Abstract
Background
CRISPR/CAS9 (epi)genome editing revolutionized the field of gene and cell therapy. Our previous study demonstrated that a rapid and robust reactivation of the HIV latent reservoir by a catalytically-deficient Cas9 (dCas9)-synergistic activation mediator (SAM) via HIV long terminal repeat (LTR)-specific MS2-mediated single guide RNAs (msgRNAs) directly induces cellular suicide without additional immunotherapy. However, potential off-target effect remains a concern for any clinical application of Cas9 genome editing and dCas9 epigenome editing. After dCas9 treatment, potential off-target responses have been analyzed through different strategies such as mRNA sequence analysis, and functional screening. In this study, a comprehensive analysis of the host transcriptome including mRNA, lncRNA, and alternative splicing was performed using human cell lines expressing dCas9-SAM and HIV-targeting msgRNAs.
Results
The control scrambled msgRNA (LTR_Zero), and two LTR-specific msgRNAs (LTR_L and LTR_O) groups show very similar expression profiles of the whole transcriptome. Among 839 identified lncRNAs, none exhibited significantly different expression in LTR_L vs. LTR_Zero group. In LTR_O group, only TERC and scaRNA2 lncRNAs were significantly decreased. Among 142,791 mRNAs, four genes were differentially expressed in LTR_L vs. LTR_Zero group. There were 21 genes significantly downregulated in LTR_O vs. either LTR_Zero or LTR_L group and one third of them are histone related. The distributions of different types of alternative splicing were very similar either within or between groups. There were no apparent changes in all the lncRNA and mRNA transcripts between the LTR_L and LTR_Zero groups.
Conclusion
This is an extremely comprehensive study demonstrating the rare off-target effects of the HIV-specific dCas9-SAM system in human cells. This finding is encouraging for the safe application of dCas9-SAM technology to induce target-specific reactivation of latent HIV for an effective “shock-and-kill” strategy.
Electronic supplementary material
The online version of this article (10.1186/s12920-018-0394-2) contains supplementary material, which is available to authorized users.
Keywords: Genome editing, CRISPR, Off-target, RNA sequencing, Transcriptome, HIV, Latency, Shock and kill
Background
Recently, CRISPR/Cas9 genome editing technology has been rapidly developed and attracted extensive attention in biomedical research, with preclinical examples and potential clinical trials in genetic diseases, cancer biology, and infectious diseases [1–7]. Simultaneously, the catalytically-deficient Cas9 (dCas9) epigenome editing technology has emerged as a novel platform for the manipulation of cellular or viral gene regulation by incorporating monoplex or multiplex transcriptional activators or repressors [8–19]. Cas9-mediated genome editing technology has been utilized to excise the HIV-1 provirus via HIV-specific multiplex single guide RNAs (sgRNAs) in cultured HIV latent cell lines [20–22], primary T cells [22, 23], and HIV transgenic rodents [24, 25]. The dCas9 epigenome editing technology [8–11, 19] is also used to reactivate the latent HIV-1 provirus using HIV long terminal repeat (LTR)-specific sgRNAs [26–29]. A rapid and robust reactivation of the HIV latent reservoir by dCas9-synergistic activation mediator (SAM) via MS2-mediated sgRNAs (msgRNAs) [30] directly induces cellular suicide without additional immunotherapy [31], which might be a novel, practical, and specific method for the “shock and kill” strategy to cure HIV/AIDS. The dCas9-SAM approach also induces specific activation of endogenous viral restriction factors that affect virus replication [32].
In addition to transcriptional activation, the dCas9 property is also extensively repurposed for transcriptional repression and DNA (de)methylation [12, 33–35]. These epigenome-editing approaches can alter the epigenetic code of the target region, and thus offer a durable manipulation of many genes important in infectious diseases, cancer, and chronic noninfectious diseases [12, 36]. Modification of an individual chromatin mark may suppress target gene expression in most cases [36]. However, permanent silencing of target genes in all cell types may require a combination of several epigenetic effectors [12].
Potential off-target effect remains a critical concern for any clinical application of this technology. Several promising strategies have been developed to mitigate any potential off-target responses, such as the sgRNA design optimization [37–42], transcriptome analysis [28, 30], and functional screening after dCas9 treatment [43]. For the parent Cas9 genome editing system, increasing experimental data suggests that the genome editing is highly specific [20, 44–48]. Newly developed unbiased profiling techniques further validate the high specificity of this Cas9/sgRNA technology [49–54]. In vivo off-target effects are expected to be low due to epigenetic protection [55, 56]. Specifically for dCas9 technology, the frequency of off-target binding to essential (functional) exons would also be very low [57]. Further mRNA-seq analysis confirmed the specificity of this dCas9-SAM technology [28, 30].
Our previous studies analyzed the exogenous viral DNA against the host genome for the best scores of efficiency and specificity [20, 21, 31]. In TZM-bI cells expressing the HIV LTR-driven luciferase reporter without the viral genome itself [58], the dCas9-SAM technology with HIV LTR-specific msgRNAs induced potent reactivation of the HIV reporter, but did not influence the cell growth/proliferation [31], supporting the absence of off-target effects by the dCas9-SAM technology [27, 28, 59]. The aim of this study is to further explore the dCas9-SAM-related potential off-target effects by generating deep sequence coverage of the entire transcriptome, comprehensively analyzing mRNAs, lncRNAs, alternative splicing, genetic mutations including single-nucleotide polymorphisms (SNPs) and indels (insertions and deletions) in TZM-bI cells stably expressing dCas9-SAM and HIV-specific msgRNAs. These analyses are important for safety considerations during the potential clinical application of dCas9 epigenome editing technology [60].
Methods
Experimental design and RNA sample preparation
The HeLa cell-derived TZM-bl cell line stably expressing higher levels of CD4 and CCR5 was obtained from Dr. John C. Kappes through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH. It was generated by introducing separate integrated copies of the luciferase and ß-galactosidase genes under control of the HIV-1 LTR promoter. To establish the dCas9-SAM stable expression cell line (designated TZMb-6465 cell line), TZM-bI cells were transduced with pMSCV-dCas9-BFP (puromycin) retroviral vector (Addgene, plasmid #46912) [10], and Lenti-MS2-p65-HSF1 (hygromycin) lentiviral vector (Addgene, plasmid #61426) [30]. After 2 days, cells were subcultured and selected with puromycin (2 μg/ml) and hygromycin (200 μg/ml). After 2 weeks of selection culture, the TZMb-6465 cells were transduced with msgRNA-expressing empty control lentiviral vector (Addgene, Plasmid #61427) [30], HIV-1 LTR_L msgRNA-expressing lentivirus or LTR_O msgRNA-expressing lentivirus. Six samples were prepared: two replicates for the LTR_L editing (LTR_L1 and LTR_L2), two replicates for the LTRO editing (LTR_O1 and LTR_O2), and two replicates for control (LTR_Zer1 and LTR_Zer2). After four days, cells were subjected to total RNA extraction using the Direct-Zol RNA MiniPrep Kit (Genesee Scientific, Catalog number: 11–330). The 4-day post-infection time point was based on the sufficient msgRNA expression and potent LTR-target reactivation [31] while minimizing the possible confounding factor resulting from the indirect downstream effects of any potential off-targets, if they existed. The RNAs were preserved with RNAstable LD (Sigma, Catalog number: 53201–013) and shipped to Novogene Bioinformatics Institute (https://en.novogene.com/) for total RNA sequencing and bioinformatics analysis. The RNA integrity was verified by 1% agarose gel electrophoresis and Agilent 2100. The RNA purity was checked using a NanoPhotometer® spectrophotometer (IMPLEN, CA, USA) and the DNA concentration was measured using Qubit® DNA Assay Kit in Qubit® 2.0 Fluorometer (Life Technologies, CA, USA).
Library construction and sequencing
The RNA quality control (QC) was done using Trimmomatic with default settings, and this step discarded less than 3% of the RNA reads, and the results were shown in Additional file 1: Table S1. After RNA QC, rRNAs were removed by using the Epicentre Ribo-Zero™ Kit. The purified RNAs were first fragmented randomly into short fragments of 150~ 200 bp by addition of a fragmentation buffer, then cDNA synthesis was performed using random hexamers. After the first strand was synthesized, a custom second strand synthesis buffer (Illumina), dNTPs (dUTP, dATP, dGTP and dCTP) and DNA polymerase I were added to synthesize the second strand, then followed by purification by AMPure XP beads, terminal repair, polyadenylation, sequencing adapter ligation, size selection, and degradation of the second strand U-contained cDNA by the USER enzyme. The strand-specific cDNA library was generated after the final PCR enrichment. The concentration of the library was first quantified by Qubit2.0, then diluted to 1 ng/ul, and the insert size was checked by Agilent 2100 and further quantified by qPCR (library concentration > 2 nM). The libraries were then subjected to HiSeq sequencing according to the concentration and the expected data volume.
Sequence analysis
About 60 GB of RNA sequencing data was generated for all six samples. Original RNA-Seq reads contain adapters and low quality reads that needed to be filtered out. To ensure the quality of the analysis, the sequence adapters (Oligonucleotide sequences for TruSeq™ RNA and DNA Sample Prep Kits) were removed from reads using Trimmomatic [61, 62]. Then all the trimmed reads with more than 10% ambiguous bases (N) were also removed. Finally, low quality reads with a Phred score less than 20 were removed. Additional file 1: Table S1 shows the distribution of quality reads across the L, O, and Zero samples. High quality sequences are mapped to the human genome (hg38) using TopHat2 with default parameters [63]. Overall, approximately 89% of the raw reads were mapped to the human genome (detailed mapping results are shown in Additional file 1: Table S2 and Additional file 2: Figure S1). Mapped reads were then assigned to known types of RNA using the program HTSeq with the union model (see Additional file 1: Table S3 for the distribution of mapped reads in different categories of known RNAs). To quantify the transcript abundance, the FPKM metric (number of fragments per kilobase of transcript sequence per million mapped reads) was used, which considers both the sequencing depth and the transcript length. In order to measure the reliability of the experiments through biological replicates, the Pearson correlation coefficient (R2) was calculated between all pairs of the L, O, and Zero samples. A correlation coefficient close to one indicates high similarity of gene expression profiles.
