Abstract
Axolotl salamanders (Ambystoma mexicanum) remain aquatic in their natural state, during which biomechanical forces on their diarthrodial limb joints are likely reduced relative to salamanders living on land. However, even as sexually mature adults, these amphibians can be induced to metamorphose into a weight‐bearing terrestrial stage by environmental stress or the exogenous administration of thyroxine hormone. In some respects, this aquatic to terrestrial transition of axolotl salamanders through metamorphosis may model developmental and changing biomechanical skeletal forces in mammals during the prenatal to postnatal transition at birth and in the early postnatal period. To assess differences in the appendicular skeleton as a function of metamorphosis, anatomical and gene expression parameters were compared in skeletal tissues between aquatic and terrestrial axolotls that were the same age and genetically full siblings. The length of long bones and area of cuboidal bones in the appendicular skeleton, as well as the cellularity of cartilaginous and interzone tissues of femorotibial joints were generally higher in aquatic axolotls compared with their metamorphosed terrestrial siblings. A comparison of steady‐state mRNA transcripts encoding aggrecan core protein (ACAN), type II collagen (COL2A1), and growth and differentiation factor 5 (GDF5) in femorotibial cartilaginous and interzone tissues did not reveal any significant differences between aquatic and terrestrial axolotls.
Keywords: axolotl salamander, metamorphosis, skeleton
Introduction
Articular cartilage and associated skeletal structures in mammals undergo a process of morphological reorganization during postnatal development (Brama et al. 2002; Hunziker et al. 2007). At birth, the cartilage has a relatively isotropic architecture with a homogenous distribution of cells and matrix biomolecules from site to site topologically across the joint surface (Mienaltowski et al. 2008). During postnatal development, cartilage undergoes significant growth and functional maturation characterized by changes in cellular and molecular components under the influence of loading. This architectural reorganization occurs from the surface to the deeper layers, as well as across the joint surface, and is associated with growth and increased biomechanical loading of the tissues as a result of ambulation after birth (Brama et al. 2000a,b, 2002). Growth and remodeling of the long bones occur concurrently in all regions including the epiphysis, being governed in part by the overlying articular cartilage layer that acts in some respects as a surface growth plate (Hunziker et al. 2007). This structural and functional adaptation to weight bearing occurs early in life, and may be inhibited by exercise restriction (Brama et al. 2002). These postnatal articular cartilage changes have been described in several mammals. Rat tibial articular cartilage undergoes a transition from an isotropic to anisotropic architecture during the first few weeks of postnatal life (Sasano et al. 1996), and similar changes have been described in the medial femoral condyle of the rabbit (Hunziker et al. 2007). Changes in proteoglycan and collagen content have been described in young dogs, guinea pigs, horses and humans that may facilitate adaptation and strengthening of articular cartilage (Brama et al. 2000a; Helminen et al. 2000). Relative levels of proteoglycan and type II collagen expression decrease during postnatal cartilage maturation (Mienaltowski et al. 2008). However, the molecular, cellular and regulatory mechanisms of these changes in mammals from prenatal to postnatal stages have not been fully elucidated.
Axolotl salamanders (Ambystoma mexicanum) exhibit paedomorphosis, wherein they retain several of their larval characteristics through adulthood and sexual maturity (Shaffer & Voss, 1996). They are aquatic in their paedomorphic state with external gills, tail fins and well‐developed limbs with webbed digits (Fig. 1A), but they can metamorphose into a terrestrial state (Fig. 1B) through environmental stress or by the exogenous administration of thyroxine hormone (Page et al. 2009). In an aquatic habitat, the biomechanical forces placed on their appendicular joints are different compared with their terrestrial counterparts. Metamorphosis is associated with the loss of external larval characteristics, as well as a change to a terrestrial habitat wherein their joints are subject to the mechanical loading forces of weight bearing and ambulation. Histological architecture in amphibian long bones varies by species and between bones (Castanet et al. 2003). In salamanders, the epiphyses remain cartilaginous throughout life and are composed of spherical chondrocytes, which have limited mitotic activity. Towards the metaphysis, however, there is an ill‐defined region between the epiphyses and diaphysis in which chondrocytes undergo several mitotic divisions and then progress through hypertrophic differentiation to enable endochondral ossification and linear growth in bone length. The diaphysis at maturity has a collar of compact bone surrounding the marrow cavity and grows appositionally from the surrounding periosteum. Changes in appendicular skeletal structure through maturation and metamorphosis vary between species, influenced in axolotls by paedomorphosis. However, the aquatic to terrestrial transition may model, to some extent, changing environmental conditions from the prenatal to postnatal period in newborn mammals and other amniotes. This study was designed with the objective of examining the morphometric, histological and gene expression changes that occur in the appendicular skeletal tissues of axolotl salamanders during metamorphosis.
Figure 1.
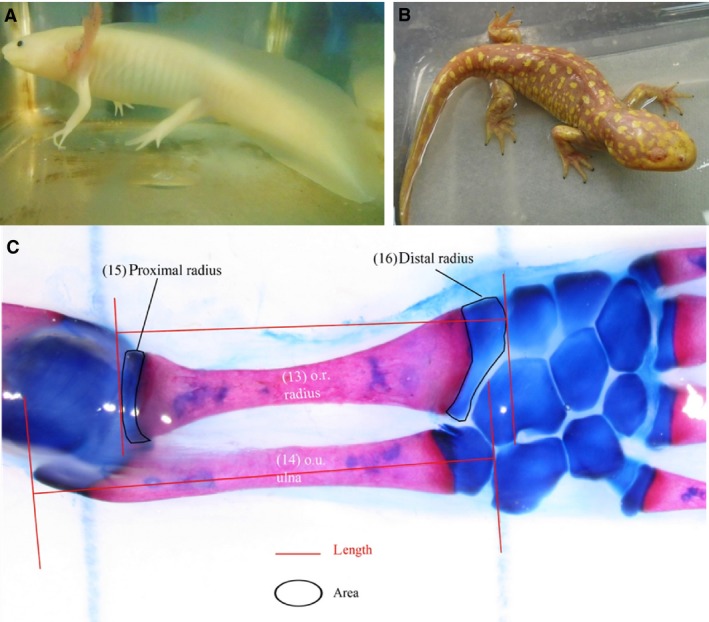
Description of samples for histomorphometric analysis. Aquatic axolotl salamanders with webbed digits, external gills and tail fins (A). Spontaneously metamorphosed terrestrial axolotl salamander, characterized by a loss of external gills, tail fins and the webbing between the digits (B). (C) Right forelimb of an aquatic axolotl salamander differentially stained for bone (red) and cartilage (blue). Framed and outlined areas illustrate the selection perimeters for bone length and area measurements.
