Abstract
Cucumber (Cucumis sativus L.) is a typical monoecism vegetable with individual male and female flowers, which has been used as a plant model for sex determination. It is well known that light is one of the most important environmental stimuli, which control the timing of the transition from vegetative growth to reproductive development. However, whether light controls sex determination remains elusive. To unravel this problem, we performed high-throughput RNA-Seq analyses, which compared the transcriptomes of shoot apices between R2B1(Red light:Blue light = 2:1)-treated and R4B1(Red light:Blue light = 4:1)-treated cucumber seedlings. Results showed that the higher proportion of blue light in the R2B1 treatment significantly induced the formation of female flowers and accelerated female flowering time in this whole study. The genes related to flowering time, such as flowering locus T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1), were up-regulated after R2B1 treatment. Furthermore, the transcriptome analysis showed that up-regulation and down-regulation of specific DEGs (the differentially expressed genes) were primarily the result of plant hormone signal transduction after treatments. The specific DEGs related with auxin formed the highest percentage of DEGs in the plant hormone signal transduction. In addition, the expression levels of transcription factors also changed after R2B1 treatment. Thus, sex differentiation affected by light quality might be induced by plant hormone signal transduction and transcription factors. These results provide a theoretical basis for further investigation of the regulatory mechanism of female flower formation under different light qualities in cucumber seedlings.
Electronic supplementary material
The online version of this article (10.1186/s40529-018-0237-7) contains supplementary material, which is available to authorized users.
Keywords: RNA-seq, Auxin, Cucumber, Flower, Sex differentiation
Background
Sex differentiation of flower buds is an important developmental process, which directly affects the product yield in plants. Cucumber (Cucumis sativus L.) is a typical monoecious plant with distinct male and female flowers. It has been served as a model system for studying physiological and molecular aspects of sex determination and differentiation in plants (Bai and Xu 2013). In the young floral buds of cucumber, both the stamen primordia and carpel primordia are initiated, with sex determination occurring just after the bisexual stage. Subsequently, male or female flowers are formed and become enlarged due to the selective arrestment of carpel or stamen development, respectively (Bai et al. 2004).
Sex differentiation of cucumber can be affected by phytohormones, such as ethylene and gibberellin. Exogenous ethylene treatment induced female flower formation in the cucumber. The ethylene content in gynoecious cucumbers is found to be higher than that of monoecious plants (Trebitsh et al. 1987; Rudich et al. 1972). Ethylene synthesis genes play an important role in the sex differentiation of the cucumber flowers, such as 1-aminocyclopropane-1-carboxylic acid oxidases (CsACO2). Organ-specific overexpression of CsACO2 driven by the organ-specific promoter P (AP3) significantly affected stamen but not carpel development, acting similarly to natural floral development of female cucumber flowers (Duan et al. 2008). A conserved-residue conversion in CsACS2 induced the formation of bisexual flowers in the cucumber (Li et al. 2009). Furthermore, ethylene signaling pathway also affects the sex differentiation. CsETR1, an ethylene receptor involved in ethylene signaling transduction, had been demonstrated to play a key role in stamen arrest in female cucumber flowers (Wang et al. 2010). Ethylene-responsive gene associated with the formation of female flowers (ERAF17), a MADS-box gene, could be induced by ethylene and might be involved in formation of female flowers in cucumbers (Ando et al. 2001). Exogenous gibberellic acid (GA3) application promoted the formation of male flowers in gynoecious plants (Pike and Peterson 1969). GA production in andromonoecious cucumbers was higher than that in gynoecious and monoecious plants (Junior et al. 1972). The GA signaling pathway is involved in stamen development in the cucumber (Zhang et al. 2014b; Fei et al. 2004). Cucumber DELLA Homolog (CsGAIP) is predominantly expressed in the male specific organs during cucumber flower development and belongs to the DELLA (the negative regulators of the GA action) family. CsGAIP inhibited stamen development through transcriptional repression of B-class floral homeotic genes APETALA3 (AP3) and PISTILLATA (PI) in Arabidopsis (Zhang et al. 2014a). CsGAMYB1, a positive regulator involved in the GA signaling pathway, also mediates the sex expression of cucumbers. Knocking out the CsGAMYB1 gene in cucumbers resulted in decreased ratios of nodes with male to female flowers (Zhang et al. 2014b). The relationship between ethylene and gibberellin in mediating sex differentiation was interpreted recently, with this study finding that gibberellin mediates sex differentiation via ethylene-dependent and ethylene-independent pathways in cucumbers (Zhang et al. 2017). Apart from ethylene and gibberellin, other hormones are also involved in the flower sex differentiation of plant (Song et al. 2013). This includes the abscisic acid (ABA) induction of male flower formation (Zhu et al. 2010); Jasmonic acid (JA) signaling pathway, which might participate in the abortion of male flowers (Acosta et al. 2009); and auxin (IAA) content, which was found to increase during female flower development and decrease during male flower development (Sakata et al. 2010). Ethylene biosynthesis has probably been induced by IAA to promote female flower formation as well (Trebitsh et al. 1987). The IAA induced CsACS1 gene expression in cucumbers, which suggested that IAA promoted female flower formation through inducing ethylene synthesis (Trebitsh et al. 1997; Mibus and Tatlioglu 2004).
Environmental cues, such as temperature, photoperiod, nutrition, also affected sex determination in many species (Golenberg and West 2013; Korpelainen 1998). Light is one of the major external factors that influence plant growth and development (Chen et al. 2004). Plants respond to light through mediating light receptors. Until now, known photoreceptors were divided into three classes: the UV-B (280–320 nm light) photoreceptors; the red/far-red reversible photoreceptors, known as the phytochromes PhyA–PhyE (Mathews and Sharrock 1997) and blue UVA photoreceptors. These blue UVA photoreceptors have three classes, which are the cryptochromes (CRY1, CRY2 and CRY3) (Mathews and Sharrock 1997; Brudler et al. 2003), phototropins (PHOT1 and PHOT2) (Briggs et al. 2001) and aureochromes (AUREO1 and AUREO2) (Ishikawa et al. 2009; Takahashi et al. 2007). There is less evidence examining the effects of light on plant sex differentiation. In the gametophytes of the Her1 mutant, blue light induced male development and red light suppressed male development (Kamachi et al. 2007). Our previous studies showed that light quality affected cucumber flower formation and sex differentiation. In our previous study, we used 4R1B (Red:Blue = 4:1), R2B1 (Red:Blue = 2:1), R6G2B1 (Red:Green:Blue = 6:2:1) ratios supplemental light to treat the cucumber seedlings. Comparing to natural light, R2B1 treatment was the best light quality ratio for improving the female flower formation, location and flowering time, but none of them has effects on male flower formation (our unpublished data). However, the mechanism of how light quality regulates flower sex differentiation still remains unclear. In our study, we aimed to understand the mechanism of light regulating cucumber sexual development. We performed RNA-Seq analyses to compare the transcriptomes of shoot apices between cucumber seedlings after R2B1 treatment and R4B1 treatment. Our results built a foundation for dissecting the molecular mechanism of flower sex differentiation in cucumbers.
Results
Effects of different LED light quality treatments on flower formation and flowering time
Comparing to natural light, R2B1 treatment was the best light quality ratio for improving the female flower formation, location and flowering time (Additional file 1: Table S1). After 20 days lighting under R2B1 and R4B1 light treatment, the formation of female flowers occurred mainly at the lower node positions under the R2B1 irradiation, as compared with R4B1 irradiation (Table 1). The number of female flowers in the 15 nodes of the cucumber plant increased markedly and flowered earlier (Table 1). All of these results implied that blue light is responsible for the high number of female flowers, early flowering time and reduction in the first female flower node.
Table 1.
Effects of different light quality on the formation of female flowers and flowering time
| Treatment | First female flower located node | Total female flower numbers in 15 nodes | Days from transplanting to first female flowering |
|---|---|---|---|
| R2B1 (Red light:blue light = 2:1) | 7.20 ± 0.23b | 2.70 ± 0.13a | 21.85 ± 0.17b |
| R4B1 (Red light:blue light = 4:1) | 8.03 ± 0.19a | 2.32 ± 0.10b | 22.53 ± 0.12a |
a,b: Statistically significant variations of mean values at different sampling points (ANOVA, p < 0.05) were indicated with different letters. Means denoted by the same letter did not differ significantly at p < 0.05
Analysis of differentially expressed genes (DEGs) under treatments with different ratios of blue and red light
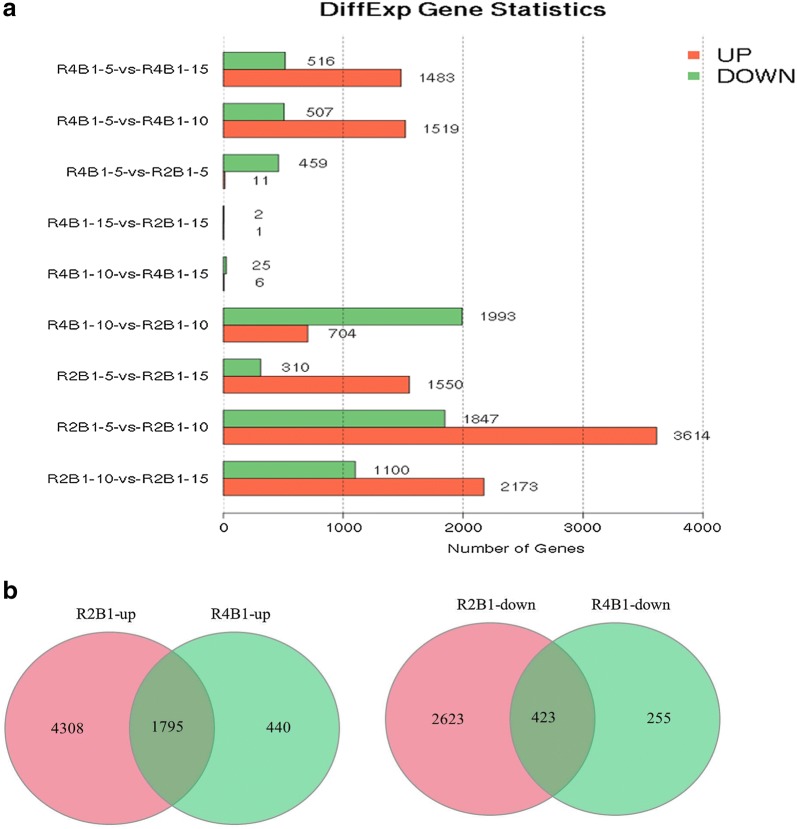
Based on deep sequencing, 23,248 unigenes were collected in 12 libraries. Genes expressed at differential stages under two treatments were listed in Table 2. After R2B1 treatment, the detected number of genes in the 5 days, 10 days and 15 days library were 18,619, 18,943 and 19,075. The mapping ratios of these libraries according to cucumber genome were 80.09, 81.48 and 82.5%. The detected new genes in these libraries were 603, 609 and 611. The relevant data for R4B1 treatment were shown in Table 2. Using |log2 (fold change)| > 1 and false discovery rate (FDR) < 0.05 as the cut-offs for significance, we identified DEGs at differential stages under two treatments (Fig. 1a). The results showed that there were 470 DEGs including 11 up-regulated genes and 459 down-regulated genes in pairwise R4B1-5-vs-R2B1-5 (5 days library comparison after R4B1 treatment and R2B1 treatment). Furthermore, we also found that 2697 DEGs were differentially expressed, with 704 genes being up-regulated and 1993 genes down-regulated in pairwise R4B1-10-vs-R2B1-10 (10 days library comparison after R4B1 treatment and R2B1 treatment). Moreover, 3 DEGs containing 1 up-regulated gene and 2 down-regulated genes were found in pairwise R4B1-15-vs-R2B1-15 (15 days library comparison after R4B1 treatment and R2B1 treatment). Most DEGs were found in the 10 days samples, which might implicate that the light quality affected flower sex differentiation mostly on the 10th day.