LncRNA analysis
The detailed workflow for identifying long noncoding RNAs (lncRNAs) is shown in Additional file 2: Figure S2b. First, cufflinks with default parameters was used to assemble the mapped reads into transcripts and quantify transcript expression (including isoforms). Candidate long noncoding RNAs (lncRNAs) were then classified into three categories (lncRNAs, intronic lncRNAs, and antisense lncRNAs) through five filtering steps (Additional file 2: Figure S2b): (1) assembled transcripts from cufflinks were merged using cuffcompare and the merged transcripts selected if they appeared in more than one sample, (2) only transcripts with more than 200 bps and two exons were kept, (3) only those transcripts that have ≥3× coverage for at least two exons were kept, (4) transcripts with high coverage were then removed if they matched known non-lncRNAs and non-mRNA (e.g., rRNA, tRNA, snRNA, snoRNA, etc), and (5) the remaining transcripts were then removed if they matched known mRNAs. The final collection of RNAs was the candidate set of lncRNAs, intronic lncRNAs, and antisense lncRNAs. Additional file 2: Figure S3 shows the number of transcripts that were filtered in each step. After all of the five filtering steps, a total of 1615 transcripts were left in the six pooled samples.
To finally determine if a transcript is a lncRNA, four popular methods for coding potential analysis were applied: (1) CPC (Coding-Potential Calculator) [64] computes the coding potential of a transcript by matching it to the NCBI nr database using BLASTX and scoring it using a support vector machine, (2) CNCI (Coding-Non-Coding Index) distinguishes protein-coding and noncoding transcripts independent of known annotations and predicts the coding or noncoding potential based solely on the features of nucleotide triplets, (3) transcripts were translated into proteins and matched to known protein domains in Pfam [65] using HMMER3 [66] where a matched sequence is considered as having coding potential, whereas others are considered as noncoding, and (4) PhyloCSF (Phylogenetic Codon Substitution Frequency) uses genome-wide mammalian sequence alignments to calculate the coding potential of transcripts.
Functions of the lncRNAs were identified by predicting their protein-coding target genes in both a cis- and trans- manner. The cis-acting target prediction assumes that the function of a lncRNA is determined by its adjacent protein coding genes, and in this study, coding genes within ±100 kb of the lncRNAs were considered as cis-acting targets. The trans-acting targets were predicted based on co-expressed genes, and only those genes that had Pearson correlation coefficients greater than 0.95 with the lncRNAs were selected.
mRNA analysis
Differentially expressed mRNAs were determined using cuffdiff with default parameters [67]. A network analysis of protein-protein interactions for the differentially expressed mRNAs was also conducted using the STRING database [68]. If the target genes (such as the expressed mRNAs) were not found in the database, a BLASTX search was done with an E-value of 1e-10 to identify potential protein-protein interactions.
SNP and indel variant calling
To examine whether the dCas9-SAM technology has an effect on genetic mutations, for example, resulting in different sets of SNPs and indel mutations due to the editing, SNPs and indels were called and compared for the six samples. Specifically, SAMtools [69] and Picard [https://broadinstitute.github.io/picard/] were used to preprocess the mapped reads. SNPs and indel variants were called using the GATK2 toolkit [70]. To quantify the similarity between the sets of SNPs and indel mutations in the samples, the Jaccard Index,
where |S| denotes the size of set S, S1 is the set of SNPs/indels in one sample, and S2 is the set of SNPs/indels in another sample, is calculated for all 15 pairs of sample comparisons. The Jaccard index ranges from 0 to 1, the higher it is, the more similarity in the sets of SNPs/indels between two samples, with 0 indicating that two samples have entirely different sets of SNPs/indels and 1 indicating that two samples have the same set of SNPs/indels.
Alternative splicing
Alternative splicing (AS) was analyzed by first classifying AS events into 12 types as illustrated in Additional file 2: Figure S4 using ASprofile [71]. Then expression levels of alternatively spliced genes were estimated using the probabilistic framework MISO (Mixture of Isoforms) [72]. MISO uses a Bayesian statistical model to give a more accurate estimate of the expression level indicated by the number of reads that covers different isoforms or exons. Differential expression of isoforms was then determined by the Bayes factor (BF) that computes the odds of differential regulation occurring. The higher the BF, the more likely the isoforms/exons are differentially regulated. A cutoff BF = 10 was applied to select the isoforms/exons that were significantly differentially regulated between conditions [72]. Five major AS events, (1) A3SS (alternative 3′ splice sites), (2) A5SS (alternative 5′ splice sites), (3) MXE (mutually exclusive exons), (4) RI (retained intron), and (5) SE (skipped exon), were analyzed.
Statistics
All the statistical tests, including Steiger’s test, two proportion z-test, and Chi-square tests were performed in R.
Results
Very similar expression profiles at the whole transcriptome level among the three conditions
In previous studies, 16 msgRNAs targeting the U3 region of the HIV LTR were screened for their efficiency in guiding dCas9-SAM to activate HIV promoter activity [31]. Two targeting sites, LTR_L (− 165/− 145 bp from the transcription start site) and LTR_O (− 112/− 92 bp from the transcription start site) surrounding the enhancer region (Fig. 1a), were identified for robust reactivation of HIV-1 provirus in various types of human cells [31]. These two hotspots were verified in other studies [26–29]. To determine if the dCas9-SAM system mediated by these two hotspots affects the host cells’ transcriptomes, the total RNAs from TZM-bI cells stably expressing the dCas9-SAM system plus msgRNA targeting LTR_L or LTR_O were prepared for lncRNA and mRNA sequencing. The empty msgRNA carrying scrambled target sequence was used as the control (LTR_Zero). The TZM-bI cell line was used because it harbors integrated HIV-1 LTR promoter but does not contain HIV-1 proviral DNA that may produce viral proteins leading to potential effects on the host transcriptome [58], complicating the analysis. A total of 600,451,484 raw reads were generated after read quality control and cleanup, of which 97.4% clean reads were kept for downstream analyses (see Additional file 1: Table S1 for details). The clean reads were then mapped to the human reference genome hg38 by Tophat2 [63]. More than 89% of the reads were mapped for all six samples (see Additional file 1: Table S2 for details) and distributions of the mapped reads in the genome are shown in Additional file 2: Figure S1.
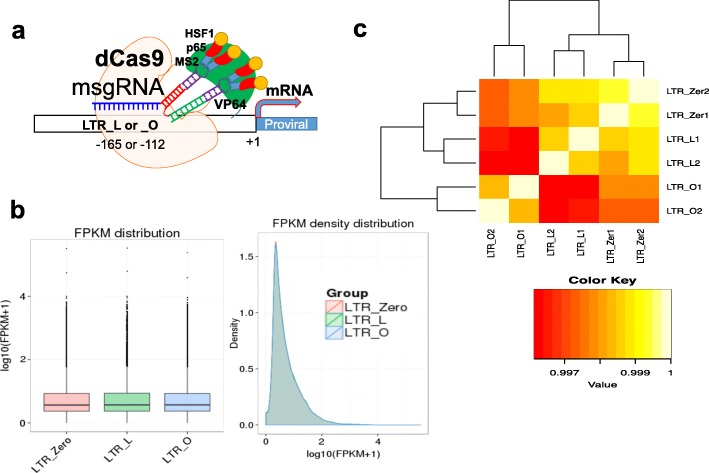
Fig. 1.
No difference in the entire RNA transcripts among the three experimental conditions. a Diagram showing the HIV proviral activation by the dCas9-SAM system with msgRNAs targeting LTR_L or LTR_O. b Box plot and density plot for the distribution of transcript expression levels measured by FPKM (averaged within replicates) of the three conditions. The plotted region of the box plot represents the maximum, upper quartile, median, lower quartile, and minimum, respectively, from top to bottom. c Hierarchical clustering of samples based on Pearson correlation coefficient of transcript expression levels for all the pairwise comparisons of the samples
The distribution of the transcript expression levels under different conditions (L, O, and Zero) was analyzed by the mean fragments per kilobase of transcript per million mapped reads (FPKM) of the two replicates for each condition (Fig. 1b). It is clear that the expression distributions of all the transcripts among the three conditions are highly similar, except for the LTR-driven reporter genes luciferase and ß-galactosidase (see Additional file 1: Table S3), which is consistent with the increased luciferase activity in the LTR-targeting groups [31]. The square of the Pearson correlation coefficient (R2) for all the transcripts among the samples and replicates was assessed, for which R2> 0.92 was considered good quality [73, 74]. Here, the correlations for all pairs of samples fell within the range of 0.9961 to 0.9993 (Fig. 1c). Samples of the same conditions (i.e., the duplicates for each condition) have significantly higher correlation coefficients than those for samples from different conditions (Steiger’s test, p < 0.05) [75].
Further analysis of the RNA types using HTSeq with the union model identified similar statistical analysis of the mapped reads (Table 1). Of all the reads that were mapped to RNAs, the majority of those reads, ranging from 88.74 to 89.42%, were mapped to protein coding regions, 1.71 to 2.03% to lncRNA, 3.59 to 4.76% to miscellaneous RNAs, 0.53 to 0.56% to processed transcripts, and 0.5 to 0.55% to antisense RNAs.
Table 1.