Materials and methods
Animal housing and care
A total of 14 axolotl salamanders (Ambystoma mexicanum), composed of seven pairs of 1‐year‐old age‐ and gender‐matched aquatic and spontaneously metamorphosed terrestrial full genetic siblings, were obtained from the Ambystoma Genetic Stock Center (University of Kentucky, Lexington, KY, USA; http://www.ambystoma.org/genetic-stock-center). Care and management of the aquatic salamanders were carried out as previously described (Cosden et al. 2011; Cosden‐Decker et al. 2012), with the spontaneously metamorphosed siblings maintained under moist terrestrial conditions. All of the salamanders were kept for an additional period of 30 days in our laboratory colony following transfer from the stock center to ensure completion of metamorphosis prior to death and sample collection (Rosenkilde & Ussing, 1996). All procedures were conducted in accordance with the guidelines of an approved University of Kentucky Institutional Animal Care and Use Committee protocol (IACUC #2008‐0282).
Sample collection and processing
Prior to tissue collection, salamanders were deeply anesthetized with 0.01% benzocaine solution (w/v, Sigma, Cat# E1501, St Louis, MO, USA) in 25% Holtfreter's solution, and killed by cervical dislocation. Fore‐ and hindlimbs were harvested, trimmed to remove skin and soft tissues, and fixed in 4% paraformaldehyde in phosphate‐buffered saline for 48 h. Alcian Blue/Alizarin Red double‐staining of bone and cartilage was performed on the fixed tissues for morphometric analysis as described previously (Wassersug, 1976; Dingerkus & Uhler, 1977). Briefly, limbs were dehydrated through increasing concentrations of ethanol after fixation and then stained with Alcian Blue solution until the cartilage appeared blue. This was followed by rehydration with decreasing concentrations of ethanol and de‐staining by incubation in a solution containing hydrogen peroxide. Limbs were enzymatically digested in trypsin, stained with Alizarin Red, and cleared in 0.05% potassium hydroxide and glycerin. Limbs processed for histological analysis were decalcified in 15–20 × volumes of ethylenediaminetetraacetic acid and hydrochloric acid (Cat# 22‐050‐130, Richard Allen Scientific, Kalamazoo, MI, USA) with periodic testing to assess the progress of decalcification. Decalcification was considered complete by the lack of precipitate formation in a test solution of 5% (v/v) ammonium hydroxide (Cat# 205840010, Thermo Fisher Scientific) and 5% (v/v) ammonium oxalate (Cat# R06840001A, Thermo Fisher Scientific) to indicate calcium leaching from the skeletal tissues. The samples were then transferred to 70% ethanol prior to paraffin embedding. All samples were sectioned at 7 μm. Hematoxylin and eosin (H&E) staining was performed using standard protocols.
Analysis of limb morphometry
Six pairs of right forelimbs and three pairs of left hindlimbs collected from seven pairs of sibling axolotl salamanders were used for histomorphometric analysis as outlined in Table 1. Digital images of Alcian Blue/Alizarin Red double‐stained joints of fore‐ and hindlimbs (Fig. S1) were obtained using a stereoscopic microscope with a 500 μm scale, and were analyzed using the line selection tools of ImageJ software (https://imagej.nih.gov/ij/download.html). Parameters assessed included length and area of bones and cartilage that were identified based on differential staining with Alcian Blue/Alizarin Red as demonstrated in Fig. 1C. In addition, length and area measurements for related bones were grouped together (Table S2) based on their functional similarity, anatomical homology and developmental origins (Shubin & Alberch, 1986; Nye et al. 2003), and compared between aquatic and terrestrial salamanders.
Table 1.
Experimental samples for the analysis of limb morphometry
| Limb | Measurement parameter | Skeletal element | Paired biological replicates (n) |
|---|---|---|---|
| Forelimb | Area | Carpal bones | 6 |
| Area | Proximal and distal radius | 6 | |
| Length | Radius | 6 | |
| Length | Ulna | 6 | |
| Length | Humerus | 6 | |
| Length | Metacarpals | 6 | |
| Hindlimb | Area | Tarsal bones | 3 |
| Area | Proximal and distal tibia | 3 | |
| Length | Metatarsals | 3 | |
| Length | Tibia | 3 | |
| Length | Fibula | 3 | |
| Length | Femur | 3 |
Each biological replicate represents a pair of genetic full sibling axolotl salamanders, one aquatic and one terrestrial.
