Abstract
Microbiota and their hosts have coevolved for millions of years. Microbiota are not only critical for optimal development of the host under normal physiological growth, but also important to ensure proper host development during nutrient scarcity or disease conditions. A large body of research has begun to detail the mechanism(s) of how microbiota cooperate with the host to maintain optimal health status. One crucial host pathway recently demonstrated to be modulated by microbiota is that of the growth factor insulin like growth factor 1 (IGF-1). Gut microbiota are capable of dynamically modulating circulating IGF-1 in the host, with the majority of data suggesting that microbiota induce host IGF-1 synthesis to influence growth. Microbiota-derived metabolites such as short chain fatty acids are sufficient to induce IGF-1. Whether microbiota induction of IGF-1 is mediated by the difference in growth hormone expression or the host sensitivity to growth hormone is still under investigation. This review summarizes the current data detailing the interaction between gut microbiota, IGF-1 and host development.
Keywords: Microbiota, Microbiome, IGF-1, Bone, Growth, SCFA
Introduction
Microbiota is the collection of microbial species that inhabit a community, while the microbiome is the genetic content, or metagenome, contributed by the entirety of this community. The intestinal microbiota is the community that is best characterized and whose impact on host physiology has been explored most deeply through experimental perturbation, primarily in mouse models, and is the focus of this review. In humans, the contribution of the intestinal metagenome to the genetic material of a host is substantial, with approximately 150 times more genes represented by the metagenome than present in the host genome [1]. Although the phyla Bacteriodetes and Firmicutes dominate the community of organisms that comprise the gut microbiome is genetically diverse at the species level and varies substantially between individuals [1, 2]. Similarly, in mouse model systems, substantial facility to facility and even cage to cage variation in microbiota has been observed [3]. Thus, correlation of particular species or defined community with microbiota’s impact on the host is challenging. Increasingly, metatranscriptomic and metabolomic approaches are employed in studying host–microbiota interactions. These approaches are likely to provide insights into microbial products or metabolic activities that can be provided by a variety of species, with the impact on the host determined by the level of the metabolite rather than on presence or absence of a particular species. Ultimately, these approaches have the potential to clarify mechanistic links between particular microbial communities and perturbations in host physiology [4, 5].
Commonly recognized microbial metabolites include short chain fatty acids (SCFA), indole derivatives including serotonin, polyamines, and ATP. In addition, microbiota are capable of biochemically modifying host metabolites, for example bile acids. These microbial metabolites are documented to have a variety of effects on the host immune system and metabolism (recently reviewed in detail by Postler and Ghosh [6]). In addition to relatively well-studied metabolites such as SCFA, numerous other metabolites in the mammalian intestinal lumen and plasma are dependent on the presence of microbiota, with approximately 10% of plasma metabolites altered by microbiota [7, 8]. Thus, microbiota generate numerous chemicals that, like hormones, can act at distal sites by entering the bloodstream. Moreover, microbiota indirectly regulate host-produced hormones including PYY, glucagon-like-peptide 1, leptin and ghrelin, as well as affecting the hypothalamic–pituitary–adrenal axis (recently reviewed by Clarke et al. [9]), leading some to refer to the gut microbiota as an endocrine organ.
Work from a number of labs, including ours, has demonstrated that microbiota also influence levels of insulin-like growth factor 1 (IGF-1) and orthologs in drosophila, zebrafish, chicken and mouse [10–15]. This work expands our understanding of microbiota’s impact on the host endocrine system.
IGF family members play an essential role in regulating skeletal development and postnatal growth as demonstrated by genetically modified mice lacking the genes encoding IGF-1 or its receptor, IGF-1R [16, 17]. A second ligand, IGF-2, also activates IGF1-R but functions predominantly in early embryogenesis [18]. Mice with germline loss of Igf1 are born runted and exhibit decreased post-natal growth rate and delayed skeletal ossification demonstrating a critical role for IGF-1 in bone [17]. Loss of IGF1 in humans has been found to similarly result in severe growth retardation [19]. Mice lacking both IGF-1 and its receptor (Igf1−/− Igf1R−/−) phenocopy Igf1R−/− mice, suggesting that IGF-1 acts solely through the IGF-1R [16].
The IGF-1R is present in many cell types within the skeleton, with tissue-specific deletion of IGF-1R in several lineages giving rise to bone phenotypes. Deletion of IGF-1R in chondrocytes (using Col II-Cre) leads to growth defects, with both growth plate abnormalities and less skeletal mineralization [20]. Deletion in IGF-1R in early osteoblasts (using Osx-Cre) impaired osteoblast proliferation and early maturation, leading to trabecular bone loss and undermineralization [21], while deletion in mature osteoblasts (using Ocn-Cre) results in an increased proportion of osteoid and decreased trabecular bone [22, 23], consistent with a role for IGF-1 promoting osteoblast proliferation, both early and late differentiation, and coupling matrix production to mineralization.
