Abstract
Background
WD40-encoding RNA antisense to p53 (Wrap53) has been implicated in cancer development. However, the role of Wrap53 remains unknown in colorectal cancer. The aim of this study was to elucidate the function of Wrap53 in colorectal cancer tumorigenesis and development.
Material/Methods
This study analyzed Wrap53 expression in colorectal cancer tissue specimens using The Cancer Genome Atlas data and tumor cell lines and assessed the effects of Wrap53 knockdown on regulation of cancer cell malignant phenotypes in vitro and in nude mouse xenografts.
Results
Wrap53 expression was upregulated in colorectal cancer tissue specimens and cell lines. Knockdown of Wrap53 expression induced colorectal cancer cell line apoptosis and cell cycle arrest in the G1 phase, but reduced tumor cell line proliferation and invasion in vitro. Knockdown of Wrap53 in a nude mouse xenograft assay inhibited tumor cell line xenograft formation and growth.
Conclusions
Wrap53 is likely a potential oncogene or possesses oncogenic activity in colorectal cancer, promoting colorectal tumorigenesis. Targeting Wrap53 expression may represent a novel strategy for the control of colorectal cancer.
MeSH Keywords: Apoptosis, Cell Cycle, Cell Proliferation, Colorectal Neoplasms, Neoplasm Invasiveness
Background
Colorectal cancer (CRC) is the fifth most often diagnosed cancer in men and the fourth most often diagnosed cancer in women, and it is the fifth leading cause of cancer-related death in both men and women in China [1]. In the USA, colorectal cancer is the third most commonly diagnosed cancer in both men and women, the second leading cause of cancer-related death in men, and the third leading cause of cancer-related death in women [2]. Colorectal cancer is a heterogeneous disease caused by the interaction of genetic and environmental factors, including hereditary, high-fat diet, alcoholism, tobacco smoke, and stress [3]. The molecular mechanisms underpinning colorectal carcinogenesis remain to be defined. Thus, further study of colorectal cancer could improve understanding of gene expression, molecular events, and novel targets for colorectal cancer therapy.
WD40-encoding RNA antisense to p53 (Wrap53) has been implicated in cancer development and is overexpressed in cancer tissues including colorectal cancer [4,5]. The Wrap53 gene is localized on chromosome 17p13 and partially overlaps the p53 tumor suppressor gene in a head-to-head direction [6]. Wrap53 is a versatile gene and can be translated into a protein product with at least 17 splicing variants [7]. Wrap53 is also the natural p53 antisense transcript regulating p53 expression via post-transcriptional interaction of Wrap53 with p53 mRNA [7]. Wrap53 protein contains a WD40 domain, an essential component for Cajal body formation and maintenance [8]. Wrap53 protein can facilitate protein-protein and protein-RNA interactions and induces other proteins to form nuclear organelles, Cajal bodies, or to telomeres and DNA double-strand breaks [6,8]. Single-nucleotide polymorphisms (SNPs) in Wrap53 have been associated with a risk of developing breast cancer [9] and ovarian cancer [10]. Wrap53 SNP (rs2287499) is localized at Wrap53 exon 1 region and produces the missense mutation R68G, resulting in altered Wrap53 protein function and cancer development [9]. Moreover, Wrap53 overexpression was observed in head and neck cancer tissues [11] and is associated with poor prognosis [12]. Wrap53 overexpression was also observed in immortalized cells and various human cancer cell lines [12]. Together, these observations suggest that Wrap53 could be an oncogene.
In this study, we first assessed Wrap53 expression in colorectal cancer tissue specimens using The Cancer Genome Atlas (TCGA) data and colorectal cancer cell lines. We then assessed the effects of Wrap53 knockdown on inhibition of colorectal cancer cell malignant phenotypes in vitro and in nude mouse xenografts in vivo. We sought to further clarify the role of Wrap53 in colorectal cancer and to validate Wrap53 as a novel therapeutic target for colorectal cancer.
Material and Methods
Extraction of RNA-seq data on Wrap53 from the TCGA database
We first extracted RNA-seq data on Wrap53 from The Cancer Genome Atlas database (TCGA; https://cancergenome.nih.gov/) and identified a total of 521 subjects with 480 cancerous tissues and 41 normal tissues. There were 247 females, 272 males, and 2 missing information about gender. We then analyzed levels of Wrap53 expression in these tissues as log2-counts-per-million (logCPM).
