Abstract
Purpose
Gene rearrangements that involve NTRK1/2/3 can generate fusion oncoproteins that contain the kinase domains of TRKA/B/C, respectively. These fusions are rare in non–small-cell lung cancer (NSCLC), with frequency previously estimated to be < 1%. Inhibition of TRK signaling has led to dramatic responses across tumor types with NTRK fusions. Despite the potential benefit of identifying these fusions, the clinicopathologic features of NTRK fusion-positive NSCLCs are not well characterized.
Methods
We compiled a database of patients with NSCLCs that harbor NTRK fusions. We characterized clinical, molecular, and histologic features with central review of histology.
Results
We identified 11 patients with NSCLCs that harbor NTRK gene fusions verified by next-generation sequencing and with available clinical and pathologic data. Fusions involved NTRK1 (n = 7) and NTRK3 (n = 4), with five and two distinct fusion partners, respectively. Fifty-five percent of cohort patients were male with a median age at diagnosis of 47.6 years (range, 25.3 to 86.0 years) and a median smoking history of 0 pack-years (range, 0 to 58 pack-years). Seventy-three percent of patients had metastatic disease at diagnosis. No concurrent alterations in KRAS, EGFR, ALK, ROS1, or other known oncogenic drivers were identified. Nine patients had adenocarcinoma, including two with invasive mucinous adenocarcinoma and one with adenocarcinoma with neuroendocrine features; one had squamous cell carcinoma; and one had neuroendocrine carcinoma. By collating data on 4,872 consecutively screened, unique patients with NSCLC, we estimate a frequency of NTRK fusions in NSCLC of 0.23% (95% CI, 0.11% to 0.40%).
Conclusion
NTRK fusions occur in NSCLCs across sexes, ages, smoking histories, and histologies. Given the potent clinical activity of TRK inhibitors, we advocate that all NSCLCs be screened for NTRK fusions by using a multiplexed next-generation sequencing–based fusion assay.
INTRODUCTION
The neurotrophin kinase (NTRK) genes NTRK1, NTRK2, and NTRK3 encode the tropomyosin receptor tyrosine kinases TRKA, TRKB, and TRKC, respectively, which function during normal neuronal development and maintenance. Gene rearrangements that involve each NTRK gene have been described in a wide variety of adult and pediatric solid tumor malignancies, and are believed to drive tumor growth and survival through expression of a constitutively active fusion protein that contains the TRK kinase domain. Although the frequency of NTRK fusions is low in common cancer types, including non–small-cell lung cancer (NSCLC), NTRK3 fusions are nearly ubiquitous among rare cancer types, such as mammary analog secretory carcinoma and infantile fibrosarcoma.1,2 In NSCLC, NTRK fusions are estimated to occur at a frequency of approximately 0.1% to 1%.1,3,4 They are less common than other oncogenic gene rearrangements that involve the anaplastic lymphoma kinase (ALK), ROS proto-oncogene 1 (ROS1), and RET proto-oncogene (RET), which occur at frequencies of approximately 4% to 6%, 1% to 2%, and 1% to 2%, respectively.5-7
Much like ALK-, ROS1-, or RET-rearranged NSCLCs, NTRK-rearranged NSCLCs seem to be oncogene dependent. Targeted inhibition of TRK signaling in preclinical models results in cell death and tumor regression.4 In early-phase clinical trials, solid tumors that harbor NTRK gene rearrangements have been highly sensitive to selective TRK tyrosine kinase inhibitors (TKIs), including larotrectinib, which is selective for TRKA/B/C, and entrectinib, which targets TRKA/B/C as well as ALK and ROS1. The objective response rate to larotrectinib across 55 adult and pediatric tumors with NTRK gene rearrangements is 75%.8 Responses have been seen across tumor types and NTRK gene partners. Among four patients with NSCLC, Response Evaluation Criteria in Solid Tumors (RECIST) responses were seen in three, and the fourth had an approximately 20% reduction in tumor size. One of four NTRK-rearranged tumors treated with entrectinib in an adult phase I trial was NSCLC, and this patient had a partial response, including a complete response in the CNS.9 Despite the potent activity of TRK inhibitors, the clinical and pathologic features of NTRK-rearranged NSCLCs are poorly defined. We show that NTRK fusions occur across age and smoking status and suggest that all patients with NSCLC be screened for fusions by using a multiplexed next-generation sequencing (NGS) assay.
METHODS
Physicians across seven institutions contributed de-identified patients with NSCLC to an NTRK fusion NSCLC database. Clinical staging was performed by treating physicians using American Joint Committee on Cancer, seventh edition, criteria. NTRK fusions were identified and validated as part of routine clinical testing at each institution. Assays used identified fusions through a variety of technologies: RNA-based fusion-targeted anchored multiplex polymerase chain reaction (PCR) and Illumina (San Diego, CA) sequencing10 (Massachusetts General Hospital [MGH] Solid Fusion Assay, Memorial Sloan Kettering [MSK] Solid Fusion Assay, ArcherDx FusionPlex performed at Caris Life Sciences [Irving, TX]); DNA hybridization capture with intron tiling and Illumina sequencing (FoundationOne11 [Foundation Medicine, Cambridge, MA], MSK-Integrated Mutation Profiling of Actionable Cancer Targets [IMPACT]12,13), total nucleic acid extraction, PCR amplification, and ion torrent sequencing (PCDx14; Paradigm, Phoenix, AZ).
The Kaplan-Meier method was used to obtain estimates of overall survival from diagnosis of stage IV disease to death or last follow-up (censored). The Clopper-Pearson exact method for the binomial distribution was used to obtain CIs for NTRK fusion frequencies.
