Summary
Fast chemical communication in the nervous system is mediated by neurotransmitter-gated ion channels. The prototypical member of this class of cell surface receptors is the cation-selective nicotinic acetylcholine receptor. As with most ligand-gated ion channels, nicotinic receptors assemble as oligomers of subunits, usually as hetero-oligomers and often with variable stoichiometries1. This intrinsic heterogeneity in protein composition provides the fine tunability in channel properties essential to brain function but frustrates structural and biophysical characterization. The α4β2 subtype of the nicotinic acetylcholine receptor is the most abundant isoform in the human brain and is the principal target in nicotine addiction. This pentameric ligand-gated ion channel assembles in two stoichiometries of α and β subunits, 2α:3β and 3α:2β. Both assemblies are functional, have distinct biophysical properties, and a misbalance in the ratio of assemblies is tied to both nicotine addiction2,3 and congenital epilepsy4,5. Here we leverage cryo-electron microscopy (cryo-EM) to obtain structures of both receptor assemblies from a single sample. Antibody fragments specific to β2 were used to ‘break’ symmetry during particle alignment and obtain high resolution reconstructions of both stoichiometries, in complex with nicotine. The results reveal principles of subunit assembly and the structural basis of the distinctive biophysical and pharmacological properties of the two different stoichiometries of this receptor.
Pseudo-symmetric heteromeric proteins, exemplified by the Cys-loop receptor family, present a challenge in cryo-EM, because particle alignment is dominated by low-resolution information6,7, and at low resolution the subunit identities may not be readily distinguishable. Heterogeneity in subunit assembly adds to the challenges in particle alignment. In the nicotinic receptor, α and β subunits have nearly identical secondary structural features, which makes them and their multiple assemblies indistinguishable at low resolution. We raised monoclonal antibodies against the recombinant α4β2 nicotinic receptor and isolated high-affinity Fab fragments to structurally distinguish α subunits from β subunits within the pseudo-symmetric pentamer. In contrast to our earlier crystallographic studies where we optimized expression conditions to produce ~exclusively the 2α:3β assembly8, here we intentionally produced a mixed population of both stoichiometries of the α4β2 receptor (2α:3β and 3α:2β). We co-purified the receptor:Fab complex, and prepared sample grids for cryo-EM (Extended Data Fig. 1a and Methods). Fab has little or no effect on [3H]-nicotine binding and activation of the channel (Extended Data Fig. 1b-d).
Single particle cryo-EM data collection and processing revealed two distinct receptor populations, one with 2 Fabs bound and one with 3 Fabs bound, corresponding to the 3α:2β and 2α:3β stoichiometries, respectively (Extended Data Fig. 2). We obtained cryo-EM maps of the two stoichiometries to ~3.5 Å resolution for the receptor and ~4 Å resolution for the entire complex including Fab (Fig. 1a-d, Extended Data Figs. 2–3), with map quality that was significantly improved over that from the crystallographic structure of the 2α:3β assembly8 (Extended Data Table 1 and Extended Data Figs. 4–6). Each receptor assembly is cylinder-shaped, with a large extracellular vestibule and funnel-shaped transmembrane channel. Fabs interact exclusively with the extracellular domain (ECD) of the β2 subunits (Extended Data Fig. 1c-d). The agonist nicotine was included throughout purification, and density for it was observed at α-β and α-α interfaces in the receptor ECD. Nicotine stabilizes the ion channel in a putative closed-desensitized conformation.
Figure 1. Fab-assisted cryo-EM resolves heterogeneity in nicotinic receptor subunit assembly.
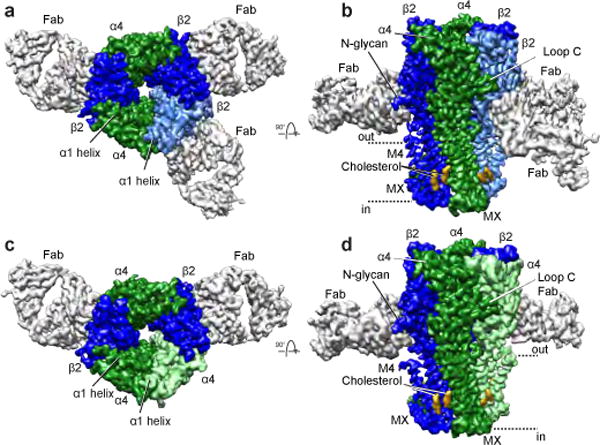
a-b, 2α:3β:Fab complex reconstruction. Fab fragments are light grey, density corresponding to cholesterol is golden orange and α4 and β2 subunits are green and blue respectively, with a lighter shade denoting the subunit position that is unique between the two stoichiometries. c-d, 3α:2β:Fab complex reconstruction. b,d, Dashed lines indicate approximate membrane position.
We sought to understand why more than two subunit assemblies are not observed3,9,10, given that our structures demonstrate the occurrence of all possible interfaces between α4 and β2 subunits. Each stoichiometry contains two α-β and two β-α interfaces, plus a third unique interface type: one α-α (3α:2β) or β-β (2α:3β) interface (Fig. 2a-d). Our reconstructions reveal that in an agonist bound, desensitized state, the α-α interface buries the most protein surface area from solvent, followed by the α-β and β-α interfaces (Fig. 2e). The β-β interface has the least buried surface area. Calculated interfacial free energies predict preferential assembly of α-β, followed by β-α as the most favorable, followed by α-α. β-β is the least favorable interface. A polar fenestration formed at the β-β interface is large enough (>5 Å) for hydrated ions to pass (Extended Data Fig. 7) and contributes to the decreased surface area buried at this interface class. These observations suggest that the more favorable α-β and β-α interfaces assemble first, followed by incorporation of the fifth subunit to create the unique interface as the final step in pentamer formation. Superposition of like subunits, both within an assembly and between assemblies, reveals no significant conformational differences among α subunits. However, there is a rigid body twist of 4.4° in the ECD relative to the TMD of the two beta subunits that compose the unique β-β interface compared to all other β2 subunits (Extended Data Fig. 6d-h). This conformational heterogeneity in β2 subunit structure likely further adds to the instability of the β-β interface. These structural observations suggest that the 3α:2β stoichiometry would be the predominant assembly in the absence of other factors, consistent with observations in rat motor cortex11 and in recombinant mammalian expression9. In our sample preparation, we exploited nicotine and low temperature during viral transduction to boost overall receptor expression, two factors that are known to shift the stoichiometry toward the 2α:3β assembly9.
Figure 2. Interface classes.
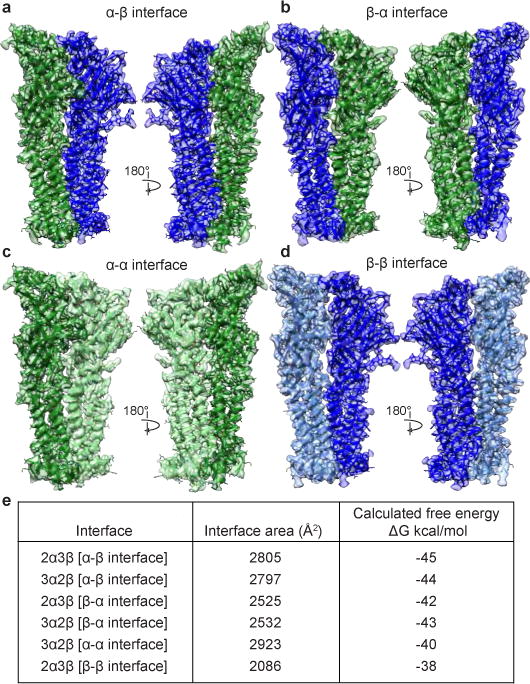
a-d, View from the periphery of the receptor toward the pore axis (left) and from the pore axis out toward the periphery of the receptor (right). α4 and β2 subunits are colored as in Fig. 1. e, Calculated interfacial buried surface areas and free energies. For interface classes for which there are two interfaces per assembly (α-β and β-α interfaces), values shown are averages.
Analysis of interfaces and subunit conformation suggested an order of subunit incorporation in the observed 2α:3β and 3α:2β assemblies driven by free energy, but did not explain why other theoretically possible subunit stoichiometries are not observed. To address this question, we superimposed the unique interface from each assembly (α-α or β-β) upon itself to build up theoretical homopentameric assemblies, wherein all interfaces except for the final interface would be equivalent to the starting, experimentally-observed interface. We then analyzed this last, non-experimental interface and found in the 5α assembly a dramatic increase in surface area buried compared to the experimental α-α interface (3849 Å2 vs. 2923 Å2) and 42 significant atomic clashes (Extended Data Fig. 8). Put simply, homopentameric α4 assemblies are unfavorable because they pack too tightly. Conversely, in the 5β assembly, at the final, non-experimental interface, we observed a gap between subunits resulting in a large decrease in surface area buried compared to the experimental β-β interface (1051 Å2 vs 2086 Å2). Thus, β-β interfaces pack such that they create too obtuse of an angle to effectively close the pentameric ring. We added one α-β interface into each of these homopentameric assemblies to examine the final theoretically possible subunit stoichiometries and observed a small decrease in the number of clashes in the 4α:1β assembly (39 clashes), and a small increase in the surface area buried at the 1α:4β assembly (1301 Å2). We conclude from these comparisons that only two stoichiometries are observed because only one α-α or one β-β interface can be accommodated in the pentamer while maintaining favorable interactions at all interfaces around the pentameric ring.
The ratio of the two observed assemblies of the α4β2 receptor in vivo is tunable and context-specific and is linked to genetic disorders and addiction2–5. Smoking, as well as nicotine added to cultured cells, preferentially upregulates the 2α:3β stoichiometry2. Mutations that shift the stoichiometry toward either the 3α:2β or 2α:3β assembly result in autosomal dominant nocturnal frontal lobe epilepsy4,5. A high-penetrance form of this syndrome is linked to a serine to leucine mutation in the α4 subunit, at the 10′ position in the M2 pore-lining helix (Extended Data Fig. 6c), which results in a shift in subunit stoichiometry toward the 3α:2β assembly4. The mechanism underlying this shift in assembly has been unclear. In the cryo-EM structures we observe this serine side chain orienting into a hydrophobic pocket in the adjacent subunit, regardless of the identity of that subunit. Mutation from the polar serine to the hydrophobic leucine should be energetically favorable given the local environment. This substitution would favor assembly of the 3α:2β stoichiometry because with three mutated α subunits, three interfaces would have improved hydrophobic interactions and fewer unsatisfied hydrogen bonding interactions, contrasted with only two such favorable interfaces in the 2α:3β assembly.