Distribution of mapped reads in different categories of RNAs in the six samples
| Sample_name | LTR_Zer1 | LTR_Zer2 | LTR_L1 | LTR_L2 | LTR_01 | LTR02 |
|---|---|---|---|---|---|---|
| 3prime_overlapping_ncrna | 159 (0.00%) | 180 (0.00%) | 160 (0.00%) | 171 (0.00%) | 180 (0.00%) | 160 (0.00%) |
| IG_C_gene | 0 (0.00%) | 2 (0.00%) | 1 (0.00%) | 0 (0.00%) | 3 (0.00%) | 1 (0.00%) |
| IG_C_pseudogene | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) |
| IG_D_gene | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) |
| IG_J_gene | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) |
| IG_J_pseudogene | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) |
| IG_V_gene | 4 (0.00%) | 2 (0.00%) | 1 (0.00%) | 3 (0.00%) | 1 (0.00%) | 4 (0.00%) |
| IG_V_pseudogene | 0 (0.00%) | 0 (0.00%) | 3 (0.00%) | 1 (0.00%) | 0 (0.00%) | 1 (0.00%) |
| Mt_rRNA | 1318 (0.00%) | 1488 (0.00%) | 1734 (0.00%) | 1496 (0.00%) | 1342 (0.00%) | 1779 (0.01%) |
| Mt_tRNA | 644 (0.00%) | 637 (0.00%) | 692 (0.00%) | 784 (0.00%) | 603 (0.00%) | 668 (0.00%) |
| TEC | 5415 (0.01%) | 5636 (0.01%) | 5036 (0.01%) | 5747 (0.01%) | 5685 (0.02%) | 5276 (0.02%) |
| TR_C_gene | 32 (0.00%) | 21 (0.00%) | 23 (0.00%) | 29 (0.00%) | 36 (0.00%) | 26 (0.00%) |
| TR_D_gene | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) |
| TR_J_gene | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) |
| TR_J_pseudogene | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) |
| TR_V_gene | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 1 (0.00%) | 0 (0.00%) | 0 (0.00%) |
| TR_V_pseudogene | 0 (0.00%) | 0 (0.00%) | 2 (0.00%) | 2 (0.00%) | 0 (0.00%) | 2 (0.00%) |
| antisense | 191,551 (0.52%) | 204,790 (0.53%) | 178,062 (0.50%) | 205,243 (0.50%) | 201,319 (0.54%) | 189,642 (0.55%) |
| known_ncrna | 0 (0.00%) | 0 (0.00%) | 1 (0.00%) | 0 (0.00%) | 1 (0.00%) | 1 (0.00%) |
| lincRNA | 738,731 (2.00%) | 761,611 (1.97%) | 706,213 (1.98%) | 702,871 (1.71%) | 742,207 (1.99%) | 702,377 (2.03%) |
| miRNA | 2479 (0.01%) | 2557 (0.01%) | 3497 (0.01%) | 3299 (0.01%) | 1525 (0.00%) | 1430 (0.00%) |
| misc_RNA | 1,612,667 (4.37%) | 1,593,547 (4.12%) | 1,627,962 (4.57%) | 1,960,500 (4.76%) | 1,343,420 (3.59%) | 1,244,791 (3.59%) |
| non_coding | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) |
| polymorphic_pseudogene | 319 (0.00%) | 355 (0.00%) | 320 (0.00%) | 369 (0.00%) | 333 (0.00%) | 312 (0.00%) |
| processed_pseudogene | 10,437 (0.03%) | 10,705 (0.03%) | 9812 (0.03%) | 11,241 (0.03%) | 10,275 (0.03%) | 8946 (0.03%) |
| processed_transcript | 196,988 (0.53%) | 213,355 (0.55%) | 194,373 (0.55%) | 229,395 (0.56%) | 203,313 (0.54%) | 192,191 (0.55%) |
| protein_coding | 32,728,357 | 34,372,393 | 31,562,319 | 36,554,719 | 33,423,746 | 30,949,051 |
| (88.74%) | (88.92%) | (88.65%) | (88.74%) | (89.42%) | (89.36%) | |
| pseudogene | 147 (0.00%) | 166 (0.00%) | 145 (0.00%) | 153 (0.00%) | 172 (0.00%) | 167 (0.00%) |
| rRNA | 32 (0.00%) | 44 (0.00%) | 38 (0.00%) | 62 (0.00%) | 46 (0.00%) | 44 (0.00%) |
| sense_intronic | 1015 (0.00%) | 1032 (0.00%) | 1070 (0.00%) | 1071 (0.00%) | 1035 (0.00%) | 1021 (0.00%) |
| sense_overlapping | 14,067 (0.04%) | 15,375 (0.04%) | 12,765 (0.04%) | 14,949 (0.04%) | 15,117 (0.04%) | 14,014 (0.04%) |
| snRNA | 3417 (0.01%) | 3214 (0.01%) | 2915 (0.01%) | 3858 (0.01%) | 3149 (0.01%) | 3236 (0.01%) |
| snoRNA | 160 (0.00%) | 172 (0.00%) | 136 (0.00%) | 149 (0.00%) | 197 (0.00%) | 170 (0.00%) |
| transcribed_processed_pseudogene | 25,420 (0.07%) | 26,038 (0.07%) | 24,532 (0.07%) | 28,196 (0.07%) | 25,315 (0.07%) | 23,318 (0.07%) |
| transcribed_unitary_pseudogene | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) |
| transcribed_unprocessed_pseudogene | 72,052 (0.20%) | 77,671 (0.20%) | 69,487 (0.20%) | 79,027 (0.19%) | 78,124 (0.21%) | 73,283 (0.21%) |
| translated_processed_pseudogene | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) |
| translated_unprocessed_pseudogene | 1 (0.00%) | 0 (0.00%) | 0 (0.00%) | 1 (0.00%) | 0 (0.00%) | 0 (0.00%) |
| unitary_pseudogene | 7892 (0.02%) | 8444 (0.02%) | 7147 (0.02%) | 8507 (0.02%) | 8233 (0.02%) | 7525 (0.02%) |
| unprocessed_pseudogene | 12,070 (0.03%) | 12,228 (0.03%) | 11,432 (0.03%) | 12,660 (0.03%) | 12,177 (0.03%) | 11,856 (0.03%) |
| Others | 1,257,539 (3.41%) | 1,342,963 (3.47%) | 1,185,376 (3.33%) | 1,370,001 (3.33%) | 1,299,971 (3.48%) | 1,201,876 (3.47%) |
Very similar expressions of lncRNAs among the three conditions
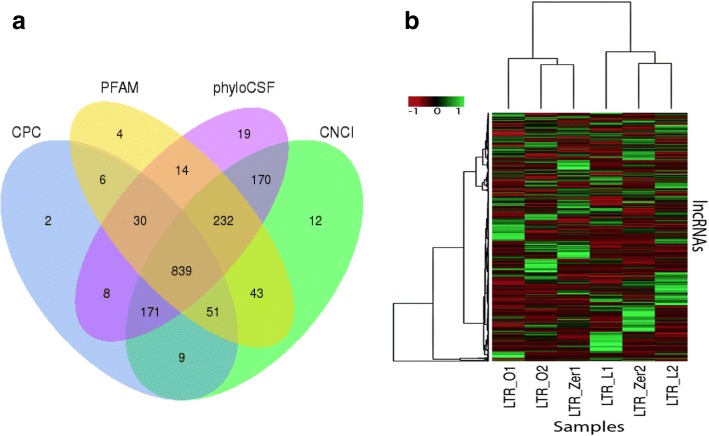
Altogether, 1615 transcripts were identified as candidate lncRNAs (see Additional file 2: Figures S2 and S4 for details). These candidate lncRNAs were then subjected to four coding potential prediction methods. A total of 839 lncRNAs were predicted by all the methods (Fig. 2a) and were therefore used in all the subsequent analyses.
Fig. 2.
No difference in the lncRNAs among the three experimental conditions. a Predicted lncRNAs based on four coding potential filtering methods. CPC, Coding-Potential Calculator; PFAM, Protein FAMily analysis; PhyloCSF, Phylogenetic Codon Substitution Frequency; CNCI, Coding-Non-Coding Index. b Expression level distribution of the 839 lncRNAs in the six samples (FPKM values are z-score normalized)
As shown in Fig. 2b, there was no clear clustering of samples from the same condition: LTR_L2 showed higher similarity to LTR_Zer2 than to LTR_L1, and LTR_O2 showed higher similarity to LTR_Zer1 than to LTR_O1. Among the 839 lncRNAs, 38 were identified to be differentially expressed for the L vs. Zero comparison at a p-value < 0.05, but none remained significant for the adjusted p-values controlling the false discovery rate (FDR) at 0.10 due to multiple testing. 40 lncRNAs were differentially expressed for the O vs. Zero comparison at p-value < 0.05, but only one lncRNA, TERC, remained statistically significant for the adjusted p-values; 53 were differentially expressed for the L vs. O comparison, but only two lncRNAs, TERC and SCARNA2, remained significant for the adjusted p-values. Interestingly, the lncRNA TERC showed differential expression levels for all pairwise comparisons of the three conditions (albeit not significant for the L vs. Zero comparison at the adjusted p-value), with the highest expression level under condition L, > 2-fold increase compared to condition O, and a 1.5-fold increase compared to the control (LTR_Zero). The lncRNA SCARNA2 showed the lowest expression level under condition O, followed by increased expression for the control condition (~ 1.4 fold), and condition L (~ 1.7 fold).