Analysis of cellularity
Four pairs of H&E‐stained femorotibial joints collected from hindlimbs were used for cell density analysis. Epiphyseal‐articular cartilage, metaphyseal cartilage and interzone were identified based on their histological characteristics as described previously (Fig. S1A and B; Cosden et al. 2011). Epiphyseal‐articular cartilage shows an isotropic distribution of chondrocytes approximately 10–20 μm in diameter, whereas the metaphyseal chondrocytes are larger, ranging from 25 µm to 35 μm in diameter and arranged in clusters. The interzone can be appreciated in the intra‐articular space as a fibrous layer with flattened cells lying close to the articular surface. Digital images of H&E‐stained interzone (20 × magnification), metaphyseal cartilage and epiphyseal/articular cartilage (4 × magnification) were analyzed using CellC and MatLab software (The Mathworks, Natick, MA, USA). The image scale was converted from pixel to micrometer prior to measurement. The area (μm2) and total cell count of a representative region of each sample was measured as demonstrated in Fig. S2C and D, respectively. The cell density was calculated as follows:
Gene expression analysis
Total RNA was isolated from whole body tissue samples of Mexican axolotl salamanders (Ambystoma mexicanum) at the following developmental stages: embryo at the tail bud stage, newly hatched larva, larva at the limb bud stage, juvenile at 8.5 cm, and adult using variations of guanidinium‐based protocols (Chomczynski & Sacchi, 1987; Macleod et al. 1996). RNA quantity, purity and integrity of both the individual samples and the resulting pool were determined with an Agilent 2100 Bioanalyzer using the Eukaryotic Total RNA nano series II analysis kit. RNA was poly‐A selected and used for an Illumina random priming directional library prep. Four lanes were sequenced only on one end providing single‐end reads, and four lanes were sequenced at both ends giving paired‐end reads. The library was sequenced on an Illumina HiSeq 2000 for 75‐bp reads producing 147 248 512 single‐end reads and 2 × 153 254 667 paired‐end reads. Base calling and processing was performed using the Illumina pipeline version 1.7. Illumina reads were filtered and trimmed using FASTX toolkit 0.0.13 (http://hannonlab.cshl.edu/fastx_toolkit/index.html) to generate 161 548 989 single‐end and 2 × 113 807 507 paired‐end high‐quality reads. RNA‐seq data have been deposited in the ArrayExpress database at EMBL‐EBI (http://www.ebi.ac.uk/arrayexpress) under accession number E‐MTAB‐6092. Then, a de novo assembly of both the single‐end and paired‐end reads was performed using Oases 0.2.07 (Schulz et al. 2012). The number of transcripts generated totaled 1 068 004 and covered 143 780 loci with a median length of 776 bp. A reference genome was not available for the axolotl salamander or a closely related species, so cDNA contigs were assembled and aligned to a BLAST database comprised of Genbank sequences for a set of targeted genes expressed in articular cartilage and joint tissues for human and the African clawed frog (Xenopus laevis) using CLC genomics workbench version 7.1 (https://www.qiagenbioinformatics.com/). An e‐value threshold of 10−7 was used. In circumstances where multiple but non‐overlapping axolotl cDNA fragments aligned to a targeted gene locus, forward and reverse primers were designed to both extend and connect these fragments as illustrated in Fig. 2. Resulting polymerase chain reaction (PCR) amplicons were sequenced (ACGT, Wheeling, IL, USA) to generate longer cDNA sequences for the 17 target genes expressed in cartilage and other joint tissues (https://www.ncbi.nlm.nih.gov/genbank/ – accession numbers MF375375 – MF375391.
Figure 2.
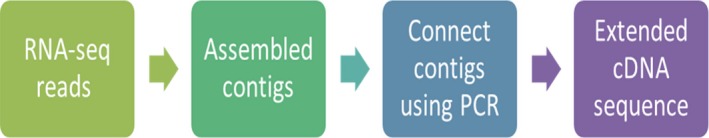
Process of sequence alignment and reconstruction of axolotl contigs for target genes.
Femorotibial samples of epiphyseal‐articular cartilage, metaphyseal cartilage and interzone tissue were dissected carefully under a stereoscopic microscope from sibling pairs of aquatic and terrestrial (metamorphosed) axolotl salamanders, snap‐frozen in liquid nitrogen, and stored at −80 °C for subsequent total RNA extraction. Total RNA was isolated using a spin column‐based kit according to the manufacturer's protocol (Norgen Biotek, Animal Tissue RNA Purification Kit, Cat# 25700). RNA samples were quantified by fluorescence using the Qubit BR Assay or Qubit HS Assay (Thermo Fisher Scientific, Cat# Q10210, Q32852). The removal of any potential contaminating genomic DNA (Thermo Fisher Scientific, DNA‐free™ DNA Removal Kit, Cat# AM 1906) followed by reverse transcription of total RNA to generate cDNA (Thermo Fisher Scientific, SuperScript® VILO™ cDNA Synthesis Kit Cat# 11754050) were performed according to the manufacturer's protocols. The resulting cDNA was uniformly diluted to 2 ng μL−1 with nuclease‐free water and stored at −20 °C. Primer sets were custom designed for the following three targeted gene sequences: aggrecan core protein (ACAN); type II collagen (COL2A1); and growth and differentiation factor 5 (GDF5), and used to quantitate steady‐state mRNA levels in the aquatic and terrestrial axolotl femorotibial tissue samples. The reactions were performed in 384‐well plates in a total reaction volume of 12.5 μL using the Power SYBR® Green PCR Master Mix (Thermo Fisher Scientific, Cat# 4367659). Nuclease‐free water and minus reverse transcriptase reaction samples served as negative controls. The calibrator sample used was total RNA from an age‐matched adult axolotl salamander. All samples were analyzed in triplicate and randomly assigned to each plate to control for any plate effects. Reactions were performed on a robotic ViiA™ 7 Real‐Time PCR System (Thermo Fisher Scientific, Cat# 4453536). Reaction amplification efficiencies were evaluated using LinRegPCR (Ramakers et al. 2003), and then used to correct cycle threshold measurements for each sample. Relative expression values (RE; relative quantity, RQ) of the transcript targets were calculated using the 2−ΔΔCt method (Livak & Schmittgen, 2001).
Statistical analysis
Data from the histomorphometric analysis were evaluated using a one‐way analysis of variance (anova) with Tukey–Kramer analysis to allow for comparison of unequally sized groups. Length and area measurements for the related bone groups were independently evaluated using a one‐way ANOVA. Analysis was performed using the SAS statistical program, version 9.1 (Statistical Analysis System, SAS Institute). Differences were considered significant at P < 0.05.
Results and discussion
Analysis of limb morphometry
Total length and area measurements of long bones and cuboidal bones of forelimbs and hindlimbs were in general higher in aquatic axolotls compared with their metamorphosed terrestrial siblings. However, not all of these differences reached statistical significance (Table 2). The second (P = 0.0061) and third (P = 0.0006) metacarpals in the forelimbs, and third (P = 0.0048) and fourth (P = 0.0214) metatarsals in the hindlimbs were significantly longer in aquatic axolotls. To carry the assessment further, ossified and non‐ossified sub‐segments of these four bones were measured separately (Table S2). Lengths of the ossified diaphyseal segments were numerically longer in each bone, but reached significance (P < 0.05) only in the metacarpals. Combined lengths of the proximal and distal cartilaginous regions were also numerically longer for all four bones in the aquatic salamanders, but the differences were not significant. Taken together, the significant shortening in metacarpal 2, metacarpal 3, metatarsal 3 and metatarsal 4 observed in the terrestrial siblings following metamorphosis appears to reflect the combined changes in both ossified and cartilaginous segments of these bones. Among the cuboidal bones, the total areas of radial carpals (P = 0.0141) and fused basal carpals of 1st and 2nd forelimb digits (P = 0.0011) were significantly higher in aquatic salamanders relative to their terrestrial siblings. In the hindlimbs, the central tarsals (P = 0.0225) and the basal tarsal of the 5th toe (P = 0.0492) were also significantly larger in aquatic axolotls (Table 3).