The 3α:2β assembly is classically described as the low-sensitivity isoform of the α4β2 receptor, with EC50 values for agonist ~100-fold higher than for the 2α:3β stoichiometry9. We sought to understand this agonist sensitivity difference in the context of receptor structure. The high-sensitivity neurotransmitter sites in the α4β2 receptor are found at the α4-β2 subunit interfaces in ECDs. The α-α interface in the 3α:2β assembly may serve as a low-sensitivity neurotransmitter site, whose occupation is necessary to obtain a full response12,13. We included 1 mM nicotine in our preparation to saturate high and low-sensitivity sites. A comparison of the four classes of interfaces reveals density for nicotine at the low-sensitivity α-α interface in the 3α:2β stoichiometry in addition to the high-sensitivity α-β interfaces in both stoichiometries of the α4β2 receptor (Fig. 3a-b and Extended Data Fig. 9). All α-β interfaces are qualitatively equivalent and are consistent with the crystal structure8 in the binding orientation of nicotine and the interactions it makes at this interface class. The binding orientation of nicotine at the α-β neurotransmitter binding sites is distinct from that observed at the α-α binding site. In the α-β interfaces, nicotine is oriented with its long axis approximately parallel to the pore axis, with its pyridine ring oriented away from the membrane. At the α-α interface (Fig. 3a and Extended Data Fig. 9), the pyridine ring of nicotine orients toward the pore axis with the molecule tilted off the membrane normal. This pose of nicotine in the α-α binding site is consistent with crystal structures of other α4β2 agonists bound to a soluble extracellular domain engineered to mimic the α-α binding site in α4β214. This trend suggests that the distinct binding orientation is a meaningful feature of the low-sensitivity α-α binding site. The difference in sensitivity between the classical α-β neurotransmitter binding site (high sensitivity) and the unique α-α binding site (low sensitivity) has been attributed to three amino acid positions on the complementary (−) face of the binding pocket: V111/F119/L121 for the β2 (−) subunit and H116/Q124/T126 for the α4 (−) subunit14,15 (Fig. 3c-d and Extended Data Fig. 9). In the α-β site (Fig. 3b and Extended Data Fig. 9), the three hydrophobic residues from the β2 (−) subunit provide a surface of van der Waals contacts that orient the pyridine ring of nicotine, locking it in the aromatic box in a vertical position. In the α-α interface, two of the three variant amino acid positions of α4 (−), T126 and Q124, by the nature of being shorter and polar amino acids, provide a less compact and more hydrophilic site that allows for the alternative binding orientation.
Figure 3. Agonist and pseudo-agonist binding sites.
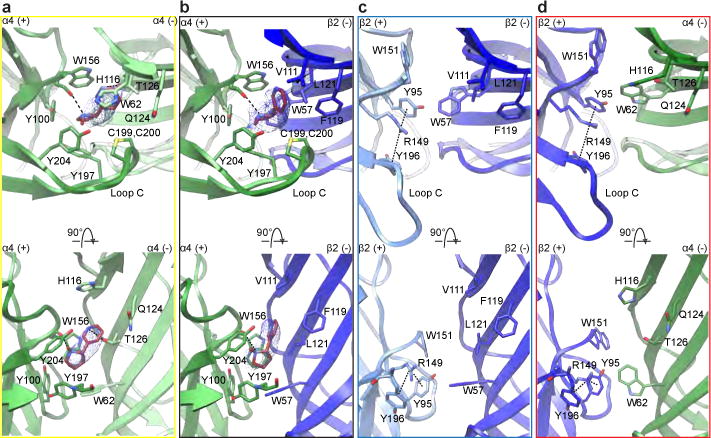
a-d, Molecular details of the α-β and α-α nicotine-binding interfaces and corresponding regions at the β-β and β-a interfaces. Top row is synaptic perspective viewed parallel to channel axis. Nicotine and interacting residues are shown as sticks. Nicotine density is displayed as blue mesh. In the bottom row, loop C backbone is hidden.
Due in part to limited high-resolution structural information for heteropentameric assemblies, the role of the pseudo-agonist sites, or interface sites in the ECD that do not bind agonist, is a relatively unexplored area of study. Our previous crystallographic work revealed the molecular architecture of the pseudo-agonist sites8, and here we extend this analysis to all interfaces found in the two stoichiometries of the α4β2 nicotinic acetylcholine receptor (Fig. 3c-d). The uniqueness of these pseudo-agonist site interfaces stems from the amino acid sidechains presented by β2 when it occupies the principal (+) side of the subunit interface. Of particular interest is R149, which would electrostatically and sterically impede agonists from binding and is positioned to form cation-π interactions with the sandwiching aromatic residues Y95 and Y196 from β2 (+) (Fig. 3c-d). This intramolecular interaction is conserved among all pseudo-agonist binding interfaces between the two assemblies; furthermore, in the closed, presumably desensitized conformations presented here, all the β-α pseudo-agonist sites in both assemblies appear to be structurally equivalent. We speculate that the guanidium group of R149, which chemically and structurally mimics the nicotinic pharmacophore, may act in part as a covalent agonist, potentiating activation.
The two α4β2 receptor assemblies have different permeation properties; the 3α:2β assembly is ~3-fold more permeable to Ca2+ than the 2α:3β assembly16 and has a higher single channel conductance9. Both permeation pathways are strongly electronegative, as would be expected for a cation-selective channel, with the 3α:2β assembly having a stronger electronegative potential throughout (Fig. 4a-b). These differences in electrostatic potential are localized to the extracellular vestibule and the junction of the ECD and transmembrane domain (TMD). The stronger electronegative potential seen in the extracellular vestibule of the 3α:2β assembly could not be predicted from sequence alone as the acidic side chains lining the extracellular vestibule that are thought to affect conductance are conserved between the α4 and β2 subunits17. The continuity of the electronegative potential is interrupted in the 2α:3β assembly at the junction between the ECD and transmembrane domain (TMD), and results from opposing charges at the 20′ position, E268 in 3α:2β and K260 in 2α:3β, in the pore-lining M2 α-helices (Fig. 4c). Point mutations at the 20′ position have demonstrated that having a negative charge at this extracellular entrance to the channel pore is an important determinant of conductance in nicotinic receptors18 and calcium permeability in cation-selective Cys-loop receptors16,19. In the α4β2 receptor specifically, mutational studies with concatemers demonstrated that increasing the proportion of negative charges at this position correlates directly with increasing permeability to Ca2+ 16. Thus, the difference in charge at the 20′ position is a principal determinant of the different permeation properties of the two receptor subunit assemblies. A caveat to these comparisons is that our EM construct lacks much of the receptor’s intracellular domain, a region that plays a role in subunit assembly and ion conductance in the structurally homologous 5-HT3 receptor19,20.
Figure 4. Pore and conductance determinants.
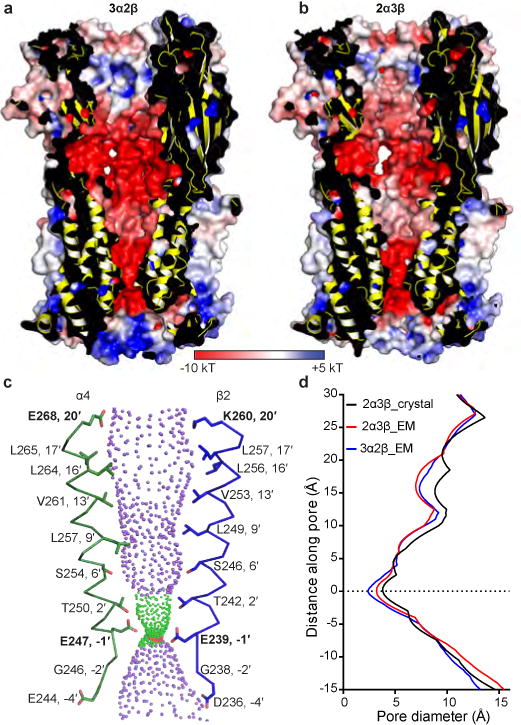
a, Cutaway of the 3α:2β assembly showing the permeation pathway colored by electrostatic potential. b, As a, but for the 2α:3β assembly. c, M2 α-helices from opposing α4 (green) and β2 (blue) subunits from the 3α:2β assembly with sidechains shown for pore-lining residues. Purple spheres indicate pore diameters >5.6Å; green are >2.8 Å and <5.6Å; red are <2.8Å. d, Pore diameter for the α4β2 receptor 2α:3β and 3α:2β assemblies from the cryo-EM study compared to the crystal structure of the 2α:3β assembly (Protein Data Bank (PDB) accession 5KXI8). Uncertainty in the position of most acidic sidechains in EM density maps adds ambiguity to the constriction point diameters defined by the −1′ glutamates.
Analysis of the transmembrane pore reveals that both α4β2 receptor assemblies taper to a constriction point created by glutamates present in all subunits, located at the −1′ position of the M2 α-helices, where the pore interfaces with the cytosol (Fig. 4c). The 3α:2β assembly has a smaller constriction point, a diameter of 2.4 Å compared to 3.2 Å (Fig. 4d and Extended Data Fig. 10), which is consistent with the more tightly packed α-α interface compared to the β-β in the 2α:3β assembly (Fig. 2e and Extended Data Figs. 7–8). Early studies on permeation through different channel classes suggested that, unlike for K+ channels, permeation through nicotinic receptors occurs with the ion at least partially hydrated21. The smallest physiologically permeant ion is Na+, with a crystal diameter of ~1.9 Å. Addition of a single equatorial water molecule (diameter of 2.8 Å) raises the diameter of the permeant species above the size of both constriction points. With the caveat that the structure was determined in the presence of detergent and is not in a native lipid membrane, the constriction diameter is consistent with the pore representing a non-conducting, agonist-bound desensitized state.
Reconstitution of nicotinic receptors into defined lipid preparations has demonstrated that cholesterol is important to nicotinic receptor function22. We tested cholesterol derivatives in thermal stabilization assays23 and found the water-soluble cholesterol analog cholesteryl-hemisuccinate (CHS) potently stabilizes the receptor; thus, we included it throughout purification of the receptor. We identified two putative cholesterol molecules per subunit in the EM density maps (Extended Data Fig. 11). These sausage-shaped densities are found in pairs located at the receptor periphery along the intracellular half of the transmembrane domain, where they flank the subunit interfaces. When viewed at the interface, each putative cholesterol molecule interacts almost exclusively with one subunit and one adjacent cholesterol molecule. Each pair of cholesterol molecules shares a single amino acid contact that bridges these intrasubunit sites (Extended Data Fig. 11 c-d). These intrasubunit sites contrast with intersubunit binding sites found for cholesterol and potentiating neurosteroids in GABAA receptor chimeras24,25, and are distinct from the intrasubunit site identified for inhibitory neurosteroids in a GABAA receptor chimera25 (Extended Data Fig. 11f). Comparison of the cholesterol positions in the α4β2 structures suggests that the orientations of the two molecules at an interface are dictated by the identity of the subunit on the principal side. When the principal subunit is β2, the cholesterols are oriented parallel to the pore axis; however, when the principal subunit is α4, the apical ends of both molecules tilt toward the principal face. The cholesterol binding site on the principal subunit is a bowl-shaped surface formed by the junction of the M3, MX and M4 helices. Close examination of this binding site reveals only a single amino acid position that is non-isosteric between the two subunit types, located on the penultimate helical turn in the M3 transmembrane helix. In β2 subunits this residue is a cysteine (C292) and in α4 subunits it is a phenylalanine (F300, Extended Data Fig. 11c-e). The bulky phenylalanine residue likely occludes access to the apical portion of the binding region observed when β2 subunits form the principal face. Analysis of human Cys-loop receptor sequences reveals that a small hydrophobic residue that would allow for this axial binding mode is conserved in neuronal β2 and β4 subunits as well as the muscle-type β1 subunit. Meanwhile, in all α subunits from cation-selective receptor types, a bulky hydrophobic residue (F, I, L or M) is conserved at this position. Our assignment of this cholesterol binding site is consistent with mapping studies using photo-activatable cholesterol analogs26,27.
