Highlights
-
•
R. pusillus encodes cellulose-, xylan- and chitin-degrading proteins.
-
•
Two putative GH9 endoglucanases were identified.
-
•
Enzyme system of R. pusillus is suited to consume easily accessible sugars.
-
•
Endoglucanase and xylanase activity detected when the fungus was grown on wheat bran and xylan.
Keywords: Carbohydrate-active enzymes, GH9, Endoglucanase, Thermophilic, CAZyme, R. miehei
Abstract
We report here the annotated draft genome sequence of the thermophilic zygomycete Rhizomucor pusillus strain FCH 5.7, isolated from compost soil in Vietnam. The genome assembly contains 25.59 Mb with an overall GC content of 44.95%, and comprises 10,898 protein coding genes. Genes encoding putative cellulose-, xylan- and chitin-degrading proteins were identified, including two putative endoglucanases (EC 3.2.1.4) from glycoside hydrolase family 9, which have so far been mostly assigned to bacteria and plants.
1. Introduction
Fast growth and rapid spore production mark the thermophilic zygomycete Rhizomucor pusillus, a member of the Mucorales, as a mycoflora pioneer species in compost [1,2]. A number of its encoded enzymes have been produced and characterised, such as polygalacturonase, glycoamylase, phytase and glucanase [[3], [4], [5], [6]], and particularly the high temperature optima of R. pusillus enzymes of around 60–70 °C [5,6] make the fungus an interesting source of biocatalysts. A lipase of a close relative, Rhizomucor miehei, is commercially available and used industrially (Palatase®, Novozymes) [7]. Therefore, R. pusillus represents a promising resource for the exploration of useful enzymes. The R. pusillus strain FCH 5.7 described in this study was isolated from 3 month old compost in Ly Nhan, Ha Nam province, Vietnam (geographical location 20°33′06.2″ N, 106°04′11.8″ E). Genome sequencing was performed to enable a systematic exploration, particularly of the strain’s carbohydrate-active enzymes (CAZymes) [8], as well as to set the basis for investigating its growth strategies at higher temperatures.
2. Materials & methods
After isolation, R. pusillus FCH 5.7 was cultivated on potato dextrose agar (PDA) at 50 °C and the species identified by internal transcribed spacer (ITS) sequencing, as described previously [9]. For whole genome sequencing, 5-day-old mycelium from PDA plates was used for the inoculation of basal liquid medium (4 g L−1 KH2PO4, 13.6 g L−1 (NH4)2SO4, 0.8 g L−1CaCl2⋅2H2O, 0.6 g L−1 MgSO4⋅7H2O, 6 g L−1 Bacto peptone, 10 mg L−1 FeSO4⋅7H2O, 3.2 mg L−1 MnSO4⋅H2O, 2.8 mg L−1 ZnSO4⋅7H2O, 4 mg L−1 CoCl2⋅6H2O, 200 mL L−1 Tween 80, pH 5.8) containing 20 g L−1 glucose as carbon source. Cultivation was carried out in 125 mL liquid medium in baffled Erlenmeyer flasks (500 mL; 50 °C, 48 h, shaking at 250 rpm). The mycelium was harvested after two days of cultivation and DNA was extracted with cetyltrimethyl ammonium bromide (CTAB) buffer (2% CTAB, 100 mM TrisHCl, pH 8.0, 20 mM EDTA, 1.4 M NaCl) and purified from the supernatant by a combination of phenol-chloroform extraction and isopropanol precipitation and DNeasy Plant Mini Kit (Qiagen), as described previously [9]. Quality of the purified DNA was verified by agarose gel electrophoresis, Nanodrop (Thermo Scientific) and Qubit (Life Technologies) before genome sequencing. RNA was extracted from cultures growing in basal liquid medium, supplemented with 1% glucose, wheat bran or beechwood xylan, respectively, after 4 h and 48 h, using TRIzol (Invitrogen) and chloroform, according to the manufacturer’s instructions. Enzymatic activities (xylanase, endoglucanase) were determined in supernatant samples from the same cultivations through measurement of produced reducing ends, using the 3,5-dinitrosalicylic acid (DNS) method [10].