IGF-1 has long been recognized as a key mediator of skeletal growth, with evidence supporting IGF-1 actions via endocrine, paracrine, as well as autocrine fashion. Circulating IGF-1 is primarily synthesized by the liver in response to growth hormone. The majority of IGF-1 in circulation is bound in a ternary complex with IGF binding protein 3 (IGFBP3) and acid labile subunit (ALS), with only a small proportion bound to other IGFBP or free. IGF-1 is also produced in peripheral tissues, including muscle and bone [24]. Initially, the importance of circulating IGF-1 for bone turnover and skeletal development was unclear, as tissue-specific deletion of IGF-1 in liver decreased circulating IGF-1 levels by 70% without impairing skeletal growth or mineralization [25]. The likely importance of autocrine/paracrine functions of IGF-1 in the skeleton was highlighted by demonstration that overexpression of IGF-1 in osteoblasts is sufficient to increase bone formation and promote mineralization, while selective loss of IGF-1 in osteocytes (using Dmp1-Cre) results in decreased bone turnover, diminished bone mineral content and loss of response to load [26, 27]. Both autocrine and paracrine functions of IGF-1 seem to be required for optimal osteoclast differentiation [28]. Although this data suggests that local IGF-1 trumps circulating IGF-1 in skeletal regulation, other data strongly support a role for circulating IGF-1 in controlling bone. Serum IGF-1 levels correlate well with bone mineral content in healthy children [29], and declining hepatic IGF-1 production has been invoked as one mechanism of osteopenia in cirrhosis [30]. A threshold level of circulating IGF-1 appears to be required for normal linear growth and bone turnover, however, as elimination of both liver IGF-1 and ALS (which is known to prolong the half-life of IGF-1) causes further decreases in IGF-1 serum levels and results in decreased bone length, periosteal circumference and bone density [31]. Moreover, mice heterozygous for Igf1 demonstrated lower serum IGF-1 levels and decreased cortical thickness and tissue bone mineral density, which were rescued by exogenous IGF-1 injection [32]. Thus, the work of a number of investigators in both mice and humans suggests that modulation of circulating IGF-1 can impact skeletal growth and bone turnover.
The major factors regulating circulating IGF-1 is the induction of liver IGF-1 production in response to GH, and regulation of half-life by ALS, which is also produced in the liver in response to GH. Little is known about other pathways by which circulating IGF-1 levels could be modulated. Recently, data from experiments using germ-free and antibiotic-treated animals in several evolutionarily divergent species have accumulated implicating gut microbiota in the regulation of IGF-1 levels.
Microbiota Induced IGF-1 Signaling in Drosophila Larval Development
The initial evidence suggesting that microbiota can affect host IGF-1 production comes from two complementary studies in the invertebrate Drosophila melanogaster [10, 14]. Insulin and insulin-like growth factor pathways are highly conserved throughout the animal kingdom with orthologs in a wide variety of species, including Drosophila. Insulin/insulin-like growth factor signaling (IIS) plays an important role in regulating growth and metabolism in Drosophila, with the Drosophila homolog of IGF-1 being Drosophila insulin-like peptides (dILPs).
Drosophila has a relatively simple gut microbiota community. The adult midgut is typically in stable contact with a symbiotic commensal community composed of 5–20 different microbial species that consist primarily members of the Acetobacter and Lactobacillus genera. Utilizing a laboratory-raised Drosophila that harbors five bacterial species, Shin et al. examined the host growth rate and body size in conventionally reared and germ-free fruit fly [14]. They found that the time to develop from larvae to puparium is longer for germ-free larvae than for conventional larvae, although body size is not affected. However, on a diet containing < 1% yeast the germ-free larvae were < 10% of the size of corresponding conventionally raised larvae and died at first instar. By colonizing germ-free larvae with individual bacterial species, they demonstrated that colonization with Acetobacter pomorum alone was sufficient to restore the host developmental rate and body size under nutrient-poor conditions to comparable to larvae harboring all of the 5 species. A similar requirement for gut microbiota under nutrient deprivation was reported by Storelli et al. [10]. They found that the Drosophila commensal Lactobacillus plantarum rescued larval lethality on nutrient-poor diet containing 0% yeast. Furthermore, monoassociation with L. plantarum but not Enterococcus faecalis or a second strain of L. plantarum, was sufficient to promote larvae growth under nutrient poor (10% yeast) conditions. These data demonstrate that a single Drosophila commensal bacterial species is sufficient to recapitulate the beneficial growth effects of convention gut microbiota under nutrient-poor conditions.
To address the mechanism by which monocolonization with a given species rescued growth, Shin et al. established a draft genome sequence for A. pomorum, generated a library of mutants via transposon-mediated random mutagenesis and screened mutants for lack of growth promotion under nutrient-free conditions [14]. They identified the pyrroloquinoline quinone-dependent alcohol dehydrogenase (PQQ-ADH)-dependent oxidative respiratory chain pathway as essential for the effects of A. pomorum on larval development under nutrient-poor conditions.
Monoassociation with an A. pomorum mutant in this pathway, P3G5, resulted in adults with smaller wings, reduced cell size and number, and smaller intestines, a phenotype similar to that of flies deficient in IIS signaling. In fact, A. pomorum induced IIS activation while P3G5 did not. Restoring IIS activity through ectopic overexpression of Drosophila insulin-like peptide 2 (DILP2) largely rescued the growth phenotype of P3G5-monoassociated flies, suggesting that a key difference between PG35 and wild-type A. pomorum is the ability to induce dILP and activate downstream pathways that enhance larval growth under nutrient-poor conditions, although it cannot be ruled out that DILP2 overexpression non-specifically bypasses the growth defect of P3G5-monoassociated flies. Interestingly, DILP2 overexpression didnot rescue developmental defects in germ-free flies, suggesting that microbiota may activate additional host pathways necessary for growth during nutrient deprivation [14]. Storelli et al. similarly identified increased dILP activity as the mechanism by which L. plantarum rescues larval development during undernutrition [10]. Using lnR gene expression as a readout of systemic dILP activity, they found that L. plantarum association correlates with increased systemic InR signaling during larval growth and L. plantarum colonization is sufficient to induce dILP activity. Taken together, the work of these two groups supports the concept that gut microbiota regulate host IGF signaling to influence growth.