Table 2.
Statistical results of expressed genes in different libraries
| library | Numbers of known genes detected | Numbers of detected new genes |
|---|---|---|
| R2B1-5 | 18,619 (80.09%) | 603 |
| R4B1-5 | 18,831 (81.00%) | 614 |
| R2B1-10 | 18,943 (81.48%) | 609 |
| R4B1-10 | 19,099 (82.15%) | 618 |
| R2B1-15 | 19,075 (82.05%) | 611 |
| R4B1-15 | 19,306 (83.04%) | 618 |
Fig. 1.
a Statistical results of differentially expressed genes (DEGs) among different stages and treatments and b Venn diagrams of DEGs that were significantly (left) up-regulated and (right) down-regulated after R2B1 and R4B1 treatment
In addition, there were more DEGs in the 5 days, 10 days and 15 days library after R2B1 irradiation compared to R4B1 irradiation. We used a Venn diagram to analyze all these up-regulated or down-regulated DEGs collected from all these treatments at different time points. The results showed that 423 DEGs were found in all three stages after both R4B1 and R2B1 treatment as well as more up-regulated DEGs (1795 DEGs) at all three stages under both treatments (Fig. 1b).
Gene ontology (GO) analysis of DEGs after different ratios of blue and red light treatments
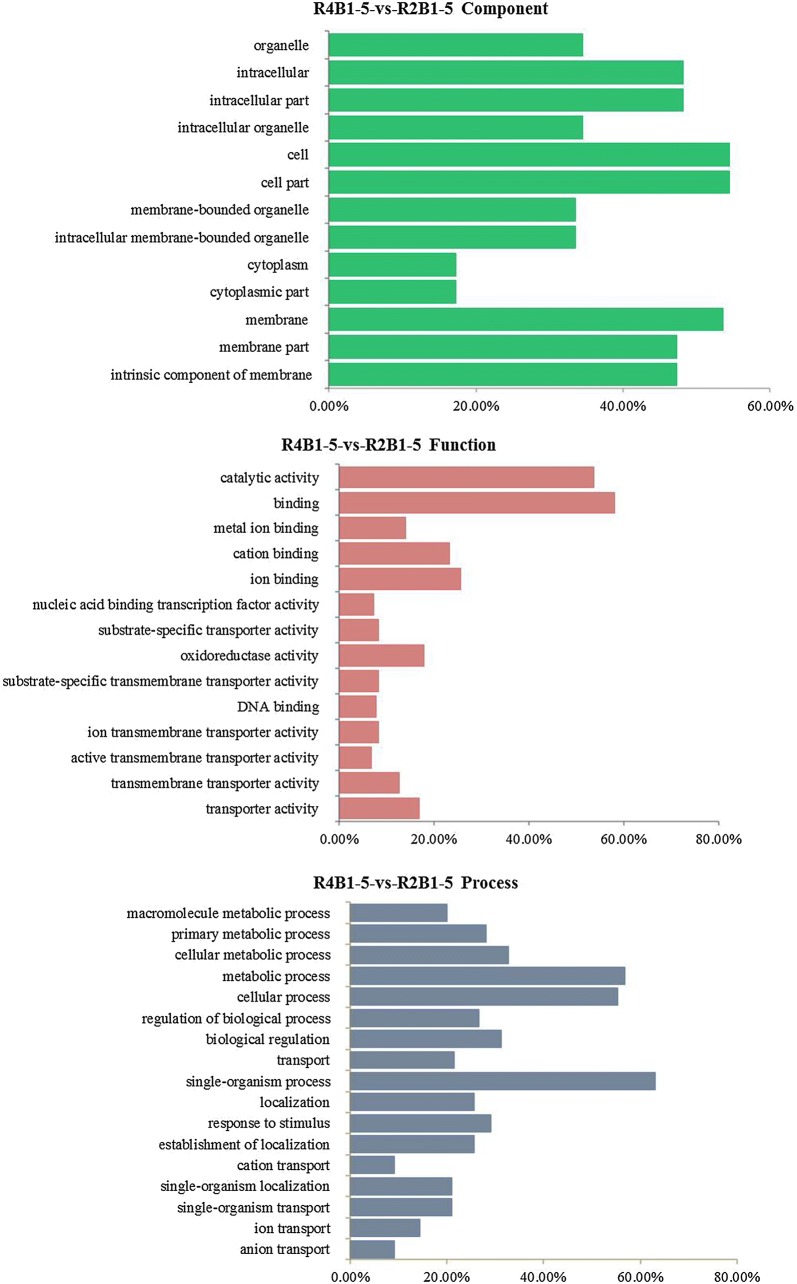
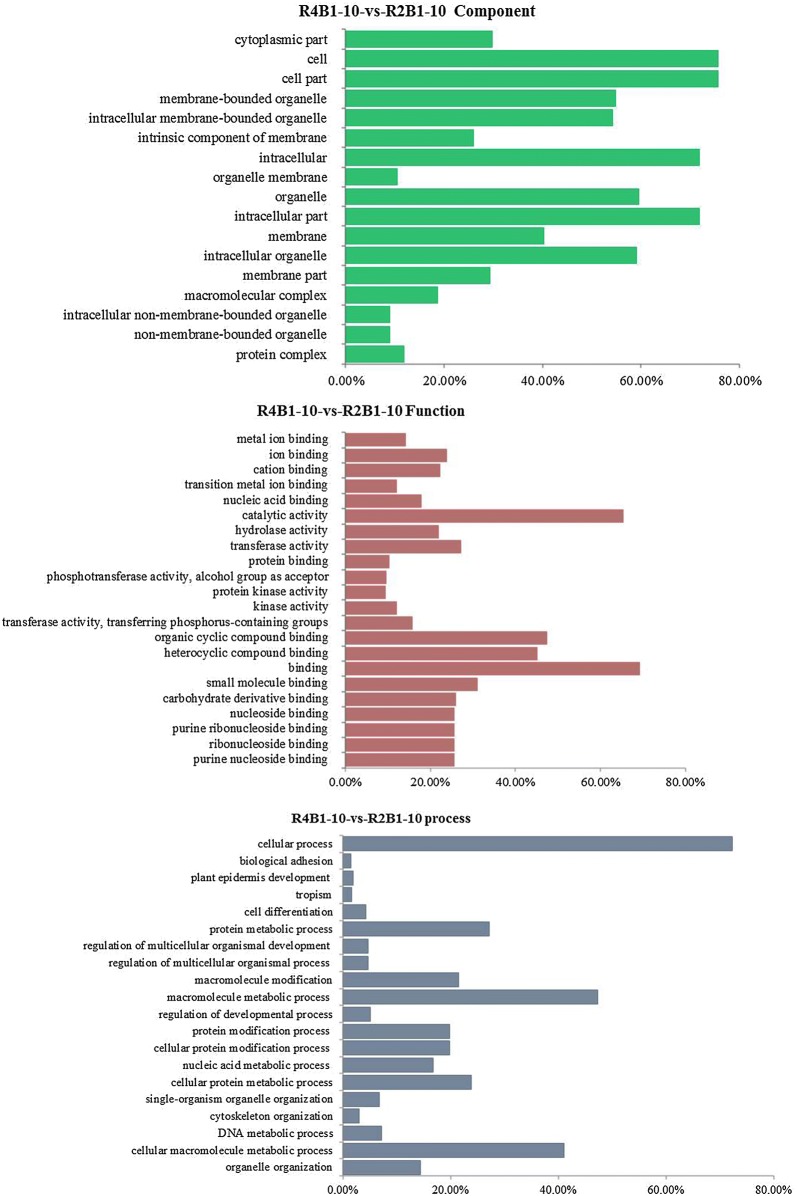
Gene ontology (GO) mainly includes three categories: molecular function, cellular component and biological process. A total of 110 DEGs in pairwise R4B1-5-vs-R2B1-5 and 745 DEGs in pairwise R4B1-10-vs-R2B1-10 were divided into cell components, which were related to the cell membrane, cytoplasm and organelle (Figs. 2 and 3). In pairwise R4B1-5-vs-R2B1-5, a total of 207 DEGs were classified into molecular functions: catalytic activity, binding, transporter activity and oxidoreductase activity (Fig. 2). A total of 195 DEGs were termed into the biology pathway, which mainly included ion transport, primary metabolic process, localization establishment of localization, biological regulation and response to stimulus (Fig. 2). In pairwise R4B1-10-vs-R2B1-10, 1060 DEGs were termed into molecular functions, which were mainly related to purine nucleoside binding, carbohydrate derivative binding, heterocyclic compound binding, organic cyclic compound binding, catalytic activity, hydrolase activity and transferase activity (Fig. 3). In pairwise R4B1-10-vs-R2B1-10, a total of 1060 genes were classified into the biology pathway. These genes mainly participated in a cellular process, cellular metabolic process, single-organism process, organic substance metabolic process, metabolic process and protein metabolic process (Fig. 3). In pairwise R4B1-15-vs-R2B1-15, only 3 DEGs were found and cannot be classified into any category.
Fig. 2.
Gene ontology (GO) categories of differently expressed genes (DEGs) in the pairwise R4B1-5-vs-R2B1-5
Fig. 3.