Table 2.
Bone length measurements of appendicular skeletal joints
| Limb | Bone | Mean ± SE in mm (Aquatic) | Mean ± SE in mm (Terrestrial) |
|---|---|---|---|
| Forelimb | Humerus | 16.33 ± 0.26 | 15.76 ± 0.26 |
| Forelimb | Radius | 7.98 ± 0.16 | 7.46 ± 0.13 |
| Forelimb | Ulna | 9.15 ± 0.28 | 8.94 ± 0.28 |
| Forelimb | Metacarpal 1 | 4.26 ± 0.11 | 3.94 ± 0.11 |
| Forelimb | Metacarpal 2 | 5.66 ± 0.06 | 5.29 ± 0.06* |
| Forelimb | Metacarpal 3 | 5.55 ± 0.04 | 5.14 ± 0.04* |
| Forelimb | Metacarpal 4 | 4.64 ± 0.09 | 4.34 ± 0.09 |
| Hindlimb | Femur | 14.26 ± 0.12 | 13.65 ± 0.12 |
| Hindlimb | Tibia | 8.47 ± 0.15 | 8.15 ± 0.15 |
| Hindlimb | Fibula | 7.99 ± 0.17 | 7.82 ± 0.17 |
| Hindlimb | Metatarsal 1 | 3.89 ± 0.10 | 3.71 ± 0.10 |
| Hindlimb | Metatarsal 2 | 5.32 ± 0.08 | 4.89 ± 0.08 |
| Hindlimb | Metatarsal 3 | 5.87 ± 0.02 | 5.45 ± 0.02* |
| Hindlimb | Metatarsal 4 | 5.64 ± 0.04 | 5.22 ± 0.04* |
| Hindlimb | Metatarsal 5 | 4.66 ± 0.09 | 4.34 ± 0.09 |
Bone length (mm) measurements from forelimbs and hindlimbs. Data are presented as mean ± standard error, with an asterisk (*) designating statistical significance.
Table 3.
Bone area measurements of appendicular skeletal joints
| Limb | Bone | Mean ± SE in mm2 (Aquatic) | Mean ± SE in mm2 (Terrestrial) |
|---|---|---|---|
| Forelimb | Cartilago prepollicis | 1.09 ± 0.06 | 1.01 ± 0.06 |
| Forelimb | Radial carpal | 1.19 ± 0.03 | 0.97 ± 0.04* |
| Forelimb | Intermediate carpal | 2.50 ± 0.19 | 2.09 ± 0.16 |
| Forelimb | Ulnar carpal | 1.73 ± 0.07 | 1.49 ± 0.06 |
| Forelimb | Central carpal | 1.63 ± 0.03 | 1.51 ± 0.03 |
| Forelimb | Fused basal carpals of 1st and 2nd fingers | 1.71 ± 0.02 | 1.51 ± 0.02* |
| Forelimb | Basal carpal of 3rd finger | 0.91 ± 0.07 | 0.92 ± 0.07 |
| Forelimb | Basal carpal of 4th finger | 1.16 ± 0.14 | 1.22 ± 0.14 |
| Hindlimb | Cartilago prehallucis | 1.57 ± 0.15 | 1.54 ± 0.15 |
| Hindlimb | Tibial tarsal | 1.29 ± 0.06 | 1.25 ± 0.06 |
| Hindlimb | Intermediate tarsal | 3.26 ± 0.18 | 3.16 ± 0.18 |
| Hindlimb | Fibular tarsal | 3.61 ± 0.19 | 3.45 ± 0.19 |
| Hindlimb | Central tarsal | 2.73 ± 0.03 | 2.41 ± 0.03* |
| Hindlimb | Fused basal tarsals of 1st and 2nd toes | 2.01 ± 0.09 | 1.66 ± 0.09 |
| Hindlimb | Basal tarsals of 3rd toe | 1.32 ± 0.06 | 1.24 ± 0.06 |
| Hindlimb | Basal tarsals of 4th toe | 1.43 ± 0.08 | 1.37 ± 0.08 |
| Hindlimb | Basal tarsals of 5th toe | 0.86 ± 0.02 | 0.72 ± 0.02 * |
Bone area (mm2) measurements from forelimbs and hindlimbs. Data are presented as mean ± standard error, with an asterisk (*) designating statistical significance.
The area of cartilage tissue at the proximal and distal ends of the radius and tibia are presented in Table 4. Although the overall area measurements of the cartilaginous ends of the radius and tibia were higher in aquatic axolotls compared with their terrestrial counterparts, only the area difference of the distal end of the tibia was statistically significant (P = 0.0433). The results of the grouped analysis were congruent with the analysis of individual bone parameters (Fig. S3; Table S1). In the forelimb, differences in the carpals (Group 1) and metacarpals (Group 3) were statistically significant, whereas in the hindlimb the metatarsals (Group 3) were significantly longer in the aquatic axolotls compared with the terrestrial siblings.
Table 4.
Cartilage area measurements of appendicular skeletal joints
| Limb | Bone | Mean ± SE in mm2 (Aquatic) | Mean ± SE in mm2 (Terrestrial) |
|---|---|---|---|
| Forelimb | Proximal radius | 0.87 ± 0.09 | 0.69 ± 0.08 |
| Forelimb | Distal radius | 1.39 ± 0.11 | 1.12 ± 0.14 |
| Hindlimb | Proximal tibia | 1.87 ± 0.19 | 1.29 ± 0.19 |
| Hindlimb | Distal tibia | 1.58 ± 0.05 | 1.23 ± 0.05* |
Cartilage area (mm2) measurements from forelimbs and hindlimbs. Data are presented as mean ± standard error, with an asterisk (*) designating statistical significance.
Analysis of cellularity
Cell density of the epiphyseal‐articular cartilage, metaphyseal cartilage and interzone of the distal femur are presented in Table 5. The cellularity of the epiphyseal‐articular cartilage (P = 0.0247) and interzone tissue (P = 0.0075) of the femorotibial joint was higher in aquatic compared with terrestrial metamorphs. No difference was observed in the cellularity of the metaphyseal cartilage.
Table 5.