Here we used single particle cryo-EM coupled with antibody fragments to determine structures of both stoichiometries of the α4β2 nicotinic receptor from one experimental preparation. Comparison of the structures reveals principles underlying subunit assembly and the differences in permeation properties between the two stoichiometries. We observed agonist binding at the non-canonical α-α subunit interface, differentially stabilized by residues unique to this interface class. The structure further allowed mapping of putative cholesterol sites on the surface of the transmembrane domain near the cytosolic junction. This study provides a framework for further biophysical analysis of the allosteric interfacial sites, therapeutic targeting of these sites with modulators akin to benzodiazepines, and more broadly, a general approach for resolving the structural biology of heterogeneous assemblies.
Methods
Nicotinic receptor expression and purification
The human α4 and β2 nicotinic receptor genes were provided by Dr. Jon Lindstrom at the University of Pennsylvania. Deletion constructs used for expression and structural analysis lack a large portion of the intracellular domain of the receptor as previously described8. These genes encode the native signal peptides, residues 1-338 and 556-601 in the α4 subunit and residues 1-330 and 417-477 in the β2 subunit (residue numbering here is for the wild-type mature, signal-peptide-cleaved protein sequence). To promote good biochemical behavior a Glu-Arg linker was inserted in the MX-M4 junction, between Phe559-Ser560 in the α4 subunit and between Gln420-Ser421 in the β2 subunit. For purification purposes a Strep-tag was inserted at the C-terminus of the β2 subunit preceded by a Ser-Ala linker. The genes were subcloned into the pEZT bacmam expression vector28 and viral titration and protein expression in GnTI- HEK cells (ATCC CRL-3022, not mycoplasma tested) was performed as previously described28 with modifications. 3.2 L of suspension GnTI- cells were transduced with multiplicities of infection (MOIs) of 0.5:0.5 for the α4 and β2 subunits, respectively, to promote expression of two receptor assemblies. Nicotine (Sigma-Aldrich) and sodium butyrate (Sigma-Aldrich) were added at the time of transduction to 0.1 mM and 3 mM, respectively. At the time of transduction, suspension cells were moved to 30 °C and 8% CO2. Cells were collected after ~72 hr by centrifugation, resuspended in 20 mM Tris, pH 7.4, 150 mM NaCl (TBS buffer), 1 mM nicotine and 1 mM phenylmethanesulfonyl fluoride (Sigma-Aldrich), and disrupted using an Avestin Emulsiflex. Lysed cells were centrifuged for 15 minutes at 10,000 g; supernatants containing membranes were centrifuged 2 hours at 186,000 g. Membrane pellets were mechanically homogenized and solubilized for 1 hour at 4 °C in a solution containing TBS, 40 mM n-dodecyl-β-D-maltopyranoside (DDM; Anatrace), 1 mM nicotine and 0.2 mM CHS (Tris salt, Anatrace). Solubilized membranes were centrifuged for 40 minutes at 186,000 g then passed over high capacity Strep-Tactin (IBA) affinity resin. The resin was washed with Size Exclusion Chromatography (SEC) buffer containing TBS, 1 mM DDM, 1 mM nicotine, 0.2 mM CHS and 1 mM TCEP (Thermo Fisher Scientific) and eluted in the same buffer containing 5 mM desthiobiotin (Sigma-Aldrich).
Generation of monoclonal antibodies and Fab fragments
The 12H2 monoclonal antibody (mAb) against the α4β2 nicotinic receptor (IgG1, κ) was raised by Dr. Daniel Cawley (Monoclonal Core, Vaccine and Gene Therapy Institute, OHSU) using standard methods. Hybridomas were generated by immunization of mice (female BALB/c) with α4β2 receptor protein purified in DDM, followed by fusion of splenocytes with mouse myeloma cells and selection of clones by enzyme-linked immunosorbent assay. The mAb was purified from hybridoma cell supernatant by cation exchange and protein A affinity chromatography. High affinity and specificity of the antibody for properly folded pentameric α4β2 receptor was assessed using Fluorescence-SEC (FSEC) with GFP-tagged receptor (shift in elution volume) and by western blot (no binding). The top candidate antibodies were then more thoroughly assessed using FSEC monitoring tryptophan fluorescence with purified α4β2 receptor and Fab fragments to ensure fidelity of the α4β2:Fab complex and estimate receptor:Fab stoichiometry. α4β2:Fab complexes were co-purified, then re-injected in analytical FSEC to test for dissociation under these dilute conditions over a Sepax SRT SEC-500 (4.6x300 mm) gel filtration column. Fabs that did not dissociate were qualitatively deemed to have sufficiently high affinity to use in EM studies. The 12H2 Fab fragment used here was generated by papain cleavage of whole antibody at a final concentration of 1 mg/mL for 2 hours at 37 °C in 50 mM NaPO4, pH 7.0, 1 mM EDTA, 10 mM cysteine and 1:100 w:w papain. Digestion was quenched using 30 mM iodoacetamide at 25 °C for 10 min. Fab was purified by cation exchange using a Hi-Trap SP column in 10 mM sodium citrate pH 5.0 and a NaCl gradient elution. Cloning and sequencing of Fab heavy and light chain genes were performed from mouse hybridoma cells.
Cryo-EM sample preparation
Purified α4β2 from affinity chromatography was mixed with Fab in a 1:2 w:w ratio and injected over a Superose 6 Increase 10/300 GL column (GE) equilibrated in SEC buffer. Peak fractions were assayed by SEC, monitoring tryptophan fluorescence. The peak fraction was concentrated four-fold to an A280 of 5.9 in SEC buffer, ultracentrifuged, and used directly to prepare EM grids. 3 μL of purified α4β2-nicotine-Fab complex was applied to glow-discharged gold R1.2/1.3 300 mesh holey carbon grids (Quantifoil) and immediately blotted for 4 s at ~100% humidity and 4 °C, then plunge-frozen into liquid ethane cooled by liquid nitrogen using a Vitrobot Mark IV (FEI).
Cryo-EM image collection and processing
Two separate data sessions were taken on a Titan Krios electron microscope (FEI) operated at 300 kV. Images were recorded on a K2 Summit direct electron detector (Gatan) equipped with GIF quantum energy filter (20 e−V) (Gatan) in super-resolution mode (super-resolution pixel size: 0.535 Å/pixel). In the first session, 3,323 images were collected and each micrograph in the first session was exposed for 15 sec with a dose rate of ~5 e−/A2/sec (total specimen dose, ~75 e−/A2), and 30 frames were captured per micrograph. In the second session, 2588 images were collected and each micrograph in the second session was exposed for 10 sec with ~5 e−/A2/sec dose rate (total specimen dose, ~50 e−/A2), and 40 frames were captured per micrograph. Images were recorded using the automated-acquisition program EPU (FEI) with defocus values varied from 0.5 μm to 4.5 μm.
Dose-fractionated images (movies) were gain normalized, 2X Fourier binned (resulting in a pixel size of 1.07 Å/pixel), aligned, dose-weighted and summed using Unblur29. Defocus values were estimated using GCTF30. A total of 5,166 micrographs were manually selected and used for further analysis (2,836 out of 3,323 from the first session and 2,330 of 2,588 from the second session). Approximately 100 particles were manually picked and subjected to reference-free 2D class averaging in Relion 2.131,32. Resulting references were then used for auto-picking in Relion 2.1 for a subset of 10 images, then the classification process was repeated to obtain good references for auto-picking on all images. Following auto-picking, micrographs were manually inspected, and false positive particles were manually removed. Curation of auto-picked particles resulted in a total of 649,773 particles. 516,450 particles were selected following 2D classification in Relion. Two hundred particles from 30 different 2D classes (6000 particles total) were used to generate an ab-initio model in Relion. 3D classification using 8 classes resulted in six good classes (403,246 particles) that could be divided into to two distinct categories based on Fab occupancy and subunit-defining features, representing the 3α:2β (139,551), and 2α:3β (263,695) stoichiometries of the α4β2 receptor. 3D refinement of these two pooled particle sets yielded reconstructions of the overall receptor-Fab complexes at ~4.0 Å resolution. The Fab molecules, in particular the peripheral constant domains, were poorly resolved, and of less biological interest to us, so we focused our effort on the receptor. Removing the Fab regions from the global resolution calculations results in reconstructions for the 3α:2β assembly at 3.7 Å and for the 2α:3β assembly at 3.4 Å resolution, consistent with side chain features throughout the density maps. All global resolutions were estimated by applying a soft mask around the protein density and the gold-standard Fourier shell correction (FSC) = 0.143 criterion. ResMap33 was used to estimate the local resolution.
Model building, refinement and validation
The higher resolution 2α:3β assembly was built first. The x-ray coordinates (PDB accession code 5KXI) were docked into the density map. Secondary structural features that are distinct between the two stoichiometries (Loop C and α1-helix, Fig. 1) were used to guide the initial rigid body fit using UCSF Chimera34. SwissModel35 was used to generate a homology model of the Fab light chain using PDB entry 4TPR and for the heavy chain using PDB entry 2ZJS. These chains were docked into the EMD density at one Fab site. Manual adjustments of the receptor-Fab structure were then performed in Coot36. The ECD and transmembrane domain (TMD) halves of each subunit, and the variable and constant halves of each Fab, were rigid body fitted into the density map. The epitope binding loops of the Fabs were rebuilt into unambiguous density at the site of interaction with the receptor, and then this first Fab molecule was copied into the two additional sites in the 2α:3β assembly. The map quality for the receptor exceeded that from the earlier x-ray structure, which allowed for more residues to be built at the N-termini and in the linkers connecting the MX amphipathic helix with the M4 helix. There was unclear density and thus a gap in the connectivity in the MX-M4 linker for 7 residues in α4 subunits and 10 residues in β2 subunits. There was improved density for side chains and better connectivity in the ECD loops, in particular loop C in the β subunits. After rebuilding in Coot, global real space coordinate and B factor refinement were performed in Phenix37.
The cryo-EM density maps drew our attention to a striking conformational difference in the loop C of β2 subunits compared to that modeled in the crystallographic structure8. We observed continuous density for this substructure in all β2 subunits in EM maps that resolve loop C in a conformation bending out and away from the subunit interface (Fig. 3 c-d and Extended Data Fig. 6a-c). This conformation was independent of whether Fab was bound to the adjacent (−) subunit or not (Extended Data Fig. 1d-e). In the crystal structure, loop C in β2 subunits was modeled in a conformation packed against the (−) subunit, roughly akin to the conformation of loop C in α4 subunits. The crystallographic modeling was based on clear electron density for loop C in one β2 subunit that was at a crystal contact (Extended Data Fig. 6a-c). We thus suggest that an open conformation of loop C in β2 subunits is the proper one.