Two central pathologists (M.S.T. and M.M.-K.) reviewed tumor histology. When used, immunohistochemistry was performed on a BOND automated system (Leica Biosystems, Buffalo Grove, IL) with the standard chromogen 3,3′-diaminobenzidine tetrahydrochloride hydrate using antigen retrieval solution ER1 (citrate buffer with surfactant, pH 6.0) or ER2 (EDTA buffer with surfactant, pH 9.0), with antibody incubated at room temperature as follows: α-TTF1 (ready-to-use [RTU] PA0364 [Leica Biosystems], ER2 for 30 minutes), α-ΔNp63 (p40, RTU API3066AA [Biocare Medical, Pacheco, CA], ER1 for 30 minutes), α-chromogranin (RTU PA0430 [Leica Biosystems], ER2 for 20 minutes), and α-synaptophysin (RTU PA0299 [Leica Biosystems], ER2 for 20 minutes). Data collection and analysis were performed under institutional review board–approved protocols.
RESULTS
We reached out to physicians at 47 institutions in the United States who were actively participating in a TRK inhibitor clinical trial that enrolled adult patients and invited them to contribute data on living or deceased patients with NSCLCs that harbored an NTRK gene rearrangement. Data on 14 patients were initially contributed from seven institutions. Candidate fusions initially were identified by using a combination of RNA- and DNA-based NGS assays, with validation by one or more of RNA-based NGS, fluorescent in situ hybridization, and reverse transcription PCR on a patient-by-patient basis. Among these patients, in-frame TRK fusions that contained the kinase domain were verified in 11, which formed the study cohort.
Of note, three patients were excluded from the study cohort for the following reasons. The first patient had an NTRK1 fusion detected by MSK-IMPACT, a DNA-based hybridization capture NGS assay,12 but not by subsequent confirmatory testing with the MSK Solid Fusion Assay, an RNA-based fusion-specific targeted NGS assay that uses anchored multiplex PCR.10 The candidate fusion contained P2RY8 exon 2 fused with NTRK1 exons 1 to 5. Because NTRK1 exons 1 to 5 lack the kinase domain, this was believed to be a nonfunctional fusion. This patient also had a concurrent KRAS G12C mutation, an established oncogenic driver. The second patient had an NTRK2 intragenic deletion that disrupted the exon 18 3′ splice site, which is predicted to disrupt the kinase domain and, therefore, to be inactivating. The third patient had an NTRK1 alteration detected by fluorescent in situ hybridization but not verified by NGS. This patient also had a concurrent HER2 L755P mutation, which is predicted to be activating.15
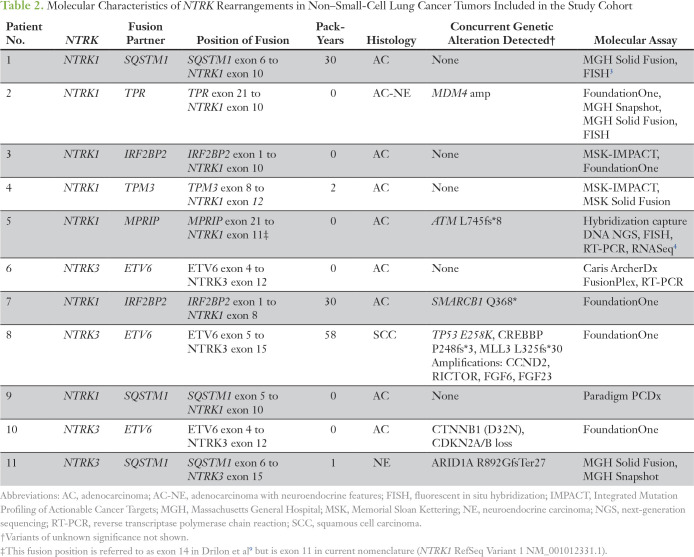
The clinical characteristics of the 11 patients in the study cohort are listed in Table 1. At the time of data analysis, six patients were living, and five were deceased. The molecular characteristics of the study cohort are listed in Table 2 and shown in Figure 1 (patients 1 to 11). Seven patients had NTRK1 fusions with five distinct fusion partners, and four had NTRK3 fusions with two distinct fusion partners. Patient 4 had a candidate NTRK1 fusion detected by MSK-IMPACT with an equivocal partner, and the correct fusion partner was determined using the MSK Solid Fusion Assay. All NTRK fusions couple the kinase domain of NTRK1 or NTRK3 (with or without the membrane-spanning helix) to an N-terminal gene fusion partner with domains known or predicted to mediate dimerization or oligomerization (Table 2; Fig 1). Two of nine patients tested had concurrent mutations in TP53. In all patients tested, potential oncogenic alterations in the following genes, when interrogated, were not detected: KRAS (zero of 10), EGFR (zero of 11), ALK (zero of 11), ROS1 (zero of 11), BRAF (zero of 11), PIK3CA (zero of 10), HER2 (zero of eight), and MET (zero of eight).