Analysis of cellularity
| Zone | Number of cells μm−2 (Aquatic) | Number of cells μm−2 (Terrestrial) |
|---|---|---|
| Epiphyseal articular cartilage | 0.000751 ± 0.00003 | 0.000575 ± 0.00003* |
| Metaphyseal cartilage | 0.000700 ± 0.000053 | 0.000727 ± 0.000053 |
| Interzone | 0.002088 ± 0.000058 | 0.001561 ± 0.000058* |
Cell density (number of cells μm−2) from the cartilaginous tissues and interzone of the distal femur. Data are presented as mean ± standard error, with an asterisk (*) designating statistical significance.
The histomorphometric parameters assessed in this study, length of appendicular long bones, area of appendicular cuboidal bones, and cellularity of femorotibial cartilaginous and interzone tissues, were broadly higher on a numerical level in aquatic axolotls compared with their metamorphosed terrestrial siblings. Not all of these differences reached the threshold for statistical significance set at P < 0.05. Yet, these data indicate that the bones of the appendicular skeleton shorten and become more compact as the axolotl transitions from an aquatic to terrestrial environment. The potential for gender‐dependent differences was controlled by gender‐matched pairings. However, potential gender‐specific differences in skeletal morphology were not investigated due to the size of the available sample set.
Size reduction of bones in the appendicular skeleton may contribute to the reduction in total body weight reported during metamorphosis in axolotl salamanders (Rosenkilde & Ussing, 1996), which contrasts to the increase in total body size observed in anurans through metamorphosis (Stebbins & Cohen, 1997; Elinson, 2013; Buchholz, 2015). The spectrum of axolotl morphological changes are all likely functionally relevant to the fundamental differences in biomechanical and kinesiology parameters imposed by aquatic vs. terrestrial environments (Karakasiliotis et al. 2013). Salamanders exhibit a wide variety of gaits in different environments broadly categorized into three types – aquatic stepping, swimming, and terrestrial stepping. Aquatic salamanders do not engage in extended periods of sustained swimming, but instead move in short bursts, separated by periods of relative inactivity (Wells, 2010). Although little has been described in the literature on differences between aquatic and terrestrial stepping, a few changes have been noted (Ashley‐Ross et al. 2009). The average speed during underwater stepping is twice the speed observed during terrestrial locomotion. Another difference is a switch from a diagonal to a lateral sequence walk when salamanders transit from an aquatic to a terrestrial environment (Ashley‐Ross, 1994). In the paper by Ashley‐Ross and colleagues, quantitative analysis of limb kinematics of the California newt, Taricha torosa, during steady‐speed walking in simulated aquatic and terrestrial environments was conducted. However, the authors did not measure changes in skeletal morphometry and, therefore, direct comparisons cannot be made. To the best of our knowledge, this is the first study comparing appendicular skeletal morphometry during metamorphosis in axolotl salamanders. Yet, it is unclear how the biomechanical loads imposed by the aquatic and terrestrial environments differ, and how components of the locomotor system, the bones, joints and supporting soft tissues respond to the different mechanical loads. These are relevant questions to be investigated in future studies.
In considering environmental influences on the appendicular skeleton during metamorphosis for axolotls at sexual maturity to the transition out of a fluid‐filled amnion in amniotes, an important difference may be the rate of skeletal growth. For example, the bones in neonatal mammals are growing rapidly through both longitudinal (growth plate) and appositional mechanisms. Environmentally associated changes in biomechanical forces with metamorphosis in axolotls have similarities, but this is not accompanied with a high rate of skeletal growth. It would be worth investigating whether the changes in skeletal morphometry during metamorphosis observed in the current study reflect an aquatic to terrestrial environmental transition. One interesting similarity with mammals, however, is the reduction in cellularity of the epiphyseal‐articular cartilage with metamorphosis. In our study, the cell density in epiphyseal‐articular cartilage was significantly lower in the terrestrial salamanders compared with their aquatic counterparts. In newborn mammals, the chondrocyte cellularity in articular cartilage is relatively high with an even cellular distribution, both of which likely facilitate a high anabolic state of tissue growth. As the articular cartilage matures postnatally, cellularity decreases overall and with depth from the articular surface. In addition, both chondrocyte and matrix organization becomes anisotropic, with cells arranged in groups separated by extracellular matrix molecules (Hunziker et al. 1997, 2002, 2007; Pearle et al. 2005; Mienaltowski et al. 2008). These remodeling events reflect the dynamics of growth and changing biomechanical forces in progression towards skeletal maturity (Jadin et al. 2007). The decrease in cellularity of the epiphyseal‐articular cartilage in axolotl salamanders during metamorphosis also correlates with substantial environmental and biomechanical changes. The reduction in cell density could also be a function of larger chondrocytes or an increase in the extracellular matrix (Cooper et al. 2013). It would be interesting to investigate potential changes in the size of chondrocytes and the molecular structure and organization of extracellular matrix components during metamorphosis, and whether any differences are concordant to the matrix changes that occur during mammalian articular cartilage maturation. This study focused on the observed appendicular skeletal parameters at a tissue, cellular and molecular level before and after metamorphosis. Additional studies examining the changes in individuals that are actively undergoing metamorphosis would allow for a better understanding of the process by which these changes occur.
Gene expression analysis
Generation of target gene sequence
Due to the absence of a reference axolotl genome, predicted cDNA sequences of the 17 targeted genes listed in Table 6 were generated using axolotl RNA‐seq reads assembled into transcript contigs and then aligned to a BLAST database comprised of Genbank sequences for human and African clawed frog (Xenopus laevis). Assembled contigs from the RNA‐seq reads included partial cDNAs for three axolotl gene loci that previously had not been reported; IHH, NOG and WNT4. For nine gene loci; BOC, CD44, CHRD, ENPP2, GDF5, TNC, ACAN, MMP13 and WNT9A, the RNA‐seq contigs extended the length of previously published axolotl cDNA sequences (Stewart et al. 2013) by an average 3.08‐fold with additional data ranging in length from 485 to 4046 nucleotides. The lengths of the cDNA sequence for the remaining five gene loci listed in Table 6; COL1A1, COL2A1, COL9A2, ERG and TGFBR2, were extended by connecting adjoining published axolotl cDNA contigs (Stewart et al. 2013) using end‐point PCR.
Table 6.