For genome sequencing, the NEBNext Ultra DNA Library Prep kit for Illumina (New England Biolabs) was used to process the DNA samples according to the manufacturer’s protocol. After fragmentation of DNA with a Covaris ultrasonicator (Thermo Scientific), quality and yield were measured with a Bioanalyzer (Agilent Technologies). Clustering and DNA sequencing with the Illumina cBot and HiSeq 2500 were performed with 8.0 pM of DNA, standard Illumina primers and HiSeq control software HCS v2.2.58. Image analysis, base calling, and quality check were done with the Illumina data analysis pipeline RTA v1.18.64 and Bcl2fastq v1.8.4. The GenomeScan in-house tool FASTQFilter v2.05 was used for adapter trimming and quality filtering of the 250 bp paired-end reads. The short-read genome assembler Abyss v1.3.7 [11] with a k-mer length of 64 was used for assembly. Scaffolds shorter than 500 bp were removed.
For RNA sequencing, total RNA was further prepared with the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs) with oligo-dT magnetic beads for mRNA enrichment. Sequencing of the resulting cDNA (16 pM) was done with an Illumina cBot and HiSeq 2500 instrument. The newly assembled genome was used as a reference to map the mRNA-Seq reads using the packages Tophat (v2.0.14. Linux_x86_64) and Bowtie (v2-2.1.0) with a default mismatch rate of 2%. Gene finding was performed combining two approaches. The HMM-based algorithm Glimmer v.3.02 [12] was trained using the genome of Mucor circinelloides (downloaded from the Joint Genome Institute (JGI), http://genome.jgi.doe.gov/). In addition, an evidence-based method of gene finding was performed using mapped mRNA-Seq reads and the software tool CodingQuarry [13]. Combining the two methods, gene models for 10,898 genes were obtained. For annotation, a BLASTp search (version 2.2.28+) (http://blast.ncbi.nlm.nih.gov) was performed on the UniprotKB/SwissProt database with default parameters. For comparison to other species, publicly accessible genomes were downloaded from JGI Mycocosm (https://genome.jgi.doe.gov/programs/fungi/index.jsf, Supplemental File 1).
The CAZyme contents of R. pusillus FCH 5.7 and the fungal species included for comparison were determined by identifying genes containing CAZyme domains using the dbCAN2 meta server (cys.bios.niu.edu/dbCAN2). Only CAZyme domains predicted by at least two of the three algorithms (DIAMOND, HMMER and Hotpep) employed by dbCAN2 were kept. It should be noted that the stringency of this approach led to slightly different numbers of identified CAZymes than has previously been reported for the respective species.
3. Results & discussion
The 25.59 Mb genome of R. pusillus FCH 5.7 was obtained with 294-fold coverage (Table 1). The whole genome assembly has been submitted to the European Nucleotide Archive and deposited at DDBJ/EMBL/GenBank under the assembly accession number FWWN00000000.
Table 1.
Genome features of R. pusillus FCH 5.7.
| Genome assembly size (Mb) | 25.59 |
| Genome coverage (x times) | 294 |
| Number of scaffolds (≥500 bp) | 618 |
| Contig N50 (bp) | 102,680 |
| Number of scaffolds > N50 | 77 |
| Max scaffold size (bp) | 534,899 |
| Number of protein coding genes | 10,898 |
| Mean gene length (bp) | 1,359 |
| GC content, overall (%) | 44.95 |
| GC content, coding (%) | 48.43 |
The analysis of CAZyme-encoding genes showed that R. pusillus FCH 5.7 possesses a similar number of total CAZymes as other zygomycetes, such as Hesseltinella vesiculosa and Phycomyces blakesleeanus, but considerably fewer than Lichtheimia corymbifera and Rhizopus oryzae (Table 2), which could be due to the reported gene expansion and genome duplication in L. corymbifera and R. oryzae, respectively [14,15]. The difference was particularly obvious for carbohydrate esterases (CE) and glycoside transferases (GT), two CAZy enzyme classes that are known to be highly abundant in R. oryzae [16]. Compared to basidiomycetes, zygomycetes generally have higher numbers of GTs, and this was also true for R. pusillus, which harbours 77 CAZy domains across 27 GT families in its genome. The number of auxiliary activity (AA) enzymes was relatively small in the investigated zygomycetes, and R. pusillus FCH 5.7 was found to only possess members of AA families 1, 5 and 6, as well as one member in family 12. None of the zygomycetes used for comparison contained AA9 lytic polysaccharide monooxygenases (LPMOs), which are abundant in ascomycetes and basidiomycetes and involved in cellulose degradation (Supplemental File 1; [17])
Table 2.