Cell lines and culture
Human colorectal cancer SW480, HT-29, HCT116, and LoVo cell lines were originally obtained from Shanghai Genechem Co., Ltd. (Shanghai, China). SW480, HCT116, and LoVo cells were cultured in Roswell Park Memorial Institute 1640 (RPMI-1640; HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; Ausbian, Australia), while HT-29 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Corning, NY, USA) with 10% FBS (Ausbian, Australia) in a humidified incubator with 5% CO2 at 37°C. All cell lines were re-authenticated with STR analysis by Shanghai Genechem Co., Ltd. (Shanghai, China) in 2017.
Lentivirus production and infection
To knock down Wrap53 expression, we constructed 3 different shWrap53 lentivirus vectors targeting Wrap53 (GenBank access #NM_001143991) and assembled them with the help of Shanghai Genechem Co., Ltd. (Shanghai, China). After DNA sequence confirmation, they were named shWrap53-A, shWrap53-B, and shWrap53-C. The shScramble lentivirus (shScramble) was used as a negative control. After lentiviruses were produced, the multiplicity of infection (MOI) was calculated. Lentiviruses carrying Wrap53 shRNA were used to infect colorectal cancer cell lines. Cells stably expressing Wrap53 shRNA were selected using puromycin (Clontech, Mountain View, CA, USA).
Quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR)
Total RNAs were isolated from cells using a Trizol regent (Pufei, Shanghai, China) and reverse-transcribed into cDNA using Moloney Murine Leukemia Virus Reverse Transcriptase (M-MLV RT; Promega, Madison, WI, USA) according to the manufacturers’ protocols (denaturation and primer annealing: mix primer and RNA, incubate at 70°C for 10 min, then chill on ice; enzyme activation and extension: incubate at 42°C for 1 h; enzyme deactivation: incubate at 70°C for 10 min). The resulting cDNA samples were amplified with qPCR using the SYBR Master Mixture (TAKARA, Dalian, China) and Wrap53 primers of 5′-TTTGGAGACTCAACCGTTAGC-3′ and 5′-CGGTGGCATCAGTTCAGAG-3′ or GAPDH primers of 5′-TGACTTCAACAGCGACACCCA-3′ and 5′-CACCCTGTTGCTGTA GCCAAA-3′ (Initial activation: 1 cycle at 95°C for 30 s; Denaturation and Annealing and Extension: 40 cycles at 95°C for 5 s and 60°C for 30 s; Final Extension: 1 cycle at 95°C for 15 s, 60°C for 30 s, and 95°C for 15 s). qPCR was quantified using the 2−ΔΔCT method [13]. GAPDH mRNA was used as an internal control.
Simple western blot
Western blot analysis was performed using a capillary-based automated system, called the Simple Western System (http://www.proteinsimple.com/simple_western_overview.html). Total cellular protein was extracted using the Radio Immunoprecipitation Assay buffer (RIPA; Beyotime, Shanghai, China) and quantified using the bicinchoninic acid (BCA) protein assay kit (TAKARA, Dalian, China). Protein samples were then run in a Wes capillary electrophoresis system (ProteinSimple, San Jose, CA, USA) using the standard manufacturer’s protocol from ProteinSimple. The quantification of protein was done using Compass software (ProteinSimple) and β-actin protein was used as an internal control. The primary antibodies were anti-Wrap53 (Abcam, Cambridge, UK) and anti-β-actin (Santa Cruz, CA, USA), and the secondary antibodies were anti-Rabbit and anti-Mouse purchased from ProteinSimple.
Tumor cell proliferation and flow cytometry apoptosis and cell cycle assays
To assess tumor cell phenotypes, we first performed tumor cell proliferation MTT assay. In brief, after transfection with Wrap53 shRNA or negative control shRNA, cells were seeded into 6-well plates at 2×103 cells per well and cultured up to 5 days. To each culture well, 20 μl MTT reagent (Genview, USA) was added and further cultured for 4 h. Optical density was measured using a Microplate reader (Tecan Infinite, Switzerland) at 490/570 mm. Cell proliferation was calculated.