The validation to test for overfitting of the model was performed as previously described38. Briefly, the atom positions of the final refined model were randomly displaced by a maximum of 0.5 Å using PDBSET in the CCP4 suite39. This perturbed model was then refined in Phenix in real space against the first half map of the reconstruction comprising 50% of the particles. A map vs. model FSC comparison was made for this model vs. the map used in its refinement (“work”), as well as the same model vs. the half map not used in refinement (“free”). The FSC curves of map vs. model agree well (Extended Data Fig. 4–5).
Sequences used in alignments were retrieved from the UniProtKB database40. Sequence alignments were made using PROMALS3D41. Subunit interfaces including free energies were analyzed using the PDBePISA server42. Pore diameters were calculated using HOLE43. Domain motions were analyzed using DynDom44. Ligand interactions were characterized by visual inspection and analysis using LIGPLOT45. Structural figures were made using UCSF-Chimera34 and PyMOL (Schrodinger, LLC) including the APBS electrostatics plugin46. Structural biology software packages were compiled by SBGrid47.
Density maps were displayed in Chimera at a threshold of 0.0306 in Figs. 1 and 2. In Fig. 3, ligand density is displayed in Chimera at a threshold of 0.0403 for the α-α interface, and 0.05 for the α-β interface.
Radioligand Binding
Experiments to test the effect of Fab on [3H]-nicotine (PerkinElmer, 79.8 Ci/mmol) binding to the α4β2 receptor were performed with protein purified as for EM (i.e., a mixture of stoichiometries) but in the absence of Fab and nicotine. The concentration of binding sites was kept at 2.5 nM after a preliminary experiment to determine optimal receptor concentration. For the +Fab samples, Fab was added in excess (5 μM). In addition to the protein, the binding assay conditions included 20 mM Tris pH 7.4, 150 mM NaCl, 1 mM DDM, and 1 mg/mL streptavidin-YiSi scintillation proximity assay beads (SPA; GE Healthcare Life Sciences). Non-specific signal was determined in the presence of 100 μM [1H]-nicotine; all data shown are from background-subtracted measurements. Experiments were performed in triplicate. All data were analyzed using Prism 6 software (GraphPad) with variable Hill slope.
Electrophysiology
The open reading frames of α4 and β2 EM constructs described above were subcloned into the pGH19 vector48. cRNAs of α4 and β2 constructs were synthesized in vitro using the MEGAscript T7 Transcription Kit (Thermo Fisher). Xenopus laevis ovaries purchased from NASCO (LM00935MX) were treated with 1-2 mg/mL of collagenase I (Gibco) in Barth’s solution (in mM, 88 NaCl, 1 KCl, 2.4 NaHCO3, 0.82 MgSO4, 0.33 Ca(NO3)2, 0.68 CaCl2, 10 Hepes, pH 7.4) for 1-1.5 hr, and washed with ND96 solution (in mM, 96 NaCl, 2 KCl, 1 MgCl2. 1.8 CaCl2, and 5 Hepes pH 7.4). Stages V–VI oocytes were collected and kept at 17 °C in ND96 solution with 25 μg/ml ampicillin and 0.1 mg/ml gentamycin. Oocytes were injected with 1-10 ng of cRNA mixture in the ratio 1:5 or 5:1 of α4:β2. Two to three days after injection of cRNA, receptor activity was measured by two-electrode voltage clamp recording in ND96 solution using Oocyte Clamp OC-725 amplifier (Warner Instruments), and the signal was digitized with a Digidata 1440A (Molecular Devices). Data were analyzed using Clamp 10.3. Pipettes were filled with 1 M KCl and showed resistances of 0.8-2 MΩ. Ligands were dissolved in ND96.
Data availability
Atomic coordinates of the two α4β2 receptor-nicotine-Fab complexes have been deposited in the PDB (2α:3β is PDB-6CNJ; 3α:2β is PDB-6CNK). The cryo-EM density maps have been deposited in the EMDB upon acceptance for publication (2α:3β is EMDB-7535; 3α:2β is EMDB-7536).
Extended Data
Extended Data Figure 1. Biochemistry, binding, electrophysiology and α4β2-Fab interactions.
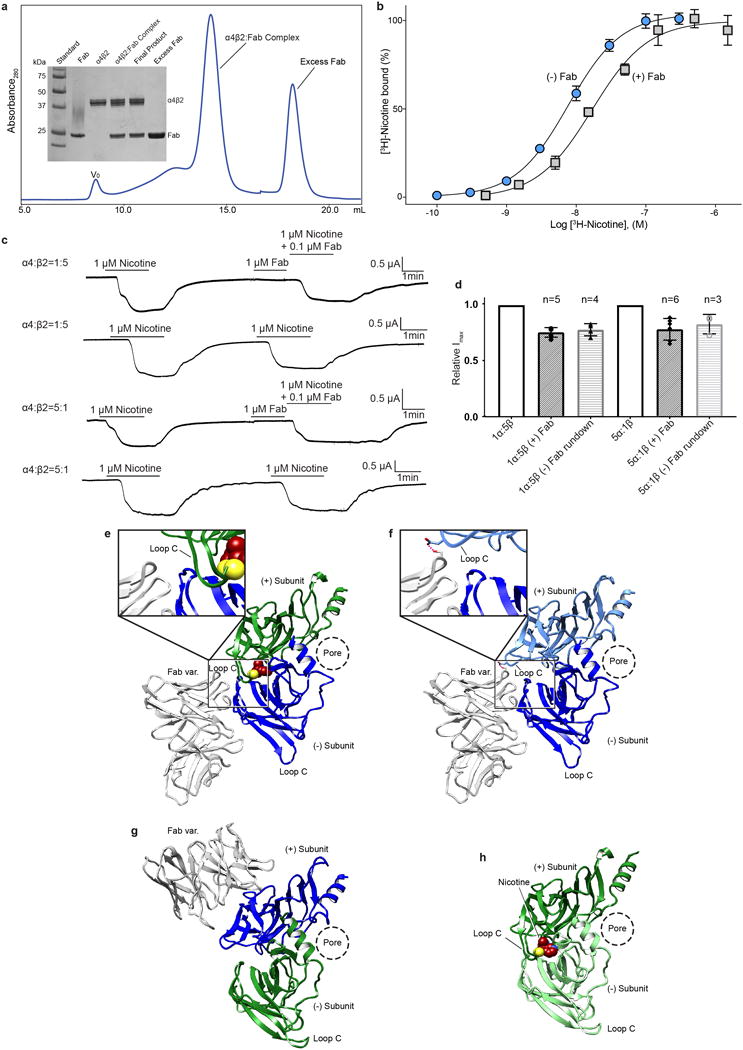
a, Size-exclusion chromatogram of α4β2:Fab complex and SDS-PAGE analysis of complex purification (V0, void volume). b, Saturation binding experiments with [3H]-nicotine and a mixture of receptor subunit stoichiometries. Grey squares denote samples with receptor plus Fab and blue circles denote samples with receptor alone. Receptor alone Kd = 7.7 nM (95% confidence interval of 6.9-8.6 nM) and receptor plus Fab Kd = 17.0 nM (95% confidence interval of 10.2-26.6 nM). Receptor alone and receptor plus Fab both exhibited a hill slope of ~1 (1.14 and 1.09 respectively). Plotted results are from a representative experiment performed in triplicate. c, Representative two-electrode voltage clamp (−60 mV) recordings of oocytes injected with cRNAs for the α4 and β2 subunits in ratios to bias assembly,1α:5β (top two traces) and 5α:1β (bottom two traces) for the 2α:3β and 3α:2β assemblies respectively. The experiments were performed in the presence and absence of Fab to assess effect of Fab on receptor gating. Each oocyte was perfused with a 1 μM nicotine solution for ~2 min, washed with bath solution for 10 min and then perfused for ~2 min with 1 μM nicotine +/− Fab. For samples were Fab was included, 1 μM final [Fab] was added directly to the oocyte bath following the first perfusion and allowed to incubate for 1.5 min before perfusing with a second solution containing 1 μM nicotine + 0.1 μM Fab. d, Bar graph quantifying peak currents before and after adding Fab. Currents were normalized to the amplitude of the first nicotine application. Appreciable current rundown was observed (assessed by applying nicotine without Fab as the second application). Change in peak current is similar in the presence and absence of Fab for both stiochiometries, suggesting no substantial effect on gating. n values are number of oocytes; error bars are standard deviation from the mean. e-h, Fab:β subunit interactions. With one exception, the Fab molecules interact exclusively with a single β subunit. The exception is at the β-β interface, where the Fab on the complementary β subunit forms one potential interaction with the preceding β subunit. This interaction is displayed as a dashed line in the panel f inset. The conformation of the principal Loop C at the exceptional β-β interface is indistinguishable from those where Fab is not interacting, suggesting that Fab does not affect the loop C conformation. e-h, Fab interactions at the α-β, β-β, α-β, and α-α interfaces, respectively. Subunits are colored as in Fig. 1.
Extended Data Figure 2. Cryo-EM image processing procedure.
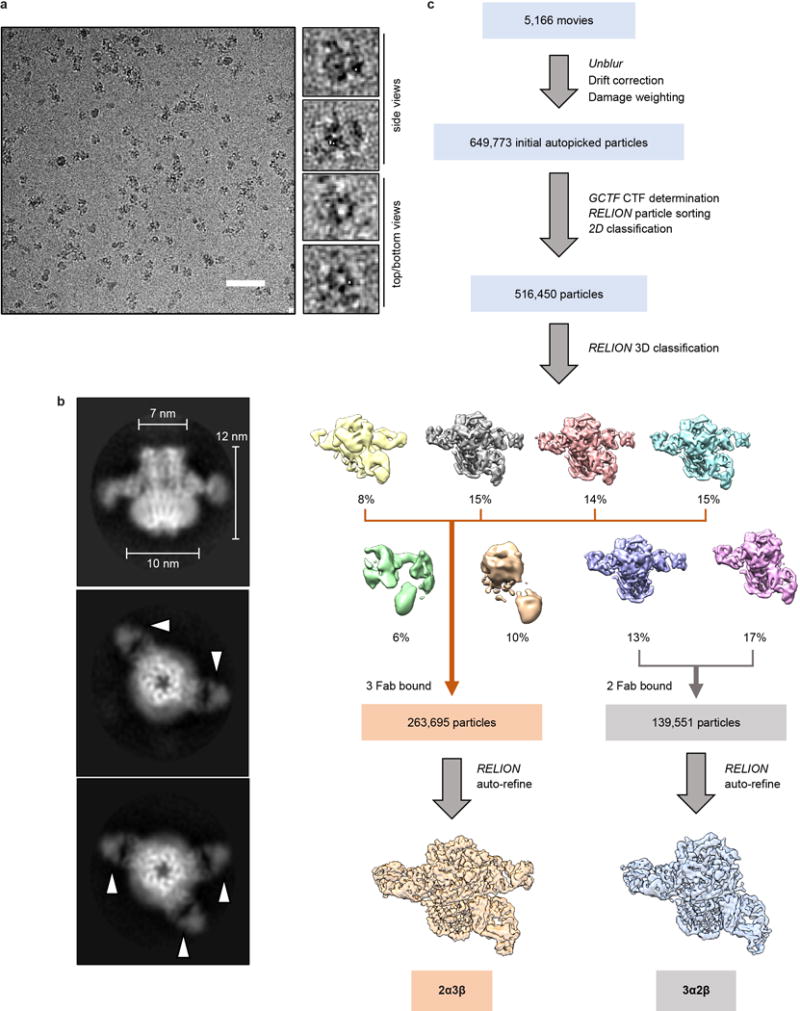
a, Representative micrograph of the α4β2:Fab complex (scale bar, 50 nm). Boxed regions on the right are magnified to show representative particle images. Top/bottom and side views are labeled. b, Images of selected two-dimensional classes from reference-free two-dimensional classification by RELION. Fab fragments are denoted by white arrowheads. c, Overview of the image processing procedure (see Methods).