Comparison of the numbers of CAZymes in R. pusillus FCH 5.7 with those in other fungi.
| total | AA | CBM | GH | CE | PL | GT | ||
|---|---|---|---|---|---|---|---|---|
| ZYG | Rhizomucor pusillus | 184 | 11 | 1 | 81 | 12 | 2 | 77 |
| Absidia repens | 236 | 11 | 0 | 78 | 19 | 2 | 126 | |
| Hesseltinella vesiculosa | 183 | 11 | 1 | 67 | 13 | 1 | 90 | |
| Lichtheimia corymbifera | 261 | 11 | 2 | 93 | 28 | 1 | 126 | |
| Mucor circinelloides | 221 | 10 | 2 | 74 | 23 | 2 | 110 | |
| Phycomyces blakesleeanus | 195 | 9 | 2 | 66 | 19 | 3 | 96 | |
| Rhizopus microsporus | 201 | 11 | 3 | 72 | 17 | 3 | 95 | |
| Rhizopus oryzae | 263 | 14 | 4 | 105 | 24 | 3 | 113 | |
| Syncephalastrum racemosum | 239 | 11 | 3 | 96 | 24 | 2 | 103 | |
| ASC | Thielavia terrestris | 405 | 65 | 28 | 202 | 23 | 4 | 83 |
| BAS | Schizophyllum commune | 331 | 56 | 6 | 175 | 19 | 15 | 60 |
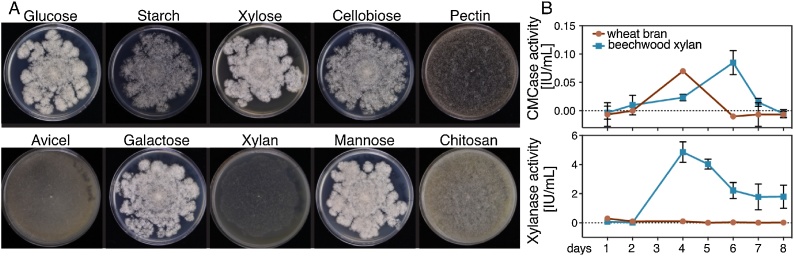
Growth on plates containing basal medium and 1% beechwood xylan was very poor, and R. pusillus FCH 5.7 grew much better on mono- and disaccharides (glucose, galactose, mannose, cellobiose), as well as starch (Fig. 1A). R. pusillus FCH 5.7 was not able to grow efficiently on cellulose (Avicel), which indicates a lack of endoglucanases and/or cellobiohydrolases to break down this polymer. Indeed, no putative GH6 or GH7 cellobiohydrolases could be found in the R. pusillus FCH 5.7 genome. The ability to degrade chitin was indicated by the presence of many chitinases (GH18) and chitin deacetylases (CE4) in the genome, and was corroborated by the ability of R. pusillus FCH 5.7 to grow on chitosan (Fig. 1A, Supplemental File 1). In contrast to a previous study [4], R. pusillus FCH 5.7 grew quite poorly on pectin, and many genes typically involved in pectin degradation, such as pectin and pectate lyases (PL1, 3, 4), were found missing in its genome. We did, however, detect sequences of three GH28s that encode putative polygalacturonases. Similar to R. miehei [18], a large number of lipases was found in the genome of R. pusillus FCH 5.7, with in total 80 putative lipases and phospholipases.