List of targeted genes expressed in axolotl cartilage and other joint tissues with their contig length, accession numbers and alignment quality scores.
| Gene name | cDNA length | Accession | ORF | Length (nt | aa) | Blastp (Species) | Blastp (Bit score) | Blastn (Species) | Blastn (Bit score) |
|---|---|---|---|---|---|---|---|---|
| ACAN | 1557 | MF375375 | ORF9 | 675 | 224 | Gallus gallus | 395 | Nanorana parkeri | 455 |
| BOC | 4593 | MF375376 | ORF4 | 3369 | 1122 | Mus musculus | 1368 | Coturnix japonica | 1319 |
| CD44 | 3852 | MF375377 | ORF1 | 2001 | 666 | Mus musculus | 204 | Ambystoma mexicanum | 2190 |
| Chordin | 2598 | MF375378 | ORF1 | 2148 | 715 | Xenopus laevis | 1121 | Cynops pyrrhogaster | 2152 |
| COL1A1 | 5235 | MF375379 | ORF6 | 1488 | 495 | Cynops pyrrhogaster | 869 | Cynops pyrrhogaster | 5963 |
| COL2A1 | 1342 | MF375380 | ORF6 | 840 | 279 | Xenopus laevis | 1127 | Cynops pyrrhogaster | 1531 |
| COL9A2 | 1081 | MF375381 | ORF1 | 984 | 327 | Mus musculus | 534 | Fopius arisanus | 52.8 |
| ENPP2 | 3328 | MF375382 | ORF17 | 963 | 320 | Homo sapiens | 568 | Alligator mississippiensis | 1040 |
| ERG | 820 | MF375383 | ORF4 | 651 | 216 | Gallus gallus | 364 | Struthio camelus australis | 628 |
| GDF5 | 2217 | MF375384 | ORF16 | 984 | 327 | Mus musculus | 355 | Parus major | 477 |
| IHH | 918 | MF375385 | ORF2 | 714 | 237 | Xenopus laevis | 387 | Coturnix japonica | 619 |
| MMP13 | 2116 | MF375386 | ORF2 | 1452 | 483 | Xenopus laevis | 723 | Ictalurus punctatus | 67.6 |
| NOG | 1603 | MF375387 | ORF7 | 684 | 227 | Gallus gallus | 352 | Crocodylus porosus | 606 |
| TGFBR2 | 1095 | MF375388 | ORF2 | 486 | 161 | Homo sapiens | 302 | Jaculus jaculus | 540 |
| TNC | 7031 | MF375389 | ORF20 | 5376 | 1791 | Gallus gallus | 2756 | Pterocles gutturalis | 1908 |
| WNT4 | 1976 | MF375390 | ORF20 | 1056 | 351 | Xenopus laevis | 699 | Nanorana parkeri | 878 |
| WNT9A | 2419 | MF375391 | ORF1 | 1083 | 360 | Mus musculus | 629 | Ambystoma mexicanum | 3014 |
Differential gene expression
Quantitative comparisons of gene expression in the paired aquatic and terrestrial axolotls were limited by RNA yields from the small microdissected femorotibial tissue samples. Sufficient RNA was isolated to compare steady‐state mRNA levels for three target genes, ACAN, COL2A1 and GDF5. Total RNA from an age‐matched adult axolotl salamander (whole body) served as the calibrator sample, the expression value for which was set to 0. ACAN mRNA expression was significantly higher in the cartilaginous tissues, both epiphyseal‐articular and metaphyseal, compared with interzone (Fig. 3a). Expression patterns of COL2A1 were similar to those of ACAN in all the tissue types tested (Fig. 3b). Type II collagen and aggrecan are matrix proteins synthesized almost exclusively by chondrocytes and well‐established biomarkers of cartilage. Their expression distinguishes chondrocytes from other cell types (Feng et al. 2010). However, the levels of expression for both genes were similar in aquatic and terrestrial across the tissue types tested. As noted, potential changes in the structural organization of these important extracellular matrix proteins as a function of metamorphosis and terrestrial ambulation will be an interesting issue to investigate.
Figure 3.
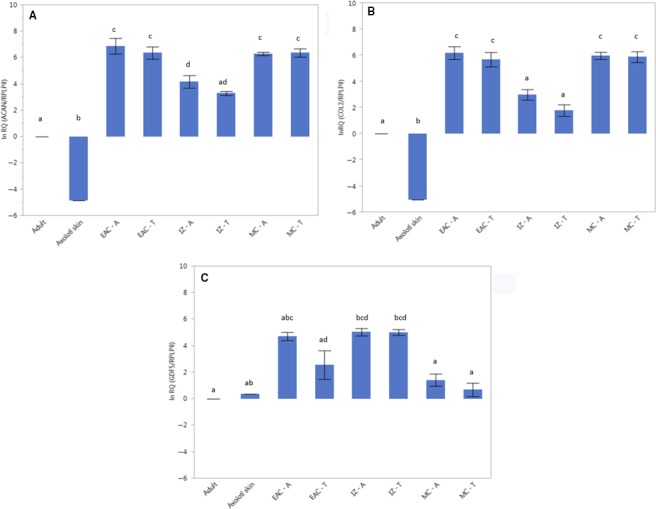
Differential gene expression. Steady‐state mRNA expression of aggrecan core protein (ACAN) (a), type II collagen (COL2) (b), growth and differentiation factor 5 (GDF5) (c) in epiphyseal articular cartilage, metaphyseal cartilage and interzone tissue samples from the femorotibial joints of aquatic and terrestrial sibling axolotls. Samples include total RNA from EAC, epiphyseal‐articular cartilage; MC, metaphyseal cartilage; and IZ, interzone from A, aquatic; and T, terrestrial axolotl salamanders. Bar graphs represent the relative expression levels relative to an age‐matched adult axolotl RNA sample.
Interzone tissue samples showed significantly higher levels of GDF5 expression compared with metaphyseal cartilage, a result consistent with studies of interzone in other species (Buxton et al. 2001; Koyama et al. 2008; Shwartz et al. 2016). As with ACAN and COL2A1, however, expression of GDF5 was not significantly different between aquatic and terrestrial individuals. Taken together, micro‐dissected cartilage and interzone tissue samples from the femorotibial joints did not identify significant differences as a function of metamorphosis in the expression of these three biomarker genes. Improved spatial resolution of gene expression patterns using techniques such as in situ hybridization and immunohistochemistry, as well as expansion of the study to more genes may identify differences between aquatic and terrestrial axolotls.