Lactobacillus Species Induce IGF-1 and Promote Growth in Vertebrates
Subtherapeutic antibiotics, which modulate the gut microbiota, have been widely used to enhance growth in livestock. With rising concerns about antibiotic resistance, modulation of livestock gut microbiota by feeding with prebiotics (non-digestible dietary fiber and oligosaccharides), probiotics (microorganisms) and combinations of both have been investigated as an alternative means to enhance productivity. Supplementing the feed of broiler chickens with either of two strains of L. plantarum along with the prebiotic inulin increased body weight and enhanced the expression of IGF-1 in the liver [13]. Although circulating IGF-1 was not directly measured, the 3–5 fold increases in liver IGF-1 transcript observed in supplemented animals would seem likely to increase serum IGF-1 levels. A growth-promoting effect of another Lactobacillus species, L. rhamnosus, a commonly used probiotic, was demonstrated in zebrafi [15]. Addition of live L. rhamnosus to tank water resulted in detection of L. rhamnosus in the gastrointestinal tract. Treated fish demonstrated increased body length, weight and enhanced calcification of the vertebrae, along with increased transcription of IGF1 and IGF2 in larvae [15]. Thus, a correlation between gut microbiota, IGF levels and growth appears to be conserved from invertebrates to vertebrates.
Microbiota Induced IGF-1 Signaling in Mammalian Post-natal Growth and Skeletal Development
IGF-1 is a critical growth factor that controls postnatal growth in mammals. Studies in mice demonstrate that circulating IGF-1 levels are significantly higher in mice with an intact gut microbiota (conventionally raised mice) compared to germ-free (GF) mice. As expected, lower IGF-1 in GF mice was correlated with decreased linear growth and body weight, suggesting that in mammals the gut microbiota is required to ensure optimal growth. Skeletal development was impacted, with decreased femur length, cortical thickness and trabecular bone seen in GF mice compared to conventionally raised animal [12]. The critical role of IGF-1 for growth promotion by gut microbiota was demonstrated in two ways. Administering recombinant IGF-1 to germ-free mice after weaning promoted the body growth and femur length, while blocking IGF-1 signaling pathway with an inhibitor of IGF-1R in conventionally raised mice diminished the growth benefit of colonization [12]. Moreover, modulating the gut microbiota can dynamically modulate circulating IGF-1 and affect bone formation in adult mice. Our group recently demonstrated that reconstitution of adult GF mice with convention microbiota led to significantly higher levels of serum IGF-1 compared to littermates that remained GF [11]. Conversely, treatment with broad-spectrum antibiotics decreased serum IGF-1 compared to the control mice. Treatment with oral vancomycin, which targets intestinal Gram-positive bacteria, as sufficient to decrease circulating IGF-1, narrowing the microbiota association with circulating IGF-1 levels to Gram-positive species. Increased serum IGF-1 after colonization of GF mice was accompanied by increases in the serum marker of bone formation marker, N-terminal propeptide of type I collage (P1NP) while antibiotic treatment decreased P1NP, suggesting that decreases in circulating IGF-1 negatively affected bone formation. Consistent with the increase in IGF-1 and P1NP, microbiota colonization increased host bone formation rate and led to increased femur length [11]. This data suggests the possibility that manipulation of gut microbiota composition could be used to promote IGF-1 production and growth.
As in flies, the gut microbiota in mice appears to be particularly important under conditions of nutrient deprivation. Mouse juvenile growth under conditions of malnutrition is significantly impaired in the absence of microbiota [12]. Similar to the observations in Drosophila, gnotobiotic mice monoassociated with L. plantarum alone can maintain mouse growth upon malnutrition, suggesting an evolutionarily conserved nutrient-sensing endocrine pathway that regulates juvenile growth [12].
Studies have not uniformly found a positive correlation between microbiota, IGF-1 and skeletal growth. A recent study found increased serum IGF-1 and bone Igf1 expression in GF compared to conventional mice, with GF mice having a corresponding higher bone formation rate and trabecular bone volume consistent with higher circulating IGF-1 [33]. In this study, it is important to note that C57BL/6 mice from Taconic were used [33], while the hybrid strain CB6F1 or BALB/c mice were used in the studies demonstrating an increase in IGF-1 in colonized mice [11, 12]. Therefore, the difference in the impact of microbiota on IGF-1 and skeletal remodeling seen in this study and those results discussed above could be due to the different strain backgrounds used and/or to differences in the composition of the gut microbial community between facilities [34]. This points to the difficulty in making conclusive statements about the effect of gut microbiota on host physiology when the identity of the microbiota or microbial product affecting the host are poorly defined.