To assess tumor cell apoptosis, we employed Annexin V-APC staining. Tumor cells were infected with Wrap53 shRNA or negative control shRNA for 5 days, after which 10 μl Annexin V-APC reagent (eBioscience, USA) was added and further incubated for 10 min. Cell apoptosis rate was analyzed using flow cytometry (Millipore, USA).
Cell cycle distribution was assessed using propidium iodide staining and flow cytometry. Specifically, cells were grown and infected with lentiviruses, then detached using trypsin and reseeded into 6-cm dishes and grown for 3 days. Next, the cells were resuspended using trypsin and stained with propidium iodide (Sigma, St. Louis, MO, USA) at 37°C for 30 min in the dark. Cell cycle distribution was then analyzed using a flow cytometer (Millipore, USA).
Transwell invasion assay
Cell invasion capacity was assessed using the Transwell invasion assay. After infection with Wrap53 shRNA or negative control shRNA, cells were trypsinized and reseeded in the upper chamber of Transwell inserts in 24-well plates. The Transwell membrane was precoated with Matrigel (Corning, NY, USA). The bottom wells were filled with DMEM (Corning, NY, USA) supplemented with 10% FBS (Ausbian, Australia). After culture for 48 h, the cells remaining on the surface of the upper well were removed using a cotton swab and the cells that crossed the filter were fixed in 10% formalin and stained with Giemsa (DingGuo, Shanghai, China). The cells were then photographed under an inverted microscope and counted.
A nude mouse tumor cell xenograft model
Four-week-old BALB/c (nu/nu) female nude mice were purchased from Shanghai SLAC Laboratory Animal Co., Ltd. (Shanghai, China). Mice were subcutaneously injected with 200 μl of cell solution containing 4×106 colorectal cancer cells infected with Wrap53 shRNA or negative control shRNA in the right groin region. Tumor xenograft formation and growth were monitored daily and length and width were recorded twice weekly. On day 21, all mice were killed via cervical dislocation. Tumor xenografts were resected and weighed. All procedures performed in studies involving animals were in accordance with the ethics guidelines of the Institutional Animal Care and Use Committee of Wannan Medical College.
Statistical analysis
Data are expressed as mean ±SD for statistical analysis with the unpaired t test. A two-sided P value <0.05 was considered to indicate statistical significance. All statistical analyses were performed using R software (version 3.3.2).
Results
Wrap53 expression is upregulated in colorectal cancer tissue specimens and cell lines
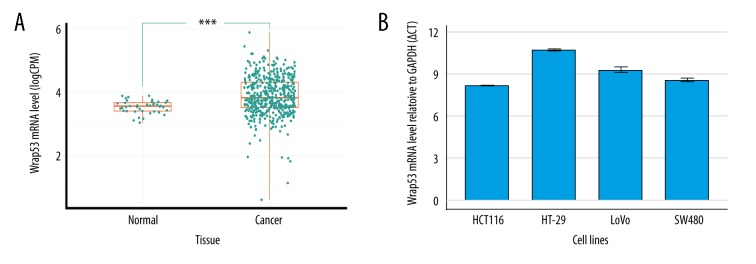
In this study, we first extracted the RNA-seq data on Wrap53 expression in colorectal tissues from the TCGA database. We identified 480 colorectal cancerous tissues and 41 normal tissues. Wrap53 expression was shown to be expressed at a higher level in colorectal cancer tissues than normal tissues (P<0.001; Figure 1A). We then used qRT-PCR to assess Wrap53 expression level in colorectal cancer cell lines and found Wrap53 mRNA to be highly expressed in SW480, HT-29, HCT116, and LoVo cell lines (Figure 1B). Wrap53 mRNA level of HCT116 was highest and we then selected the HCT116 cell line for subsequent Wrap53 knockdown experiments.
Figure 1.
Wrap53 expression is upregulated in colorectal cancer tissues and cell lines.(A) TCGA data. The analysis shows higher Wrap53 expression in colorectal cancer tissues than normal tissues (*** P<0.001; t test). (B) qRT-PCR. Colorectal cancer cell lines were grown and subjected to RNA isolation and qRT-PCR. All Δ;CT ≤12 indicated that Wrap53 was highly expressed in 4 cell lines. The experiments were carried out in triplicate.