Differentially expressed mRNAs
Altogether, 142,791 mRNAs were compared for differential expression among groups. With a false discovery rate of 0.10, four genes (DSC3, EGF, TRIM26, FHDC1, see Additional file 1: Table S5) were differentially expressed between the L and Zero samples, 24 genes were differentially expressed between the O and Zero samples (Additional file 1: Table S5), and 63 genes were differentially expressed between the L and O samples (Additional file 1: Table S5). Gene Ontology analysis revealed no statistically significant enrichment of any specific categories (results not shown). Comparison of the genes across these three lists of differentially expressed genes for the three pairwise comparisons showed that only one gene, TRIM26, was more robustly down regulated in the L samples (FPKM = ~ 1.4) than in both the O (FPKM = ~ 4.5) and Zero (FPKM = ~ 3.9) samples (all pairwise comparisons are statistically significant). REPS2 was significantly upregulated in both the O and L samples compared to the Zero control, but only showed a statistical significance in the O vs. Zero sample comparison for the adjusted p-value; in the L vs. Zero sample comparison, although the p-value was significant, the adjusted p-value was not. There were 21 genes differentially expressed in the O samples compared with either the L or Zero samples (but not between the L and Zero samples, Table 2). Interestingly, all these 21 genes were significantly downregulated in the O samples as compared to those in both the L and Zero samples. Also interesting was that one third of these genes were histone related: HIST1H2AB, HIST1H2AD, HIST1H2AM, HIST1H4J, HIST2H2AC, HIST2H2BF, HIST2H3D. This result suggestsed that there were no apparent upregulated changes from Zero to LTR_L in all mRNA transcripts. However, LTR_O significantly downregulated some genes. Since the dCas9-SAM was expected to activate the mRNA expression of any potential off-target genes, these downregulated genes might not be directly related to the action of the dCas9-SAM activation system. However, these downregulated genes were specific for the msgRNA LTR_O, and histone-related genes were the most striking, perhaps implying that LTR_O-mediated LTR transcription activation may exhaust some histone proteins. It was unlikely that LTR_O induced direct suppression of several histone genes, unless the enriched transcriptional activator (VP64, p65, HSF1) by the dCas9-SAM via LTR_O msgRNA might suppress histone genes by interacting with their transcriptional complex. It was also possible that LTR_O affected some genes such as TERC and REPS2 that might negatively regulate the expression of these histone genes.
Table 2.
21 genes that are significantly downregulated in the O samples as compared to the Zero and L samples
| Genes | LTR_O_FPKM | LTR_Zero_FPKM | log2(fold) | LTR_L_FPKM | log2(fold) |
|---|---|---|---|---|---|
| HNRNPAB | 6.55 | 38.66 | − 2.56 | 43.94 | −2.75 |
| PTP4A2 | 3.62 | 20.44 | −2.50 | 23.02 | −2.67 |
| B4GALT2 | 2.07 | 6.02 | −1.54 | 6.07 | −1.55 |
| C4orf48 | 4.99 | 11.81 | −1.24 | 11.15 | −1.16 |
| TPGS1 | 3.36 | 7.16 | −1.09 | 8.80 | −1.39 |
| HPCAL1 | 4.01 | 8.33 | −1.05 | 8.39 | −1.06 |
| SLBP | 10.55 | 20.53 | −0.96 | 20.64 | −0.97 |
| CITED4 | 3.56 | 6.79 | −0.93 | 7.66 | −1.11 |
| HIST2H2BF | 97.90 | 175.32 | − 0.84 | 176.52 | − 0.85 |
| TMEM160 | 8.40 | 14.67 | −0.80 | 16.45 | −0.97 |
| HIST2H2AC | 444.08 | 750.59 | −0.76 | 850.05 | −0.94 |
| C17orf89 | 57.82 | 95.66 | −0.73 | 109.36 | −0.92 |
| IER5L | 6.61 | 10.87 | −0.72 | 13.23 | −1.00 |
| CEBPD | 16.45 | 26.54 | −0.69 | 29.57 | −0.85 |
| HIST2H3D | 248.87 | 400.31 | −0.69 | 425.10 | −0.77 |
| HIST1H2AB | 336.25 | 536.04 | −0.67 | 587.46 | −0.80 |
| HIST1H2AM | 468.32 | 743.47 | −0.67 | 809.48 | −0.79 |
| MIF | 128.97 | 200.45 | −0.64 | 247.69 | −0.94 |
| HIST1H4J | 1102.07 | 1656.42 | −0.59 | 1819.34 | −0.72 |
| CYBA | 179.61 | 268.40 | −0.58 | 308.07 | −0.78 |
| HIST1H2AD | 722.53 | 1057.74 | −0.55 | 1177.36 | −0.70 |
SNP and indel analysis
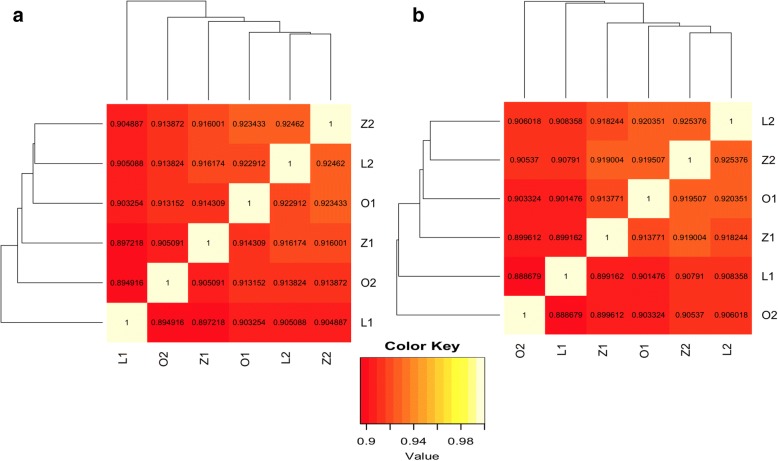
To examine whether the dCas9-SAM epigenome editing had an effect on the rate of genetic mutations, SNPs and indel variants in all the samples were identified using GATK2 [70]. Totally, there were 733,334 SNPs and 36,715 indels identified in the six samples. The Jaccard index was computed for each pair of samples where the number of reads that supported the called SNPs and indels was greater than or equal to 20. Figure 3 showed the Jaccard index matrix and clustering result of the six samples for both SNPs and indels. The Jaccard index was high for all sample comparisons, ranging from 0.895 (O2 vs. L1) to 0.925 (Z2 vs. L2) for SNPs, and from 0.889 (O2 vs. L1) to 0.925 (Z2 vs. L2) for indels. The clustering result revealed no clear grouping within the same conditions (that is, L samples grouped together, O samples grouped together, or control samples grouped together), suggesting that there were no systematic differences in SNP and indel variations between different editing conditions.
Fig. 3.
Hierarchical clustering of the six samples based on the Jaccard index for SNPs (a) and indels (b)
Very similar distribution of alternative splicing events among the three groups
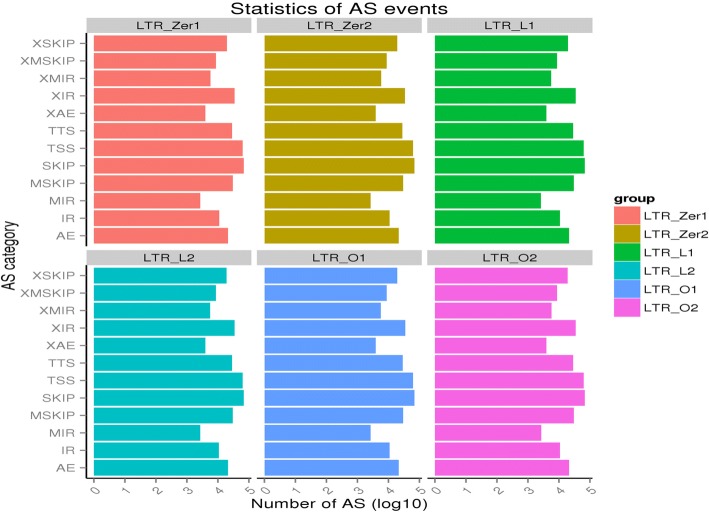
Alternative splicing is an important means for increasing the diversity of transcripts and proteins. In fact, a majority of mammalian genes have around 2~ 12 mRNA isoforms, with some having a few thousand isoforms [76]. Therefore, characterizing the off-target effects of dCas9 epigenome editing is incomplete without considering how alternative splicing might be affected among different groups as compared to the control. To investigate in detail how isoforms or exons might be affected, alternative splicing events were first classified into 12 types as illustrated in Additional file 2: Figure S4 using ASprofile [71]. The number of each type of alternative splicing event for the six samples was shown in Fig. 4 (also see Additional file 1: Table S6). The total number of alternative splicing events ranged from 297,334 to 298,098 with the two LTR_O samples (O1: 298, 098; O2: 297,999) having the highest number of alternative splicing events, followed by LTR_Zer2 (297,789), LTR_L2 (297,763), LTR_Zer1 (297,580), and LTR_L1 (297,334). The distribution of different types of alternative splicing was very similar among the six samples, and there was no significant difference either within or between groups (all the pairwise Chi-square tests’ p-values are greater than 0.98).
Fig. 4.
