Abstract
The progression of age-related sarcopenia can be accelerated by impaired recovery of muscle mass following periods of disuse due to illness or immobilization. However, the mechanisms underlying poor recovery of aged muscle following disuse remain to be delineated. Recent evidence suggests that mitochondrial energetics play an important role in regulation of muscle mass. Here, we report that 22- to 24-month-old mice with low muscle mass and low glucose clearance rate also display poor early recovery of muscle mass following 10 days of hind limb unloading. We used unbiased and targeted approaches to identify changes in energy metabolism gene expression, metabolite pools and mitochondrial phenotype, and show for the first time that persistent mitochondrial dysfunction, dysregulated fatty acid β-oxidation, and elevated H2O2 emission occur concomitantly with poor early recovery of muscle mass following a period of disuse in old mice. Importantly, this is linked to more severe whole-body insulin resistance, as determined by insulin tolerance test. The findings suggest that muscle fuel metabolism and mitochondrial energetics could be a focus for mining therapeutic targets to improve recovery of muscle mass following periods of disuse in older animals.
Keywords: Disuse atrophy, Skeletal muscle, Fatty acid oxidation, Insulin resistance
Skeletal muscle is a highly plastic tissue that can adapt its mass, structure, and metabolic capacity in response to changes in mechanical load. During aging, skeletal muscle exhibits a progressive loss of mass, strength, and function also called sarcopenia (1), a condition that can increase risk of frailty, morbidity, and loss of independence (2). In addition, there is an attenuated adaptive recovery in aging following atrophy induced by: disuse, immobilization, starvation, and glucocorticoid treatment (3–7). Poor recovery of muscle mass and strength, even with exercise training, a potent anabolic stimuli has also been reported in older humans following unilateral leg immobilization (8). Indeed, the lack of recovery from periods of disuse has been suggested to accelerate sarcopenia in older adults (9). Currently, there are no effective therapies to counteract disuse-induced muscle atrophy and poor recovery of muscle mass, in part because a detailed understanding of the mechanisms underlying loss of muscle plasticity with aging are unknown. In addition, the older population demographic is growing and tends to experience more frequent and prolonged periods of inactivity, increasing the need for the development of safe and effective therapeutics.
Maintenance of skeletal muscle mass reflects a finely controlled balance between protein synthesis and degradation. Hypertrophy results from a shift to protein synthesis, and the primary anabolic pathway involves the mammalian target of rapamycin complex 1 (mTORC1). mTORC1 is principally regulated by nutritional cues, insulin/growth factor signaling cascades, and mechanotransduction (10). Contractile inactivity–induced muscle atrophy is due to reduced basal and postprandial protein synthesis, also known as anabolic resistance, and elevated protein degradation by ubiquitin/proteasome and autophagy/lysosome pathways (11). Protein breakdown is initiated by inflammatory, glucocorticoid, and myostatin/transforming growth factor beta β pathways (12). Although the protein degradation/synthesis pathways are relatively well characterized, upstream regulation, particularly in the context of muscle plasticity in aging, is less well understood.
Separate lines of investigation have shown that mitochondrial dysfunction occurs in aging muscle, as evidenced by decreased content, mitochondrial protein synthesis, elevated reactive oxygen species (ROS) emission and apoptosis, and reduced adenosine triphosphate (ATP) production, calcium handling (13). Recent evidence also points to mitochondria as an important nexus of control for acute muscle atrophy during disuse (14). Rodent studies using mitochondrial-targeted antioxidants support a crucial role for mitochondrial ROS in mediating muscle atrophy, through upregulation of ubiquitin-protease enzymes (15,16). In addition, a model of increased ROS production, the superoxide dismutase (Sod1) knockout, shows exacerbated muscle atrophy during aging (17). Elevated mitochondrial oxidative stress is purported to stimulate muscle protein breakdown by activating lysosome-autophagy and ubiquitin-proteasome systems (18–21) and energetic stress (reduced ATP production) may also activate the adenosine monophosphate-activated protein kinase (AMPK)-FoxO3 pathways leading to increased protein degradation (22,23). Moreover, overexpression of PGC-1α, a key driver of mitochondrial biogenesis, can protect muscle mass from acute atrophy due to immobilization or disuse (24–26). Key regulators of autophagy and mitochondrial fission also regulate muscle mass (22,27).