Knockdown of Wrap53 expression in colorectal cancer cell line using lentivirus carrying Wrap53 shRNA
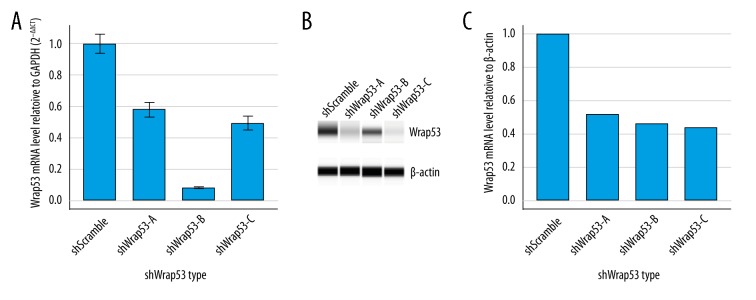
To knock down Wrap53 expression, we designed 3 different shWrap53 lentivirus products (shWrap53-A, shWrap53-B, and shWrap53-C) and shScramble lentivirus (shScramble) to infect HCT116 cells. Both qRT-PCR and Western blot analysis indicated that shWrap53-B most effectively knocked down Wrap53 expression in colorectal cell line HCT116 (Figure 2A–2C). Thus, this shWrap53-B was used in subsequent experiments.
Figure 2.
Knockdown of Wrap53 in colorectal cancer cell using lentivirus. (A) qRT-PCR. Colorectal cancer cells were transfected with Wrap53 shRNAs and then subjected to qRT-PCR analysis. The experiments were performed in triplicate. (B) Western blot analysis. Total cellular protein was extracted and subjected to Western blot analysis. (C) Relative quantification of Western blot analysis corrected by shScramble.
Wrap53 knockdown reduced colorectal cancer cell line proliferation in vitro
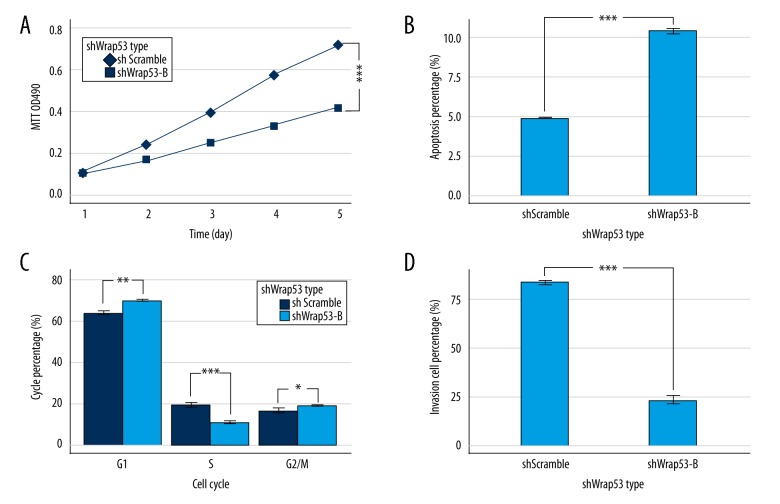
We then assessed the effects of Wrap53 knockdown on regulation of colorectal cancer phenotype in proliferation assays. The cell growth curve revealed that HCT116 cells infected with shWrap53-B proliferated significantly less than control group cells (P<0.001; Figure 3A), demonstrating that Wrap53 is required for colorectal cancer cell proliferation.
Figure 3.
Knockdown of Wrap53 in colorectal cancer HCT116 cells promotes apoptosis and arrest at the G1 stage but inhibits tumor cell proliferation and invasion. (A) Cell proliferation assay. HCT116 cells were transfected with Wrap53 shRNA or negative control shRNA, and proliferation was assessed by MTT assay (*** P<0.001; t test at day 5). (B) Cell apoptosis assay. HCT116 cells were transfected with Wrap53 shRNA or negative control shRNA and Annexin V staining was assessed by flow cytometry (*** P<0.001; t test). (C) Cell cycle assay. HCT116 cells were transfected with Wrap53 shRNA or negative control shRNA and PI staining was assessed by flow cytometry (* P<0.05, ** P<0.01, *** P<0.001; t test). (D) Transwell invasion assay. HCT116 cells were transfected with Wrap53 shRNA or negative control shRNA and invasion was assessed by Transwell invasion assay (*** P<0.001; t test). All experiments were performed in triplicate.