Summary statistics of the 12 types of alternative splicing in the six samples. The number of events for each type is log10 transformed
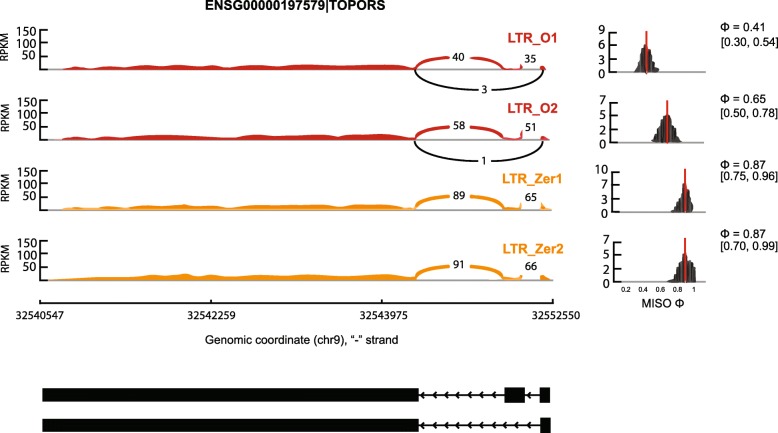
To further examine whether isoforms produced by alternative splicing differed in expression level among the three groups, the MISO (mixture-of-isoforms) model [72] was used to determine the isoforms that differentiate the groups. MISO uses a Bayesian statistical model to estimate the expression level of different isoforms/exons and identifies differentially regulated isoforms by the Bayes factor (BF) that calculates the odds of differential regulation of isoforms or exons. Five major types of alternative splicing events, alternative 3′ splice sites (A3SS), alternative 5′ splice sites (A5SS), mutually exclusive exons (MXE), retained intron (RI), and skipped exon (SE), were analyzed and compared among the three groups. Table 3 showed the genes that exhibited significant differential isoform regulation between the group comparisons. Figure 5 showed an example of the TOPORS gene exhibiting significant differential exon skipping in LTR_O samples compared to the Zero samples. Altogether, there were not many differential isoform regulations between the groups. For example, of the 7244 A3SS events compared between the L samples and Zero samples, only seven (< 0.1%) had significant differential isoform regulation. In fact, the percentage of significant differential isoform regulations between groups for the three pairwise comparisons (L vs. Zero, O vs. Zero, L vs. O) ranged from 0.097 to 0.111% for A3SS, from 0.130 to 0.2% for A5SS, from 0.180 to 0.181% for MXE, from 0.122 to 0.197% for RI, and from 0.081 to 0.112% for SE. Taken together, less than 0.2% of the alternative splicing events considered showed differential isoform regulations between the groups, suggesting no genome-wide systematic alternative splicing changes occurred due to the dCas9 editing. Moreover, comparison of the list of genes with differential isoform regulation to the list of differentially expressed genes (Additional file 1: Table S5) showed that only DSC3 had differential exon regulation between the L and Zero samples, and DSC3 was also significantly downregulated in the L samples compared to the Zero samples.
Table 3.
Comparison of differential isoform regulation between the three groups. The genes in bold font are those shared by two pairwise comparisons. The numbers in parenthesis are the number of events considered for the particular group comparison
| AS types | L vs. Zero | O vs. Zero | L vs. O |
|---|---|---|---|
| A3SS (# of events) | (7244) | (7143) | (7239) |
| C8orf22 | ANKRD11 | BMP1 | |
| CLSPN | C11orf48 | NFAT5 | |
| COBLL1 | CNOT2 | OCEL1 | |
| DAK | GNB2L1 | ORMDL1 | |
| JOSD1 | SETMAR | ST5 | |
| OCEL1 | YWHAB | ZNF84 | |
| PIH1D1 | ZNF587 | ||
| A5SS (# of events) | (5399) | (5350) | (5407) |
| C17orf70 | ANGPT1 | HILPDA | |
| MTMR2 | CLEC2D | NBPF11 | |
| NOC2L | LAMA4 | NDUFV2 | |
| RP5-1198O20.4 | NAA60 | NT5C | |
| SMARCC2 | SLC50A1 | OXLD1 | |
| TWF1 | TYSND1 | RP4-583P15.15 | |
| ZNF30 | SRRM1 | ||
| TBC1D7 | |||
| TYSND1 | |||
| VPS52 | |||
| chr1:32336239:32335947 | |||
| MXE (# of events) | (4959) | (4946) | (5006) |
| CNOT1 | CNOT1 | AKIRIN1 | |
| DBNL | EIF4G2 | DDHD2 | |
| DPY30 | HMGN1 | DEK | |
| MPPE1 | PTRH1 | DPP3 | |
| PLA2G6 | RPS6KC1 | ELMOD3 | |
| SPDL1 | TMBIM4 | MPV17L | |
| TMBIM6 | TMEM116 | RPS6KC1 | |
| UQCC1 | chr7:143284899:143284974:+@chr7:143285348 | TCTN1 | |
| WBP1 | TMEM116 | ||
| RI (# of events) | (4109) | (4057) | (4084) |
| CENPV | BAX | BAX | |
| MRRF | CAPRIN2 | FANCI | |
| RP11-5A19.5 | CDK5RAP3 | HSD17B4 | |
| RPRD2 | CENPV | MTA1 | |
| SMTN | GPS2 | TAB3 | |
| IMPDH2 | |||
| QARS | |||
| SERAC1 | |||
| SE (# of events) | (25,942) | (25,835) | (25,969) |
| C2CD5 | AC013394.2 | AC013394.2 | |
| CDC42BPA | AGPAT2 | AC124789.1 | |
| DCTD | ATG7 | ARID1B | |
| DSC3 | B3GALNT2 | ATG10 | |
| GRB10 | BCL2L12 | ATG7 | |
| HMGN1 | CENPU | B3GALNT2 | |
| KCTD17 | CMTR2 | BBS1 | |
| LINC00570 | GABPB2 | BCL2L12 | |
| MIPOL1 | HMGN1 | BTBD7 | |
| MRPL52 | IMMP1L | CD320 | |
| NCSTN | KDM6A | CD59 | |
| NUMB | KLHL5 | DCTD | |
| PDE4DIP | MAPK9 | LINC00472 | |
| PXK | MRPL52 | MAPK9 | |
| RAB40B | PTK2 | MIR4435-1HG | |
| SCMH1 | SETD8 | PTK2 | |
| SPATA20 | SMURF2P1 | RHBDD1 | |
| TMEM139 | SP3 | RP4-717I23.3 | |
| TTC23 | TBL1XR1 | RPS6KB2 | |
| ZNF138 | TINF2 | SP3 | |
| ZSCAN21 | TMEM139 | ST20-MTHFS | |
| TMEM189 | TBL1XR1 | ||
| TOPORS | TOPORS | ||
| TRIP6 | UBE2I | ||
| UBE2I | YDJC | ||
| VWA9 | ZNF639 | ||
| ZNF584 | ZNF678 | ||
| chr7:143284899:143284974 |
Fig. 5.
The sashimi plot showing exon skipping in TOPORS that exhibits significant differential regulation between the LTR_O group and the control group. The top left panel shows the FPKM of reads that supports the corresponding exons and exon junctions in the two LTR_O samples and two control samples, respectively. The top right panel shows the posterior distribution of Ψ (the fraction of inclusive isoform), with the red line denoting the estimated Ψ and grey lines the 95% confidence interval of Ψ. The bottom panel shows the two transcripts due to exon skipping in the bottom transcript
Discussion
Determining off-target effects from CRISPR/Cas9-based genome editing in a thorough and highly sensitive manner has been a great challenge in the field [6, 77–79]. Apart from ongoing extensive work in optimizing the technology to minimize off-target cleavage [39, 80–82], serious effort has also been devoted to examining the off-target effects resulting in changes at the levels of genomes and transcriptomes [50, 52, 83–89]. In particular, the specificity of the dCas9-SAM system itself has been validated by mRNA-seq analysis [17, 28, 30], although the dCas9-VP160 alone (in the absence of sgRNA) has been shown to reactivate latent HIV-1 in U1 cells [90]. Here, deep sequencing of transcriptomes of human cells after epigenome (transcriptional) editing by HIV-specific msgRNA/dCas9-SAM was performed, and a comprehensive analysis was done to examine any potential off-target effects of the HIV-targeted msgRNA/dCas9-SAM on the mRNA transcription, lncRNA expression, alternative splicing, as well as genetic mutations including SNPs and indels.
Off-target effect on the overall mRNA expression level
In terms of mRNA expression, if there were significant off-target effects, many genes would be upregulated in the O and L samples compared to the control group (the genes that are upregulated could differ between the O and L samples), but only a handful of the host genes showed significant difference, most of which were actually downregulated (Additional file 1: Table S5). Specifically, of the 28 genes showing a statistically significant difference, only two, HDGF and REPS2, were significantly upregulated in the O samples compared to the control group. Four genes were found differentially expressed in the L group vs. Zero group comparison, but all of them were downregulated in the L group compared to the Zero group (the control group). It is puzzling that most of the differentially expressed genes were significantly downregulated in the dCas9-SAM editing system (O and L samples) compared to the control group. This phenomenon has not yet been reported anywhere in the literature.
The 12~ 14-bp target sequence near the protospacer-adjacent motif (PAM) region (NGG) is critical for the specificity of Cas9 genome editing [91, 92]. In silico off-target effect prediction for LTR_L and LTR_O was done by blasting > 14-bp target + NGG against the human genome/transcripts as we described previously [20, 21, 23], then comparing the list of potential off-target gene locations with the genes identified in Additional file 1: Table S5. There is no overlap between the two lists, suggesting that genes that show significant expression difference between the two dCas9-SAM edited groups and the control group may not be the direct result of the potential off-target effect.