Fig. 1.
Growth on plates containing 0.1 g L−1 of a single carbon source (A), and enzyme assays evaluating endoglucanase (CMCase) and xylanase activities (B). For plates, fungus was grown at 50 °C for 3 days. For enzyme assays, fungus was cultivated on wheat bran (red) or beechwood xylan (blue) over the course of 8 days.
Despite the poor growth on plates containing Avicel or xylan, xylanase and low levels of endoglucanase (CMCase) activities were detected in liquid cultures of R. pusillus FCH 5.7 grown on wheat bran or beechwood xylan as carbon sources (Fig. 1A). Cultivation on wheat bran and beechwood xylan resulted in almost identical levels of CMCase activity. Xylanase activity, on the other hand, was much higher during growth on xylan compared to growth on wheat bran. This indicates the presence of xylanases that are induced more by hardwood glucuronoxylans, as found in beechwood, rather than the grass-specific arabinoxylans contained in wheat bran.
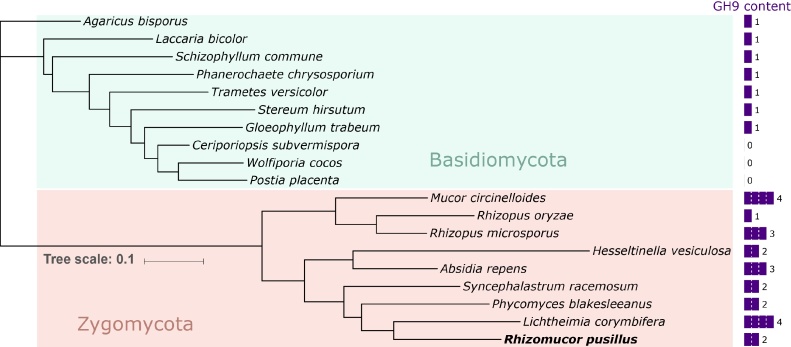
Several putative endoglucanases from family GH45 were detected in the genome, which could explain the low enzyme activity observed on CMC (Fig. 1B). Interestingly, R. pusillus FCH 5.7 also encodes two GH9 members, which is a family containing endoglucanases typically found in bacteria, plants and occasionally animals [19,20], but completely absent in ascomycetes fungi. We compared the genomes of a selection of fungi to determine how widespread the occurrence of GH9 genes is. Many of the basidiomycetes in our selection were shown to encode a single putative GH9 protein, while up to four were detected in zygomycetes (Fig. 2). Zygomycete GH9s are, to our knowledge, completely unexplored proteins and the function of fungal GH9s and the importance for, for instance, growth on polysaccharides is unclear and has not been described yet. One study indicated an involvement in degradation of crystalline cellulose, as a putative GH9 gene was found to be upregulated in the basidiomycete P. chrysosporium grown on cellulose [21], but conclusive functional evidence is still missing.
Fig. 2.
Maximum-likelihood phylogenetic tree depicting the relationships among selected fungal species and the GH9 content of their respective genomes. The tree was constructed based on protein sequences of GT33 proteins, which are present as a single copy in each of the selected species, in MEGA7 [23] and was annotated with iTOL [24].
The lifestyle and habitat, and therefore the repertoire of CAZymes of zygomycetes is quite different to ascomycetes and basidiomycetes. The enzyme system of R. pusillus FCH 5.7 seems to be better suited to consume easily accessible, simple sugars found in plant biomass [22] as well as lipid-containing compounds. As seen in the growth studies (Fig. 1A) and also shown for R. oryzae, zygomycetes are in general unable to degrade complex plant cell wall polysaccharides [16]. The lack of genes encoding enzymes of the CAZy families GH6, GH7 and AA9 in the R. pusillus genome further supports this statement. However, the low endoglucanase and xylanase activities detected in R. pusillus FCH 5.7 culture supernatants, as well as the presence of putative xylanases and endoglucanases (GH9, GH45) in the genome indicate that this fungus may be able to degrade oligo- and/or polysaccharide to a certain extent.