Extended Data Figure 3. Three-dimensional reconstructions of the 2α:3β and 3α:2β assemblies.
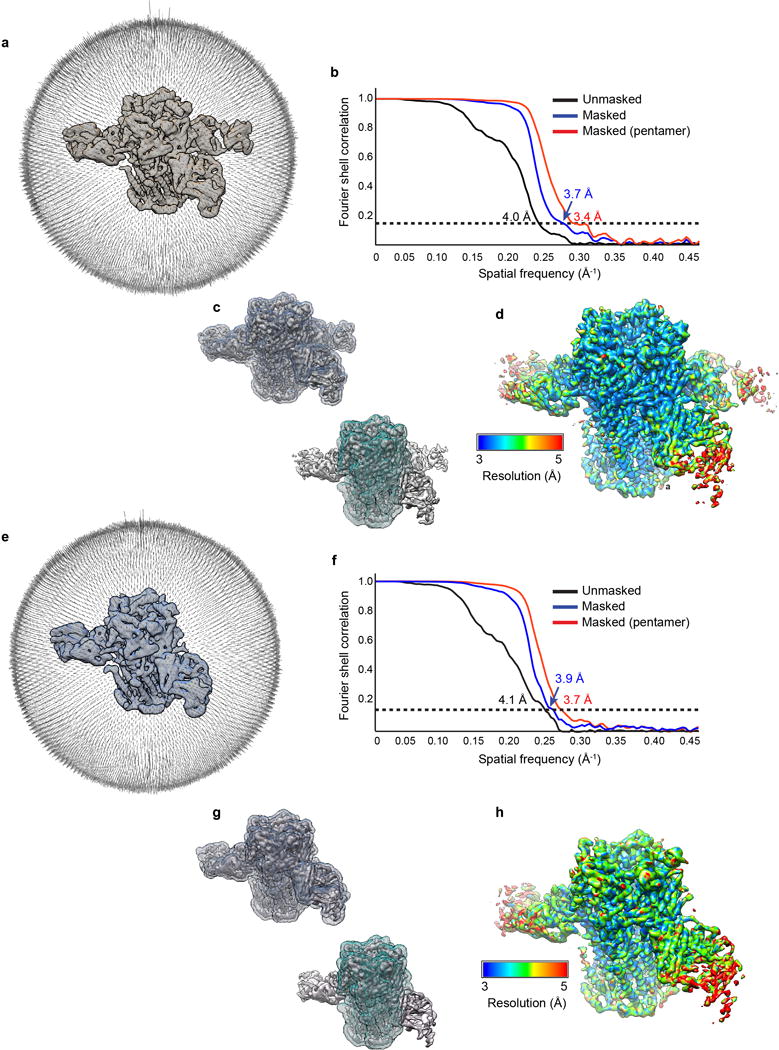
a, Angular distribution histogram of 2α:3β assembly projections. b, Fourier shell correlation (FSC) of 2α:3β assembly maps before (black) and after (blue and red) masking. Two soft masks were used: one (red) mask that included only the receptor and one (blue) mask that included the whole 2α:3β:Fab complex. When a mask was used, the FSC curve was corrected for masking effects during the RELION post-processing procedure. Masks used in FSC calculations are shown in c, for the whole 2α:3β:Fab complex (top), and receptor only (bottom), respectively, superimposed on the map in question. d, Local resolution of the 2α3β:Fab reconstruction estimated by ResMap33. Shown is the combined map, which has not been sharpened or filtered. e-h, as in a-d but for the 3α2β assembly.
Extended Data Figure 4. 3α:2β assembly model-map validation.
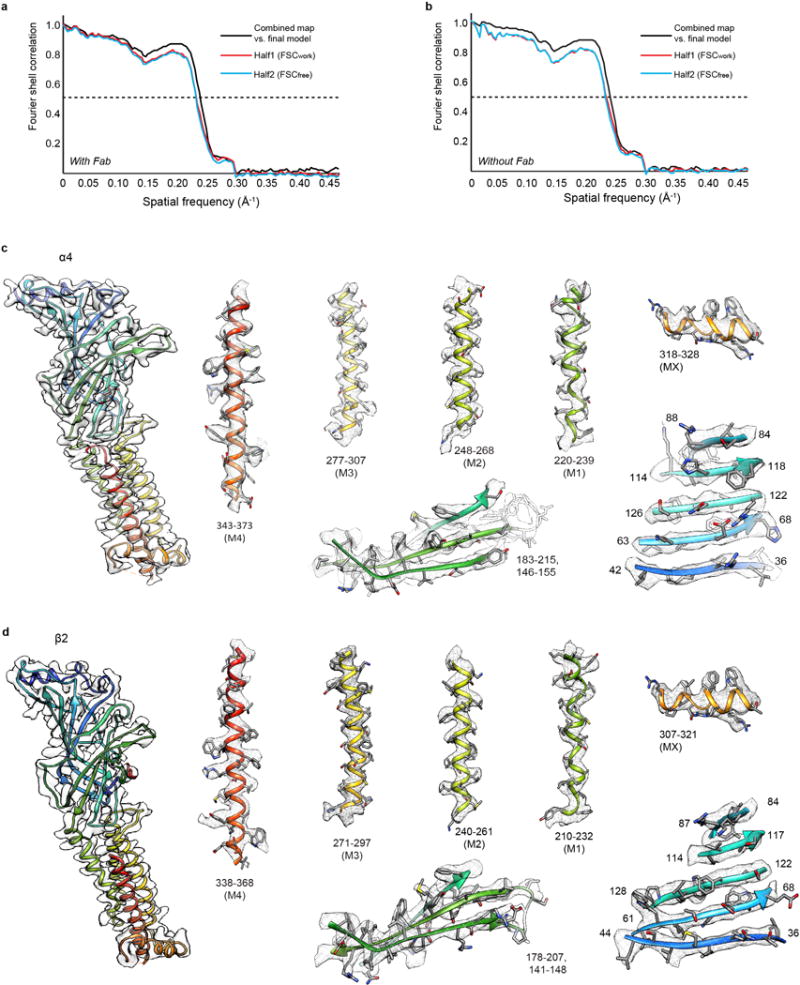
a-b, FSC curves for cross-validation between the maps and the models with (a) and without Fab fragments (b) for the 3α:2β assembly. Curves for final model versus summed map (full) in black, for model versus half map in red (work), and for model versus half map not used for refinement in blue (free). For validation of receptor alone (without Fab), maps were segmented to exclude the Fab fragments. c, EM density segments of the 3α:2β assembly for a representative α4 subunit. Density map and model for α4 subunit (left). Representative density for extracellular and transmembrane spanning regions (right). Regions are numbered and helices are labeled. Maps were sharpened with a single B factor at −150 Å2. d, as in c, but for a representative β2 subunit.
Extended Data Figure 5. 2α:3β assembly model-map validation.
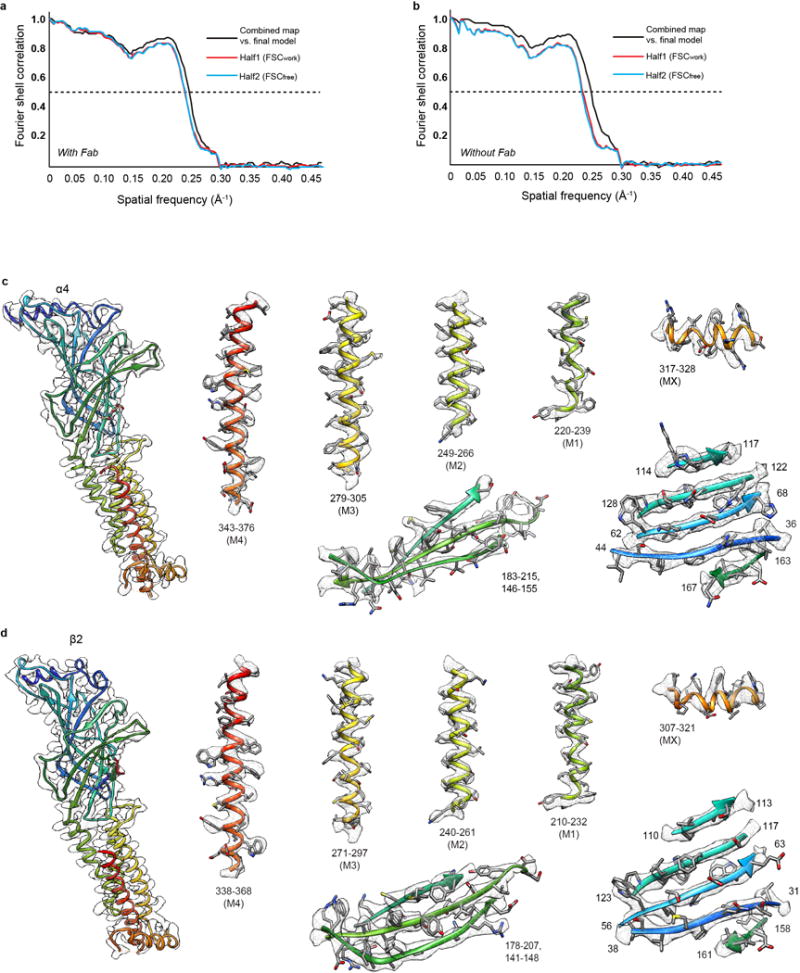
a-b, FSC curves for cross-validation between the maps and the models with (a) and without fab fragments (b) for the 2α:3β assembly. Curves for final model versus summed map (full) in black, for model versus half map in red (work), and for model versus half map not used for refinement in blue (free). For validation of receptor alone (without fab) maps segmented to exclude the Fab fragments were used. c, EM density segments of the 2α:3β assembly for a representative α4 subunit. Density map and model for α4 subunit (left). Representative density for extracellular and transmembrane spanning regions (right). Regions are numbered and helices are labeled. Maps were sharpened with a single B-factor at −170 Å2. d, as in c, but for a representative β2 subunit.
Extended Data Figure 6. Comparison of map and model of 2α3β by x-ray crystallography at 3.9 Å resolution and by cryo-EM at 3.4 Å resolution, and comparison of subunit backbone conformations.