Wrap53 knockdown induced colorectal cancer cell line apoptosis in vitro
To assess the mechanism underpinning Wrap53 knockdown-induced reduced proliferation, we assessed Annexin V-APC staining by flow cytometry. The rate of apoptosis was significantly higher in HCT116 cells with shWrap53-B than shScramble (P<0.001; Figure 3B). This data supports the anti-apoptosis role of Wrap53 in colorectal cancer.
Wrap53 knockdown induced colorectal cancer cell line G1 cell cycle arrest in vitro
To investigate the cell cycle distribution after Wrap53 knockdown, we assessed PI staining by flow cytometry. Knockdown of Wrap53 expression significantly increased the percentage of cells in the G1 phase but reduced the percentage of cells in the S phase (P<0.01; Figure 3C). Taken together, these results suggest that Wrap53 promotes colorectal cancer cell growth but prevents apoptosis.
Wrap53 knockdown inhibited colorectal cancer cell line invasion in vitro
Tumor cell invasion leads to cancer metastasis and progression. Thus, we examined the invasion ability of HCT116 cells in which Wrap53 was knocked down in a Transwell invasion assay. HCT116 cells infected with shWrap53-B invaded more slowly than negative control cells (P<0.001; Figure 3D), suggesting that knockdown of Wrap53 expression suppressed tumor cell invasion capacity.
Wrap53 knockdown suppressed colorectal cancer cell xenograft formation and growth in vivo
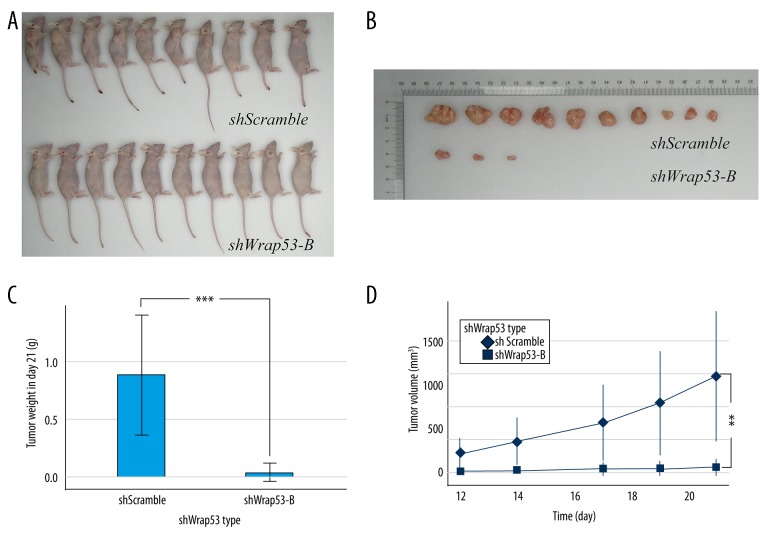
To better illustrate the oncogenic potential of Wrap53 in colorectal cancer in vivo, we first established a nude mouse colorectal cancer cell xenograft model (Figure 4A). On day 21, mice were killed, and tumor xenografts were resected and weighed (Figure 4B). We found that knockdown of Wrap53 expression suppressed colorectal cancer cell xenograft formation and growth in vivo (P<0.001; Figure 4C). The tumor volume growth curve also showed that the tumor volume of shWrap53-B was approximately 5% that of the control group on day 21 (P<0.01; Figure 4D). These data show that silencing of Wrap53 expression inhibited tumor growth in vivo.
Figure 4.