Gene ontology (GO) categories of differently expressed genes (DEGs) in the pairwise R4B1-10-vs-R2B1-10
Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis of DEGs after different ratios of blue and red light treatments
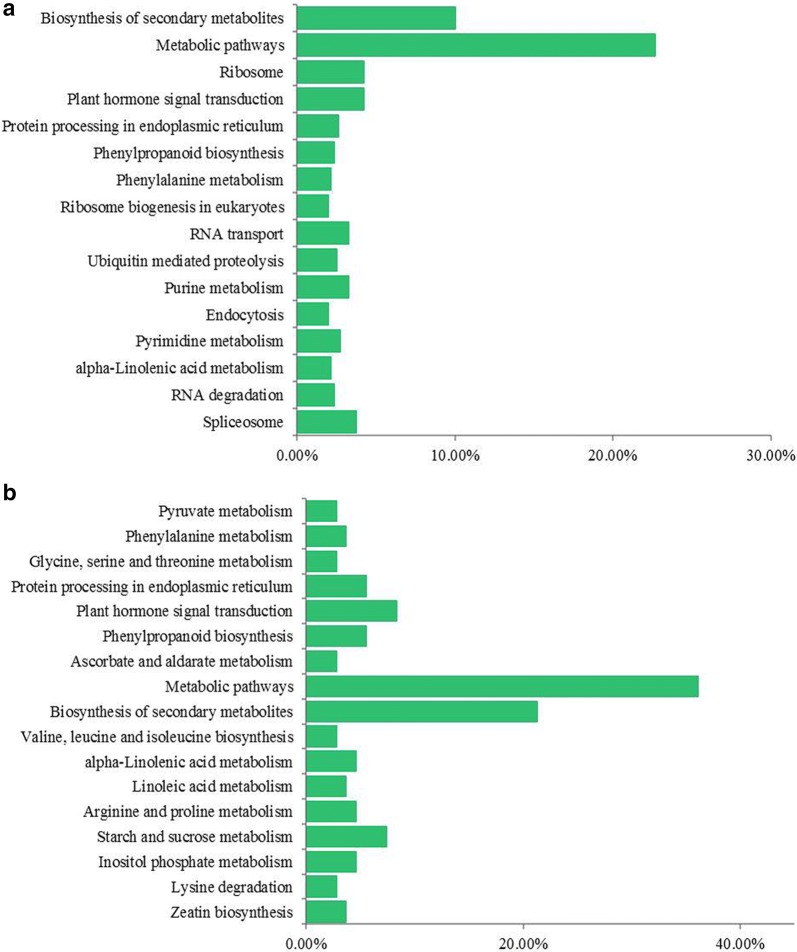
To further explore the biological pathways associated with sex differentiation development, DEGs in each comparison were mapped to the reference canonical pathways in the KEGG database. In pairwise R4B1-5-vs-R2B1-5, 108 DEGs were assigned into 52 KEGG pathways. They were significantly enriched in the following pathways: metabolic pathways, biosynthesis of secondary metabolites, starch and sucrose metabolism and plant hormone signal transduction (Fig. 4a). In pairwise R4B1-10-vs-R2B1-10, a total of 798 DEGs were assigned into 106 KEGG pathways. They were significantly enriched in the following pathways: metabolic pathways, biosynthesis of secondary metabolites and plant hormone signal transduction (Fig. 4b).
Fig. 4.
Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment of differently expressed genes (DEGs) in a pairwise R4B1-5-vs-R2B1-5 and b pairwise R4B1-10-vs-R2B1-10
All specifically up-regulated or down-regulated DEGs after R2B1 and R4B1 treatment were also mapped to the reference canonical pathway in the KEGG database. The results showed that 543 specifically up-regulated DEGs from R2B1 were mostly enriched in the following KEGG pathways: plant hormone signal transduction, secondary protein processing in endoplasmic reticulum, RNA transport and spliceosome pathway (Table 3). A total of 382 specifically down-regulated DEGs from R2B1 were grouped into the following KEGG pathways: biosynthesis of amino acids, protein processing in endoplasmic reticulum and RNA transport pathway (Table 3). The relevant data for R4B1 treatment were listed in Table 3.
Table 3.
Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment of significantly up/down-regulated DEGs in R2B1/R4B1
| Pathway | DEGs with pathway annotation (543) | P-value | |
|---|---|---|---|
| Plant hormone signal transduction | 49 | 0.000917 | Up-regulated DEGs in R2B1 |
| Protein processing in endoplasmic reticulum | 40 | 0.000665 | |
| RNA transport | 33 | 0.012593 | |
| Spliceosome | 31 | 0.078185 | |
| Endocytosis | 25 | 0.010854 | |
| Pyrimidine metabolism | 24 | 0.029783 | |
| RNA degradation | 22 | 0.001054 | |
| Ubiquitin mediated proteolysis | 21 | 0.0133 | |
| Starch and sucrose metabolism | 6 | 0.024124 | |
| Alanine, aspartate and glutamate metabolism | 4 | 0.002149 | Up-regulated DEGs in R4B1 |
| Diterpenoid biosynthesis | 3 | 0.005284 | |
| Insulin resistance | 3 | 0.010482 | |
| Biosynthesis of amino acids | 34 | 0.000934 | Down-regulated DEGs in R2B1 |
| Protein processing in endoplasmic reticulum | 29 | 0.002805 | |
| RNA transport | 29 | 0.000742 | |
| Spliceosome | 28 | 0.004517 | |
| Purine metabolism | 25 | 0.00642 | |
| Aminoacyl-tRNA biosynthesis | 23 | 0 | |
| Pyrimidine metabolism | 22 | 0.001743 | |
| RNA polymerase | 10 | 0.034698 | |
| Plant hormone signal transduction | 16 | 0.955624 | |
| Ribosome | 16 | 0 | Down-regulated DEGs in R4B1 |
| Porphyrin and chlorophyll metabolism | 5 | 0.000222 | |
| Photosynthesis | 4 | 0.002138 |
Auxin plays an important role in the regulation of cucumber sex differentiation under different ratios of blue and red light treatments
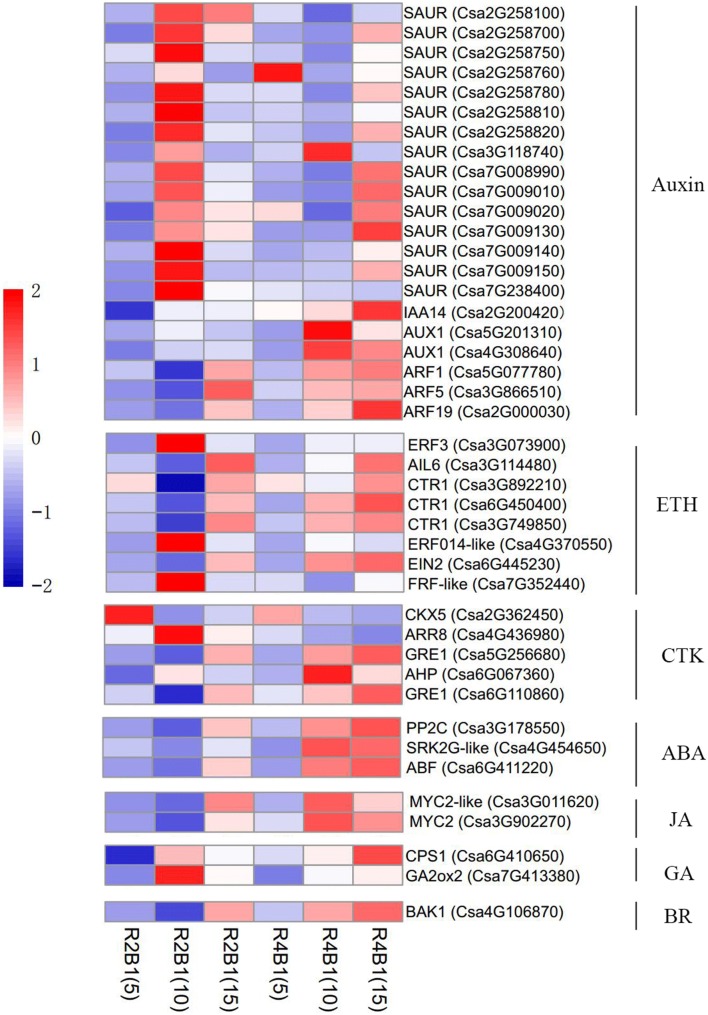
Since cucumber sex differentiation is closely related to all types of endogenous hormones, we further explored the role of endogenous hormones regulating the sex differentiation process under different light treatments. Thus, DEGs enriched in plant hormone signal transduction were further analyzed. In pairwise R4B1-5-vs-R2B1-5, 33.33% of the DEGs were related to auxin, 33.33% to cytokines, 22.22% to SA and 11.11% to ABA. In pairwise R4B1-10-vs-R2B1-10, 55.88% of the DEGs were related to auxin, 14.71% to ethylene, 11.76% to CTK, 8.82% to ABA, 5.88% to JA and 2.94% to SA (Fig. 5a, b). In addition, there were 49 up-regulated DEGs related to the plant hormone signaling pathway in R2B1, including 53.06% related to IAA, 8.16% to ETH, 12.24% to CTK and 14.29% to ABA (Fig. 5c). IAA plays an important role during the flower sex differentiation of cucumber seedlings under light irradiation. As most of the DEGs were related to plant hormone, 16 DEGs were found to be involved in auxin biosynthesis and signaling pathway, with most of them showing a higher expression level at R2B1-10 stage (Fig. 6). Five DEGs involved in CTK signaling pathway, eight DEGs in ETH signal transduction pathway and three DEGs in ABA signaling pathway were also identified from the transcriptomes of cucumber shoot apices (Fig. 6). There was no DEGs involved in the GA signal transduction, while only two genes, CsCPS1 (Csa6G410650) and CsGA2ox2 (Csa7G413380), involved in GA biosynthesis were identified. CsGA2ox2 (Csa7G413380) accumulated more transcripts at the R2B1-10 stage, while CsCPS1 (Csa6G410650) expressed more at the R4B1-15 stage (Fig. 6). In conclusion, light regulated cucumber flower sex differentiation through meditating multiple hormone signals: auxin plays an important role; CTK, ABA and ETH function as supplementary regulators; and gibberellin is not involved.
Fig. 5.
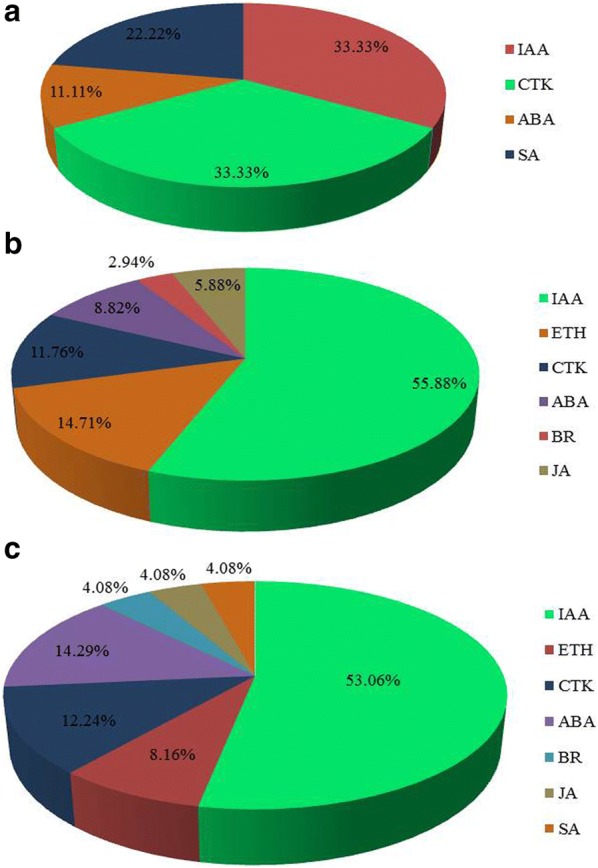
The proportion of differently expressed genes (DEGs) related to plant hormone signaling pathways in a pairwise R4B1-5-vs-R2B1-5, b pairwise R4B1-10-vs-R2B1-10 and c R2B1 library. IAA auxin, CTK cytokine, ABA abscisic acid, SA salicylic acid, ETH ethylene, BR brassinolide, JA jasmonic acid
Fig. 6.
