollak MN, Missmer SA. A prospective study of insulin-like growth factor 1, its binding protein 3, and risk of endometriosis. Am J Epidemiol. 2015;182:148–156. doi: 10.1093/aje/kwv037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim H, Ku SY, Kim SH, Lee GH, Choi YM, Kim JM, Lee TH, Moon SY. Endothelial nitric oxide synthase gene Glu298Asp polymorphism is associated with advanced stage endometriosis. Hum Reprod. 2009;24:2656–2659. doi: 10.1093/humrep/dep212. [DOI] [PubMed] [Google Scholar]
- 35.Heinosalo T, Gabriel M, Kallio L, Adhikari P, Huhtinen K, Laajala TD, Kaikkonen E, Mehmood A, Suvitie P, Kujari H, et al. Secreted frizzled-related protein 2 (SFRP2) expression promotes lesion proliferation via canonical WNT signaling and indicates lesion borders in extraovarian endometriosis. Hum Reprod. 2018;33:817–831. doi: 10.1093/humrep/dey026. [DOI] [PubMed] [Google Scholar]
- 36.Greaves E, Collins F, Esnal-Zufiaurre A, Giakoumelou S, Horne AW, Saunders PT. Estrogen receptor (ER) agonists differentially regulate neuroangiogenesis in peritoneal endometriosis via the repellent factor SLIT3. Endocrinology. 2014;155:4015–4026. doi: 10.1210/en.2014-1086. [DOI] [PubMed] [Google Scholar]
- 37.Yang Y, Jiang Y, Wan Y, Zhang L, Qiu J, Zhou S, Cheng W. UCA1 functions as a competing endogenous RNA to suppress epithelial ovarian cancer metastasis. Tumour Biol. 2016;37:10633–10641. doi: 10.1007/s13277-016-4917-1. [DOI] [PubMed] [Google Scholar]
- 38.Sun M, Nie F, Wang Y, Zhang Z, Hou J, He D, Xie M, Xu L, De W, Wang Z, Wang J. LncRNA HOXA11-as promotes proliferation and invasion of gastric cancer by scaffolding the chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76:6299–6310. doi: 10.1158/0008-5472.CAN-16-0356. [DOI] [PubMed] [Google Scholar]
- 39.Yu F, Zheng J, Mao Y, Dong P, Lu Z, Li G, Guo C, Liu Z6, Fan X. Long non-coding RNA growth arrest-specific transcript 5 (GAS5) inhibits liver fibrogenesis through a mechanism of competing endogenous RNA. J Biol Chem. 2015;290:28286–28298. doi: 10.1074/jbc.M115.683813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding J, Xie M, Lian Y, Zhu Y, Peng P, Wang J, Wang L, Wang K. Long noncoding RNA HOXA-AS2 represses P21 and KLF2 expression transcription by binding with EZH2, LSD1 in colorectal cancer. Oncogenesis. 2017;6:e288. doi: 10.1038/oncsis.2016.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao J, Zhou B, Zhang J, Geng P, Liu K, Zhu Y, Zhu W. A new tumor suppressor LncRNA ADAMTS9-AS2 is regulated by DNMT1 and inhibits migration of glioma cells. Tumour Biol. 2014;35:7935–7944. doi: 10.1007/s13277-014-1949-2. [DOI] [PubMed] [Google Scholar]
- 42.Ledermann J, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, Scott C, Meier W, Shapira-Frommer R, Safra T, et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N Engl J Med. 2012;366:1382–1392. doi: 10.1056/NEJMoa1105535. [DOI] [PubMed] [Google Scholar]
- 43.Kim M, Lee SH, Kim J, Lee SE, Kim YJ, Min CK. Copy number variations could predict the outcome of bortezomib plus melphalan and prednisone for initial treatment of multiple myeloma. Genes Chromosomes Cancer. 2015;54:20–27. doi: 10.1002/gcc.22213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.