Knockdown of Wrap53 suppresses nude mouse colorectal cancer cell xenograft formation and growth in vivo. (A) Stable Wrap53-B knocked down HCT116 cells or negative control cells were subcutaneously injected into the right flank region of nude mice (n=10 each of group). Twenty-one days later, the mice were killed and photographed. (B) Tumor xenografts. Tumor xenografts were resected and weighted. (C) The weight of shWrap53-B tumor xenografts was significantly less than negative control xenografts (*** P<0.001; t test). (D) Tumor xenograft growth curve. The volume of shWrap53-B tumor xenografts was significantly lower than that of negative control xenografts (** P<0.01; t test at day 21).
Discussion
Wrap53 can be transcribed into at least 17 variants via 3 alternative start exons. Exon 1α, 1β, and 1γ lead to transcription into Wrap53α, Wrap53β, and Wrap53γ transcripts, respectively [7]. Previous studies reported that Wrap53 possesses dual functions as a natural p53 antisense transcript and a protein-encoding transcript [6,7,14]. However, only Wrap53α is a natural p53 antisense transcript and overlaps the first exon of p53 in an antisense fashion for up to 227 base pairs [7]. Several lines of evidence indicate that Wrap53α does not encode Wrap53 protein but is merely involved in regulation of p53 rather than Wrap53β, Wrap53γ, and Wrap53 protein [6,7,14]. Wrap53 protein contains a WD40 domain and is mainly produced from Wrap53β transcript, while WD40 domain proteins function to regulate a large variety of cell processes via protein-protein, protein-peptide, and protein-DNA interactions [15,16]. The Wrap53 protein can be detected in the cytoplasm and is highly enriched in Cajal bodies and nuclear organelles [6]. Cajal bodies are characterized by the marker protein coilin, which serves as a platform for the assembly of Cajal body components, including the splice variant small nuclear RNPs (snRNPs), small Cajal body-specific RNPs (scaRNPs), and small nucleolar RNPs (snoRNPs) [17–19]. In the present study, we found that Wrap53 was highly expressed in colorectal cancer tissues and cell lines, which is consistent with the expression pattern found in head and neck cancer cell lines and tissue samples [11]. Moreover, Wrap53 expression was previously reported to be a novel biomarker in early detection of rectal cancer since Wrap53 expression was higher in primary rectal cancer tissues than in normal mucosa, while expression of Wrap53 did not differ significantly between primary and metastasized tumors [5].
Furthermore, to explore the potential oncogenic role of Wrap53 in colorectal tumorigenesis, we knocked down Wrap53 expression using shWrap53 in HCT116 cells. Wrap53 shRNA significantly reduced Wrap53 mRNA and protein levels, which subsequently impaired HCT116 cell proliferation capacity, which was consistent with previous observations in head and neck cancer Cal-27, ACC2, and HSC-3 cell lines [11]. Moreover, we also found that knockdown of Wrap53 expression induced HCT116 cell apoptosis. Indeed, a previous study have reported that Wrap53-depletion triggered apoptosis of HCT116 and cervical cancer HeLa and osteosarcoma U2OS cell lines, but not in normal cells [12]. This process may result from activation of the mitochondria-induced apoptosis pathway after Wrap53 silencing [12]. This finding indicates that Wrap53 expression promotes cancer cell survival and proliferation.
The growth and survival of many cancer cells is dependent on telomerase activity and its recruitment to the telomere. The latter prevents the chromosomes from undergoing end-to-end fusion and degradation [20]. Despite this protective mechanism, the telomere is usually progressively shortened by each DNA replication, and a DNA damage response is triggered when it becomes too short to provoke cell senescence or apoptosis [20,21]. Such a reaction or activity is the most important mechanism restricting proliferation. However, cancer cells can avoid telomere shortening by expressing telomerase to maintain telomere length, promoting proliferative immortality [22]. Telomerase is a ribonucleoprotein complex, containing a RNA component (TR), telomerase reverse transcriptase (TERT), and the accessory proteins dyskerin, NOP10, and NHP2 [23]. Telomerase is assembled and matures in the Cajal body, and telomerase recruitment to telomere also requires the Cajal body, which is regulated and driven by Wrap53 protein by specifically binding to dyskerin and TR [24,25]. Dyskerin is a RNA-binding protein and can bind to numerous small nucleolar RNAs during telomerase assembly via its PUA domain [24]. Therefore, Wrap53 protein is essential for telomerase trafficking. If Wrap53 is mutated, telomerase deficiency and dyskeratosis congenital will occur [25,26]. Moreover, Wrap53 protein also plays a role in transportation of telomerase in a Cajal body-independent manner [27]. In addition, Wrap53 protein is also a subunit of the telomerase holoenzyme and is important for telomere maintenance in human cancer cells [25]. Taken together, knockdown of Wrap53 expression can alter telomerase activity and reduce tumor cell proliferation, but can also induce tumor cell apoptosis. Indeed, the present study confirmed this pattern in colorectal cancer cells in nude mouse xenografts. In cell cycle assay, the percentage of G1 phase HCT116 cells increased and the percentage of S phase HCT116 cells decreased after Wrap53 knockdown. The recruitment of telomerase to telomere occurs in S phase [26]. As mentioned above, Wrap53 is involved in telomerase transport to the telomere. Therefore, Wrap53 depletion inhibits telomerase trafficking, causing G1 cell cycle arrest. In general, apoptosis and cell cycle arrest are the most common inhibitors of cellular proliferation.