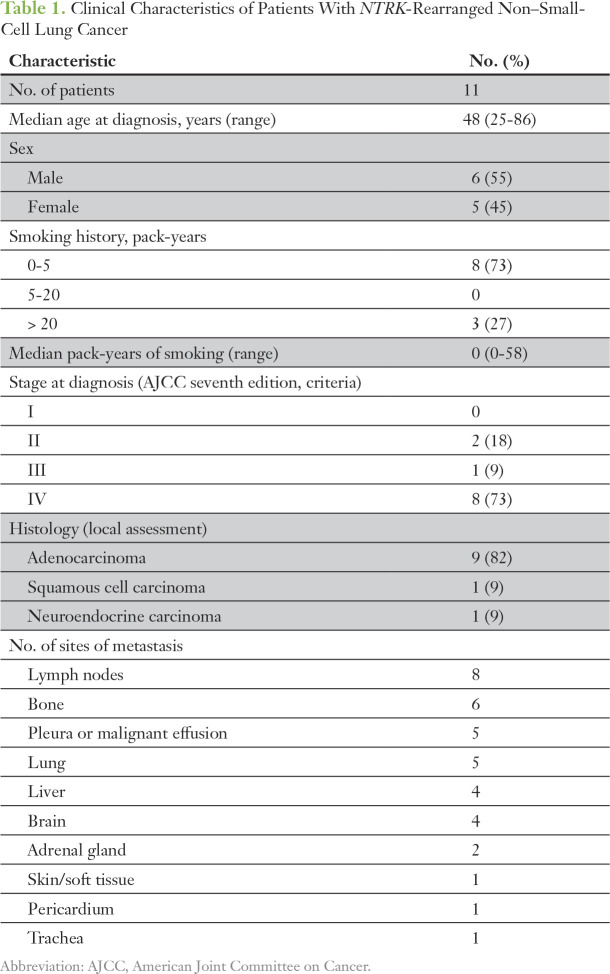
Table 1.
Clinical Characteristics of Patients With NTRK-Rearranged Non–Small-Cell Lung Cancer
Table 2.
Molecular Characteristics of NTRK Rearrangements in Non–Small-Cell Lung Cancer Tumors Included in the Study Cohort
Fig 1.
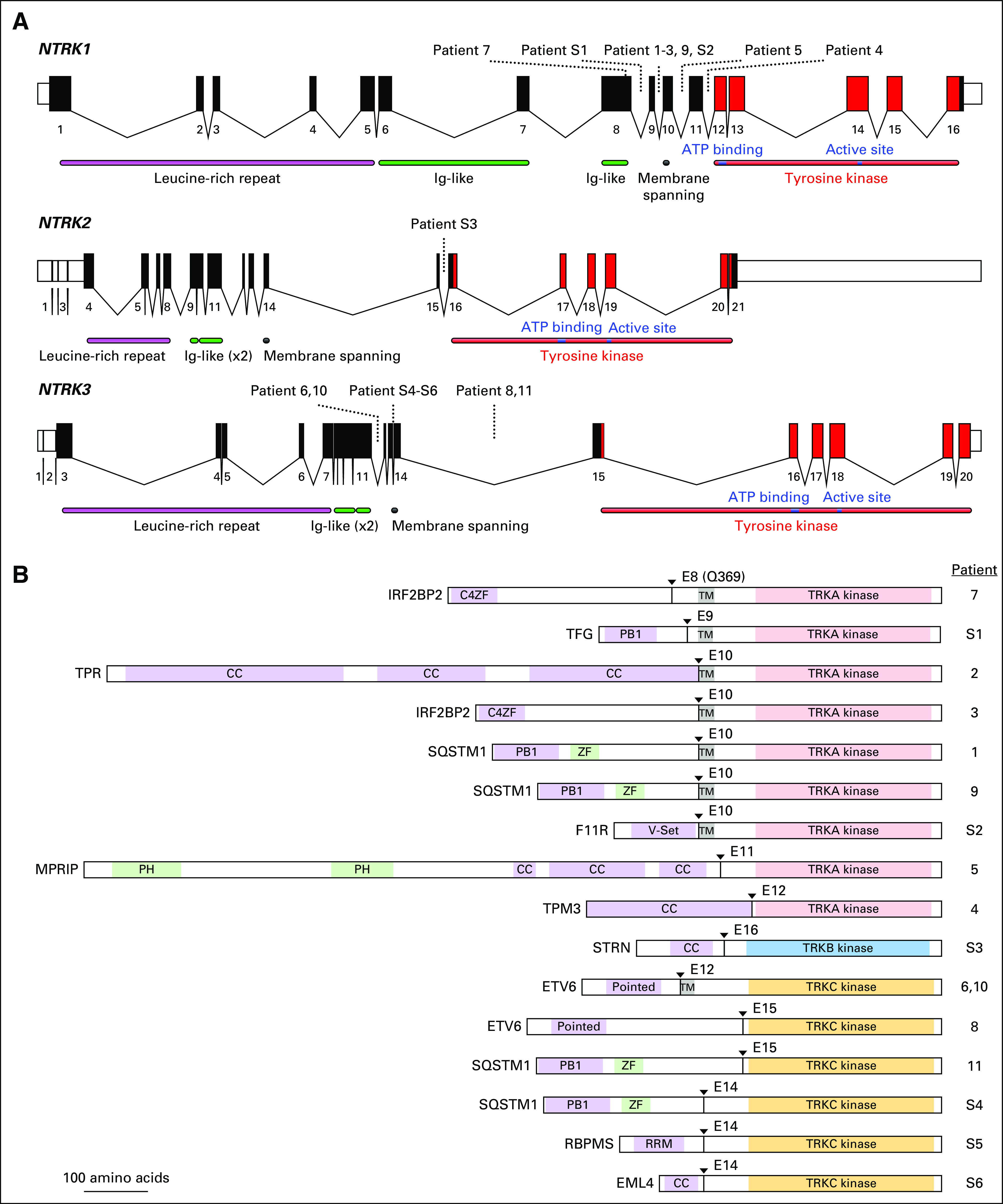
(A) Schematic of the human NTRK loci. Exon numbers are shown below their respective boxes for reference sequence NTRK1 transcript variant 1 (NM_001012331.1), NTRK2 transcript variant a (NM_006180.4), and NTRK3 transcript variant 1 (NM_001012338.2). Fusion breakpoints are shown as dotted lines for the indicated patients. Patient 7 has an exonic breakpoint; all other breakpoints are intronic. Note that exons are drawn at a larger scale than introns and that introns are not drawn to the same scale for each gene (NTRK1 locus is approximately 21 kilobases [kb], NTRK2 is approximately 358 kb, and NTRK3 is approximately 384 kb). (B) Schematic of predicted fusion protein products (see also Tables 2 and 3). Triangles and E notation indicate the fusion breakpoints and subsequent TRK exon. Purple-shaded domains are those predicted or shown to induce dimerization in the fusion partner (C4ZF, C4 zinc finger; PB1, Phox and Bem1p interaction domain; CC, coiled coil; Pointed, sterile alpha motif [SAM]/helix loop helix [HLH] oligomerization domain; RRM, RNA recognition motif). Green-shading indicates domains are other annotated sequence features (ZF, zinc finger; PH, pleckstrin homology). Gray shading indicates the transmembrane domain (TM). The kinase domains of TRKA/B/C are indicated and shown in light red, blue, and gold, respectively. Proteins are drawn to scale (MPRIP-NTRK1 fusion = 1,332 amino acids). Ig, immunoglobulin.