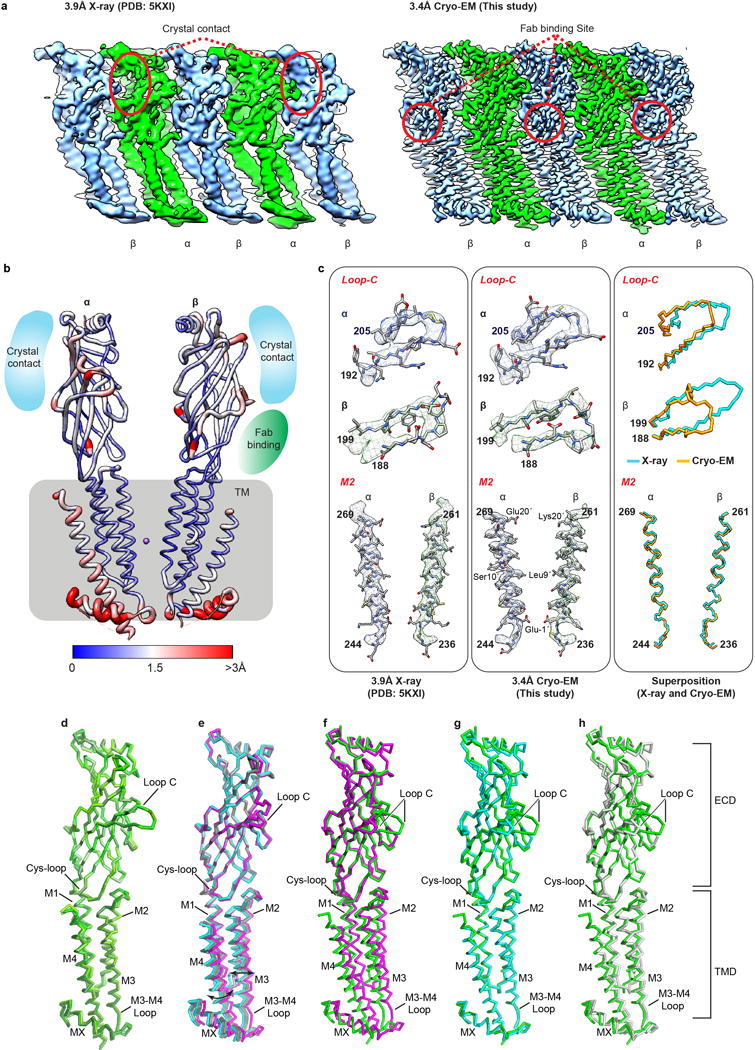
a, The x-ray electron density map (left panel) and cryo-EM map (right panel) are displayed in a linear representation (unwrapped) to illustrate the overall resolvability of all five subunits: two α4 subunits (green) and three β2 subunits (blue). Red circles indicate the location of crystal contacts on the x-ray electron density map (left panel) or Fab binding sites on cryo-EM density map (right panel). b, x-ray structure of the 2α:3β assembly (PDB accession 5KXI8) after alignment to the cryo-EM structure (from this study) shown in ribbon representation and colored according to Cα r.m.s.d. values [ranging from blue (low) to red (high)]. While the areas involved in crystal contacts displayed relatively higher r.m.s.d. values for Cα, over 2 Å, Fab binding location shows r.m.s.d. values for Cα of less than 1 Å. This result implies that zones of protein-protein contacts in the crystal lattice may significantly affect local structure on the pentamer, but Fab binding in cryo-EM study did not result in significant structural change, consistent with the binding assays and electrophysiological results (Extended Data Fig. 1). c, Fit of x-ray model (5KXI) to x-ray electron density map (left column) and cryo-EM model to cryo-EM density map (middle column) are shown as selected areas for high r.m.s.d. (Loop C) and low r.m.s.d. (M2 helix), respectively. Right column shows superposition of x-ray and cryo-EM model at the selected areas. d-h, Subunit superpositions. d, Comparison of all α4 subunits in the 2α:3β and 3α:2β assemblies reveals there are no significant conformational differences in α-carbon backbones. e, Superposition of all β2 subunits in the two assemblies reveals a domain motion of the TMD relative to the ECD of the subunits that comprise the β-β interface. The rotation of 4.4° between the two beta subunits that compose the β-β interface is denoted by black double headed arrows. The principal beta subunit (β2 +) is colored magenta while the complementary beta subunit (β2 -) is shown in cyan. f-h, Superposition of α4 (green) with the three distinct β2 conformations: principal β-β (magenta) complementary β-β (cyan) and α-β-α (grey). The conformation of the principal β-β (magenta) subunit is distinct from every other subunit in the 2α:3β and 3α:2β assemblies. The conformation of the complementary (−) subunit of the β-β interface is rotated towards the pore axis, accommodating the dramatic conformational change of the principal β2 subunit away from the pore axis. Interestingly this conformational change results in the complementary β-β (cyan) subunit adopting a backbone conformation similar to α4 subunits.
Extended Data Figure 7. Fenestration at β-β interface.
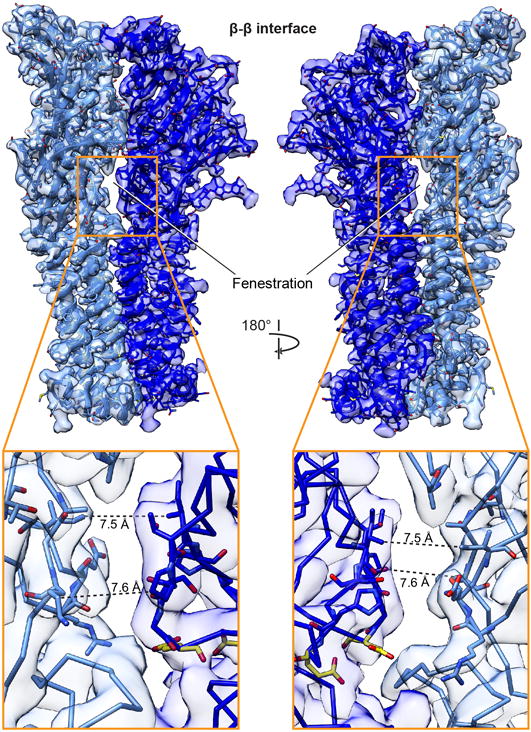
Extracellular fenestration unique to this class of interface. Inset of fenestration with distances indicated by dashed lines and side chains surrounding fenestration shown as sticks. Two conserved glutamic acid residues in close proximity to this gap at the β-β interface, shown in yellow, are implicated in Ca2+ potentiation of nicotinic receptors49.
Extended Data Figure 8. Basis of α4β2 heteromeric assembly.
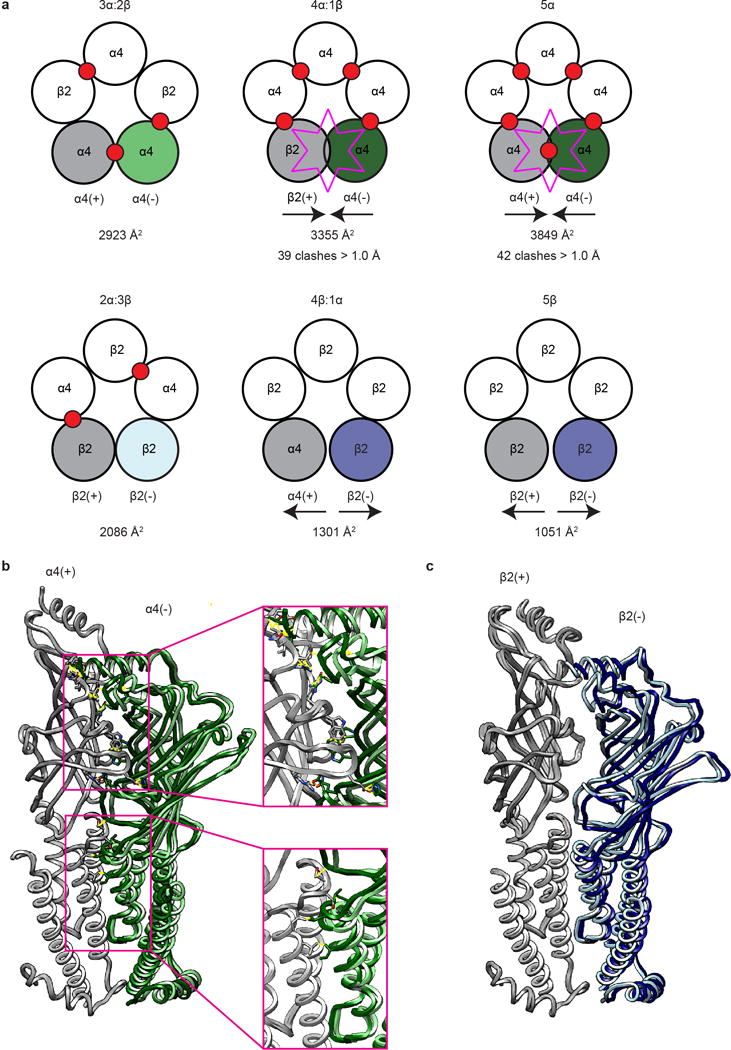
a, Cartoon representation of top view of observed (2α:3β and 3α:2β) and computational (4α:1β, 1α:4β, 5α and 5β) α4β2 pentameric assemblies. Assemblies on the top row (3α:2β, 4α:1β, and 5α) are arranged by increasing α4 composition. Assemblies on the bottom row (2α:3β, 1α:4β and 5β) are arranged by increasing β2 composition. Agonist binding sites are denoted by red circles. Buried interface areas (Å2) for the interfaces analyzed in b-c are listed below each pentameric assembly. Subunits are colored in a as they are in b-c, as described below. b, Superposition of α-α from 3α:2β and final α-α interface in 5α homopentamer. Principal subunits (grey) were superimposed to highlight differences at the interface. Complementary (−) subunits are colored light green for α-α from 3α:2β and dark green for the α-α interface in the 5α homopentamer. Sticks are displayed for amino acid clashes that are greater than 1.5 Å overlap assessed by Molprobity. c, Superposition of the β-β interface from 2α:3β and final β-β interface in the 5β homopentamer. Principal subunits (grey) were superimposed to highlight differences at the interface. Complementary (−) subunits are colored light blue for the of β-β interface from the 2α:3β assembly and dark blue for the final β-β interface in the 5β homopentamer.
Extended Data Figure 9. Ligand binding site comparisons.