Approximately 90% of colorectal cancer-related mortality is caused by tumor distant metastases [28]. Suppression of tumor invasion can effectively inhibit tumor cell spread and infiltration into distant organs. In the present study, we further assessed the effects of Wrap53 knockdown on colorectal cancer cell invasion. The observed inhibition of colorectal cancer cell invasion by Wrap53 knockdown is consistent with a previous study of head and neck cancer [11], suggesting that Wrap53 is involved in cell mobility, which warrants further investigation. Furthermore, the effects of Wrap53 knockdown in vitro need to be confirmed in vivo. Thus, we performed a tumor cell nude mouse xenograft assay and confirmed our in vitro data, consistent with a previous study of head and neck cancer [11]. We did not record survival of colorectal cancer patients, which could be crucially important to determine the role of Wrap53 in colorectal cancer progression. However, at least 2 studies have reported that Wrap53 expression is associated with poor prognosis of colorectal cancer [4,5].
Conclusions
In conclusion, our results show Wrap53 overexpression in colorectal cancer tissues and cell lines. Knockdown of Wrap53 expression reduced tumor cell proliferation and invasion, but induced tumor cell apoptosis and G1 cell cycle arrest in vitro. In a nude mouse tumor cell xenograft assay, we confirmed our in vitro data. Thus, we conclude that Wrap53 is likely a potential oncogene or possesses oncogenic activity in colorectal cancer.
Footnotes
Conflict of interest
None.
Source of support: The study was supported by grants from the Natural Science Fund of the Education Department of Anhui Province (KJ2016A740) and the Education Department of Anhui Province (gxfx2017068)
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. Cancer J Clin. 2016;66:115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Lizarbe MA, Calle-Espinosa J, Fernández-Lizarbe E, et al. Colorectal cancer: From the genetic model to posttranscriptional regulation by noncoding RNAs. Biomed Res Int. 2017;2017 doi: 10.1155/2017/7354260. 7354260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang MJ, Ping J, Li Y, et al. The prognostic factors and multiple biomarkers in young patients with colorectal cancer. Sci Rep. 2015;5:10645. doi: 10.1038/srep10645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang H, Wang DW, Adell G, Sun XF. WRAP53 is an independent prognostic factor in rectal cancer – a study of Swedish clinical trial of preoperative radiotherapy in rectal cancer patients. BMC Cancer. 2012;12:294. doi: 10.1186/1471-2407-12-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henriksson S, Farnebo M. On the road with Wrap53β: guardian of Cajal bodies and genome integrity. Front Genet. 2015;6:91. doi: 10.3389/fgene.2015.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahmoudi S, Henriksson S, Corcoran M, et al. Wrap53, a natural p53 antisense transcript required for p53 induction upon DNA damage. Mol Cell. 2016;64:1009. doi: 10.1016/j.molcel.2016.11.027. [DOI] [PubMed] [Google Scholar]
- 8.Mahmoudi S, Henriksson S, Weibrecht I, et al. Wrap53 is essential for Cajal body formation and for targeting the survival of motor neuron complex to Cajal bodies. PLoS Biol. 2010;8:e1000521. doi: 10.1371/journal.pbio.1000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Closas M, Kristensen V, Langerød A, et al. Common genetic variation in TP53 and its flanking genes, WDR79 and ATP1B2, and susceptibility to breast cancer. Int J Cancer. 2007;121:2532–38. doi: 10.1002/ijc.22985. [DOI] [PubMed] [Google Scholar]