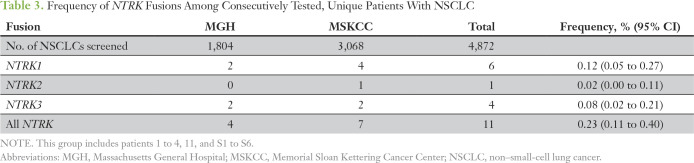
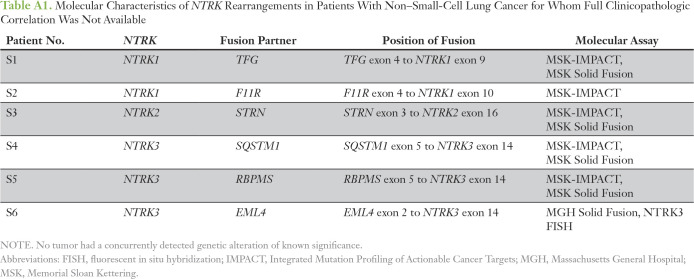
To estimate the overall frequency of NTRK fusions in NSCLC, we reviewed consecutively tested patients with NSCLC from MGH and the MSK Cancer Center, where NGS screening of 4,872 unique patients identified 11 NTRK fusions (0.23%; Table 3). The frequencies of NTRK1, NTRK2, and NTRK3 fusions were 0.12%, 0.02%, and 0.08%, respectively. Five of these patients had available clinical and pathologic data for inclusion in the study cohort, and we report the molecular details of the additional six patients (S1 to S6; Appendix Table A1). We diagram the fusion positions of all 17 of these patients (study cohort patients 1 to 11 plus patients S1 to S6) in Figure 1.
Table 3.
Frequency of NTRK Fusions Among Consecutively Tested, Unique Patients With NSCLC
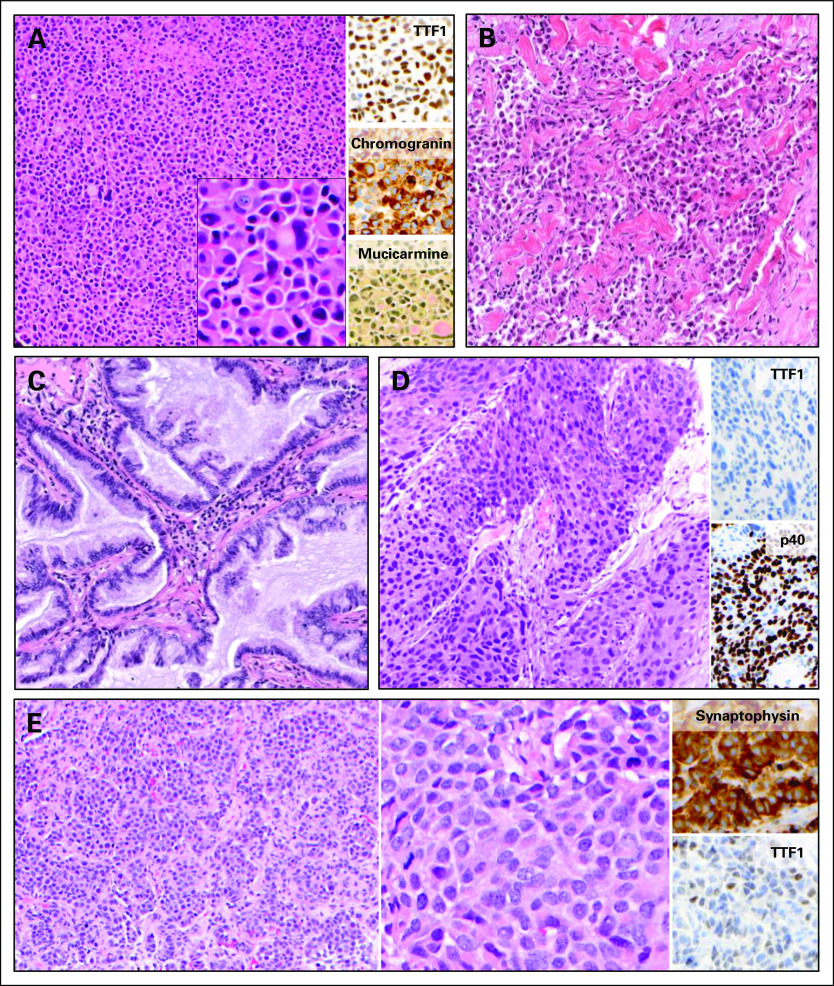
We next examined the histologic features of the 11 patients who formed the study cohort. Nine were adenocarcinoma, one was squamous cell carcinoma, and one was neuroendocrine carcinoma. Among the patients with adenocarcinoma, we observed a range of histologic subtypes, including adenocarcinoma with neuroendocrine features (patient 2; Fig 2A), poorly differentiated adenocarcinoma with solid pattern and signet ring cells (patient 1; Fig 2B), and invasive mucinous adenocarcinoma (patients 4 and 6; Fig 2C). Squamous cell histology was observed in patient 8 and was confirmed with adequate sampling and by immunohistochemical expression of p40 and absence of TTF1 (patient 8; Fig 2D). Patient 11 (Fig 2E) had a morphologically well-differentiated neuroendocrine tumor (equivalent to atypical carcinoid) with an increased mitotic index of 12 per 10 high-power fields and a brain metastasis; this tumor was classified as large-cell neuroendocrine carcinoma in accordance with current WHO criteria.16
Fig 2.