The expression of a number of genes altered in cucumber shoot apices under different light quality treatments: genes involved in plant hormone signaling pathway
Validation of RNA-Seq results by real-time quantitative PCR (qRT-PCR)
Twenty-six genes with different expression profiles were randomly selected to verify the RNA-Seq results by qRT-PCR. α-Tubulin (TUA) was used as the internal reference control to standardize the results (Wan et al. 2010). As shown in Additional file 2: Figure S1, twenty-three genes had the same expression pattern. The Pearson correlation coefficient between the RNA-Seq and qRT-PCR data were high, confirming the accuracy of the RNA-Seq data (Additional file 3: Figure S2 and Additional file 4: Figure S3).
Effect of different ratios of blue and red light treatments on light signal transduction of cucumber seedling leaves
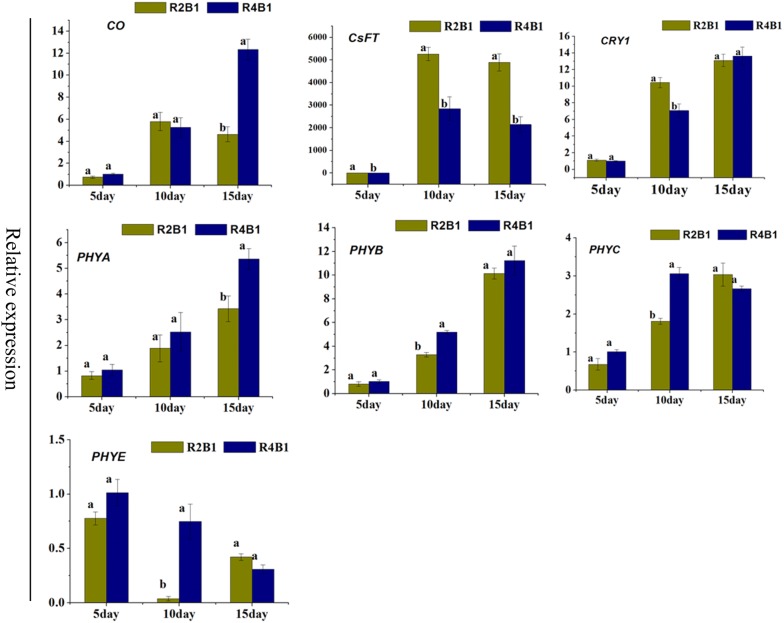
Photoperiodic induction affects flower meristem differentiation and number of flowers. As leaves are the site for photoperiodic induction, genes related to light signal transduction and flowering were checked by qRT-PCR on 5-day, 10-day and 15-day cucumber seedling leaves after R2B1 and R4B1 light irradiation. There were two blue light receptors, named CRY1 (Csa3M889670) and CRY2 (Csa3M88160). Only CRY1(Csa3M889670) was found in cucumber leaves. In the 10 days treatment samples, the expression of CRY1 was higher in R2B1 comparing to R4B1 (Fig. 7). Four red light receptors (PHYA, PHYB, PHYC and PHYE) were found in cucumber leaves. Compared to R2B1 treatment, there was a greater expression of PHYA in 10 days samples under R4B1 treatment, while the others showed a higher expression in the 15 days samples after R4B1 treatment (Fig. 7). CONSTANS (CO) is responsible for turning the light transduction into a flowering signal. It showed a higher expression in 15 days samples after R4B1 treatment than after R2B1 treatment (Fig. 7). FLOWERING LOCUST (FT) is a key gene in regulating flowering time. FT was expressed more in 10 and 15 days samples after R2B1 treatment than after R4B1 treatment (Fig. 7). All of these results suggested that the high proportion of blue light promoted the early flowering of cucumbers by regulating these flowering genes.
Fig. 7.
The expression of a number of flowering and light signal transduction related genes were altered in cucumber seedlings leaves under different light quality treatments. Error bars represented standard deviations of the means of three independent replicates. Statistically significant variations of expression and mean values at different sampling points (ANOVA, p < 0.05) were indicated with different letters
Analysis of differentially expressed genes related to flower sex differentiation of cucumber seedlings under different ratios of blue and red light treatments
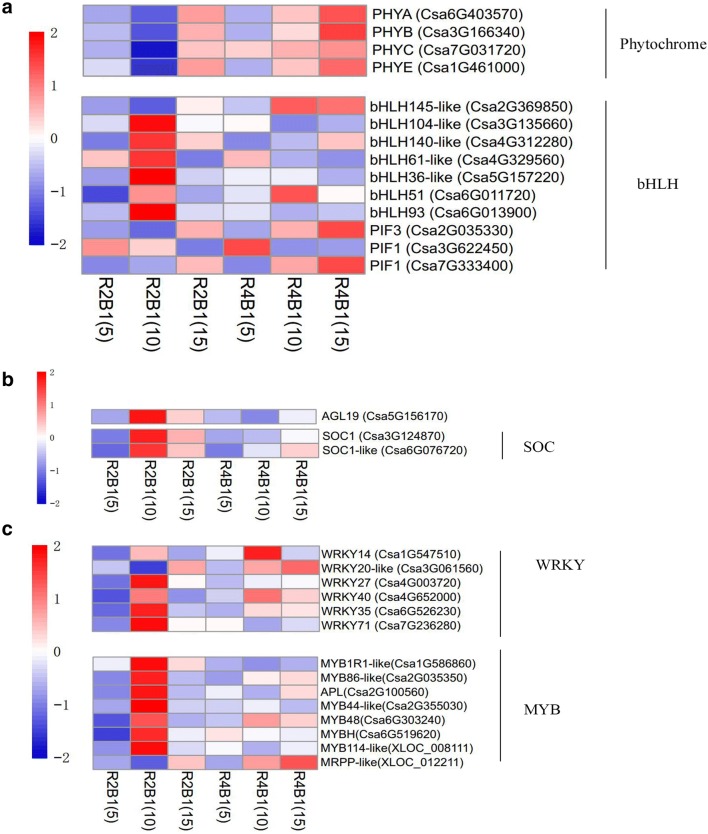
Plant sex differentiation is closely related to endogenous hormone regulation, while flowering time is related to the photoperiod pathway, vernalization and gibberellin pathway. Aiming to identify genes related to flower sex differentiation and promoting flowering under light irradiation, the expression pattern analysis of DEGs was performed in pairwise R4B1-5-vs-R2B1-5, R4B1-10-vs-R2B1-10 and R4B1-15-vs-R2B1-15. Four light receptors (Csa6G403570 (PHYA), Csa3G166340 (PHYB), Csa7G031720 (PHYC) and Csa1G461000 (PHYE)) responding to red light were identified from the shoot apices of cucumber seedlings. These genes showed a higher expression level after R4B1 treatment compared to R2B1 treatment (Fig. 8a). The (basic helix-loop-helix) bHLH transcription factor is involved in light signal transduction, hormone signal transduction and flowering pathway. Seven bHLH DEGs were identified in the RNA-seq transcriptome, including 6 up-regulated and 1 down-regulated genes in R2B1-10 samples. PIF, phytochrome interaction factor, initiates light signal transduction through interacting with the activated phytochrome. PIF belongs to bHLH family. Its expression levels increased at the R4B1-10 stage, while these levels decreased at the R2B1-10 stage (Fig. 8a). The WKRY (transcription factors with WRKYGQK peptide) family transcription factors participate in multiple metabolic pathways. Six differently expressed WKRY transcription factors were identified in cucumber shoot apices, including 3 (Csa4G003720, Csa6G526230 and Csa7G236280) up-regulated genes and 2 (Csa1G547510 and Csa3G061560) down-regulated genes at R2B1-10 stage (Fig. 8c). The MYB transcription factor is related to plant hormone and flower organ development. Nine MYB DEGs were identified in shoot apices transcriptome of cucumber seedlings, including 2 new genes (XLOC_008111 and XLOC_012211), 7 (Csa1G586860, Csa2G035350, Csa2G100560, Csa2G355030, Csa6G303240, Csa6G519620 and XLOC_012211) up-regulated genes and 2 (Csa4G338950 and XLOC_012211) down-regulated genes at the R2B1-10 stage (Fig. 8c) (AGAMOUS [AG]-like MADS box protein) AGL and (SUPPRESSOR OF OVEREXPRESSION OF CO1) SOC1 belong to MADS-box gene, which both could regulate flowering time. Seven AGLs and two SOC1 s were identified from the shoot apices of cucumber seedlings. AGL (Csa5G156170) and SOC1 (Csa3G124870, Csa6G076720) accumulated more transcripts at the R2B1-10 stage (Fig. 8b, c).
Fig. 8.
The expression of a number of genes altered in cucumber shoot apices under different light quality treatments: a genes involved in light signaling transduction; b genes related to flowering time regulation and c transcription factors involved in plant hormone and flower organ development
Discussion
Light quality affects cucumber flowering time
Blue light is a strong signal in floral bud formation. In Chrysanthemum, flower budding was formed even after a longer photoperiod than a critical day length under blue light illumination (Jerzy et al. 2011). In petunia plants, floral bud formation and flowering occurred earlier under blue light treatment, while no floral buds were observed under low red irradiance. High red irradiance and temporal switching to blue light during long-term low red irradiance induced floral development (Fukuda et al. 2016). In our experiment, we found that the first female flower opened earlier and occurred mainly at the lower node positions under a higher proportion of blue light irradiance (Table 1). This suggested that blue light promoted flower bud formation and accelerated cucumber flowering.
Cryptochromes (CRY) are flavo-proteins that direct a diverse array of developmental processes in response to blue light in plants (Liu et al. 2016). CRY1 and CRY2 function as major blue light receptors regulating blue light-induced de-etiolation and photo-periodic flowering (Guo and Li 1998). In Arabidopsis, CRY1 and CRY2 serve both distinct and partially overlapping functions in regulating photomorphogenic responses and photoperiodic flowering. The gain-of-function mutant alleles of CRY1 exhibited an early flowering phenotype after several days (Exner et al. 2010). In our study, we detected a higher expression of CRY1 in cucumber seedling leaves after R2B1 treatment than R4B1 treatment (Fig. 7), which was consistent with the earlier flowering phenotype after R2B1 irradiation. The phytochrome (phy) family of sensory photoreceptors (phyA to phyE in Arabidopsis thaliana) also responds to inducing floral budding. Low red/far-red ratio promotes flowering in Arabidopsis through PHYA inhibiting PHYB (Chory 2003; Halliday et al. 2003). In our study, we detected 4 phytochrome orthologous genes in cucumber seedling leaves and shoot apices. Expression analysis showed that PHYA accumulated more in leaves at the R4B1-15 stage than R2B1-15 stage (Fig. 7). In comparison, the other genes displayed a higher expression in leaves at R4B1-10 stage than R2B1-10 stage (Fig. 7). Similar results were found in shoot apices, with PHYA, PHYB, PHYC, PHYE expressed more in the shoot apices of R4B1-10 sample than R2B1-10 sample (Fig. 7). The expression of CRY2 and PHYD were not detected in both cucumber leaves and shoot apices.