- 10.Schildkraut JM, Goode EL, Clyde MA, et al. Single nucleotide polymorphisms in the TP53 region and susceptibility to invasive epithelial ovarian cancer. Cancer Res. 2009;69:2349–57. doi: 10.1158/0008-5472.CAN-08-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun CK, Luo XB, Gou YP, et al. TCAB1: A potential target for diagnosis and therapy of head and neck carcinomas. Mol Cancer. 2014;13:180. doi: 10.1186/1476-4598-13-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahmoudi S, Henriksson S, Farnebo L, et al. WRAP53 promotes cancer cell survival and is a potential target for cancer therapy. Cell Death Dis. 2011;2:e114. doi: 10.1038/cddis.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Farnebo M. Wrap53, a novel regulator of p53. Cell Cycle. 2009;8:2343–46. doi: 10.4161/cc.8.15.9223. [DOI] [PubMed] [Google Scholar]
- 15.Stirnimann CU, Petsalaki E, Russell RB, Müller CW. WD40 proteins propel cellular networks. Trends Biochem Sci. 2010;35:565–74. doi: 10.1016/j.tibs.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Xu C, Min J. Structure and function of WD40 domain proteins. Protein Cell. 2011;2:202–14. doi: 10.1007/s13238-011-1018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nizami Z, Deryusheva S, Gall JG. The Cajal body and histone locus body. Cold Spring Harb Perspect Biol. 2010;2:a000653. doi: 10.1101/cshperspect.a000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cioce M, Lamond AI. Cajal bodies: A long history of discovery. Annu Rev Cell Dev Biol. 2005;21:105–31. doi: 10.1146/annurev.cellbio.20.010403.103738. [DOI] [PubMed] [Google Scholar]
- 19.Machyna M, Heyn P, Neugebauer KM. Cajal bodies: Where form meets function. Wiley Interdiscip Rev RNA. 2013;4:17–34. doi: 10.1002/wrna.1139. [DOI] [PubMed] [Google Scholar]
- 20.Calado RT, Young NS. Telomere diseases. N Engl J Med. 2009;361:2353–65. doi: 10.1056/NEJMra0903373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stanley SE, Armanios M. The short and long telomere syndromes: Paired paradigms for molecular medicine. Curr Opin Genet Dev. 2015;33:1–9. doi: 10.1016/j.gde.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sandin S, Rhodes D. Telomerase structure. Curr Opin Struct Biol. 2014;25:104–10. doi: 10.1016/j.sbi.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gomez DL, Armando RG, Cerrudo CS, et al. Telomerase as a cancer target. Development of new molecules. Curr Top Med Chem. 2016;16:2432–40. doi: 10.2174/1568026616666160212122425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cerrudo CS, Ghiringhelli PD, Gomez DE. Protein universe containing a PUA RNA-binding domain. FEBS J. 2014;281:74–87. doi: 10.1111/febs.12602. [DOI] [PubMed] [Google Scholar]
- 25.Venteicher AS, Abreu EB, Meng Z, et al. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science. 2009;323:644–48. doi: 10.1126/science.1165357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt JC, Cech TR. Human telomerase: Biogenesis, trafficking, recruitment, and activation. Genes Dev. 2015;29:1095–105. doi: 10.1101/gad.263863.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stern JL, Zyner KG, Pickett HA, et al. Telomerase recruitment requires both TCAB1 and Cajal bodies independently. Mol Cell Biol. 2012;32:2384–95. doi: 10.1128/MCB.00379-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu C, Zhu X, Liu W, et al. Hedgehog signaling pathway in colorectal cancer: Function, mechanism, and therapy. Onco Targets Ther. 2017;10:3249–59. doi: 10.2147/OTT.S139639. [DOI] [PMC free article] [PubMed] [Google Scholar]