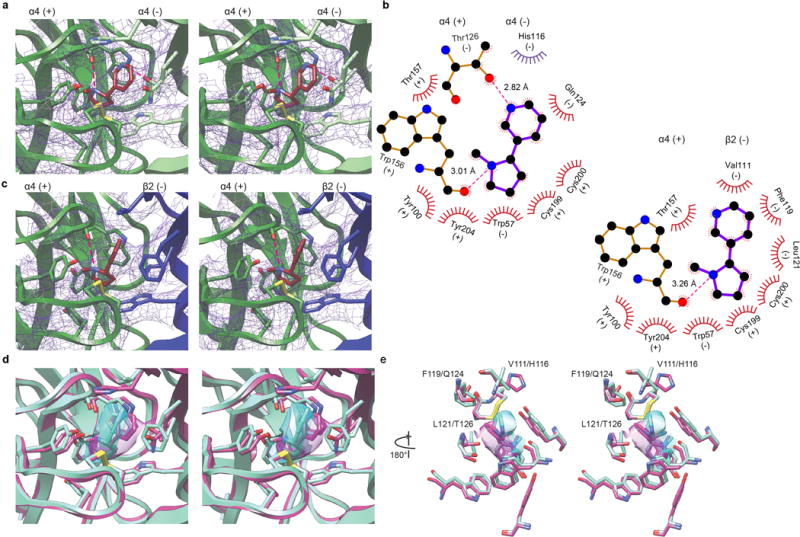
a, Stereo images of nicotine bound at the α-α interface. Subunits are colored as in Fig. 1. Density (purple mesh) is displayed at a threshold of 0.0306 in Chimera. Electrostatic interactions denoted as dashed magenta lines. Binding pocket residues and nicotine displayed as sticks. b, Ligplot of nicotine bound at the α-α and α-β interface, top left and bottom right respectively. Residue contacts within 4.5 Å are displayed in red, residue displayed in purple (His116) is within 5.5 Å. c, Stereo images of nicotine bound at the α-β interface. Density (purple mesh) is displayed at a threshold of 0.0306 in Chimera. Hydrogen bonds denoted as dashed magenta lines. Binding pocket residues and nicotine displayed as sticks. d-e, Stereo image overlays of nicotine bound at the α-α (magenta) and α-β interface (cyan). Ligand density is displayed as transparent surfaces. Density is displayed at a threshold of 0.406 for the α-α (magenta) and 0.500 for the α-β (cyan) interface in Chimera. e, ribbon removed for clarity, rotated 180° relative to d. Key residues implicated in the difference in sensitivity between the classical α-β neurotransmitter binding site (high sensitivity V111/F119/L121) and the unique α-α binding site (low sensitivity H116/Q124/T126) are labeled.
Extended Data Figure 10. Comparison of ions located in the pore above the constriction point.
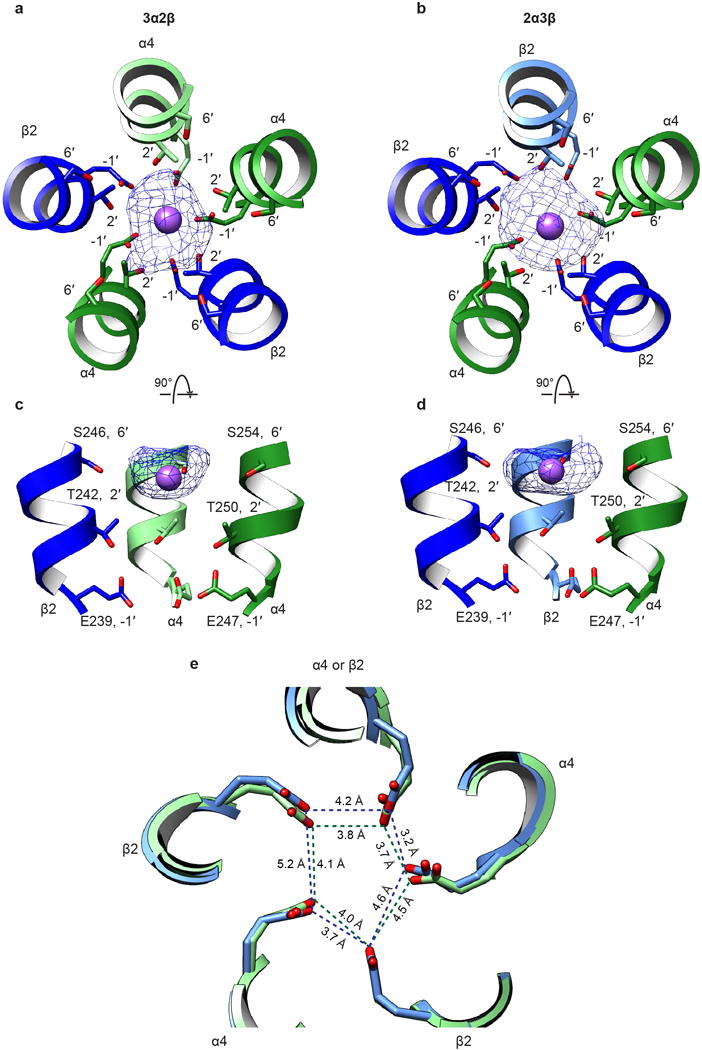
a, Top view of EM density map of ion in the pore of the 3α:2β assembly. Density (blue mesh) is displayed at a threshold of 0.027 in Chimera. Subunits are colored as in Fig. 1. Modeled Na+ ion is represented as a purple sphere. Nearest residues on the M2 α-helices are indicated. b, As a, but for the 2α:3β assembly. c, Side view of EM density map of ion in the pore of the 3α:2β assembly. Two subunits (one α and one β) removed for clarity. Colors and residues indicated as in a. d, As c, but for the 2α:3β assembly. e, Superposition of constriction region of 2α:3β (light blue) and 3α:2β (light green) assemblies.
Extended Data Figure 11. Comparison of putative cholesterol binding orientations.
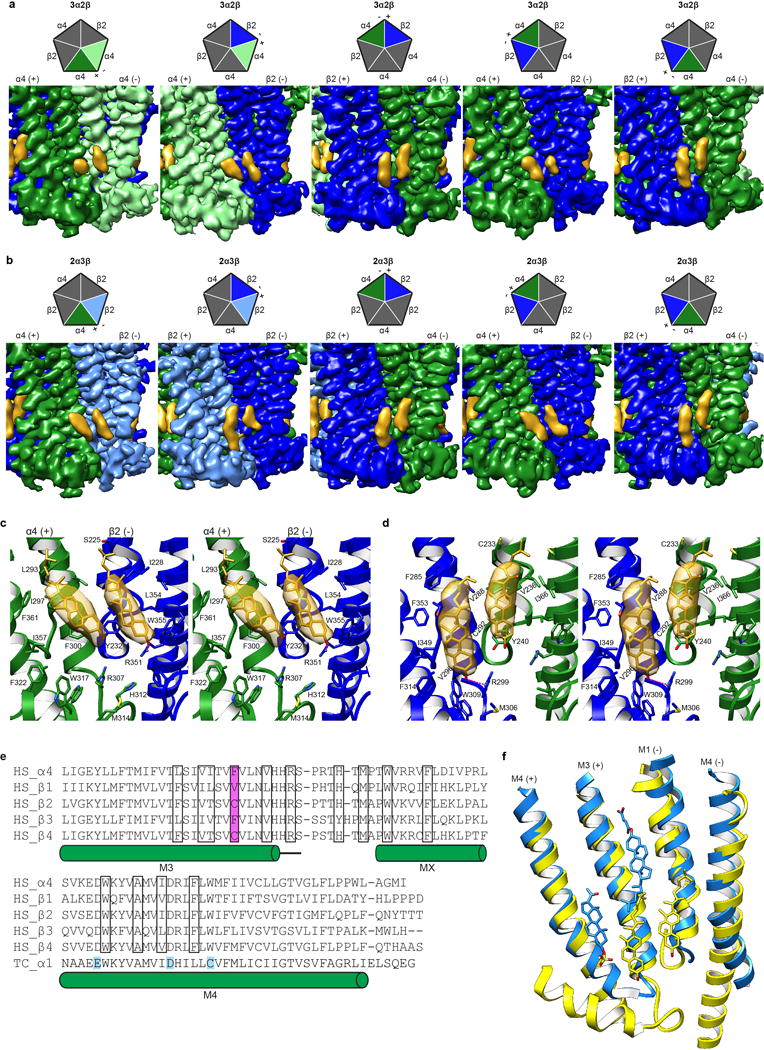
a-b, EM density map showing cholesterol sites from all TMD interfaces in the 3α:2β assembly and 2α:3β assembly respectively. Cartoon pentagons (top) are colored to illustrate subunits composing the displayed interface. c, Stereo image of Representative cholesterol binding site with α on principal side (+). When the principal side (+) is an α4 subunit both molecules tilt toward the principal face. One residue from the complementary side β2 (−), Y232, contacts both cholesterol molecules at the α-β interface (at the α-α interface the equivalent residue Y240 in the complementary (−) α4 contacts both cholesterol molecules at the interface; not shown). d, Stereo image of Representative cholesterol binding site with β on principal side (+). When the principal side (+) is a β2 subunit both cholesterol molecules are oriented orthogonal to the plane of the membrane. One residue from the principal (+) β2 subunit, V288, makes contacts with both cholesterol molecules at the β-α and the β-β (not shown) interface. Cholesterol and interacting side chains are shown as sticks. Density maps in panels a-d are displayed at a threshold of 0.025 in Chimera. e, Alignment of nicotinic receptor subunits for region encompassing putative cholesterol sites. Residues in cholesterol binding pocket are boxed and amino acid position implicated in differential cholesterol binding is highlighted in magenta. Uniprot accession IDs are provided40. Residues mapped using a photoreactive cholesterol analog in Torpedo californica (TC P02710)26,27 are highlighted in cyan. All other sequences are from Homo sapiens (HS P43681, P11230, P17787, Q05901, P30926 for α4 and β1-4 respectively). f, Superposition of β-α (yellow) from α4β2 and α1-α1 (Dodger blue) GABAA-GLIC chimera (PDB accession 5OSC25) TMD interface to compare putative binding sites. Pregnenolone sulfate, CHS and cholesterol are shown as sticks.
Extended Data Table 1.
Cryo-EM data collection, refinement and validation statistics.
| Dataset | 2α3β stoichiometry EMDB-7535 PDB-6CNJ |
3α2β stoichiometry EMDB-7536 PDB-6CNK |
|---|---|---|
| Data collection and processing | ||
| Magnification | 46,730 | |
| Voltage (kV) | 300 | |
| Electron exposure (e-/Å2) | 50 (2,588 images) or 75 (3,323 images) | |
| Defocus range (μm) | −0.5 to −4.5 | |
| Pixel size (Å) | 1.07 | |
| Symmetry Imposed | C1 | |
| Initial number of particles | 649,773 | |
| Final number of particles | 263,695 | 139,551 |
| Map resolution (Å) | 3.4/3.7* | 3.7/3.9* |
| FSC threshold | 0.143 | 0.143 |
| Refinement | ||
| Intial model used (PDB code) | 5KXI | 5KXI |
| Model resolution (Å) | 4.2 | 4.3 |
| FSC threshold | 0.5 | 0.5 |
| Map-sharpening B factor (Å2) | −150 | −170 |
| Model composition | ||
| Non-H atoms | 25,347 | 22,082 |
| Protein residues | 3,183 | 2,757 |
| B factors (Å2) | ||
| Receptor | 128 | 115 |
| Fab | 233 | 201 |
| Glycosylation | 135 | 117 |
| Ligand | 107 | 101 |
| Cholesterol | 142 | 140 |
| Ions | 117 | 27 |
| Validation | ||
| MolProbity score | 1.9 (100th percentile) | 1.98 (100th percentile) |
| Clashscore | 11.8 | 13.6 |
| Poor rotamers (%) | 0.25 | 0.16 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.005 | 0.007 |
| Bond angles (°) | 0.68 | 0.69 |
| Ramachandran plot | ||
| Favored (%) | 95.5 | 95.1 |
| Allowed (%) | 4.5 | 4.9 |
| Disallowed (%) | 0 | 0 |
Mask including Fab.