Microbiota Induced IGF-1 Ameliorates Muscle Wasting in DSS Colitis
Microbiota induction of host IGF-1 could have many benefits for the host, including protection during disease challenges. The microbial community present in the mouse intestine is well documented to vary between different animal facilities, and multiple examples have been reported where disease phenotype is modulated by the different microbiota across facilities. Treatment with dextran sulfate sodium (DSS) is used to induce inflammatory bowel disease in mice. DSS treated C57BL/6 mice from Jackson exhibit muscle and fat wasting with weight loss, and treatment with broad-spectrum antibiotic cocktail has no significant impact on disease severity. In contrast, antibiotic treatment protects DSS treated C57BL/6 mice maintained at UC Berkeley from weight loss, consistent with decreased disease severity. Co-housing the antibiotic-treated Jackson mice with Berkeley mice protected the Jackson mice from wasting, strongly suggesting that the microbiota composition may account for the difference between mouse colonies [35]. The investigators then used 16S rRNA sequencing to identify an E. coli O21:H+ strain that is absent in antibiotic-treated Jackson but not Berkeley mice. Monoassociation of germ-free mice with this E. coli O21:H+ strain protected DSS treated mice from wasting, including muscle wasting. Like DSS treatment, S. typhimurium and B. thailandensis infections causes muscle wasting in Jackson C57BL/6 mice that is rescued by colonization with E. coli O21:H+ [35]. To understand the protective mechanism, the investigators performed RNA sequencing analysis of leg muscles after S. typhimurium infection and found that E.coli O21:H+ colonized mice had evidence of increased IGF-1 signaling compared with infected control mice. In fact, the decrease in circulating IGF-1 after infection was ameliorated by colonization. The causal role of increased IGF-1 in protection from muscle wasting was demonstrated by the loss of the E.coli O21:H+ protective effect in mice treated with an IGF-1 neutralizing antibody [35]. Thus, the effect of pathogenic infection on host circulating IGF-1 can be modulated by the presence of specific microbiota to alter the expression of disease.
Multiple Host Organs/Tissues Coordinate to Produce IGF-1 in Response to Microbiota
In Drosophila, dILPs are produced by the larval fat body, the Drosophila counterpart of liver and adipose tissue. While liver is the primary source of circulating IGF-1 in mammals, as discussed above, bone and muscle cells can also produce IGF-1 locally to elicit autocrine and paracrine effects on postnatal growth. Additionally, white-adipose-tissue (WAT) has been reported to be capable of producing IGF-1 in sufficient amounts to impact circulating IGF-1 [36].
The effect of gut microbiota on host IGF-1 production by various tissues has been examined by a number of investigators. Different microbiota or host conditions may impact IGF-1 production by individual tissues more or less, though comparisons between studies are limited by the limitations in which tissues were assessed and differences in method of assessment (transcript level versus protein level per gram of tissue versus protein produced per organ).
Expression of Igf1 is higher in the liver from conventional juvenile mice compared to GF juveniles [12]. Similarly, liver Igf1 expression was higher in chickens fed L. plantarum with the probiotic inulin [13] and colonization of adult GF mice with conventional microbiota significantly increased IGF-1 production by the liver [11]. Colonization of GF mice also increased IGF-1 production in WAT, as measured in the abdominal fat pad [11]. IGF-1 mRNA and protein levels in WAT were substantially and significantly higher in E. coli O21:H+ colonized animals after infection, though liver Igf1 expression was unchanged [35]. Muscle is also a source of IGF-1, and muscle Igf1 expression was higher in conventional compared to GF juvenile mice, but was not altered by colonization of adult GF mice with conventional microbiota or by E. coli O21:H+ colonization [11, 12, 35].
Microbiota-mediated changes in local IGF-1 could also play an important role in regulating bone remodeling. In our recent study, we proposed that microbiota promote bone growth through not only systemic but also local IGF-1 production. We found that upon colonization with conventional microbiota there is a significant increase in bone marrow Igf1 expression. The expression of Runx2, a downstream target gene of IGF-1 was also significantly increased in bone from colonized mice [11]. In contrast, in the report which showed that there is a negative association between microbiota and bone growth, Igf1 expression was found to decrease in SPF compared to GF bone marrow and calvaria [33]. Together, these data indicate that changes in local Igf1 expression correlates with the effect of microbiota on bone.
The Mechanism of IGF-1 Induction by Microbiota
How microbiota induce IGF-1 systemically and locally is still under investigation. IGF-1 is downstream of growth hormone during postnatal development [24]. Whether microbiota-mediated IGF-1 production is dependent on growth hormone is unclear. Circulating growth hormone was similar in germ-free mice, conventional raised mice and colonized mice, despite the substantial differences in IGF-1 levels [11, 12]. Similarly, serum growth hormone in B. thailandensis infected mice is not altered by E. coli O21:H+ colonization, suggesting growth hormone independent modulation of IGF-1 levels by E. coli colonization. Although growth hormone levels of GF and L. plantarum monoassociated mice are similar, the tissue response to growth hormone, as assessed by expression of Ghr and transcriptional targets of growth hormone signaling, is impaired. Thus, in the absence of microbiota the activity of the somatrotropic axis may be impaired due to a growth hormone resistance state. Together, these data suggest that alterations in IGF-1 production and function in response to microbiota are not mediated solely by changes in growth hormone levels.