Off-target effects on alternative splicing
Comparison with 12 types of alternative splicing events reveals no statistically significant differences between the edited groups (L and O) and the control group (Fig. 4). Moreover, a detailed expression analysis of isoforms caused by five major types of alternative splicing shows only a small number of differential isoform regulations between groups (< 0.2%, Table 3), further suggesting that there are no pronounced genome-wide alternative splicing changes occurring due to the dCas9-SAM editing. DSC3 is the only gene that shows both significant differential exon regulation and expression level differences between the edited group (L) and the control group, but contrary to expectations, is significantly downregulated. Previous studies show about 47~ 74% of alterative splicing events show variation among different human tissues and 10~ 30% of alternative splicing events show variation among individuals [93]. Therefore, comparatively, the level of variation in alternative splicing detected among the three groups (L, O, and control) is 2~ 3 orders of magnitude lower. Although the level of genetic variation among the samples is also lower (less than one order of magnitude, see results on SNPs and indel comparison), these comparisons nonetheless suggest that the off-target effect due to the dCas9 epigenome editing does not include any noticeable changes at the genome-wide alternative splicing level. Since alternative splicing is an important mechanism for increasing transcript and protein diversity [76, 94], and fine-tuning gene expression and function, any off-target effect caused by dCas9 editing could conceivably create undesirable consequences that in turn limit dCas9 usage. The current finding is thus very encouraging for the safe application of dCas9 epigenome editing to reactivate the silent HIVs for their ultimate elimination.
Off-target effect on lncRNAs
Long noncoding RNA (lncRNA), transcripts longer than 200 nucleotides that cannot be translated into proteins, are derived from 70~ 90% of the mammalian genome while mRNAs are transcribed from only 1% of the genome [95]. These lncRNAs have been shown to play important regulatory roles in chromatin reprogramming and pre- and post-mRNA processing [96–98]. Therefore, any off-target effects on lncRNA expression is also important to consider. Using the pipeline shown in Additional file 2: Figure S2b, 839 lncRNAs (Fig. 2a) were identified in the transcripts and their expression compared in six samples. Results (Fig. 2b) reveal no clear clustering of samples within the same groups and no clear separation among groups. There is no significant lncRNA expression difference between the L group and the control group. Only one lncRNA, TERC, is significantly downregulated in the O samples compared to the control samples. In fact, TERC has the highest expression level under condition L, followed by the control condition, and then condition O. This expression difference does not seem to be directly linked to any off-target effect, as one would expect TERC lncRNA to have higher expressions in both edited groups (O and L groups) compared to the control group. The observation for lncRNA expression is similar to the observation for mRNA expression, because the handful of mRNAs and lncRNAs tend to be downregulated, contrary to an expectation of elevated expressions in the edited groups due to the potential off-target transcriptional activation effect. It is therefore concluded that there is little, if any, detectable off-target effects on lncRNA transcription. As more studies have shown the involvement of lncRNAs in various diseases and cancer [99–102], our current finding is reassuring, and further supports the safe application of dCas9-SAM epigenome editing. Note that the current finding does not preclude the possibility that the off-target effects could upregulate some unknown genetic elements/factors, which in turn suppress/reduce the expression of the mRNA and lncRNAs identified in the current study.
Off-target effect on SNPs and indels
Off-target-induced mutations are also another important consideration for the safe application of dCas9-SAM system in clinical settings. Although dCas9 itself does not induce indels or SNPs directly due to its lack of endonuclease activity, it is possible that the dCas9-SAM system induces indels indirectly through potential off-target effects on some mutagenic genes. Results (Fig. 3) comparing both SNPs and indels in the six samples did not show any significant off-target effects. Although previous studies have shown that RNA-guided endonuclease mediated genome editing can induce off-target indel mutations [92, 103–106], numerous studies have also shown that off-target mutations can be effectively reduced and possibly eliminated by careful selection of unique target sequences and guide RNA and Cas9 variant optimization [107]. One cautionary note is that since SNPs and indels were identified using RNA-seq data, the current study cannot address whether there is any significant mutagenic effect due to the dCas9 epigenome editing in non-transcribed regions.
Conclusion
To the authors’ knowledge, this study is the most comprehensive and exhaustive characterization of the off-target effects on transcriptomes after HIV-targeted dCas9-SAM epigenome editing. Analysis of known types of RNAs reveals no significant difference between transcriptomes of HIV-targeted and non-targeted msgRNA-treated human cells, supporting the contention that msgRNA-directed dCas9-based SAM technology can be safely used to reactivate dormant HIV for an effective “shock-and-kill” strategy to finally eliminate the virus [108]. One caveat with the current study is that there were only two replicates for each group, which limits the statistical power of the study. Future work needs to include more replicates. Additionally, further assessment of the potential off-target effects with the dCas9-SAM system in human primary cells and preclinical animal models is warranted.
Additional files
Table S1. Statistics of RNA-Seq quality reads. Table S2. Mapping results. Table S3. Validation of dCas9-SAM mRNA and sgRNA expression (transcripts per million). Table S4. Distribution of reads in known types of RNAs. Table S5. Differentially expressed mRNA transcripts for all the three pairwise comparisons of the samples (O vs Zero, L vs Zero, and O vs L). Table S6. Distribution of the 12 types of alternative splicing events across samples. (XLSX 30 kb)
Figure S1. Distributions of the mapped reads in the genome for the six samples. Figure S2. Workflow charts for RNA-seq analysis. (a) Library construction. (b) lncRNA filtering by four pipelines to predict candidate lncRNAs based on their structures and noncoding features. Figure S3. Statistics of lncRNA filtering. Horizontal axis represents the filtering step and vertical axis represents the number of remaining transcripts after the filtering step. Figure S4. Illustration of 12 types of alternative splicing events analyzed by ASprofile (Picture taken from Florea L, Song L, Salzberg SL: Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Res 2013, 2:188). (PDF 2000 kb)
Acknowledgements
We acknowledge Dr. Xiaoxue Jiang and Dr. Wenjie Wei for bioinformatics analysis.
Funding
This work was supported by National Institutes of Health (R01NS087971 and R01DK075964 to W.H. and VT open access subvention fund to L.Z.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available from the sequence read archive (SRA): https://www.ncbi.nlm.nih.gov/bioproject/PRJNA477008 and the raw data: https://www.ncbi.nlm.nih.gov/Traces/study/?acc=SRP150955.
Abbreviations
- A3SS
Alternative 3′ splice sites
- AS
Alternative splicing
- BF
Bayes factor
- dCas9
dead CRISPR-associated protein 9
- FDR
False discovery rate
- FPKM
Fragments per kilobase of transcript per million mapped reads
- lncRNA
long noncoding RNA
- LTR
Long terminal repeat
- MISO
Mixture of isoforms
- msgRNA
MS2-mediated single guide RNA
- MXE
Mutually exclusive exons
- PAM
Protospacer-adjacent motif
- RI
Retained intron
- SAM
Synergistic activation mediator
- SE
Skipped exon
- SNP
Single-nucleotide polymorphism
Authors’ contributions
WH, YZ, LZ and HW conceived and designed the experiments. YZ, FL, XX, RP and JY performed the experiments, acquired/discussed the data and reviewed/edited the manuscript. GA, WH, LZ, LTW and XY analyzed/interpreted the data, prepared figures and extensively edited the manuscript. WH, LZ, YZ, HW and LTW supervised the study, drafted and revised the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yonggang Zhang, Email: biozyg@163.com.
Gustavo Arango, Email: gustavo1@vt.edu.
Fang Li, Email: flihu@temple.edu.
Xiao Xiao, Email: ken_023@126.com.
Raj Putatunda, Email: tuf26817@temple.edu.
Jun Yu, Email: jun.yu@temple.edu.
Xiao-Feng Yang, Email: xiao-feng.yang@temple.edu.
Hong Wang, Email: hong.wang@temple.edu.
Layne T. Watson, Email: ltw@vt.edu
Liqing Zhang, Phone: 540-231-9413, Email: lqzhang@cs.vt.edu.
Wenhui Hu, Phone: 1-215-707-5164, Email: whu@temple.edu.