Histology of select patient tumors; original magnification x100 unless otherwise specified. (A) Patient 2 had an adenocarcinoma with solid growth pattern, diffuse neuroendocrine differentiation, and signet ring cells; inset shows high magnification (x400). (B) Patient 1 had poorly differentiated adenocarcinoma with solid and single-cell growth patterns. (C) Patient 4 had mucinous adenocarcinoma, and patient 6 had similar histology (data not shown). (D) Patient 8 had squamous cell carcinoma. (E) Patient 11 had neuroendocrine carcinoma with well-differentiated morphology and increased mitotic activity (left); high magnification (x400) shown in middle.
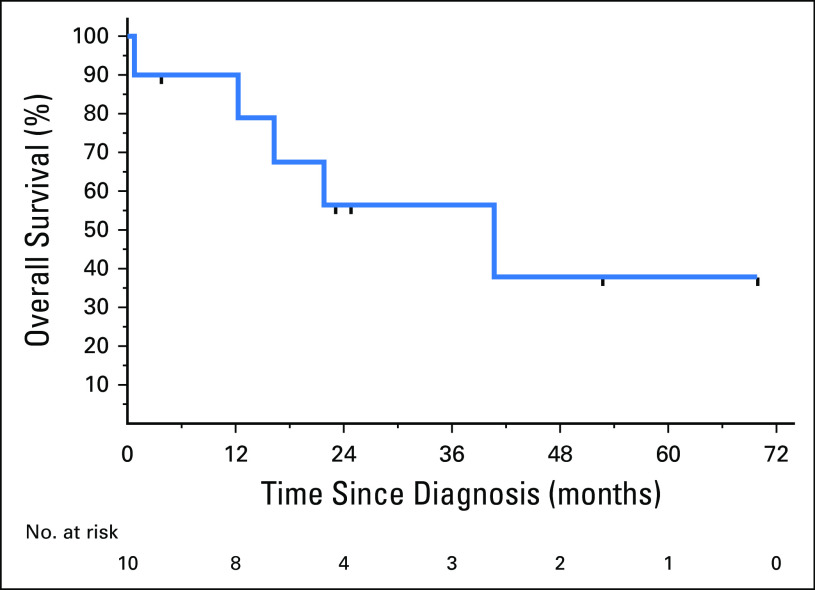
Although analysis of the cohort is limited by size and the fact that this review is retrospective across multiple institutions, we sought to describe clinical outcomes in these patients. Across the cohort of 11 patients, eight (73%) received at least one TRK TKI at some point in their treatment course, and 10 (91%) received a platinum doublet. One patient (9%) received no treatment. The median overall survival of the 10 patients with metastatic disease was 40.8 months (95% CI, 0.79 months to not reported), with a median follow-up of 52.8 months (Fig 3).
Fig 3.
Overall survival of the 10 patients with metastatic disease measured from the date of stage IV diagnosis to date of death or last known follow-up. Censored patients are marked on the curve.
Three patients had early-stage disease at the time of diagnosis. Patient 4 had stage IIB (T3N0) disease at diagnosis, was treated with surgery followed by adjuvant cisplatin and pemetrexed, and remained recurrence free at the most recent follow-up 30.0 months after initial diagnosis. Patient 6 had stage IIA (T1bN1) disease at diagnosis, was treated with surgery followed by cisplatin and pemetrexed, and developed metastatic disease 24.5 months after initial diagnosis. Patient 10 had stage IIIB (T4N2M0) disease at diagnosis, was treated with chemotherapy and radiation, and developed metastatic disease 10 months after initial diagnosis. The remaining eight patients had metastatic disease at the time of diagnosis.