Taken together, this body of evidence implicates a central role for mitochondrial energetics in regulating muscle mass and as such, it may also play a role in recovery of muscle mass following a period of disuse. The goal of this study was to determine whether mitochondrial function and gene/metabolic profile are linked with responses to hind limb unloading–induced atrophy and recovery during reloading in adult (6 months) and old mice (22–24 months). Our data suggest that dysregulated fatty acid β-oxidation (FAO) and elevated H2O2 emission from mitochondria may contribute to compromised recovery of soleus muscle mass following unloading in old mice.
Methods
Animals
All animal experiments and euthanasia protocols were conducted in strict accordance with the National Institute of Health guidelines for humane treatment of animals and approved by the Institutional Animal Care and Use Committee at the Sanford Burnham Prebys Medical Discovery Institute at Lake Nona (protocol #2013-0102). Six- and 22- to 24-month-old male C57BL/6J mice were obtained from the National Institute on Aging rodent colony (Charles River Laboratories, Madison, Wisconsin). Young and aged animals were divided into the following experimental groups: 10 days of hind limb unloading (UN), 10 days of hind limb unloading followed by 3 days of reloading recovery (RL), and control (CON). Mice were fed ad libitum with a standard chow diet (#2916, Harlan-Teklad, Houston, Texas) and water/hydrogel (ClearH2O, Westbrook, Maine) and housed at 22°C with a 12-hour light/dark cycle.
Tail Suspension Hind Limb Unloading
Tail suspension is a most commonly used animal models of musculoskeletal nonweight bearing. Prior to tail suspension experiments, the individual mice acclimated to single-cage housing and routine handling for 3 days. Tail suspension was performed using a modified version of the Morey-Holton and Globus protocol (28). Briefly, a small metallic hook is taped to the base of the tail using nonabrasive adhesive tape wrapped in a helical pattern. The hook is then attached to a small swivel key chain that is attached to a metal rod that runs the length of the microisolator cage. The mice could move on a y-axis and rotate 360 degrees and so had access to all areas of the cage. The hind limbs are maintained just off the cage floor with the body of the mouse at approximately 30° angle from the cage floor. The mice can move freely and the angle and height of the mice are checked daily. The control mice were separated into the individual cages with the exactly the same conditions as unloading groups, but without tail suspension.
After 10 days of UN, RL, or CON, the mice were fasted for 4 hours and euthanized by CO2 asphyxiation and cervical dislocation, or anesthetized with sodium pentobarbital (IP administration, 50 mg/kg) for muscle harvested for metabolomics analysis. The soleus, gastrocnemius, and quadriceps femoris muscle groups were immediately harvested and weighed. The soleus muscle from right hind limb was used for fresh tissue assay immediately and other muscles were snap-frozen in liquid nitrogen for other analysis. Separate experiments were conducted for cardiometabolic phenotyping, permeabilized fiber bundle assays, RNA analysis, immunoblots, and metabolomics/lipidomics.
Cardiometabolic Phenotyping
Body Composition. Measurement of fat and lean mass (g) is accomplished using a LF90II TD-NMR (Bruker, Madison, WI). Conscious mice are placed in a plastic restraint tube that is inserted into the instrument. Measurements are obtained in less than 1 minute. Ambulatory cage activity. Locomotor activity measurements are obtained using a Promethion Mouse Multiplexed Metabolic System (Sable Systems International, Las Vegas, NV). Locomotor activity along the X, Y, and Z planes is measured using infrared beams that span the home cage. Data are collected by a host computer. Insulin-stimulated Tissue Glucose Uptake, was assessed as described in the Supplementary Material section. Processing of tissues for glucose uptake was determined as previously described (29), and as detailed in the Supplementary Material section.