CONSTANS (CO) is a key transcription factor regulating flowering time. CO positively regulates two floral integrators. In Arabidopsis, CO activated SOC1 through FT to promote flowering (Rosas et al. 2014; Lee and Lee 2010). In this study, compared to R2B1-15, CO showed a higher expression in R4B1-15 (Fig. 7). Although CO transcription regulation was affected by blue light treatment, CO was not only positively regulated by CRY2 and PHYA, but also negatively regulated by PHYB (Valverde et al. 2004). The lower expression under a high proportion of blue irradiation might result from the co-regulation of the cryptochrome receptor and phytochrome receptor. In Arabidopsis, blue light could induce SOC1 and FT expression to promote flowering (Hori et al. 2011). In petunia plants, blue light induces FBP28 expression, the orthologous gene of SOC1 (Fukuda et al. 2011). SOC1 is one of the direct targets of AGL24 and is up-regulated by AGL24. In Arabidopsis, SOC1 and AGL24 are able to up-regulate each other’s expressions, both of them are MADS-domain-containing transcription factors that determine flowering time (Michaels et al. 2003). In this study, FT showed a higher expression in R2B1-10 and R2B1-15 than in R4B1-10 and R4B1-15 (Fig. 8b), which suggested that higher proportion of blue light irradiation accelerated cucumber flowering through inducing FT expression. Two SOC1 orthologous genes and seven AGLs (only AGL19 showed different expression) orthologous genes were found in cucumber shoot apices. SOC1 and AGL19 displayed the same expression pattern, with increased expression in R2B1-10 and R2B1-15 treatments (Fig. 8b). All these results suggested that a higher proportion of blue light induced cucumber flower bud formation and promoted flowering time through up-regulating expression of these flowering regulation factors.
Light quality affects flower sex differentiation of cucumber seedlings through regulating plant hormone signaling transduction
In this study, we found that under a higher proportion of blue light (R2B1) treatment, the first female flower node declined significantly and the number of female flowers within fifteen plant nodes increased markedly (Table 1). Transcriptome analysis of cucumber seedlings under different light treatment by RNA-seq technology showed that there were more DEGs under R2B1 treatment. These DEGs were mostly involved in the hormone signaling pathway (Table 3). This suggested that a higher proportion of blue light regulated female flower formation through mediating plant hormone signaling pathways.
Plant sex differentiation is closely related to plant hormones, such as ethylene, which is considered as a potent sex hormone in cucumbers that can induce formation of female flowers (Trebitsh et al. 1987; Rudich et al. 1972; Li et al. 2017). Another example is the gibberellins, which can promote the formation of male flowers via ethylene-dependent and ethylene-independent pathways in cucumber (Hao et al. 2003). ABA can promote male flower development (Zhu et al. 2010), while auxin is able to promote the female tendency of vegetables and induce female flower development through triggering ethylene synthesis (Atsmon and Tabbak 1979; Galun et al. 1962). Exogenous CTK application can turn the grape male flower into a female flower (Chang et al. 1999; Negi and Olmo 1966). JA can regulate gynoecium development (Figueroa and Browse 2015), while BR induces androecium development in maize and inhibits gynoecium development (Hartwig et al. 2011; Makarevitch et al. 2012). In this study, we found that among these DEGs related to plant hormone transduction pathways, the specific DEGs related with IAA formed the higher proportion of DEGs in plant hormone signal transduction, which was followed by the specific DEGs related with CTK, ABA, ETH, BR, JA and SA (Fig. 5).
Ethylene is considered as a potent sex hormone in cucumber that can induce formation of female flowers (Malepszy and Niemirowicz-Szczytt 1991). Ethylene content in shoot apices of gynoecious cucumbers is higher than that of monoecious plants (Rudich et al. 1972; Fujita and Fujieda 1981; Trebitsh et al. 1987). Treatment with exogenous ethylene or ethylene-releasing reagents can increase the numbers of female and bisexual flowers in monoecious and andromonoecious lines, respectively (Mcmurray and Miller 1968; Iwahori et al. 1969). Until now, the molecular mechanism of ethylene-regulated sex determination of cucumber has been well understood. Ethylene biosynthetic genes and ethylene signal transduction genes are involved in the sex expression of cucumber (Wang et al. 2010). In the ethylene signal transduction pathway, the receptors, such as ETRs, function as negative regulators, while ERFs (downstream components of receptors) act as positive transcription factors to regulate sex determination (Tao et al. 2018; Prescott et al. 2016). In our study, the expression levels of some ERFs were increased under R2B1 treatment compared to R4B1 treatment (Fig. 6a). They were probably involved in cucumber sex expression through inhibiting the differentiation into males and promoting the differentiation into females. However, this understanding was based on bioinformatics analysis. The precise roles of ERFs in cucumber flower development remained unclear and should be verified in future studies using advanced physiological and molecular techniques. CTR1 is a negative regulator of ethylene signaling, while CsCTR1 expression gradually declined during male flower development and increased during female flower development (Bie et al. 2014). In our study, CsCTR1 expression increased on the 5th day after a higher proportion of blue light treatment.
Ethylene plays a key role in plant sex determination, with several research results proving that there is communication between auxin and ethylene during plant development (Trebitsh et al. 1987; Gao et al. 2015; Wang et al. 2018). In our study, we found a considerably number of transcription factors involving in auxin signaling pathway altered their expression levels under R2B1 treatment (Fig. 6). IAA may play a key role in effect of light quality on sex differentiation in cucumber. IAA has been considered as a transcription factor activated by phytochrome, which participates in signaling transduction. During the early development of plants, there are several reactions between phytochrome and auxin, such as auxin homeostasis promoted by light (Zhang et al. 2014c). Auxin is an important regulator in the development of male flowers (Sakata et al. 2010). The auxin synthesis pathway can be regulated by aldehyde oxidases, which have been identified in cucumber (Brown and Purves 1978). Aldehyde oxidases are involved in the conversions of indole-3-acetaldehyde to IAA (Mano and Nemoto 2012). Higher activity of aldehyde oxidase has also been measured in the auxin-overproducing superroot1 (sur1) mutant of Arabidopsis (Seo et al. 1998). Aldehyde oxidase was reported to regulate auxin and ABA biosynthesis pathway (Koshiba et al. 1996; Seo et al. 1998, 2000). In our study, the expression of Aldehyde oxidase (Csa4G269130) was down-regulated in R2B1 samples (Additional file 5: Table S2), which suggested that the auxin synthesis or ABA synthesis in cucumber seedlings flowers was low under a higher proportion of blue light. This result was contradicted against the finding that more female flower were formed in the R2B1 samples which might result from higher auxin content. Meanwhile, it was also contrary to the most up-regulated auxin responsive genes SAURs (Fig. 6). While the expression of auxin transporting gene LAX1 and PIN1 were also altered indicating that the auxin transport was changed inside the cucumber flower. Even though the low biosynthesis might be found in the flowers, the higher transportation might also cause a high auxin content in the flower tissues. However, auxin content detection and gene function analysis experiments should be performed to confirm this hypothesis.
Gibberellins, a class of tetracyclic diterpenoid phytohormones, can promote the differentiation of cucumbers into male flowers. GA production in andromonoecious cucumbers is higher than that in gynoecious and monoecious plants (Junior et al. 1972). Exogenous GA3 application can increase the ratio of males to females in monoecious cucumbers and induce the formation of male flowers in gynoecious plants (Wittwer and Bukovac 1962; Pike and Peterson 1969). In our study, we identified two DEGs involved in the GA synthesis pathway. Ent-copalyl diphosphate synthase (CPS) is limiting for ent-kaurene production in the first portion of the GA synthesis pathway (Prisic and Peters 2007). CsCPS1 was down-regulated in R2B1-5 and up-regulated in R4B1-15 (Fig. 6), which suggested that a high proportion of blue light reduces levels of GA in cucumber seedling flowers through breaking down the CPS1 gene expression. GA2-oxidase (GA2ox) is also a key gene for GA synthesis pathway. It catalyzes catabolism and inactivation of bioactive GAs or their precursors (Schomburg and Amasino 2003; Lester et al. 1999; Thomas et al. 1999). In our study, CsGA2ox2 was up-regulated in R2B1-10 samples (Fig. 6), which suggested that a high proportion of blue light reduces active GA levels through inducing the expression of CsGA2ox2. In addition, the GA signaling pathway is involved in stamen and anther development in hermaphroditic plants, such as Arabidopsis and rice (Oryza sativa) (Aya et al. 2009; Plackett et al. 2011; Sun 2011). However, in this study, we did not identify any DEGs involved in GA signaling pathway.
PIF, phytochrome interaction factor, plays a central role in light signal transduction mediated by phytochrome (De and Prat 2014). PIF3 can activate the transcription factor EIN3 directly, which was involved in the ethylene signaling pathway (Zhong et al. 2012). COI1, JAZ, MYC2 and JAR1 involved in the JA signaling pathway affects the light signal response (Kazan and Manners 2011). PHYB negatively regulated ABA accumulation through mediating light signals, while PHYA positively regulated ABA signaling pathway under far-red light treatment (Gu et al. 2012; Mach 2014). Thus, it was seen that light could regulate plant sex differentiation through mediating hormones by light receptors. The phytohormones, auxin, BR, CK, ETH, GA and JA, influenced sex differentiation in flowering plants (Durand and Durand 2010; Louis and Durand 1978; Chailakhyan 1979; Irish and Nelson 1989). Our results showed that the expression of some key genes involved in ABA, auxin, CK, ETH, GA and JA biosynthesis pathways significantly changed during development and sex expression, which suggesting that these hormones might participate in these processes. Thus, it is interesting to investigate how these hormones interact with one another to regulate the abortion of male flowers in gynoecious plants.
Light quality affects flowering time of cucumber seedlings through regulating transcription factors
A total of 433 transcription factors have been identified by RNA-seq technology. Among them, ethylene transcription factor accounted for 24.02%, MYB transcription factor accounted for 20.79%, bHLH transcription factor accounted for 16.17% and WRKY transcription factor accounted for 10.62%.