DISCUSSION
TRK TKIs have shown tremendous promise in NTRK fusion-positive solid tumors across cancer types,8,9 which follows the paradigm now well established for EGFR mutant and ALK or ROS1 fusion-positive NSCLCs. Although NTRK fusions are rare in NSCLC, uncertainty remains about which patients should undergo testing for these alterations. We describe the clinicopathologic features of a cohort of 11 patients with NSCLCs harboring NTRK gene rearrangements that resulted in the fusion of the TRK tyrosine kinase domain with a dimerization-inducing partner. These are predicted or previously have been reported to be activating.4,17 The current cohort includes both men and women across a range of ages, histologies, and smoking histories. Although the cohort is small, the only defining pattern of clinical characteristics that emerges is the lack of an alternate canonical driver mutation in all patients, similar to others with kinase fusion-positive NSCLCs.18 Of note, NTRK rearrangements were identified in patients with and without a history of smoking; although the majority of patients (eight [73%] of 11) had a minimal to never smoking history, three of the 11 had a history of ≥ 30 pack-years. Similarly, ALK-, ROS1-, and RET-driven NSCLCs are enriched in never-smokers but can be seen in current and former smokers as well.5,19,20 Nine of the 11 patients had adenocarcinoma that tended to be mucinous or poorly differentiated, including one with a TPR-NTRK1 fusion with neuroendocrine differentiation. However, other histologies also were observed, including one squamous cell carcinoma with an ETV6-NTRK3 fusion and one neuroendocrine carcinoma with an SQSTM1-NTRK3 fusion.
Ascertainment bias as a result of selective testing has historically limited an accurate assessment of frequency of NTRK fusions in NSCLC. We have combined the clinical experience from multiplexed targeted NGS screening of 4,872 unique, consecutive patients with NSCLC at both MGH and MSK Cancer Center to estimate an NTRK fusion frequency of 0.23% in NSCLC. These assays generally are used at the time of tissue diagnosis in both institutions; therefore, this population likely represents a previously unscreened group in which patients were not already selected to be negative for other known driver mutations in lung cancer. We note that cancers selected for molecular testing may be enriched for patients with metastatic disease because no established role exists for targeted therapies in early-stage lung cancer to date. Although NTRK fusions are rare in lung cancer, we estimate that with approximately 234,000 new NSCLC diagnoses annually in the United States, > 500 of these patients may be candidates for highly effective TRK inhibitor therapy. Significantly more patients with NTRK fusion NSCLC may exist when considering the global incidence of lung cancer.
The natural history of NTRK fusion NSCLCs, compared with NSCLCs in general, is not well established. Although we observed a median overall survival of 40.8 months among the 10 patients with metastatic disease, we acknowledge the small size of this retrospective cohort, among whom eight received at least one TRK TKI. The observation that one of two patients diagnosed at stage II and one at stage III developed relapsed metastatic disease is consistent with the natural history of NSCLCs in general, although selection bias may have existed against screening patients with early-stage cancer who did not develop metastatic disease.
Because there seems to be no uniform defining clinical or pathologic feature of NTRK fusion-positive NSCLCs, we recommend screening all NSCLCs for NTRK gene rearrangements. In our experience, RNA-based fusion assays, such as the MGH or MSK Solid Fusion Assays or related ArcherDx FusionPlex, have a number of advantages over DNA-based methods, including high sensitivity, confident identification of breakpoints and in-frame fusions, and deeper coverage.10 Three patients with predicted nonfunctional NTRK alterations also were identified in this study, which emphasizes the added value of NGS-based sequencing and attention to the breakpoints. Although immunohistochemical assays for the detection of TRK expression are in development,21 allocation of an unstained slide for TRK immunohistochemistry may be impractical given the need to test for a wide range of molecular alterations on often-limited tissue samples. Similarly, given the seeming lack of concurrent canonical driver mutations in these patients, consideration of an initial DNA-based NGS for mutational profiling may be reasonable, with reflex multiplexed fusion-targeted RNA-based NGS in tumors that lack such a driver. However, sequential testing for possible gene alterations can delay the ultimate molecular diagnosis, may be problematic for small samples, and relies on mutual exclusivity of a kinase fusion and oncogenic driver mutation. Therefore, we favor concurrent NGS-based mutational analysis with multiplexed NGS-based targeted RNA sequencing for the identification of gene fusions in NSCLC rather than sequential mutation testing or immunohistochemistry, which consumes more time and tissue. Ultimately, we anticipate that more widespread and comprehensive NTRK fusion testing in patients with NSCLC will lead to expanded treatment options for NTRK fusion–positive patients.
ACKNOWLEDGMENT
We thank Roxana Azimi for administrative support and the patients and families who contributed to this study.
Appendix
Table A1.
Molecular Characteristics of NTRK Rearrangements in Patients With Non–Small-Cell Lung Cancer for Whom Full Clinicopathologic Correlation Was Not Available
Footnotes
Supported by the President and Fellows of Harvard College (Catalyst Medical Research Investigator Training to A.F.F.), the National Cancer Institute (Chabner K12CA087723 to A.F.F.), and the National Cancer Institute University of Colorado Lung Cancer Specialized Program of Research Excellence (P50 CA058187 to R.C.D.).
Presented at the 2017 Annual Meeting of the American Society of Clinical Oncology, Chicago, IL, June 2-6, 2017.