As gut microbiota are affecting IGF-1 production in distant tissues, and given the many different bacterial species implicated in modulating IGF-1 levels, it seemed likely that a microbial metabolite produced in common by each of these species could provide the mechanistic link between gut microbiota and regulation of host IGF-1. An excellent candidate was SCFA (acetate, propionate and butyrate), abundant microbial metabolites produced by fermentation of non-digestible dietary fibers. Butyrate is a major energy source for enterocytes, and SCFA act both locally in the gut but also enter the circulation and can act distantly. SCFA can modulate host cell function either by activation of their cognate G protein-coupled receptors (GPR41, GPR43 and GPR109) or by acting as inhibitors of histone deacetylase [37]. Cecal concentrations of SCFA are higher in conventionally raised compared to GF mice [38, 39]. We found that colonization of GF mice did result, as expected, in increases in cecal SCFA, which were decreased by broad-spectrum antibiotics and vancomycin treatment of conventional mice [11]. As cecal SCFA concentration roughly correlated with trends in serum IGF-1, we tested if SCFA supplementation was sufficient to increase circulating IGF-1 in antibiotic-treated mice. After 4 weeks of supplementation with SCFA in the drinking water, serum IGF-1 levels were higher in SCFA supplemented antibiotic-treated mice compared to antibiotic only controls. Similar to colonization of GF mice, SCFA supplementation increased adipose tissue IGF-1 production and resulted in a trend toward increased liver IGF-1 production [11]. Thus, we hypothesize that microbiota produced SCFA act either directly or indirectly on host liver and adipose tissue to increase circulating IGF-1 levels and promote growth and skeletal development (see Fig. 1). However, despite broad-spectrum antibiotics, some gut microbiota species remain and it is not possible to definitively conclude that SCFA are sufficient to induce IGF-1, and it is likely that additional microbiota–host interactions contribute to the increased IGF-1 production by host tissues.
Fig. 1.
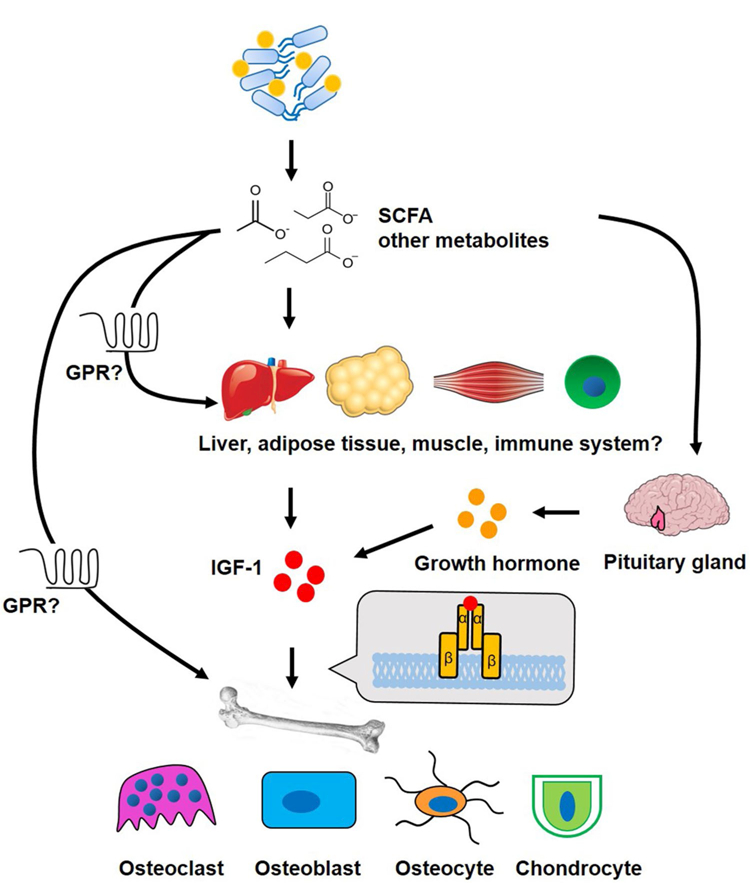
Schematic of the connection of gut microbiota, host IGF-1 and effects on bone. Gut microbiota produced short chain fatty acids (SCFA), and possibly other yet to be identified microbial metabolites, act on liver and adipose tissue to induce production of insulin-like growth factor 1 (IGF-1), which in turn acts on bone cells to influence linear growth, bone mass and mineralization. Additionally, metabolites could act directly on bone and muscle, inducing local IGF-1 production. Whether the effect of SCFA on host IGF-1 is mediated by their G-protein coupled receptors (GPR) is not known. Microbial metabolites including SCFA may also have as yet unappreciated effect on growth hormone (GH) release from the pituitary, promoting liver IGF-1 production. Further, it is possible that the well-documented effects of SCFA on the immune system indirectly influence host growth and skeletal development
Work from other groups in a variety of species supports a connection between microbiota produced SCFA and either circulating IGF-1 or skeletal health. In Drosophila, the mutant P3G5 monoassociated flies showed impaired production of acetate, and supplementation with acetic acid reversed the growth defects in P3G5 colonized flies suggesting a role for SCFA in the growth promotion and increased dILP signaling [14]. In chickens, feeding with the combination of probiotics and L. plantarum significantly increased fecal SCFA, particularly acetate [13]. In mice,supplementation with either L. rhamnosus, L. reuteri or a defined community including a number of Lactobacillus species, prevented sex-steroid deficiency induced bone loss [40, 41]. Additionally, there are many reports that feeding with probiotics, non-digestible fiber and oligosaccharides that are fermented into SCFA, promotes skeletal health, a topic recently thoroughly reviewed by McCabe et al. [42]. Although the presence of microbiota and production of SCFA are correlated with growth and/or induction of IGF-1 in a number of species and across a variety of experimental manipulations, existing data suggest that other mechanism(s) likely exist by which microbiota regulate IGF-1, growth and skeletal development. In fact, in the case of E. coli O21:H+ colonization, this species colonized WAT after host infection with B. thailandensis [35], suggesting that microbiota may directly induce IGF-1 production by adipose tissue. Furthermore, supplementation with acetic acid, a precursor for acetate, could not rescue the growth phenotype of GF flies, in contrast to P3G5 monoassociated fl [14], suggesting that other microbiota–host interactions contribute to growth phenotypes. For example, undernutrition has been linked to low IGF-1 levels and GH resistance (reviewed by Fazeli and Klibanski [43]), and it is possible that the composition of microbiota alters either the ability to extract nutrients or the host response to nutrients, which could indirectly impact IGF-1 levels.