Analyzing the impact, both structural and functional, of biomechanical changes imposed by birth and ambulation on the skeletal elements of neonatal mammals is confounded by concurrent rapid growth. Metamorphosis of adult axolotl salamanders shares the substantial environmental transition, but without growth as a confounding variable. Therefore, how anatomical changes observed in the appendicular skeleton during axolotl metamorphosis specifically relate to an aquatic to terrestrial environmental transition is an important question.
Conclusions
In this study, we examined morphometric, histological and gene expression changes that occur in the appendicular skeletal tissues of axolotl salamanders during metamorphosis. The histomorphometric data indicated that the bones of the appendicular skeleton shorten and become more compact as axolotls transition from an aquatic to terrestrial environment. However, a comparison of steady‐state mRNA transcripts for ACAN, COL2A1 and GDF5 biomarker genes in cartilaginous and interzone tissues did not identify significant differences as a function of metamorphosis.
Author contributions
P.T., J.L. and J.M. made substantial contributions to study conception and design, as well as the acquisition, analysis, and interpretation of samples and data. Z.Z. contributed to the analysis of RNA sequencing data and its submission to public data repositories. Drafting of the manuscript was performed by P.T. and J.M. All authors participated in revising the manuscript and approving the final submission.
Supporting information
Fig. S1. Experimental samples for the analysis of limb morphometry. Forelimbs and hindlimbs of an aquatic axolotl salamander differentially stained for bone (red) and cartilage (blue) showing the skeletal elements used for histomorphometric analysis.
Fig. S2. Description of samples for the analysis of limb morphometry. Representative image of a histological section of the femorotibial joint (knee) illustrating the demarcation of metaphyseal cartilage, epiphyseal‐articular cartilage and the interzone for histomorphometric analysis (A) at 4 × and (B) at 20 × magnification. Representative images of a histological section of the distal end of femur illustrating the procedure of area (C) and total cell count (D) measurements for analysis of cellularity.
Fig. S3. Grouped analysis of limb morphometry. (a) Forelimb grouped analysis: the average bone area or length in aquatic axolotls was significantly higher than in the terrestrial axolotls groups 1 and 3 (mean ± SD). (b) Hindlimb grouped analysis: the average bone area or length in aquatic axolotls was significantly higher than in the terrestrial axolotls in group 3 (mean ± SD). Components of each group are defined in Table S1.
Table S1. Description of skeletal tissue groups from forelimbs and hindlimbs.
Table S2. Bone length measurements from ossified and non‐ossified sub‐segments of metacarpals and metatarsals.
Acknowledgements
The authors thank Illumina for generating the RNA‐sequencing data. The authors are grateful to Dr David Horohov and John Stewart for use of the ViiA™ 7 Real‐Time PCR System. The authors are grateful to Dr Rebekah Cosden‐Decker, D. Taylor Lundeen and Travis Trisko for assistance with axolotl care. Financial support was received from Gluck Equine Research Foundation and Geoffrey C. Hughes Foundation. The authors declare no conflict of interest.
References
- Ashley‐Ross M (1994) Hindlimb kinematics during terrestrial locomotion in a salamander (Dicamtodon tenebrosus). J Exp Biol 193, 255–283. [DOI] [PubMed] [Google Scholar]
- Ashley‐Ross MA, Lundin R, Johnson KL (2009) Kinematics of level terrestrial and underwater walking in the California newt, Taricha torosa. J Exp Zool A Ecol Genet Physiol 311, 240–257. [DOI] [PubMed] [Google Scholar]
- Brama PA, Tekoppele JM, Bank RA, et al. (2000a) Functional adaptation of equine articular cartilage: the formation of regional biochemical characteristics up to age one year. Equine Vet J 32, 217–221. [DOI] [PubMed] [Google Scholar]
- Brama PA, Tekoppele JM, Bank RA, et al. (2000b) Topographical mapping of biochemical properties of articular cartilage in the equine fetlock joint. Equine Vet J 32, 19–26. [DOI] [PubMed] [Google Scholar]
- Brama PA, Tekoppele JM, Bank RA, et al. (2002) Development of biochemical heterogeneity of articular cartilage: influences of age and exercise. Equine Vet J 34, 265–269. [DOI] [PubMed] [Google Scholar]
- Buchholz DR (2015) More similar than you think: frog metamorphosis as a model of human perinatal endocrinology. Dev Biol 408, 188–195. [DOI] [PubMed] [Google Scholar]
- Buxton P, Edwards C, Archer CW, et al. (2001) Growth/differentiation factor‐5 (GDF‐5) and skeletal development. J Bone Joint Surg Am 83, S23–S30. [PubMed] [Google Scholar]
- Castanet J, Francillon‐Vieillot H, De Ricqles A, et al. (2003) The skeletal histology of the Amphibia In: Amphibian Biology (ed. Heatwole H, DaviesM).Chipping Norton, Australia: Surrey Beatty. [Google Scholar]
- Chomczynski P, Sacchi N (1987) Single‐step method of RNA isolation by acid guanidinium thiocyanate‐phenol‐chloroform extraction. Anal Biochem 162, 156–159. [DOI] [PubMed] [Google Scholar]
- Cooper KL, Oh S, Sung Y, et al. (2013) Multiple phases of chondrocyte enlargement underlie differences in skeletal proportions. Nature 495, 375–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosden RS, Lattermann C, Romine S, et al. (2011) Intrinsic repair of full‐thickness articular cartilage defects in the axolotl salamander. Osteoarthritis Cartilage 19, 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosden‐Decker RS, Bickett MM, Lattermann C, et al. (2012) Structural and functional analysis of intra‐articular interzone tissue in axolotl salamanders. Osteoarthritis Cartilage 20, 1347–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingerkus G, Uhler LD (1977) Enzyme clearing of alcian blue stained whole small vertebrates for demonstration of cartilage. Stain Technol 52, 229–232. [DOI] [PubMed] [Google Scholar]
- Elinson RP (2013) Chapter Nine – Metamorphosis in a frog that does not have a tadpole In: Current Topics in Developmental Biology (ed. Shi Y‐B.). Cambridge, MA: Academic Press. [DOI] [PubMed] [Google Scholar]