3.1. Nucleotide sequence accession number(s)
The R. pusillus FCH 5.7 whole genome sequence has been submitted to GenBank and deposited at DDBJ/EMBL/GenBank under the accession No. FWWN00000000.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Acknowledgments
This research was financially supported by the European Union OPTIBIOCAT project (FP7 KBBE. 2013.3.3-04; grant agreement no. 613868), by the Swedish Research Council (Vetenskapsrådet) grant no. 2014-3523, and by the Wallenberg Wood Science Center, granted by the Knut and Alice Wallenberg Foundation.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.btre.2018.e00279.
Contributor Information
Silvia Hüttner, Email: huttner@chalmers.se.
Zoraide Granchi, Email: z.granchi@genomescan.nl.
Thanh Thuy Nguyen, Email: thuyn0106@gmail.com.
Sake van Pelt, Email: s.vanpelt@genomescan.nl.
Johan Larsbrink, Email: johan.larsbrink@chalmers.se.
Vu Nguyen Thanh, Email: thanh@firi.ac.vn.
Lisbeth Olsson, Email: lisbeth.olsson@chalmers.se.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Satyanarayana T., Littlechild J., Kawarabayasi Y. 2013. Thermophilic Microbes in Environmental and Industrial Biotechnology. [Google Scholar]
- 2.Morgenstern I., Powlowski J., Ishmael N., Darmond C., Marqueteau S., Moisan M.C., Quenneville G., Tsang A. A molecular phylogeny of thermophilic fungi. Fungal Biol. 2012;116:489–502. doi: 10.1016/j.funbio.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 3.Viikari L., Alapuranen M., Puranen T., Vehmaanperä J., Siika-Aho M. Thermostable enzymes in lignocellulose hydrolysis. Adv. Biochem. Eng. Biotechnol. 2007;108:121–145. doi: 10.1007/10_2007_065. [DOI] [PubMed] [Google Scholar]
- 4.Siddiqui M.A., Pande V., Arif M. Production, purification, and characterization of polygalacturonase from Rhizomucor pusillus isolated from decomposting orange peels. Enzyme Res. 2012;2012:138634. doi: 10.1155/2012/138634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chadha B.S., Harmeet G., Mandeep M., Saini H.S., Singh N. Phytase production by the thermophilic fungus Rhizomucor pusillus. World J. Microbiol. Biotechnol. 2004;20:105–109. [Google Scholar]
- 6.He Z., Zhang L., Mao Y., Gu J., Pan Q., Zhou S., Gao B., Wei D. Cloning of a novel thermostable glucoamylase from thermophilic fungus Rhizomucor pusillus and high-level co-expression with α-amylase in Pichia pastoris. BMC Biotechnol. 2014;14:1–10. doi: 10.1186/s12896-014-0114-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodrigues R.C., Fernandez-Lafuente R. Lipase from Rhizomucor miehei as an industrial biocatalyst in chemical process. J. Mol. Catal., B Enzym. 2010;64:1–22. [Google Scholar]
- 8.Lombard V., Golaconda Ramulu H., Drula E., Coutinho P.M., Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hüttner S., Nguyen T.T., Granchi Z., Chin-A-Woeng T., Ahrén D., Larsbrink J., Thanh V.N., Olsson L. Combined genome and transcriptome sequencing to investigate the plant cell wall degrading enzyme system in the thermophilic fungus Malbranchea cinnamomea. Biotechnol. Biofuels. 2017;10 doi: 10.1186/s13068-017-0956-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao Z., Storms R., Tsang A. Microplate-based carboxymethylcellulose assay for endoglucanase activity. Anal. Biochem. 2005;342:176–178. doi: 10.1016/j.ab.2005.01.052. [DOI] [PubMed] [Google Scholar]