References
- 1.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32(4):347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vasileva EA, Shuvalov OU, Garabadgiu AV, Melino G, Barlev NA. Genome-editing tools for stem cell biology. Cell Death Dis. 2015;6:e1831. doi: 10.1038/cddis.2015.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanchez-Rivera FJ, Jacks T. Applications of the CRISPR-Cas9 system in cancer biology. Nat Rev Cancer. 2015;15(7):387–395. doi: 10.1038/nrc3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riordan SM, Heruth DP, Zhang LQ, Ye SQ. Application of CRISPR/Cas9 for biomedical discoveries. Cell Bioscience. 2015;5:33. doi: 10.1186/s13578-015-0027-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saayman S, Ali SA, Morris KV, Weinberg MS. The therapeutic application of CRISPR/Cas9 technologies for HIV. Expert Opin Biol Ther. 2015;15(6):819–830. doi: 10.1517/14712598.2015.1036736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thakore PI, Black JB, Hilton IB, Gersbach CA. Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nat Methods. 2016;13(2):127–137. doi: 10.1038/nmeth.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agne M, Blank I, Emhardt AJ, Gabelein CG, Gawlas F, Gillich N, Gonschorek P, Juretschke TJ, Kramer SD, Louis N, et al. Modularized CRISPR/dCas9 effector toolkit for target-specific gene regulation. ACS Synth Biol. 2014;3(12):986–989. doi: 10.1021/sb500035y. [DOI] [PubMed] [Google Scholar]
- 9.Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013;10(10):977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154(2):442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng AW, Wang H, Yang H, Shi L, Katz Y, Theunissen TW, Rangarajan S, Shivalila CS, Dadon DB, Jaenisch R. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23(10):1163–1171. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amabile A, Migliara A, Capasso P, Biffi M, Cittaro D, Naldini L, Lombardo A. Inheritable silencing of endogenous genes by hit-and-run targeted epigenetic editing. Cell. 2016;167(1):219–232. doi: 10.1016/j.cell.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chavez A, Tuttle M, Pruitt BW, Ewen-Campen B, Chari R, Ter-Ovanesyan D, Haque SJ, Cecchi RJ, Kowal EJ, Buchthal J, et al. Comparison of Cas9 activators in multiple species. Nat Methods. 2016;13(7):563–567. doi: 10.1038/nmeth.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, E PRI, Lin S, Kiani S, Guzman CD, Wiegand DJ, et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 2015;12(4):326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Black JB, Adler AF, Wang HG, D'Ippolito AM, Hutchinson HA, Reddy TE, Pitt GS, Leong KW, Gersbach CA. Targeted epigenetic remodeling of endogenous loci by CRISPR/Cas9-based transcriptional activators directly converts fibroblasts to neuronal cells. Cell Stem Cell. 2016;19(3):406–414. doi: 10.1016/j.stem.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thakore PI, D'Ippolito AM, Song L, Safi A, Shivakumar NK, Kabadi AM, Reddy TE, Crawford GE, Gersbach CA. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Methods. 2015;12(12):1143–1149. doi: 10.1038/nmeth.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hilton IB, D'Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33(5):510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013;10(10):973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu W, Kaminski R, Yang F, Zhang Y, Cosentino L, Li F, Luo B, Alvarez-Carbonell D, Garcia-Mesa Y, Karn J, et al. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc Natl Acad Sci U S A. 2014;111(31):11461–11466. doi: 10.1073/pnas.1405186111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin C, Zhang T, Li F, Yang F, Putatunda R, Young WB, Khalili K, Hu W, Zhang Y. Functional screening of guide RNAs targeting the regulatory and structural HIV-1 viral genome for a cure of AIDS. AIDS. 2016;30(8):1163–1174. doi: 10.1097/QAD.0000000000001079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaminski R, Chen Y, Salkind J, Bella R, Young WB, Ferrante P, Karn J, Malcolm T, Hu W, Khalili K. Negative feedback regulation of HIV-1 by gene editing strategy. Sci Rep. 2016;6:31527. doi: 10.1038/srep31527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaminski R, Chen Y, Fischer T, Tedaldi E, Napoli A, Zhang Y, Karn J, Hu W, Khalili K. Elimination of HIV-1 genomes from human T-lymphoid cells by CRISPR/Cas9 gene editing. Sci Rep. 2016;6:22555. doi: 10.1038/srep22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaminski R, Bella R, Yin C, Otte J, Ferrante P, Gendelman HE, Li H, Booze R, Gordon J, Hu W, et al. Excision of HIV-1 DNA by gene editing: a proof-of-concept in vivo study. Gene Ther. 2016;23(8–9):690–695. doi: 10.1038/gt.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin C, Zhang T, Qu X, Zhang Y, Putatunda R, Xiao X, Li F, Xiao W, Zhao H, Dai S, et al. In Vivo Excision of HIV-1 Provirus by saCas9 and Multiplex Single-Guide RNAs in Animal Models. Mol Ther. 2017;25:1168–1186. doi: 10.1016/j.ymthe.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bialek JK, Dunay GA, Voges M, Schafer C, Spohn M, Stucka R, Hauber J, Lange UC. Targeted HIV-1 latency reversal using CRISPR/Cas9-derived transcriptional activator systems. PLoS One. 2016;11(6):e0158294. doi: 10.1371/journal.pone.0158294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Limsirichai P, Gaj T, Schaffer DV. CRISPR-mediated activation of latent HIV-1 expression. Mol Ther. 2016;24(3):499–507. doi: 10.1038/mt.2015.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saayman SM, Lazar DC, Scott TA, Hart JR, Takahashi M, Burnett JC, Planelles V, Morris KV, Weinberg MS. Potent and targeted activation of latent HIV-1 using the CRISPR/dCas9 activator complex. Mol Ther. 2016;24(3):488–498. doi: 10.1038/mt.2015.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji H, Jiang Z, Lu P, Ma L, Li C, Pan H, Fu Z, Qu X, Wang P, Deng J, et al. Specific reactivation of latent HIV-1 by dCas9-SunTag-VP64-mediated guide RNA targeting the HIV-1 promoter. Mol Ther. 2016;24(3):508–521. doi: 10.1038/mt.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517(7536):583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Yin C, Zhang T, Li F, Yang W, Kaminski R, Fagan PR, Putatunda R, Young WB, Khalili K, et al. CRISPR/gRNA-directed synergistic activation mediator (SAM) induces specific, persistent and robust reactivation of the HIV-1 latent reservoirs. Sci Rep. 2015;5:16277. doi: 10.1038/srep16277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bogerd HP, Kornepati AV, Marshall JB, Kennedy EM, Cullen BR. Specific induction of endogenous viral restriction factors using CRISPR/Cas-derived transcriptional activators. Proc Natl Acad Sci U S A. 2015;112(52):E7249–E7256. doi: 10.1073/pnas.1516305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA, Jaenisch R. Editing DNA methylation in the mammalian genome. Cell. 2016;167(1):233–247. doi: 10.1016/j.cell.2016.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choudhury SR, Cui Y, Lubecka K, Stefanska B, Irudayaraj J. CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter. Oncotarget. 2016;7:46545–46556. doi: 10.18632/oncotarget.10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDonald JI, Celik H, Rois LE, Fishberger G, Fowler T, Rees R, Kramer A, Martens A, Edwards JR, Challen GA. Reprogrammable CRISPR/Cas9-based system for inducing site-specific DNA methylation. Biol Open. 2016;5(6):866–874. doi: 10.1242/bio.019067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kungulovski G, Jeltsch A. Epigenome editing: state of the art, concepts, and perspectives. Trends Genet. 2016;32(2):101–113. doi: 10.1016/j.tig.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 37.Wolt JD, Wang K, Sashital D, Lawrence-Dill CJ. Achieving plant CRISPR targeting that limits off-target effects. Plant Genome. 2016;9(3) [DOI] [PubMed]
- 38.Ma J, Koster J, Qin Q, Hu S, Li W, Chen C, Cao Q, Wang J, Mei S, Liu Q, et al. CRISPR-DO for genome-wide CRISPR design and optimization. Bioinformatics. 2016;32(21):3336–3338. doi: 10.1093/bioinformatics/btw476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chari R, Yeo NC, Chavez A, Church GM. sgRNA scorer 2.0: a species-independent model to predict CRISPR/Cas9 activity. ACS Synth Biol. 2017;6:902–904. doi: 10.1021/acssynbio.6b00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cradick TJ, Qiu P, Lee CM, Fine EJ, Bao G. COSMID: a web-based tool for identifying and validating CRISPR/Cas off-target sites. Mol Ther Nucleic Acids. 2014;3:e214. doi: 10.1038/mtna.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y, Liu KI, Sutrisnoh NB, Srinivasan H, Zhang J, Li J, Zhang F, Lalith CRJ, Xing H, Shanmugam R, et al. Systematic evaluation of CRISPR-Cas systems reveals design principles for genome editing in human cells. Genome Biol. 2018;19(1):62. doi: 10.1186/s13059-018-1445-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bae S, Park J, Kim JS. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30(10):1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai SQ, Joung JK. Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nat Rev Genet. 2016;17(5):300–312. doi: 10.1038/nrg.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zuckermann M, Hovestadt V, Knobbe-Thomsen CB, Zapatka M, Northcott PA, Schramm K, Belic J, Jones DT, Tschida B, Moriarity B, et al. Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nat Commun. 2015;6:7391. doi: 10.1038/ncomms8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith C, Gore A, Yan W, Abalde-Atristain L, Li Z, He C, Wang Y, Brodsky RA, Zhang K, Cheng L, et al. Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell. 2014;15(1):12–13. doi: 10.1016/j.stem.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Veres A, Gosis BS, Ding Q, Collins R, Ragavendran A, Brand H, Erdin S, Cowan CA, Talkowski ME, Musunuru K. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell. 2014;15(1):27–30. doi: 10.1016/j.stem.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang L, Grishin D, Wang G, Aach J, Zhang CZ, Chari R, Homsy J, Cai X, Zhao Y, Fan JB, et al. Targeted and genome-wide sequencing reveal single nucleotide variations impacting specificity of Cas9 in human stem cells. Nat Commun. 2014;5:5507. doi: 10.1038/ncomms6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sung K, Park J, Kim Y, Lee NK, Kim SK. Target specificity of Cas9 nuclease via DNA rearrangement regulated by the REC2 domain. J Am Chem Soc. 2018;140:7778–7781. doi: 10.1021/jacs.8b03102. [DOI] [PubMed] [Google Scholar]
- 49.Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520(7546):186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33(2):187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frock RL, Hu J, Meyers RM, Ho YJ, Kii E, Alt FW. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol. 2015;33(2):179–186. doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martin F, Sanchez-Hernandez S, Gutierrez-Guerrero A, Pinedo-Gomez J, Benabdellah K. Biased and unbiased methods for the detection of off-target cleavage by CRISPR/Cas9: an overview. Int J Mol Sci. 2016;17(9):1507. [DOI] [PMC free article] [PubMed]