Future Directions
The idea that manipulation of the gut microbial community by supplementing with specific bacterial species or defined microbial communities, with or without probiotics, could be used to treat osteopenia or growth deficits in children is extremely attractive as it would take advantage of endogenous host regulatory circuits. Additionally, it is likely to have high acceptability to patients because of the perception that it is a “natural” treatment and the general popularity of probiotic supplementation with the public. While pre-clinical data suggesting that this could be possible is exciting, a substantial number of unknowns remain to be answered before this type of therapy could be rationally proposed.
At the most basic level, it is not yet clear whether particular species of microbes are required to induce host IGF-1, or if the key factor is the capacity of an individual’s gut microbiota to produce certain metabolites. Metagenomic and metabolomic approaches could be extremely informative as to the common downstream effects of the many microbiota species shown to affect IGF-1 levels in a variety of species. While SCFA are clearly implicated in the mechanism by which microbiota modulate host IGF-1, elucidation of the mechanism by which SCFA modulate host IGF-1 production is an area requiring further investigation. Whether all three SCFA are equally important in this pathway is not clear. Further, it is not known if SCFA alters liver and adipose IGF-1 production through direct effects on liver and fat or indirectly through actions on enterocytes or immune cells. Mechanistically, whether the relevant activity of SCFA is inhibition of histone deacetylases or activation of their G protein-coupled receptors is not clear, though circulating IGF-1 in GPR109-deficient mice is similar to that of wild-type littermate controls suggesting that GPR109 at least is dispensable for host IGF-1 production [44]. Perhaps the most significant outstanding question is whether SCFA are sufficient to induce host IGF-1 in the absence of any gut microbiota or whether other microbiota–host interactions influencing IGF-1 remain to be discovered. The data from flies would suggest that SCFA may not be sufficient. Whether SCFA supplementation was not sufficient to rescue growth defects in GF flies because supplementation was solely with acetate precursors, because microbiota effects on gut maturation are required for host response to SCFA or whether this reflects a requirement for activation of a second microbe–host pathway to promote IGF-1 production and host growth is unclear. Future studies incorporating metabolomics and metagenomic tools, as well as a more detailed examination of host response to colonization with defined microbial communities or treatment with microbial metabolites will hopefully answer these questions and raise new and exciting areas for additional investigation.
Acknowledgments
Funding This work was supported by NIH Grants AG046257 from the NIA, AR062590 from NIAMS, the Rheumatology Research Foundation and a Faculty Career Development Award from the Brigham and Women’s Hospital.
Footnotes
Conflict of interest Neither Dr. Jing Yan nor Dr. Julia Charles have anything to disclose.
References
- 1.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, Meta HITC, Bork P, Ehrlich SD, Wang J (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464(7285):59–65. 10.1038/nature08821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morgan XC, Segata N, Huttenhower C (2013) Biodiversity and functional genomics in the human microbiome. Trends Genet 29(1):51–58. 10.1016/j.tig.2012.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hildebrand F, Nguyen TL, Brinkman B, Yunta RG, Cauwe B, Vandenabeele P, Liston A, Raes J (2013) Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol 14(1):R4 10.1186/gb-2013-14-1-r4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bashiardes S, Zilberman-Schapira G, Elinav E (2016) Use of metatranscriptomics in microbiome research. Bioinform Biol Insights 10:19–25. 10.4137/BBI.S34610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luan H, Wang X, Cai Z (2017) Mass spectrometry-based metabolomics: targeting the crosstalk between gut microbiota and brain in neurodegenerative disorders. Mass Spectrom Rev 10.1002/mas.21553 [DOI] [PubMed] [Google Scholar]
- 6.Postler TS, Ghosh S (2017) Understanding the holobiont: how microbial metabolites affect human health and shape the immune system. Cell Metab 26(1):110–130. https://doi.org/10.1016/j. cmet.2017.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsumoto M, Kibe R, Ooga T, Aiba Y, Kurihara S, Sawaki E, Koga Y, Benno Y (2012) Impact of intestinal microbiota on intestinal luminal metabolome. Sci Rep 2:233 https://doi.org/10.1038/ srep00233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G (2009) Metabolomics analysis reveals large effects of gut microfl a on mammalian blood metabolites. Proc Natl Acad Sci USA 106(10):3698–3703. https://doi.org/10.1073/ pnas.0812874106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke G, Stilling RM, Kennedy PJ, Stanton C, Cryan JF, Dinan TG (2014) Minireview: gut microbiota: the neglected endocrine organ. Mol Endocrinol 28(8):1221–1238. https://doi.org/10.1210/ me.2014-1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Storelli G, Defaye A, Erkosar B, Hols P, Royet J, Leulier F (2011) Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab 14(3):403–414. https://doi.org/10.1016/j. cmet.2011.07.012 [DOI] [PubMed] [Google Scholar]