Supplementary Material
Acknowledgments
We thank D. Cawley at OHSU for production of monoclonal antibodies, X. Bai for EM discussion, Y. Jiang for the use of the oocyte rig, C. Noviello for assistance in EM data collection and all members of the Hibbs Lab and M. Horvath for discussion. Cryo-EM data were collected at the UT Southwestern Medical Center Cryo-Electron Microscopy Facility that is funded in part by the CPRIT Core Facility Support Award RP170644. We thank D. Nicastro and Z. Chen for support in facility access and data acquisition. We thank W. Chiu for cryo-EM training and resources in the National Center for Macromolecular Imaging (NCMI) at Baylor College of Medicine. NCMI is supported by NIH Grants P41GM103832 and U24GM116787. R.W. acknowledges support from the Sara and Frank McKnight Fund for Biochemical Research and the NIH (T32GM008203). R.E.H. is supported by a McKnight Scholar Award, The Welch Foundation (I-1812) and the NIH (DA037492, DA042072, and NS095899).
Footnotes
Author Contributions: RW expressed and purified the protein, made the EM samples, collected the EM data, processed the EM data with assistance from SR, built and refined the models and performed the binding assays. AG processed an initial EM dataset revealing the presence of two stoichiometries. CLMP performed the mAb and Fab characterization. JT performed the electrophysiology. RW and RH wrote the manuscript with assistance from SR; RH oversaw all aspects of the project. All authors gave feedback on the manuscript.
The authors declare no competing financial interests.
References
- 1.Nemecz A, Prevost MS, Menny A, Corringer PJ. Emerging Molecular Mechanisms of Signal Transduction in Pentameric Ligand-Gated Ion Channels. Neuron. 2016;90:452–470. doi: 10.1016/j.neuron.2016.03.032. [DOI] [PubMed] [Google Scholar]
- 2.Lester HA, et al. Nicotine is a selective pharmacological chaperone of acetylcholine receptor number and stoichiometry. Implications for drug discovery. Aaps J. 2009;11:167–177. doi: 10.1208/s12248-009-9090-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moroni M, Zwart R, Sher E, Cassels BK, Bermudez I. alpha4beta2 nicotinic receptors with high and low acetylcholine sensitivity: pharmacology, stoichiometry, and sensitivity to long-term exposure to nicotine. Molecular pharmacology. 2006;70:755–768. doi: 10.1124/mol.106.023044. [DOI] [PubMed] [Google Scholar]
- 4.Son CD, Moss FJ, Cohen BN, Lester HA. Nicotine normalizes intracellular subunit stoichiometry of nicotinic receptors carrying mutations linked to autosomal dominant nocturnal frontal lobe epilepsy. Molecular pharmacology. 2009;75:1137–1148. doi: 10.1124/mol.108.054494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weltzin MM, Lindstrom JM, Lukas RJ, Whiteaker P. Distinctive effects of nicotinic receptor intracellular-loop mutations associated with nocturnal frontal lobe epilepsy. Neuropharmacology. 2016;102:158–173. doi: 10.1016/j.neuropharm.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henderson R, et al. Tilt-pair analysis of images from a range of different specimens in single-particle electron cryomicroscopy. Journal of molecular biology. 2011;413:1028–1046. doi: 10.1016/j.jmb.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheres SH. Processing of Structurally Heterogeneous Cryo-EM Data in RELION. Methods in enzymology. 2016;579:125–157. doi: 10.1016/bs.mie.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Morales-Perez CL, Noviello CM, Hibbs RE. X-ray structure of the human alpha4beta2 nicotinic receptor. Nature. 2016;538:411–415. doi: 10.1038/nature19785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson ME, Kuryatov A, Choi CH, Zhou Y, Lindstrom J. Alternate stoichiometries of alpha4beta2 nicotinic acetylcholine receptors. Mol Pharmacol. 2003;63:332–341. doi: 10.1124/mol.63.2.332. [DOI] [PubMed] [Google Scholar]
- 10.Carbone AL, Moroni M, Groot-Kormelink PJ, Bermudez I. Pentameric concatenated (alpha4)(2)(beta2)(3) and (alpha4)(3)(beta2)(2) nicotinic acetylcholine receptors: subunit arrangement determines functional expression. British journal of pharmacology. 2009;156:970–981. doi: 10.1111/j.1476-5381.2008.00104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeDominicis KE, et al. The (alpha4)3(beta2)2 Stoichiometry of the Nicotinic Acetylcholine Receptor Predominates in the Rat Motor Cortex. Molecular pharmacology. 2017;92:327–337. doi: 10.1124/mol.116.106880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harpsoe K, et al. Unraveling the high- and low-sensitivity agonist responses of nicotinic acetylcholine receptors. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2011;31:10759–10766. doi: 10.1523/JNEUROSCI.1509-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazzaferro S, et al. Additional acetylcholine (ACh) binding site at alpha4/alpha4 interface of (alpha4beta2)2alpha4 nicotinic receptor influences agonist sensitivity. The Journal of biological chemistry. 2011;286:31043–31054. doi: 10.1074/jbc.M111.262014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shahsavar A, et al. Acetylcholine-Binding Protein Engineered to Mimic the alpha4-alpha4 Binding Pocket in alpha4beta2 Nicotinic Acetylcholine Receptors Reveals Interface Specific Interactions Important for Binding and Activity. Molecular pharmacology. 2015;88:697–707. doi: 10.1124/mol.115.098061. [DOI] [PubMed] [Google Scholar]
- 15.Ahring PK, et al. Engineered alpha4beta2 nicotinic acetylcholine receptors as models for measuring agonist binding and effect at the orthosteric low-affinity alpha4-alpha4 interface. Neuropharmacology. 2015;92:135–145. doi: 10.1016/j.neuropharm.2014.12.035. [DOI] [PubMed] [Google Scholar]
- 16.Tapia L, Kuryatov A, Lindstrom J. Ca2+ permeability of the (alpha4)3(beta2)2 stoichiometry greatly exceeds that of (alpha4)2(beta2)3 human acetylcholine receptors. Molecular pharmacology. 2007;71:769–776. doi: 10.1124/mol.106.030445. [DOI] [PubMed] [Google Scholar]
- 17.Hansen SB, Wang HL, Taylor P, Sine SM. An ion selectivity filter in the extracellular domain of Cys-loop receptors reveals determinants for ion conductance. The Journal of biological chemistry. 2008;283:36066–36070. doi: 10.1074/jbc.C800194200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imoto K, et al. Rings of negatively charged amino acids determine the acetylcholine receptor channel conductance. Nature. 1988;335:645–648. doi: 10.1038/335645a0. [DOI] [PubMed] [Google Scholar]
- 19.Livesey MR, et al. Structural determinants of Ca2+ permeability and conduction in the human 5-hydroxytryptamine type 3A receptor. The Journal of biological chemistry. 2008;283:19301–19313. doi: 10.1074/jbc.M802406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelley SP, Dunlop JI, Kirkness EF, Lambert JJ, Peters JA. A cytoplasmic region determines single-channel conductance in 5-HT3 receptors. Nature. 2003;424:321–324. doi: 10.1038/nature01788. [DOI] [PubMed] [Google Scholar]
- 21.Dwyer TM, Adams DJ, Hille B. The permeability of the endplate channel to organic cations in frog muscle. The Journal of general physiology. 1980;75:469–492. doi: 10.1085/jgp.75.5.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baenziger JE, Domville JA, Therien JPD. The Role of Cholesterol in the Activation of Nicotinic Acetylcholine Receptors. Current topics in membranes. 2017;80:95–137. doi: 10.1016/bs.ctm.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 23.Hattori M, Hibbs RE, Gouaux E. A fluorescence-detection size-exclusion chromatography-based thermostability assay for membrane protein precrystallization screening. Structure. 2012;20:1293–1299. doi: 10.1016/j.str.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller PS, et al. Structural basis for GABAA receptor potentiation by neurosteroids. Nature structural & molecular biology. 2017;24:986–992. doi: 10.1038/nsmb.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laverty D, et al. Crystal structures of a GABAA-receptor chimera reveal new endogenous neurosteroid-binding sites. Nature structural & molecular biology. 2017;24:977–985. doi: 10.1038/nsmb.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamouda AK, Chiara DC, Sauls D, Cohen JB, Blanton MP. Cholesterol interacts with transmembrane alpha-helices M1, M3, and M4 of the Torpedo nicotinic acetylcholine receptor: photolabeling studies using [3H]Azicholesterol. Biochemistry. 2006;45:976–986. doi: 10.1021/bi051978h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corbin J, Wang HH, Blanton MP. Identifying the cholesterol binding domain in the nicotinic acetylcholine receptor with [125I]azido-cholesterol. Biochimica et biophysica acta. 1998;1414:65–74. doi: 10.1016/s0005-2736(98)00153-9. [DOI] [PubMed] [Google Scholar]
- 28.Morales-Perez CL, Noviello CM, Hibbs RE. Manipulation of Subunit Stoichiometry in Heteromeric Membrane Proteins. Structure. 2016;24:797–805. doi: 10.1016/j.str.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grant T, Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 A reconstruction of rotavirus VP6. eLife. 2015;4:e06980. doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang K. Gctf: Real-time CTF determination and correction. Journal of structural biology. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernandez-Leiro R, Scheres SHW. A pipeline approach to single-particle processing in RELION. Acta crystallographica Section D, Structural biology. 2017;73:496–502. doi: 10.1107/S2059798316019276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheres SHA. Bayesian view on cryo-EM structure determination. Journal of molecular biology. 2012;415:406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nature methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 35.Biasini M, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic acids research. 2014;42:W252–258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica Section D, Biological crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amunts A, et al. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343:1485–1489. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta crystallographica Section D, Biological crystallography. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.The UniProt, C. UniProt: the universal protein knowledgebase. Nucleic acids research. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pei J, Kim BH, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. Journal of molecular biology. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 43.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. Journal of molecular graphics. 1996;14376:354–360. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 44.Hayward S, Lee RA. Improvements in the analysis of domain motions in proteins from conformational change: DynDom version 1.50. J Mol Graph Model. 2002;21:181–183. doi: 10.1016/s1093-3263(02)00140-7. [DOI] [PubMed] [Google Scholar]
- 45.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein engineering. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 46.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morin A, et al. Collaboration gets the most out of software. eLife. 2013;2:e01456. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Venkatachalan SP, et al. Optimized expression vector for ion channel studies in Xenopus oocytes and mammalian cells using alfalfa mosaic virus. Pflugers Archiv: European journal of physiology. 2007;454:155–163. doi: 10.1007/s00424-006-0183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Galzi JL, Bertrand S, Corringer PJ, Changeux JP, Bertrand D. Identification of calcium binding sites that regulate potentiation of a neuronal nicotinic acetylcholine receptor. The EMBO journal. 1996;15:5824–5832. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates of the two α4β2 receptor-nicotine-Fab complexes have been deposited in the PDB (2α:3β is PDB-6CNJ; 3α:2β is PDB-6CNK). The cryo-EM density maps have been deposited in the EMDB upon acceptance for publication (2α:3β is EMDB-7535; 3α:2β is EMDB-7536).