AUTHOR CONTRIBUTIONS
Conception and design: Anna F. Farago, Martin S. Taylor, Shivaani Kummar, Maria E. Arcila, Sai-Hong I. Ou, Alice T. Shaw, Alexander Drilon
Financial support: Anna F. Farago, A. John Iafrate
Administrative support: Anna F. Farago, Dara L. Aisner, Jochen K. Lennerz, A. John Iafrate, Sai-Hong I. Ou, Mari Mino-Kenudson
Provision of study materials or patients: Anna F. Farago, Robert C. Doebele, Viola W. Zhu, Theresa A. Boyle, Eric B. Haura, A. John Iafrate, Sai-Hong I. Ou
Collection and assembly of data: Anna F. Farago, Martin S. Taylor, Robert C. Doebele, Viola W. Zhu, Shivaani Kummar, Alexander I. Spira, Theresa A. Boyle, Eric B. Haura, Maria E. Arcila, Ryma Benayed, Dara L. Aisner, Jochen K. Lennerz, A. John Iafrate, Sai-Hong I. Ou, Alice T. Shaw, Mari Mino-Kenudson, Alexander Drilon
Data analysis and interpretation: Anna F. Farago, Martin S. Taylor, Robert C. Doebele, Shivaani Kummar, Theresa A. Boyle, Maria E. Arcila, Ryma Benayed, Nora K. Horick, Jochen K. Lennerz, Long P. Le, A. John Iafrate, Sai-Hong I. Ou, Alice T. Shaw, Mari Mino-Kenudson, Alexander Drilon
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Anna F. Farago
Honoraria: Foundation Medicine
Consulting or Advisory Role: PharmaMar, Takeda Pharmaceuticals, AbbVie, Loxo Oncology, Stemcentrx
Research Funding: PharmaMar (Inst), AbbVie (Inst), Bristol-Myers Squibb (Inst), Merck (Inst), Loxo Oncology (Inst), Ignyta (Inst)
Travel, Accommodations, Expenses: PharmaMar, AbbVie, Stemcentrx
Martin S. Taylor
No relationship to disclose
Robert C. Doebele
Stock and Other Ownership Interests: Rain Therapeutics
Honoraria: Pfizer, AstraZeneca, ARIAD Pharmaceuticals, Guardant Health, Takeda Pharmaceuticals, Spectrum Pharmaceuticals, Trovagene
Consulting or Advisory Role: Pfizer, OncoMed Pharmaceuticals, Trovagene, Ignyta, GreenPeptide, AstraZeneca
Research Funding: Ignyta (Inst)
Patents, Royalties, Other Intellectual Property: Licensing fees from Abbott Molecular for patent PCT/US2013/057495, licensing fees from Ignyta for biologic materials (Inst)
Travel, Accommodations, Expenses: Ignyta, ARIAD Pharmaceuticals, Guardant Health
Viola W. Zhu
Stock and Other Ownership Interests: TP Therapeutics (I)
Honoraria: AstraZeneca
Consulting or Advisory Role: AstraZeneca, Bayer AG, TP Therapeutics, Biocept
Speakers’ Bureau: AstraZeneca, Roche, Genentech, Takeda Pharmaceuticals
Travel, Accommodations, Expenses: AstraZeneca, Roche, Genentech, Takeda Pharmaceuticals, TP Therapeutics
Shivaani Kummar
Consulting or Advisory Role: Corvus Pharmaceuticals, MedTree (I), Nodus Therapeutics
Research Funding: Bristol-Myers Squibb (Inst), Dynavax Technologies (Inst), Pfizer (Inst), Loxo Oncology (Inst), Corvus Pharmaceuticals (Inst), Plexxikon (Inst), Jounce Therapeutics (Inst), ADC Therapeutics (Inst), Advenchen Laboratories (Inst)
Alexander I. Spira
Research Funding: Roche (Inst), AstraZeneca (Inst), Boehringer Ingelheim (Inst), Astellas Pharma (Inst), MedImmune (Inst), Novartis (Inst), NewLink Genetics (Inst), Incyte (Inst), AbbVie (Inst), Ignyta (Inst), LAM Therapeutics (Inst), Trovagene (Inst), Takeda Pharmaceuticals (Inst), MacroGenics (Inst), CytomX Therapeutics (Inst)
Theresa A. Boyle
Other Relationship: Bristol-Myers Squibb
Eric B. Haura
Consulting or Advisory Role: Bristol-Myers Squibb, Janssen Pharmaceuticals
Research Funding: Eli Lilly, Ignyta, Janssen Pharmaceuticals, Boehringer Ingelheim, Forma Therapeutics
Patents, Royalties, Other Intellectual Property: Patent pending on proximity ligation assay technology related to kinase inhibitor sensitivity biomarkers
Travel, Accommodations, Expenses: Bristol-Myers Squibb, Roche
Maria E. Arcila
Consulting or Advisory Role: AstraZeneca
Travel, Accommodations, Expenses: AstraZeneca, Invivoscribe, Raindance Technologies
Ryma Benayed
No relationship to disclose
Dara L. Aisner
Honoraria: AstraZeneca, Clovis Oncology
Consulting or Advisory Role: Inivata, AbbVie, Bristol-Myers Squibb
Research Funding: Genentech (Inst), Roche (Inst)
Patents, Royalties, Other Intellectual Property: Patent pending for pneumatic cell collection device
Travel, Accommodations, Expenses: SeraCare Life Sciences
Nora K. Horick
No relationship to disclose
Jochen K. Lennerz
No relationship to disclose
Long P. Le
Stock and Other Ownership Interests: ArcherDx
Consulting or Advisory Role: ArcherDx
Patents, Royalties, Other Intellectual Property: Co-inventor of the anchored multiplex PCR technology, which is licensed to ArcherDx and receive royalty payments for this patent
Travel, Accommodations, Expenses: ArcherDx
A. John Iafrate
Stock and Other Ownership Interests: ArcherDx
Consulting or Advisory Role: Debiopharm Group, Chugai Pharmaceutical, Roche
Research Funding: Blueprint Medicines
Patents, Royalties, Other Intellectual Property: ArcherDx exclusive license to AMP technology
Sai-Hong I. Ou
Stock and Other Ownership Interests: TP Therapeutics
Honoraria: Pfizer, Roche, Genentech, ARIAD Pharmaceuticals, Takeda Pharmaceuticals, Novartis, AstraZeneca, Foundation Medicine
Consulting or Advisory Role: Pfizer, Roche, Genentech, Novartis, AstraZeneca, Takeda Pharmaceuticals, Foundation Medicine, TP Therapeutics, Ignyta