- 53.Shi L, Tang X, Tang G. GUIDE-Seq to detect genome-wide double-stranded breaks in plants. Trends Plant Sci. 2016;21(10):815–818. doi: 10.1016/j.tplants.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 54.Cho GY, Schaefer KA, Bassuk AG, Tsang SH, Mahajan VB. Crispr Genome Surgery in the Retina in Light of Off-Targeting. Retina. 2018;38:1443–1455. doi: 10.1097/IAE.0000000000002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hay EA, Khalaf AR, Marini P, Brown A, Heath K, Sheppard D, MacKenzie A. An analysis of possible off target effects following CAS9/CRISPR targeted deletions of neuropeptide gene enhancers from the mouse genome. Neuropeptides. 2017;64:101–7. [DOI] [PMC free article] [PubMed]
- 56.Cao J, Wu L, Zhang SM, Lu M, Cheung WK, Cai W, Gale M, Xu Q, Yan Q. An easy and efficient inducible CRISPR/Cas9 platform with improved specificity for multiple gene targeting. Nucleic Acids Res. 2016;44(19):e149. doi: 10.1093/nar/gkw660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boyle EA, Andreasson JOL, Chircus LM, Sternberg SH, Wu MJ, Guegler CK, Doudna JA, Greenleaf WJ. High-throughput biochemical profiling reveals sequence determinants of dCas9 off-target binding and unbinding. Proc Natl Acad Sci U S A. 2017;114(21):5461–5466. doi: 10.1073/pnas.1700557114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Geonnotti AR, Bilska M, Yuan X, Ochsenbauer C, Edmonds TG, Kappes JC, Liao HX, Haynes BF, Montefiori DC. Differential inhibition of human immunodeficiency virus type 1 in peripheral blood mononuclear cells and TZM-bl cells by endotoxin-mediated chemokine and gamma interferon production. AIDS Res Hum Retrovir. 2010;26(3):279–291. doi: 10.1089/aid.2009.0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hui L, Rao WW, Yu Q, Kou C, Wu JQ, He JC, Ye MJ, Liu JH, Xu XJ, Zheng K, et al. TCF4 gene polymorphism is associated with cognition in patients with schizophrenia and healthy controls. J Psychiatr Res. 2015;69:95–101. doi: 10.1016/j.jpsychires.2015.07.022. [DOI] [PubMed] [Google Scholar]
- 60.Brocken DJW, Tark-Dame M, Dame RT. dCas9: a versatile tool for epigenome editing. Curr Issues Mol Biol. 2017;26:15–32. doi: 10.21775/cimb.026.015. [DOI] [PubMed] [Google Scholar]
- 61.Williams CR, Baccarella A, Parrish JZ, Kim CC. Trimming of sequence reads alters RNA-Seq gene expression estimates. BMC Bioinformatics. 2016;17:103. doi: 10.1186/s12859-016-0956-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35(Web Server issue):W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44(D1):D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yap CK, Eisenhaber B, Eisenhaber F, Wong WC. xHMMER3x2: utilizing HMMER3's speed and HMMER2's sensitivity and specificity in the glocal alignment mode for improved large-scale protein domain annotation. Biol Direct. 2016;11(1):63. doi: 10.1186/s13062-016-0163-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.von Mering C, Jensen LJ, Snel B, Hooper SD, Krupp M, Foglierini M, Jouffre N, Huynen MA, Bork P. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33(Database issue):D433–D437. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. Genome project data processing S: the sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Florea L, Song L, Salzberg SL. Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Res. 2013;2:188. doi: 10.12688/f1000research.2-188.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7(12):1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kang Y, Norris MH, Zarzycki-Siek J, Nierman WC, Donachie SP, Hoang TT. Transcript amplification from single bacterium for transcriptome analysis. Genome Res. 2011;21(6):925–935. doi: 10.1101/gr.116103.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li W, Turner A, Aggarwal P, Matter A, Storvick E, Arnett DK, Broeckel U. Comprehensive evaluation of AmpliSeq transcriptome, a novel targeted whole transcriptome RNA sequencing methodology for global gene expression analysis. BMC Genomics. 2015;16:1069. doi: 10.1186/s12864-015-2270-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Steiger JH. Tests for comparing elements of a correlation matrix. Psychol Bull. 1980;87(2):245. doi: 10.1037/0033-2909.87.2.245. [DOI] [Google Scholar]
- 76.Roy B, Haupt LM, Griffiths LR. Review: alternative splicing (AS) of genes as an approach for generating protein complexity. Curr Genomics. 2013;14(3):182–194. doi: 10.2174/1389202911314030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Havlicek S, Shen Y, Alpagu Y, Bruntraeger MB, Zufir NB, Phuah ZY, Fu Z, Dunn NR, Stanton LW. Re-engineered RNA-guided FokI-nucleases for improved genome editing in human cells. Mol Ther. 2017;25(2):342–355. doi: 10.1016/j.ymthe.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529(7587):490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maggio I, Goncalves MA. Genome editing at the crossroads of delivery, specificity, and fidelity. Trends Biotechnol. 2015;33(5):280–291. doi: 10.1016/j.tibtech.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 83.Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. 2014;32(7):677–683. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- 84.Kim D, Bae S, Park J, Kim E, Kim S, Yu HR, Hwang J, Kim JI, Kim JS. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015;12(3):237–243. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- 85.Wang X, Wang Y, Wu X, Wang J, Wang Y, Qiu Z, Chang T, Huang H, Lin RJ, Yee JK. Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nat Biotechnol. 2015;33(2):175–178. doi: 10.1038/nbt.3127. [DOI] [PubMed] [Google Scholar]
- 86.Gaj T, Staahl BT, Rodrigues GM, Limsirichai P, Ekman FK, Doudna JA, Schaffer DV. Targeted gene knock-in by homology-directed genome editing using Cas9 ribonucleoprotein and AAV donor delivery. Nucleic Acids Res. 2017;45:e98. doi: 10.1093/nar/gkx154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Polstein LR, Perez-Pinera P, Kocak DD, Vockley CM, Bledsoe P, Song L, Safi A, Crawford GE, Reddy TE, Gersbach CA. Genome-wide specificity of DNA binding, gene regulation, and chromatin remodeling by TALE- and CRISPR/Cas9-based transcriptional activators. Genome Res. 2015;25(8):1158–1169. doi: 10.1101/gr.179044.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liszczak GP, Brown ZZ, Kim SH, Oslund RC, David Y, Muir TW. Genomic targeting of epigenetic probes using a chemically tailored Cas9 system. Proc Natl Acad Sci U S A. 2017;114(4):681–686. doi: 10.1073/pnas.1615723114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim D, Kim J, Hur JK, Been KW, Yoon SH, Kim JS. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat Biotechnol. 2016;34(8):863–868. doi: 10.1038/nbt.3609. [DOI] [PubMed] [Google Scholar]
- 90.Kim V, Mears BM, Powell BH, Witwer KW. Mutant Cas9-transcriptional activator activates HIV-1 in U1 cells in the presence and absence of LTR-specific guide RNAs. Matters (Zur). 2017;2017 [DOI] [PMC free article] [PubMed]
- 91.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6(5):386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 95.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338(6113):1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 96.Affymetrix ETP, Cold Spring Harbor laboratory ETP: post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature 2009, 457(7232):1028–1032. [DOI] [PMC free article] [PubMed]
- 97.Millan MJ. Linking deregulation of non-coding RNA to the core pathophysiology of Alzheimer's disease: an integrative review. Prog Neurobiol. 2017;156:1–68. doi: 10.1016/j.pneurobio.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 98.Matsui M, Corey DR. Non-coding RNAs as drug targets. Nat Rev Drug Discov. 2017;16(3):167–179. doi: 10.1038/nrd.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G, 3rd, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14(7):723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ronchetti D, Manzoni M, Agnelli L, Vinci C, Fabris S, Cutrona G, Matis S, Colombo M, Galletti S, Taiana E, et al. lncRNA profiling in early-stage chronic lymphocytic leukemia identifies transcriptional fingerprints with relevance in clinical outcome. Blood Cancer J. 2016;6(9):e468. doi: 10.1038/bcj.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Malik B, Feng FY. Long noncoding RNAs in prostate cancer: overview and clinical implications. Asian J Androl. 2016;18(4):568–574. doi: 10.4103/1008-682X.177123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Niknafs YS, Han S, Ma T, Speers C, Zhang C, Wilder-Romans K, Iyer MK, Pitchiaya S, Malik R, Hosono Y, et al. The lncRNA landscape of breast cancer reveals a role for DSCAM-AS1 in breast cancer progression. Nat Commun. 2016;7:12791. doi: 10.1038/ncomms12791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31(9):822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tycko J, Myer VE, Hsu PD. Methods for optimizing CRISPR-Cas9 genome editing specificity. Mol Cell. 2016;63(3):355–370. doi: 10.1016/j.molcel.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim D, Kim S, Kim S, Park J, Kim JS. Genome-wide target specificities of CRISPR-Cas9 nucleases revealed by multiplex Digenome-seq. Genome Res. 2016;26(3):406–415. doi: 10.1101/gr.199588.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31(9):839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim JS. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24(1):132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Darcis G, Van Driessche B, Van Lint C. HIV latency: should we shock or lock? Trends Immunol. 2017;38(3):217–228. doi: 10.1016/j.it.2016.12.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Statistics of RNA-Seq quality reads. Table S2. Mapping results. Table S3. Validation of dCas9-SAM mRNA and sgRNA expression (transcripts per million). Table S4. Distribution of reads in known types of RNAs. Table S5. Differentially expressed mRNA transcripts for all the three pairwise comparisons of the samples (O vs Zero, L vs Zero, and O vs L). Table S6. Distribution of the 12 types of alternative splicing events across samples. (XLSX 30 kb)
Figure S1. Distributions of the mapped reads in the genome for the six samples. Figure S2. Workflow charts for RNA-seq analysis. (a) Library construction. (b) lncRNA filtering by four pipelines to predict candidate lncRNAs based on their structures and noncoding features. Figure S3. Statistics of lncRNA filtering. Horizontal axis represents the filtering step and vertical axis represents the number of remaining transcripts after the filtering step. Figure S4. Illustration of 12 types of alternative splicing events analyzed by ASprofile (Picture taken from Florea L, Song L, Salzberg SL: Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Res 2013, 2:188). (PDF 2000 kb)
Data Availability Statement
The datasets generated and/or analyzed during the current study are available from the sequence read archive (SRA): https://www.ncbi.nlm.nih.gov/bioproject/PRJNA477008 and the raw data: https://www.ncbi.nlm.nih.gov/Traces/study/?acc=SRP150955.