- 11.Simpson J.T., Wong K., Jackman S.D., Simpson J.T., Wong K., Jackman S.D., Schein J.E., Jones S.J.M. ABySS : a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majoros W.H., Pertea M., Salzberg S.L. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics. 2004;20:2878–2879. doi: 10.1093/bioinformatics/bth315. [DOI] [PubMed] [Google Scholar]
- 13.Testa A.C., Hane J.K., Ellwood S.R., Oliver R.P. CodingQuarry: highly accurate hidden Markov model gene prediction in fungal genomes using RNA-seq transcripts. BMC Genomics. 2015;16(170) doi: 10.1186/s12864-015-1344-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linde J., Schwartze V., Binder U., Voigt K., Horn F. De novo whole-genome sequence and genome annotation of Lichtheimia ramosa. Genome Announc. 2014;2:e0088–14. doi: 10.1128/genomeA.00888-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma L.J., Ibrahim A.S., Skory C., Grabherr M.G., Burger G., Butler M., Elias M., Idnurm A., Lang B.F., Sone T., Abe A., Calvo S.E., Corrochano L.M., Engels R., Fu J., Hansberg W., Kim J.M., Kodira C.D., Koehrsen M.J., Liu B., Miranda-Saavedra D., O’Leary S., Ortiz-Castellanos L., Poulter R., Rodriguez-Romero J., Ruiz-Herrera J., Shen Y.Q., Zeng Q., Galagan J., Birren B.W., Cuomo C.A., Wickes B.L. Genomic analysis of the basal lineage fungus Rhizopus oryzae reveals a whole-genome duplication. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Battaglia E., Benoit I., van den Brink J., Wiebenga A., Coutinho P.M., Henrissat B., de Vries R.P. Carbohydrate-active enzymes from the zygomycete fungus Rhizopus oryzae: a highly specialized approach to carbohydrate degradation depicted at genome level. BMC Genomics. 2011;12:38. doi: 10.1186/1471-2164-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monclaro A.V., Filho E.X.F. Fungal lytic polysaccharide monooxygenases from family AA9: recent developments and application in lignocelullose breakdown. Int. J. Biol. Macromol. 2017;102:771–778. doi: 10.1016/j.ijbiomac.2017.04.077. [DOI] [PubMed] [Google Scholar]
- 18.Zhou P., Zhang G., Chen S., Jiang Z., Tang Y., Henrissat B., Yan Q., Yang S., Chen C.-F., Zhang B., Du Z. Genome sequence and transcriptome analyses of the thermophilic zygomycete fungus Rhizomucor miehei. BMC Genomics. 2014;15(294) doi: 10.1186/1471-2164-15-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ni J., Takehara M., Watanabe H. Identification of activity related amino acid mutations of a GH9 termite cellulase. Bioresour. Technol. 2010;101:6438–6443. doi: 10.1016/j.biortech.2010.03.045. [DOI] [PubMed] [Google Scholar]
- 20.Davison A., Blaxter M. Ancient origin of glycosyl hydrolase family 9 cellulase genes. Mol. Biol. Evol. 2005;22:1273–1284. doi: 10.1093/molbev/msi107. [DOI] [PubMed] [Google Scholar]
- 21.Vanden Wymelenberg A., Denman S., Dietrich D., Bassett J., Yu X., Atalla R., Predki P., Rudsander U., Teeri T.T., Cullen D. Transcript analysis of genes encoding a family 61 endoglucanase and a putative membrane-anchored family 9 glycosyl hydrolase from Phanerochaete chrysosporium. Appl. Environ. Microbiol. 2002;68:5765–5768. doi: 10.1128/AEM.68.11.5765-5768.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richardson M. The ecology of the zygomycetes and its impact on environmental exposure. Clin. Microbiol. Infect. 2009;15:2–9. doi: 10.1111/j.1469-0691.2009.02972.x. [DOI] [PubMed] [Google Scholar]
- 23.Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016 doi: 10.1093/molbev/msw054. msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Letunic I., Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.