- Feng J, Gu Z, Lin X, et al. (2010) Postnatal development of type II collagen and aggrecan mRNA expression in a rabbit craniomandibular joint. Anat Rec (Hoboken) 293, 1574–1580. [DOI] [PubMed] [Google Scholar]
- Helminen HJ, Hyttinen MM, Lammi MJ, et al. (2000) Regular joint loading in youth assists in the establishment and strengthening of the collagen network of articular cartilage and contributes to the prevention of osteoarthrosis later in life: a hypothesis. J Bone Miner Metab 18, 245–257. [DOI] [PubMed] [Google Scholar]
- Hunziker EB, Michel M, Studer D (1997) Ultrastructure of adult human articular cartilage matrix after cryotechnical processing. Microsc Res Tech 37, 271–284. [DOI] [PubMed] [Google Scholar]
- Hunziker EB, Quinn TM, Hauselmann HJ (2002) Quantitative structural organization of normal adult human articular cartilage. Osteoarthritis Cartilage 10, 564–572. [DOI] [PubMed] [Google Scholar]
- Hunziker EB, Kapfinger E, Geiss J (2007) The structural architecture of adult mammalian articular cartilage evolves by a synchronized process of tissue resorption and neoformation during postnatal development. Osteoarthritis Cartilage 15, 403–413. [DOI] [PubMed] [Google Scholar]
- Jadin KD, Bae WC, Schumacher BL, et al. (2007) Three‐dimensional (3‐D) imaging of chondrocytes in articular cartilage: growth‐associated changes in cell organization. Biomaterials 28, 230–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakasiliotis K, Schilling N, Cabelguen JM, et al. (2013) Where are we in understanding salamander locomotion: biological and robotic perspectives on kinematics. Biol Cybern 107, 529–544. [DOI] [PubMed] [Google Scholar]
- Koyama E, Shibukawa Y, Nagayama M, et al. (2008) A distinct cohort of progenitor cells participates in synovial joint and articular cartilage formation during mouse limb skeletogenesis. Dev Biol 316, 62–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2− ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Macleod JN, Burton‐Wurster N, Gu DN, et al. (1996) Fibronectin mRNA splice variant in articular cartilage lacks bases encoding the V, III‐15, and I‐10 protein segments. J Biol Chem 271, 18 954–18 960. [DOI] [PubMed] [Google Scholar]
- Mienaltowski MJ, Huang L, Stromberg AJ, et al. (2008) Differential gene expression associated with postnatal equine articular cartilage maturation. BMC Musculoskelet Disord 9, 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nye HL, Cameron JA, Chernoff EA, et al. (2003) Extending the table of stages of normal development of the axolotl: limb development. Dev Dyn 226, 555–560. [DOI] [PubMed] [Google Scholar]
- Page RB, Monaghan JR, Walker JA, et al. (2009) A model of transcriptional and morphological changes during thyroid hormone‐induced metamorphosis of the axolotl. Gen Comp Endocrinol 162, 219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearle AD, Warren RF, Rodeo SA (2005) Basic science of articular cartilage and osteoarthritis. Clin Sports Med 24, 1–12. [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RHL, et al. (2003) Assumption‐free analysis of quantitative real‐time polymerase chain reaction (PCR) data. Neurosci Lett 339, 62–66. [DOI] [PubMed] [Google Scholar]
- Rosenkilde P, Ussing AP (1996) What mechanisms control neoteny and regulate induced metamorphosis in urodeles? Int J Dev Biol 40, 665–673. [PubMed] [Google Scholar]
- Sasano Y, Furusawa M, Ohtani H, et al. (1996) Chondrocytes synthesize type I collagen and accumulate the protein in the matrix during development of rat tibial articular cartilage. Anat Embryol (Berl) 194, 247–252. [DOI] [PubMed] [Google Scholar]
- Schulz MH, Zerbino DR, Vingron M, et al. (2012) Oases: robust de novo RNA‐seq assembly across the dynamic range of expression levels. Bioinformatics 28, 1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer HB, Voss SR (1996) Phylogenetic and mechanistic analysis of a developmentally integrated character complex: alternate life history modes in ambystomatid salamanders1. Am Zool 36, 24–35. [Google Scholar]
- Shubin NH, Alberch P (1986) A morphogenetic approach to the origin and basic organization of the tetrapod limb. Evol Biol 20, 319–387. [Google Scholar]
- Shwartz Y, Viukov S, Krief S, et al. (2016) Joint development involves a continuous influx of Gdf5‐positive cells. Cell Rep 15, 2577–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins RC, Cohen NW (1997) A Natural History of Amphibians. Princeton: Princeton University Press. [Google Scholar]
- Stewart R, Rascon CA, Tian S, et al. (2013) Comparative RNA‐seq analysis in the unsequenced axolotl: the oncogene burst highlights early gene expression in the blastema. PLoS Comput Biol 9, e1002936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassersug RJ (1976) A procedure for differential staining of cartilage and bone in whole formalin‐fixed vertebrates. Stain Technol 51, 131–134. [DOI] [PubMed] [Google Scholar]
- Wells KD (2010) The Ecology and Behavior of Amphibians. Chicago: University of Chicago Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Experimental samples for the analysis of limb morphometry. Forelimbs and hindlimbs of an aquatic axolotl salamander differentially stained for bone (red) and cartilage (blue) showing the skeletal elements used for histomorphometric analysis.
Fig. S2. Description of samples for the analysis of limb morphometry. Representative image of a histological section of the femorotibial joint (knee) illustrating the demarcation of metaphyseal cartilage, epiphyseal‐articular cartilage and the interzone for histomorphometric analysis (A) at 4 × and (B) at 20 × magnification. Representative images of a histological section of the distal end of femur illustrating the procedure of area (C) and total cell count (D) measurements for analysis of cellularity.
Fig. S3. Grouped analysis of limb morphometry. (a) Forelimb grouped analysis: the average bone area or length in aquatic axolotls was significantly higher than in the terrestrial axolotls groups 1 and 3 (mean ± SD). (b) Hindlimb grouped analysis: the average bone area or length in aquatic axolotls was significantly higher than in the terrestrial axolotls in group 3 (mean ± SD). Components of each group are defined in Table S1.
Table S1. Description of skeletal tissue groups from forelimbs and hindlimbs.
Table S2. Bone length measurements from ossified and non‐ossified sub‐segments of metacarpals and metatarsals.