Most studies have shown that MYB, bHLH and WRKY transcription factors play an important role in biotic stress and abiotic stress (Oh et al. 2012; Li et al. 2010; Jiang and Yu 2009), but some studies proved that they were also related to flower development (Wang et al. 2011; Luo et al. 2013; Zhang et al. 2014b). In Arabidopsis, JA mediates the transcription factors MYB21 and MYB24 to regulate stamen development (Song et al. 2011), while MYC5 regulated stamen development by activating JA signaling pathway through inducing MYB21 expression. PIFs belongs to bHLH family of transcription factors, which interact with activated Pr to initiate phytochrome signaling transduction. In Arabidopsis, PIF4 accelerated flowering by activating the FT gene (Kumar et al. 2016). WRKY family members are key factors for the ABA response pathway (Rushton et al. 2012). It had been shown that WRKY transcription factors regulated plant flowering. WRKY25 accelerated flowering through negatively regulating AP1 directly or indirectly (Wang et al. 2011). In pea, GsWRKY20 promoted flowering by regulating the flowering related genes FT, SOC1 and CO (Luo et al. 2013).
Materials and methods
Plant materials and treatments
LED light produced by Kedao technology corporation (Huizhou, China) (Red light: 650–660 nm and Blue light: 450–460 nm) was chosen for this study. Cucumber (Cucumis sativus L. cv. Huaqing No. 5) seeds were cultivated on a sponge block with a Yamasaki culture solution. When cotyledon appeared, we placed these seedlings under light conditions as follows: 300 μmol/m2/s, 12/12 (light dark) at 25 °C and a relative humidity of 70–80%. There were 2 treatments with ratios of red to blue being 4:1 (R4B1) and 2:1 (R21B), respectively. Each treatment lasted 20 days. As the flower primordium appeared 10 days after light treatment, the shoot apices and leaves after 5, 10 and 15 days of light treatments were collected immediately flash frozen in liquid nitrogen and stored at − 80 °C until use. Two biological replicates of shoot apices were used for RNA-seq analysis and real-time RT-PCR. After 20 days of treatment, 30–40 seedlings from each treatment were transplanted into the greenhouse of South China Agriculture University with a relative humidity ranging from 70 to 85% and 28 °C/20 °C (14 h/10 h) day/night temperature for further flowering time analysis.
cDNA library construction and illumina sequencing
The cucumber shoot apices stored at − 80 °C were provided to Guangzhou Gene Denovo Biological Technology Co., Ltd. (Guangzhou, China). RNA isolation and RNA-Seq library preparation and sequencing were carried out by Guangzhou Gene Denovo Biological Technology Co., Ltd. (Guangzhou, China). In brief, the workflow of library construction for transcriptome analysis was as follows: After RNA was collected, poly(A)-containing mRNA was purified using oligo(dT) magnetic beads. Following this, the mRNAs were fragmented and cDNA was synthesized using a random hexamer, DNA polymerase I and RNase H. The double-stranded cDNAs were purified and ligated to adaptors for Illumina paired-end sequencing. PCR amplication was done on the purified cDNAs with ligated adaptors. PCR products were separated by 2% agarose gel. The final PCR product chosen for sequencing is 400–500 bp band, which were cut and recovered from the gel. The quality and quantity of the library were verified using an Agilent 2100 Bioanalyzer and ABI real time RT-PCR, respectively (Additional file 6: Table S3 and Additional file 2: Figure S1). The cDNA libraries were sequenced using Illumina HiSeqTM 2500 by Gene Denovo Co. (Guangzhou, China). The libraries of two biological replicates of shoot apices were prepared independently.
Sequence read mapping and assembly
The original image data produced by sequencing were transferred into sequences (raw reads). The calculating methods for raw reads was conducted according to Cock (2010). To obtain clean reads for further analysis, the raw reads were filtered by removing adaptor sequences, low-quality reads and reads with a percentage of unknown bases (N) of more than 10%. Following this, we used Bowtie to remove the ribosome reads (Langmead et al. 2009). The clean reads were mapped to the cucumber genome assembly (Chinese long) v2 (http://www.icugi.org/cgi-bin/ICuGI/index.cgi) according to Kim et al. (2013) and Trapnell et al. (2012).
Quantification and differential expression analysis of transcripts
The expression level of each gene was determined by the numbers of reads, which were uniquely mapped to the specific gene and the total number of uniquely mapped reads in the sample. Reads per kilobase of exon model per million mapped reads (RPKM) were calculated according to Mortazavi to estimate gene expression levels. The FDR was used to determine the threshold of the P-value in multiple tests. In our study, a FDR < 0.05 and an absolute value of |log2 (fold change)| > 1 were used as the threshold to determine the significant DEGs.
GO and KEGG enrichment analysis
To identify putative biological functions and pathways for the DEGs, the Gene ontology (GO) and Kyoto encyclopedia of gene and genomes (KEGG) database were searched for annotation. The methods for GO and KEGG enrichment analysis were according to Young and Mao respectively (Young et al. 2010; Mao et al. 2005).
Real-time RT-PCR
Total RNA was isolated from 100 mg cucumber leaves and shoot apices using Huayueyang reagent kit (with DNase I step: Pipet the DNase I onto the membrane containing RNA. Incubate at 37 °C for 30 min) according to the manufacturer’s instructions. The RNA quality and purity were verified by Nanodrop 2000 and electrophoresis on 1.0% agarose gels. The first-strand cDNA was synthesized from aliquots of 1 μg of total RNA using PrimeScriptTM RT Reagent Kit with gDNA Eraser (TaKaRa, RR047A) in a reaction volume of 20 μL. The synthesized cDNA was diluted 20 times with sterile water and used as the template for real-time PCR. The reactions were carried out in a Roche LightCycler 480 system with SYBR Premix Ex Taq II (Tli RNaseH Plus) (Takara Bio, Dalian, China). The reactions reagent mix was 5 μL SYBR Premix Ex Taq II, 1.5 μL cDNA template, 0.4 μL each primer (10 μmol/μL), and 2.7 μL nuclease-free water. The amplification program was 95 °C for 30 s and 40 cycles of 95 °C for 5 s and 60 °C for 30 s. Melting curve analyses were performed at the end of 40 cycles (95 °C for 5 s followed by a constant increase from 60 to 95 °C). In addition, TUA was used to normalize expression levels (Wan et al. 2010), and the relative expression of genes was calculated using the 2−∆∆Ct method (Livak and Schmittgen 2001). The results were analyzed by Excel 2010. qPCR reactions in leaves were prepared in biological triplicate. qPCR reactions in shoot apices for RNA-seq validation were prepared in three biological replicates.
Genes used for Real-Time RT-PCR were designed on the NCBI primer blast website (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). The primer quality and efficiency were checked by qPCR on series of diluting cDNA. The expression of 26 genes chosen from RNA-seq were analyzed by real-time RT-PCR in shoot apex samples (R2B1 and R4B1 at 5 days, 10 days and 15 days). Meanwhile, 9 genes were used for checking light signal transduction in leaves (Additional file 7: Table S4).
Additional files
Additional file 1: Table S1. Effects of different light quality on the formation of female flowers and flowering time.
Additional file 2: Figure S1. 2% agrose gel result of PCR product for library sequencing.
Additional file 3: Figure S2. Correlation between gene expression levels of two biological replicates for each stage.
Additional file 4: Figure S3. Expression pattern of 26 genes validated by real-time quantitative PCR (qPCR).
Additional file 5: Table S2. Expression data of all the genes in R2B1 and R4B1 samples.
Additional file 6: Table S3. List of primers used in the expression studies.
Additional file 7: Table S4. Real-time quantitative PCR (qPCR) quality control (QC).
Authors’ contributions
JS and YZ carried out the experiment, participated in the analysis. YH drafted the manuscript. WS carried out the qRT-PCR analysis. SS and GS performed the statistical analysis. HL and YH conceived of the study, and participated in its design. RC and HL acquired of funding and helped to draft the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The raw RNA-seq reads have been deposited in NCBI Sequence Read Archive (SRA) under Accession Number of SRR7345617; SRR7345616; SRR7345615; SRR7345614; SRR7345621; SRR7345620; SRR7345619; SRR7345618; SRR7345623; SRR7345622; SRR7345625; SRR7345624.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Fundings
This work was supported by Teamwork Projects Funded by Guangdong Natural Science Foundation (No. S2013030012842), and the Guangzhou Science & Technology Project (201605030005).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jiali Song, Email: Song_46@126.com.
Yiting Zhang, Email: YitingZhang@scau.edu.cn.
Shiwei Song, Email: swsong@scau.edu.cn.
Wei Su, Email: susan_l@scau.edu.cn.
Riyuan Chen, Email: rychen@scau.edu.cn.
Guangwen Sun, Email: sungw1968@scau.edu.cn.
Yanwei Hao, Phone: +020-85280228, Email: yanweihao@scau.edu.cn, Email: yanweihaogbf@gmail.com.
Houcheng Liu, Phone: +020-85280228, Email: liuhch@scau.edu.cn.