- 11.Yan J, Herzog JW, Tsang K, Brennan CA, Bower MA, Garrett WS, Sartor BR, Aliprantis AO, Charles JF (2016) Gut microbiota induce IGF-1 and promote bone formation and growth. Proc Natl Acad Sci U S A 113(47):E7554–E7563. https://doi.org/10.1073/ pnas.1607235113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwarzer M, Makki K, Storelli G, Machuca-Gayet I, Srutkova D, Hermanova P, Martino ME, Balmand S, Hudcovic T, Heddi A, Rieusset J, Kozakova H, Vidal H, Leulier F (2016) Lactobacillus plantarum strain maintains growth of infant mice during chronic undernutrition. Science 351(6275):854–857. 10.1126/science.aad8588 [DOI] [PubMed] [Google Scholar]
- 13.Kareem KY, Loh TC, Foo HL, Akit H, Samsudin AA (2016) Effects of dietary postbiotic and inulin on growth performance, IGF1 and GHR mRNA expression, faecal microbiota and volatile fatty acids in broilers. BMC Vet Res 12(1):163 10.1186/s12917-016-0790-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shin SC, Kim SH, You H, Kim B, Kim AC, Lee KA, Yoon JH, Ryu JH, Lee WJ (2011) Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334(6056):670–674. https://doi.org/10.1126/ science.1212782 [DOI] [PubMed] [Google Scholar]
- 15.Avella MA, Place A, Du SJ, Williams E, Silvi S, Zohar Y, Carnevali O (2012) Lactobacillus rhamnosus accelerates zebrafi backbone calcification and gonadal differentiation through effects on the GnRH and IGF systems. PLoS ONE 7(9):e45572 https:// doi.org/10.1371/journal.pone.0045572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu JP, Baker J, Perkins AS, Robertson EJ, Efstratiadis A (1993) Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r). Cell 75(1):59–72 [PubMed] [Google Scholar]
- 17.Baker J, Liu JP, Robertson EJ, Efstratiadis A (1993) Role of insulin-like growth factors in embryonic and postnatal growth. Cell 75(1):73–82 [PubMed] [Google Scholar]
- 18.DeChiara TM, Efstratiadis A, Robertson EJ (1990) A growth-deficiency phenotype in heterozygous mice carrying an insulin-like growth factor II gene disrupted by targeting. Nature 345(6270):78–80. 10.1038/345078a0 [DOI] [PubMed] [Google Scholar]
- 19.Camacho-Hubner C, Woods KA, Miraki-Moud F, Clark A, Savage MO (1999) Insulin-like growth factor-I deficiency caused by a partial deletion of the IGF-I gene: effects of rhIGF-I therapy. Growth Horm IGF Res 9(Suppl B):47–51 (discussion 51–42) [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Cheng Z, Elalieh HZ, Nakamura E, Nguyen MT, Mackem S, Clemens TL, Bikle DD, Chang W (2011) IGF-1R signaling in chondrocytes modulates growth plate development by interacting with the PTHrP/Ihh pathway. J Bone Miner Res 26(7):1437–1446. 10.1002/jbmr.359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Menendez A, Fong C, ElAlieh HZ, Kubota T, Long R, Bikle DD (2015) IGF-I signaling in osterix-expressing cells regulates secondary ossification center formation, growth plate maturation, and metaphyseal formation during postnatal bone development. J Bone Miner Res 30(12):2239–2248. 10.1002/jbmr.2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Nishida S, Boudignon BM, Burghardt A, Elalieh HZ, Hamilton MM, Majumdar S, Halloran BP, Clemens TL, Bikle DD (2007) IGF-I receptor is required for the anabolic actions of parathyroid hormone on bone. J Bone Miner Res 22(9):1329–1337. 10.1359/jbmr.070517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang M, Xuan S, Bouxsein ML, von Stechow D, Akeno N, Faugere MC, Malluche H, Zhao G, Rosen CJ, Efstratiadis A,Clemens TL (2002) Osteoblast-specific knockout of the insulin-like growth factor (IGF) receptor gene reveals an essential role of IGF signaling in bone matrix mineralization. J Biol Chem 277(46):44005–44012. https://doi.org/10.1074/jbc. M208265200 [DOI] [PubMed] [Google Scholar]
- 24.Van Wyk JJ, Smith EP (1999) Insulin-like growth factors and skeletal growth: possibilities for therapeutic interventions. J Clin Endocrinol Metab 84(12):4349–4354. https://doi.org/10.1210/ jcem.84.12.6201 [DOI] [PubMed] [Google Scholar]
- 25.Yakar S, Liu JL, Stannard B, Butler A, Accili D, Sauer B, LeRoith D (1999) Normal growth and development in the absence of hepatic insulin-like growth factor I. Proc Natl Acad Sci USA 96(13):7324–7329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao G, Monier-Faugere MC, Langub MC, Geng Z, Nakayama T, Pike JW, Chernausek SD, Rosen CJ, Donahue LR, Malluche HH, Fagin JA, Clemens TL (2000) Targeted overexpression of insulin-like growth factor I to osteoblasts of transgenic mice: increased trabecular bone volume without increased osteoblast proliferation. Endocrinology 141(7):2674–2682. https://doi.org/10.1210/ endo.141.7.7585 [DOI] [PubMed] [Google Scholar]