Speakers’ Bureau: Genentech, AstraZeneca, Takeda Pharmaceuticals
Research Funding: Pfizer (Inst), Roche (Inst), AstraZeneca (Inst), MedImmune (Inst), Clovis Oncology (Inst), ARIAD Pharmaceuticals (Inst), Ignyta (Inst), Peregrine Pharmaceuticals (Inst), GlaxoSmithKline (Inst), Astellas Pharma (Inst), Chugai Pharmaceutical (Inst)
Alice T. Shaw
Honoraria: Pfizer, Novartis, Roche, Genentech, Foundation Medicine
Consulting or Advisory Role: Pfizer, Novartis, Genentech, Roche, ARIAD Pharmaceuticals, Ignyta, Blueprint Medicines, Daiichi Sankyo, EMD Serono, Taiho Pharmaceutical, KSQ Therapeutics, Natera, Loxo Oncology, Takeda Pharmaceuticals
Research Funding: Pfizer, Novartis, Roche, Genentech
Mari Mino-Kenudson
Consulting or Advisory Role: Merrimack Pharmaceuticals, H3 Biomedicine
Alexander Drilon
Consulting or Advisory Role: Ignyta, Loxo Oncology, TP Therapeutics, AstraZeneca, Pfizer, Blueprint Medicines, Genentech, Roche, Takeda Pharmaceuticals, Helsinn Group, BeiGene
REFERENCES
- 1.Vaishnavi A, Le AT, Doebele RC. TRKing down an old oncogene in a new era of targeted therapy. Cancer Discov. 2015;5:25–34. doi: 10.1158/2159-8290.CD-14-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farago AF, Azzoli CG. Beyond ALK and ROS1: RET, NTRK, EGFR and BRAF gene rearrangements in non-small cell lung cancer. Transl Lung Cancer Res. 2017;6:550–559. doi: 10.21037/tlcr.2017.08.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farago AF, Le LP, Zheng Z, et al. Durable clinical response to entrectinib in NTRK1-rearranged non-small cell lung cancer. J Thorac Oncol. 2015;10:1670–1674. doi: 10.1097/01.JTO.0000473485.38553.f0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaishnavi A, Capelletti M, Le AT, et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med. 2013;19:1469–1472. doi: 10.1038/nm.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaw AT, Yeap BY, Mino-Kenudson M, et al. Clinical features and outcome of patients with non–small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27:4247–4253. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang R, Hu H, Pan Y, et al. RET fusions define a unique molecular and clinicopathologic subtype of non-small-cell lung cancer. J Clin Oncol. 2012;30:4352–4359. doi: 10.1200/JCO.2012.44.1477. [DOI] [PubMed] [Google Scholar]
- 7.Bergethon K, Shaw AT, Ou SH, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012;30:863–870. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drilon A, Laetsch TW, Kummar S, et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med. 2018;378:731–739. doi: 10.1056/NEJMoa1714448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drilon A, Siena S, Ou SI, et al. Safety and antitumor activity of the multitargeted pan-TRK, ROS1, and ALK inhibitor entrectinib: Combined results from two phase I trials (ALKA-372-001 and STARTRK-1) Cancer Discov. 2017;7:400–409. doi: 10.1158/2159-8290.CD-16-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng Z, Liebers M, Zhelyazkova B, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014;20:1479–1484. doi: 10.1038/nm.3729. [DOI] [PubMed] [Google Scholar]
- 11.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weiss GJ, Hoff BR, Whitehead RP, et al. Evaluation and comparison of two commercially available targeted next-generation sequencing platforms to assist oncology decision making. OncoTargets Ther. 2015;8:959–967. doi: 10.2147/OTT.S81995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mishra R, Hanker AB, Garrett JT. Genomic alterations of ERBB receptors in cancer: Clinical implications. Oncotarget. 2017;8:114371–114392. doi: 10.18632/oncotarget.22825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rekhtman N, Pietanza MC, Hellmann MD, et al. Next-generation sequencing of pulmonary large cell neuroendocrine carcinoma reveals small cell carcinoma-like and non-small cell carcinoma-like subsets. Clin Cancer Res. 2016;22:3618–3629. doi: 10.1158/1078-0432.CCR-15-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ardini E, Bosotti R, Borgia AL, et al. The TPM3-NTRK1 rearrangement is a recurring event in colorectal carcinoma and is associated with tumor sensitivity to TRKA kinase inhibition. Mol Oncol. 2014;8:1495–1507. doi: 10.1016/j.molonc.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gainor JF, Varghese AM, Ou SH, et al. ALK rearrangements are mutually exclusive with mutations in EGFR or KRAS: An analysis of 1,683 patients with non-small cell lung cancer. Clin Cancer Res. 2013;19:4273–4281. doi: 10.1158/1078-0432.CCR-13-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gautschi O, Milia J, Filleron T, et al. Targeting RET in patients with RET-rearranged lung cancers: Results from the global, multicenter RET registry. J Clin Oncol. 2017;35:1403–1410. doi: 10.1200/JCO.2016.70.9352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med. 2014;371:1963–1971. doi: 10.1056/NEJMoa1406766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hyman DM, Laetsch TW, Kummar S, et al. The efficacy of larotrectinib (LOXO-101), a selective tropomyosin receptor kinase (TRK) inhibitor, in adult and pediatric TRK fusion cancers. J Clin Oncol. 2017;35 (suppl; abstr LBA2501) [Google Scholar]