References
- Acosta IF, Laparra H, Romero SP, Schmelz E, Hamberg M, Mottinger JP, Moreno MA, Dellaporta SL. Tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science. 2009;323:262–265. doi: 10.1126/science.1164645. [DOI] [PubMed] [Google Scholar]
- Ando S, Sato Y, Kamachi S, Sakai S. Isolation of a MADS-box gene (ERAF17) and correlation of its expression with the induction of formation of female flowers by ethylene in cucumber plants (Cucumis sativus L.) Planta. 2001;213:943–952. doi: 10.1007/s004250100571. [DOI] [PubMed] [Google Scholar]
- Atsmon D, Tabbak C. Comparative effects of gibberellin, silver nitrate and aminoethoxyvinyl glycine on sexual tendency and ethylene evolution in the cucumber plant (Cucumis sativus L.) Plant Cell Physiol. 1979;20:1547–1555. [Google Scholar]
- Aya K, Ueguchitanak M, Kondo M, Hamada K, Yano K, Nishimura M, Matsuoka M. Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell. 2009;21:1453–1472. doi: 10.1105/tpc.108.062935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai N, Xu H. Chapter one-unisexual cucumber flowers, sex and sex differentiation. Int Rev Cel Mol Blo. 2013;304:1–55. doi: 10.1016/B978-0-12-407696-9.00001-4. [DOI] [PubMed] [Google Scholar]
- Bai L, Peng B, Cui X, Gu T, Xu Y, Li Q, Xu H, Bai N. Developmental analyses reveal early arrests of the spore-bearing parts of reproductive organs in unisexual flowers of cucumber (Cucumis sativus L.) Planta. 2004;220:230–240. doi: 10.1007/s00425-004-1342-2. [DOI] [PubMed] [Google Scholar]
- Bie B, Sun J, Pa J, He H, Cai R. Ectopic expression of CsCTR1, a cucumber CTR-like gene, attenuates constitutive ethylene signaling in an Arabidopsis ctr1-1 mutant and expression pattern analysis of CsCTR1 in cucumber (Cucumis sativus) Int J Mol Sci. 2014;15:16331–16350. doi: 10.3390/ijms150916331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs WR, Beck CF, Cashmore AR, Christie JM, Hughes J, Jarillo JA, Kagawa T, Kanegae H, Liscum E, Nagatani A. The phototropin family of photoreceptors. Plant Cell. 2001;13:993–997. doi: 10.1105/tpc.13.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown HM, Purves WK. Cucumber seedling indoleacetaldehyde oxidase. Plant Physiol. 1978;61:107–110. doi: 10.1104/pp.61.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudler R, Hitomi K, Daiyasu H, Toh H, Kucho KI, Ishiura M, Kanehisa M, Roberts VA, Todo T, Tainer JA, Getzoff ED. Identification of a new cryptochrome class: structure, function, and evolution. Mol Cell. 2003;11:59–67. doi: 10.1016/s1097-2765(03)00008-x. [DOI] [PubMed] [Google Scholar]
- Chailakhyan MK. Genetic and hormonal regulation of growth, flowering, and sex expression in plants. Am J Bot. 1979;66:717–736. [Google Scholar]
- Chang T, Chen S, Hsu Y, Yu C, Du S, Huang L. Changes in cytokinin activities before, during and after floral initiation in Polianthes tuberosa. Plant Physiol Bioch. 1999;37:679–684. [Google Scholar]
- Chen M, Chory J, Fankhauser C. Light signal transduction in higher plants. Annu Rev Genet. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
- Chory J. Regulation of flowering time by light quality. Nature. 2003;423:881–885. doi: 10.1038/nature01636. [DOI] [PubMed] [Google Scholar]
- Cock J, Fields J, Goto N, Heuer L, Rice M. The sanger fastq file format for sequences with quality scores, and the solexa/illumina fastq variants. Nucleic Acids Res. 2010;38:1767–1771. doi: 10.1093/nar/gkp1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De M, Prat S. Pifs get BR right: phytochrome interacting factors as integrators of light and hormonal signals. New Phytol. 2014;202:1126–1141. doi: 10.1111/nph.12725. [DOI] [PubMed] [Google Scholar]
- Duan H, Wang H, Xu H, Bai N. Stamen development in Arabidopsis is arrested by organ-specific overexpression of a cucumber ethylene synthesis gene CsACO2. Planta. 2008;228:537–543. doi: 10.1007/s00425-008-0756-7. [DOI] [PubMed] [Google Scholar]
- Durand R, Durand B. Sexual differentiation in higher plants. Physiol Plantarum. 2010;60:267–274. [Google Scholar]
- Exner V, Alexandre C, Rosenfeldt G, Alfarano P, Nater M, Caflisch A, Gruissem W, Batschauer A, Hennig L. A gain-of-function mutation of Arabidopsis cryptochrome1 promotes flowering. Plant Physiol. 2010;154:1633–1645. doi: 10.1104/pp.110.160895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei H, Zhang R, Pharis P, Sawhney K. Pleiotropic effects of the male sterile33 (ms33) mutation in Arabidopsis are associated with modifications in endogenous gibberellins, indole-3-acetic acid and abscisic acid. Planta. 2004;219:649–660. doi: 10.1007/s00425-004-1270-1. [DOI] [PubMed] [Google Scholar]
- Figueroa P, Browse J. Male sterility in Arabidopsis induced by overexpression of a MYC5-SRDX chimeric repressor. Plant J. 2015;81:849–860. doi: 10.1111/tpj.12776. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Fujieda K. Relation between sex expression types and cotyledon etiolation of cucumber in vitro I. on the role of ethylene evolved from seedlings. Plant Cell Physiol. 1981;22:667–674. [Google Scholar]
- Fukuda N, Ishii Y, Ezura H, Olsen E. Effects of light quality under red and blue light emitting diodes on growth and expression of FBP28 in petunia. Acta Hortic. 2011;907:361–366. [Google Scholar]
- Fukuda N, Ajima C, Yukawa T, Olsen JE. Antagonistic action of blue and red light on shoot elongation in petunia depends on gibberellin, but the effects on flowering are not generally linked to gibberellin. Environ Exp Bot. 2016;121:102–111. [Google Scholar]
- Galun E, Jung Y, Lang A. Culture and sex modification of male cucumber buds in vitro. Nature. 1962;194:596–598. [Google Scholar]
- Gao P, Sheng Y, Luan F. RNA-Seq transcriptome profiling reveals differentially expressed genes involved in sex expression in melon. Crop Sci. 2015;55:1686–1695. [Google Scholar]
- Golenberg EM, West NW. Hormonal interactions and gene regulation can link monoecy and environmental plasticity to the evolution of dioecy in plants. Am J Bot. 2013;100:1022–1037. doi: 10.3732/ajb.1200544. [DOI] [PubMed] [Google Scholar]
- Gu J, Fang Z, Jie Z, Zhou J, Qian F, Yan L. Light signals mediated by phytochrome B affect abscisic acid pathway in rice. Chin Sci Bull. 2012;57:2371. [Google Scholar]
- Guo H, Li C. Regulation of flowering time by Arabidopsis photoreceptors. Science. 1998;279:1360–1363. doi: 10.1126/science.279.5355.1360. [DOI] [PubMed] [Google Scholar]
- Halliday KJ, Salter MG, Thingnaes E, Whitelam GC. Phytochrome control of flowering is temperature sensitive and correlates with expression of the floral integrator FT. Plant J. 2003;33:875–885. doi: 10.1046/j.1365-313x.2003.01674.x. [DOI] [PubMed] [Google Scholar]
- Hao J, Wang H, Peng B, Bai L, Xu Y, Li Q, Xu H, Bai N. DNA damage in the early primordial anther is closely correlated with stamen arrest in the female flower of cucumber (Cucumis sativus L.) Planta. 2003;217:888–895. doi: 10.1007/s00425-003-1064-x. [DOI] [PubMed] [Google Scholar]
- Hartwig T, Chuck GS, Fujioka S, Klempien A, Weizbauer R, Potluri DP, Choe S, Johal GS, Schulz B. Brassinosteroid control of sex determination in maize. Proc Natl Acad Sci USA. 2011;108:19814–19819. doi: 10.1073/pnas.1108359108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori Y, Nishidate K, Nishiyama M, Kanahama K, Kanayama Y. Flowering and expression of flowering-related genes under long-day conditions with light-emitting diodes. Planta. 2011;234:321–330. doi: 10.1007/s00425-011-1397-9. [DOI] [PubMed] [Google Scholar]
- Irish EE, Nelson T. Sex determination in monoecious and dioecious plants. Plant Cell. 1989;1:737–744. doi: 10.1105/tpc.1.8.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa M, Takahashi F, Nozaki H, Nagasato C, Motomura T, Kataoka H. Distribution and phylogeny of the blue light receptors aureochromes in eukaryotes. Planta. 2009;230:543–552. doi: 10.1007/s00425-009-0967-6. [DOI] [PubMed] [Google Scholar]
- Iwahori S, Lyons JM, Sims WL. Induced femaleness in cucumber by 2-chloroethanephosphonic acid. Nature. 1969;222:271–272. doi: 10.1038/222271a0. [DOI] [PubMed] [Google Scholar]
- Jerzy M, Zakrzewski P, Schroeterzakrzewska A. Effect of colour of light on the opening of inflorescence buds and post-harvest longevity of pot chrysanthemums (chrysanthemum × grandiflorum (ramat.) kitam) Acta Agrobot. 2011;64:13–18. [Google Scholar]
- Jiang W, Yu D. Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol. 2009;9:20. doi: 10.1186/1471-2229-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junior H, Baker R, Sell HM. Different sex phenotypes of Cucumis sativus L. and C. melo L. and their endogenous gibberellin activity. Euphytica. 1972;21:285–291. [Google Scholar]
- Kamachi H, Iwasawa O, Hickok LG, Nakayama M, Noguchi M, Inoue H. The effects of light on sex determination in gametophytes of the fern Ceratopteris richardii. J Plant Res. 2007;120:629–634. doi: 10.1007/s10265-007-0106-z. [DOI] [PubMed] [Google Scholar]
- Kazan K, Manners JM. The interplay between light and jasmonate signalling during defence and development. J Exp Bot. 2011;62:4087–4100. doi: 10.1093/jxb/err142. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. Tophat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R364. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpelainen H. Labile sex expression in plants. Biol Rev. 1998;73:157–180. [Google Scholar]
- Koshiba T, Saito E, Ono N, et al. Purification and properties of flavin- and molybdenum-containing aldehyde oxidase from coleoptiles of Maize. Plant Physiol. 1996;110:781–789. doi: 10.1104/pp.110.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SV, Lucyshyn D, Jaeger KE, Alós E, Alvey E, Harberd NP, Wigge PA. Phytochrome interacting factor 4 controls the thermosensory activation of flowering. Nature. 2016;484:242–245. doi: 10.1038/nature10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:1–10. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Lee I. Regulation and function of SOC1, a flowering pathway integrator. J Exp Bot. 2010;61:2247–2254. doi: 10.1093/jxb/erq098. [DOI] [PubMed] [Google Scholar]
- Lester DR, Ross JJ, Smith JJ, Elliott RC, Reid JB. Gibberellin 2-oxidation and the SLN gene of Pisum sativum. Plant J. 1999;19:65–73. doi: 10.1046/j.1365-313x.1999.00501.x. [DOI] [PubMed] [Google Scholar]
- Li Z, Huang S, Liu S, Pan J, Zhang Z, Tao Q, Shi Q, Jia Z, Zhang W, Chen H, Si L, Zhu L, Cai R. Molecular isolation of the M gene suggests that a conserved-residue conversion induces the formation of bisexual flowers in cucumber plants. Genetics. 2009;182:1381–1385. doi: 10.1534/genetics.109.104737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Guo S, Zhao Y, Chen D, Chong K, Xu Y. Overexpression of a homopeptide repeat-containing bHLH protein gene (OrbHLH001) from dongxiang wild rice confers freezing and salt tolerance in transgenic Arabidopsis. Plant Cell Rep. 2010;29:977–986. doi: 10.1007/s00299-010-0883-z. [DOI] [PubMed] [Google Scholar]