- 27.Sheng MH, Zhou XD, Bonewald LF, Baylink DJ, Lau KH (2013) Disruption of the insulin-like growth factor-1 gene in osteocytes impairs developmental bone growth in mice. Bone 52(1):133–144. 10.1016/j.bone.2012.09.027 [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Nishida S, Elalieh HZ, Long RK, Halloran BP, Bikle DD (2006) Role of IGF-I signaling in regulating osteoclastogenesis. J Bone Miner Res 21(9):1350–1358. https://doi.org/10.1359/ jbmr.060610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Coeverden SC, Netelenbos JC, de Ridder CM, Roos JC, Popp-Snijders C, Delemarre-van de Waal HA (2002) Bone metabolism markers and bone mass in healthy pubertal boys and girls. Clin Endocrinol 57(1):107–116 [DOI] [PubMed] [Google Scholar]
- 30.Cemborain A, Castilla-Cortazar I, Garcia M, Quiroga J, Muguerza B, Picardi A, Santidrian S, Prieto J (1998) Osteopenia in rats with liver cirrhosis: beneficial effects of IGF-I treatment. J Hepatol 28(1):122–131 [DOI] [PubMed] [Google Scholar]
- 31.Yakar S, Rosen CJ, Beamer WG, Ackert-Bicknell CL, Wu Y, Liu JL, Ooi GT, Setser J, Frystyk J, Boisclair YR, LeRoith D (2002) Circulating levels of IGF-1 directly regulate bone growth and density. J Clin Invest 110(6):771–781. https://doi.org/10.1172/ JCI15463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guerra-Menendez L, Sadaba MC, Puche JE, Lavandera JL, de Castro LF, de Gortazar AR, Castilla-Cortazar I (2013) IGF-I increases markers of osteoblastic activity and reduces bone resorption via osteoprotegerin and RANK-ligand. J Transl Med 11:271 10.1186/1479-5876-11-271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novince CM, Whittow CR, Aartun JD, Hathaway JD, Poulides N, Chavez MB, Steinkamp HM, Kirkwood KA, Huang E, Westwater C, Kirkwood KL (2017) Commensal gut microbiota immunomodulatory actions in bone marrow and liver have catabolic effects on skeletal homeostasis in health. Sci Rep 7(1):5747 10.1038/s41598-017-06126-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan J, Charles JF (2017) Gut microbiome and bone: to build, destroy, or both? Curr Osteoporos Rep 15(4):376–384. https:// doi.org/10.1007/s11914-017-0382-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schieber AM, Lee YM, Chang MW, Leblanc M, Collins B, Downes M, Evans RM, Ayres JS (2015) Disease tolerance mediated by microbiome E. coli involves inflammasome and IGF-1 signaling. Science 350(6260):558–563. https://doi.org/10.1126/ science.aac6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kloting N, Koch L, Wunderlich T, Kern M, Ruschke K, Krone W, Bruning JC, Bluher M (2008) Autocrine IGF-1 action in adipocytes controls systemic IGF-1 concentrations and growth. Diabetes 57(8):2074–2082. 10.2337/db07-1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koh A, De Vadder F, Kovatcheva-Datchary P, Backhed F (2016) From dietary fiber to host physiology: short-chain fatty acids askey bacterial metabolites. Cell 165(6):1332–1345. 10.1016/j.cell.2016.05.041 [DOI] [PubMed] [Google Scholar]
- 38.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS (2013) The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341(6145):569–573. https://doi.org/10.1126/ science.1241165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coff PJ, Rudensky AY (2013) Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504(7480):451–455. https:// doi.org/10.1038/nature12726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li JY, Chassaing B, Tyagi AM, Vaccaro C, Luo T, Adams J, Darby TM, Weitzmann MN, Mulle JG, Gewirtz AT, Jones RM, Pacifici R (2016) Sex steroid deficiency-associated bone loss is microbiota dependent and prevented by probiotics. J Clin Invest 126(6):2049–2063. 10.1172/JCI86062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Britton RA, Irwin R, Quach D, Schaefer L, Zhang J, Lee T, Parameswaran N, McCabe LR (2014) Probiotic L. reuteri treatment prevents bone loss in a menopausal ovariectomized mouse model. J Cell Physiol 229(11):1822–1830. https://doi.org/10.1002/ jcp.24636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCabe L, Britton RA, Parameswaran N (2015) Prebiotic and probiotic regulation of bone health: role of the intestine and its microbiome. Curr Osteoporos Rep 13(6):363–371. 10.1007/s11914-015-0292-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fazeli PK, Klibanski A (2014) Determinants of GH resistance in malnutrition. J Endocrinol 220(3):R57–65. 10.1530/JOE-13-0477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan J, Takakura A, Zandi-Nejad K, Charles JF (2017) Mechanisms of gut microbiota-mediated bone remodeling. Gut Microbes 10.1080/19490976.2017.1371893 [DOI] [PMC free article] [PubMed] [Google Scholar]