- Li SF, Zhang GJ, Zhang XJ, Yuan JH, Deng CL, Gao WJ. Comparative transcriptome analysis reveals differentially expressed genes associated with sex expression in garden asparagus (asparagus officinalis) BMC Plant Biol. 2017;17:143. doi: 10.1186/s12870-017-1091-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Yang Z, Gomez A, Liu B, Lin C, Oka Y. Signaling mechanisms of plant cryptochromes in Arabidopsis thaliana. J Plant Res. 2016;129:137–148. doi: 10.1007/s10265-015-0782-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Louis JP, Durand B. Studies with the dioecious angiosperm Mercurialis annua L. (2n = 16): correlation between genic and cytoplasmic male sterility, sex segregation and feminizing hormones (cytokinins) Mol Gen Genet. 1978;165:309–322. [Google Scholar]
- Luo X, Sun X, Liu B, Zhu D, Bai X, Cai H, Ji W, Cao L, Wu J. Ectopic expression of a WRKY homolog from glycine soja alters flowering time in Arabidopsis. PLoS ONE. 2013;8:e73295. doi: 10.1371/journal.pone.0073295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach J. Walk into the light response: direct targets of phytochrome A include genes that respond to light, stress, and hormones. Plant Cell. 2014;26:1832. doi: 10.1105/tpc.114.127019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevitch I, Thompson A, Muehlbauer GJ, Springer NM. Brd1 gene in maize encodes a brassinosteroid C-6 oxidase. PLoS ONE. 2012;7:e30798. doi: 10.1371/journal.pone.0030798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malepszy S, Niemirowicz-Szczytt K. Sex determination in cucumber (Cucumis sativus) as a model system for molecular biology. Plant Sci. 1991;80:39–47. [Google Scholar]
- Mano Y, Nemoto K. The pathway of auxin biosynthesis in plants. J Exp Bot. 2012;63:2853–2872. doi: 10.1093/jxb/ers091. [DOI] [PubMed] [Google Scholar]
- Mao X, Cai T, Olyarchuk JG, Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21:3787–3793. doi: 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
- Mathews S, Sharrock RA. Phytochrome gene diversity. Plant, Cell Environ. 1997;20:666–671. [Google Scholar]
- Mcmurray AL, Miller CH. Cucumber sex expression modified by 2-chloroethanephosphonic acid. Science. 1968;162:1397–1398. doi: 10.1126/science.162.3860.1397. [DOI] [PubMed] [Google Scholar]
- Mibus H, Tatlioglu T. Molecular characterization and isolation of the f/f gene for femaleness in cucumber (Cucumis sativus L.) Theor Appl Genet. 2004;109:1669–1676. doi: 10.1007/s00122-004-1793-7. [DOI] [PubMed] [Google Scholar]
- Michaels SD, Ditta G, Gustafson-Brown C, Pelaz S, Yanofsky M, Amasino RM. AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant J. 2003;33:867–874. doi: 10.1046/j.1365-313x.2003.01671.x. [DOI] [PubMed] [Google Scholar]
- Negi SS, Olmo HP. Sex conversion in a male Vitis vinifera L. by a Kinin. Science. 1966;152:1624–1625. doi: 10.1126/science.152.3729.1624. [DOI] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol. 2012;14:802–809. doi: 10.1038/ncb2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike LM, Peterson CE. Gibberellin A4/A7, for induction of staminate flowers on the gynoecious cucumber (Cucumis sativus L.) Euphytica. 1969;18:106–109. [Google Scholar]
- Plackett AR, Thomas SG, Wilson ZA, Hedden P. Gibberellin control of stamen development: a fertile field. Trends Plant Sci. 2011;16:568–578. doi: 10.1016/j.tplants.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Prescott AM, Mccollough FW, Eldreth BL, Binder BM, Abel SM. Analysis of network topologies underlying ethylene growth response kinetics. Front Plant Sci. 2016;7:1308. doi: 10.3389/fpls.2016.01308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prisic S, Peters RJ. Synergistic substrate inhibition of ent-copalyl diphosphate synthase: a potential feed-forward inhibition mechanism limiting gibberellin metabolism. Plant Physiol. 2007;144:445–454. doi: 10.1104/pp.106.095208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas U, Mei Y, Xie Q, Banta JA, Zhou RW, Seufferheld G, Gerard S, Chou L, Bhambhra N, Parks JD, Flowers JM, Mcclung CR, Hanzawa Y, Purugganan MD. Variation in Arabidopsis flowering time associated with cis -regulatory variation in CONSTANS. Nat Commun. 2014;5:3651. doi: 10.1038/ncomms4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudich J, Halevy AH, Kedar N. The level of phytohormones in monoecious and gynoecious cucumbers as affected by photoperiod and ethephon. Plant Physiol. 1972;50:585–590. doi: 10.1104/pp.50.5.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton DL, Tripathi P, Rabara RC, Lin J, Ringler P, Boken AK, Langum J, Smidt L, Boomsma DD, Emme NJ, Chen X, Finer JJ, Shen J, Rushton PJ. WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol J. 2012;10:2–11. doi: 10.1111/j.1467-7652.2011.00634.x. [DOI] [PubMed] [Google Scholar]
- Sakata T, Oshino T, Miura S, Tomabechi M, Tsunaga Y, Higashitani N, Miyazawa Y, Takahashi H, Watanabe M, Higashitani A. Auxins reverse plant male sterility caused by high temperatures. Proc Natl Acad Sci USA. 2010;107:8569–8574. doi: 10.1073/pnas.1000869107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schomburg FM, Amasino RM. Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell. 2003;15:151–163. doi: 10.1105/tpc.005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Akaba S, Oritani T, et al. Higher activity of an aldehyde oxidase in the auxin-overproducing superroot1 mutant of Arabidopsis thaliana. Plant Physiol. 1998;116:687–693. doi: 10.1104/pp.116.2.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Peeters AJM, Koiwai H, et al. The arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc Natl Acad Sci USA. 2000;97:12908–12913. doi: 10.1073/pnas.220426197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Qi T, Huang H, Ren Q, Wu D, Chang C, Peng W, Liu Y, Peng J, Xie D. The jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell. 2011;23:1000–1013. doi: 10.1105/tpc.111.083089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Qi T, Huang H, Xie D. Regulation of stamen development by coordinated actions of jasmonate, auxin, and gibberellin in Arabidopsis. Mol Plant. 2013;6:1065–1073. doi: 10.1093/mp/sst054. [DOI] [PubMed] [Google Scholar]
- Sun TP. The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr Biol Cb. 2011;21:R338–R345. doi: 10.1016/j.cub.2011.02.036. [DOI] [PubMed] [Google Scholar]
- Takahashi F, Yamagata D, Ishikawa M, Fukamatsu Y, Ogura Y, Kasahara M, Kiyosue T, Kikuyama M, Wada M, Kataoka H. AUREOCHROME, a photoreceptor required for photomorphogenesis in stramenopiles. P Natl Acad Sci USA. 2007;104:19625–19630. doi: 10.1073/pnas.0707692104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Q, Niu H, Wang Z, Li Z. Ethylene responsive factor ERF110 mediates ethylene-regulated transcription of a sex determination-related orthologous gene in two cucumis species. J Exp Bot. 2018;69:12. doi: 10.1093/jxb/ery128. [DOI] [PubMed] [Google Scholar]
- Thomas SG, Phillips AL, Hedden P. Molecular cloning and functional expression of gibberellin 2- oxidases, multifunctional enzymes involved in gibberellin deactivation. P Natl Acad Sci USA. 1999;96:4698–4703. doi: 10.1073/pnas.96.8.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebitsh T, Rudich J, Riov J. Auxin, biosynthesis of ethylene and sex expression in cucumber (Cucumis sativus) Plant Growth Regul. 1987;5:105–113. [Google Scholar]
- Trebitsh T, Staub JE, O’Neill SD. Identification of a 1-aminocyclopropane-1-carboxylic acid synthase gene linked to the female (f) locus that enhances female sex expression in cucumber. Plant Physiol. 1997;113:987–995. doi: 10.1104/pp.113.3.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;629:1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- Wan H, Zhao Z, Qian C, Sui Y, Malik AA, Chen J. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal Biochem. 2010;399:257–261. doi: 10.1016/j.ab.2009.12.008. [DOI] [PubMed] [Google Scholar]
- Wang DH, Li F, Duan QH, Han T, Xu ZH, Bai SN. Ethylene perception is involved in female cucumber flower development. Plant J. 2010;61:862–872. doi: 10.1111/j.1365-313X.2009.04114.x. [DOI] [PubMed] [Google Scholar]
- Wang FX, Li SJ, Yu DQ. Overexpression of WRKY25 causes early flowering in Arabidopsis under long-day conditions. Plant Divers. 2011;33:653–659. [Google Scholar]
- Wang C, Xin M, Zhou X. Transcriptome profiling reveals candidate genes associated with sex differentiation induced by night temperature in cucumber. Sci Hortic. 2018;232:162–169. [Google Scholar]
- Wittwer SH, Bukovac MJ. Staminate flower formation on gynoecious cucumbers as influenced by the various gibberellins. Sci Nat. 1962;49:305–306. [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A. Method gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu B, Yang S, An J, Chen C, Zhang X, Ren H. A cucumber DELLA homolog CsGAIP may inhibit staminate development through transcriptional repression of B class floral homeotic genes. PLoS ONE. 2014;9:e91804. doi: 10.1371/journal.pone.0091804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang X, Liu B, Wang W, Liu X, Chen C, Liu X, Yang S, Ren H. A GAMYB homologue CsGAMYB1 regulates sex expression of cucumber via an ethylene-independent pathway. J Exp Bot. 2014;65:3201–3213. doi: 10.1093/jxb/eru176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Ji R, Li H, Zhao T, Liu J, Li C, Liu B. CONSTANS-LIKE 7 (COL7) is involved in phytochrome B (phyB)-mediated light-quality regulation of auxin homeostasis. Mol Plant. 2014;7:1429–1440. doi: 10.1093/mp/ssu058. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhao G, Li Y, Mo N, Zhang J, Liang Y. Transcriptomic analysis implies that GA regulates sex expression via ethylene-dependent and ethylene-independent pathways in cucumber (Cucumis sativus L.). Front. Plant Sci. 2017;8:10. doi: 10.3389/fpls.2017.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Shi H, Xue C, Wang L, Xi Y, Li J, Quail PH, Deng XW, Guo H. A molecular framework of light-controlled phytohormone action in Arabidopsis. Curr Biol Cb. 2012;2:1530–1535. doi: 10.1016/j.cub.2012.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Dun X, Zhou ZF, Xia SQ, Yi B, Wen J, Shen J, Ma C, Tu J, Fu T. A separation defect of tapetum cells and microspore mother cells results in male sterility in Brassica napus: the role of abscisic acid in early anther development. Plant Mol Biol. 2010;72:111–123. doi: 10.1007/s11103-009-9556-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Effects of different light quality on the formation of female flowers and flowering time.
Additional file 2: Figure S1. 2% agrose gel result of PCR product for library sequencing.
Additional file 3: Figure S2. Correlation between gene expression levels of two biological replicates for each stage.
Additional file 4: Figure S3. Expression pattern of 26 genes validated by real-time quantitative PCR (qPCR).
Additional file 5: Table S2. Expression data of all the genes in R2B1 and R4B1 samples.
Additional file 6: Table S3. List of primers used in the expression studies.
Additional file 7: Table S4. Real-time quantitative PCR (qPCR) quality control (QC).
Data Availability Statement
The raw RNA-seq reads have been deposited in NCBI Sequence Read Archive (SRA) under Accession Number of SRR7345617; SRR7345616; SRR7345615; SRR7345614; SRR7345621; SRR7345620; SRR7345619; SRR7345618; SRR7345623; SRR7345622; SRR7345625; SRR7345624.