Myofiber Bundle Preparation
Permeabilized fiber bundles (1–3 mg) were prepared immediately following tissue harvest, as previously described (30,31) and as detailed in the Supplementary Material section.
Mitochondrial Respiration
Respirometry assays were conducted using an Oxygraph-2k (Oroboros Instruments, Innsbruck, Austria). The myofiber bundles were gently placed into the respirometer chambers and after a stable baseline was reached, the assay protocol was run in duplicate at 37°C and between 350 and 200 nmol of O2 in Buffer Z with blebbistatin (25 µM). The assay protocol was described in the Supplementary Material section.
Mitochondrial H2O2 Emission
H2O2 emission was measured with Amplex Red reagent which reacts with H2O2 to produce the stable fluorescent compound resorufin. Resorufin fluorescence was monitored using a Fluorescence Spectrometer (Perkin Elmer LS50B, Waltham, Massachusetts) with temperature control (37°C) and magnetic stirring at excitation/emission 563/587 nm. The assay was run with buffer Z containing 5,000 U/ml CuZn-SOD, 25 µM blebbistatin, 50 µM Amplex Ultra-Red, and 6 U/ml horseradish peroxidase. Briefly, the muscle fiber bundles were added to the reaction buffer with 10 µg/ml oligomycin, 10 mM glutamate, 2 mM malate, and 10 mM succinate. The rate of emission of H2O2 (pmol) was calculated from previously established fluorescence intensity standard curves with known concentrations of H2O2, after correcting for the rate of change in background fluorescence. Following each experiment, myofiber bundles were dried and weighed. Rate of H2O2 emission was expressed as pmol/min/mg dry weight.
Mitochondrial Calcium Retention Capacity
A continuous, spectrophotometric assay was utilized to measure mitochondrial calcium retention capacity (CRC) within soleus fiber bundles, as detailed in the Supplementary Material section.
RNA Sequencing and Informatics
Details of sample preparation, library preparation, sequencing, mapping, and differential expression analysis are described in the Supplementary Material section.
KEGG Pathway and Upstream Regulator Analysis
The database for annotation, visualization, and integrated discovery (DAVID) v6.8 was used to identify enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Ingenuity Pathway Analysis (IPA) Upstream Regulator was used to inferring and scoring regulator networks upstream of gene-expression data to identify putative upstream transcriptional regulators. The gene-expression data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE102284.
Real-Time Quantitative-PCR
Details of sample preparation and gene-expression analysis by polymerase chain reaction (PCR) are described in the Supplementary Information section.
Primer Sequences for qRT-PCR Analysis
Primers were designed using cDNA sequences from Genebank (http://www.ncbi.nlm.nih.gov/genbank/) and using NCBI Primer-BLAST online tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). The following primer sequences were used:
Ppargc1a,F-TGCCATTGTTAAGACCGAG;R-ACCC TTGGGGTCATTTGG.
Esrrg,F-TGACTTGGCTGACCGAG;R-CCGAGGATCAGAATCTCC.
Esrra,F-AGGAGT ACGTCCTGCTG;R-CCTCAGCATCTTCAATG
Rplp0, F-TGGAAGTCCAACTACTTCCTCAA;R-ATCT GCTGCATCTGCTTGGAG.
Immunoblot
Frozen soleus was prepared for immunoblot as described previously (32) and as detailed in the Supplementary Material section.
Lipidomics
Multidimensional mass spectrometry-based shotgun lipidomics was employed to measure and characterize the lipid patterns in mouse soleus muscle. Briefly, the lipids were extracted from soleus muscle by modified Bligh and Dyer procedure with LiCl solution (33). The lipid extracts were finally flushed with nitrogen, capped, and stored at −20°C for electrospray ionization mass spectrometry as described previously. 4-Hydroxyalkenal species (4-HNE) were measured by derivatization with 125 mM carnosine by incubating at 37°C for 24 hours (34). The derivatization of PE was performed by incubating with 4 mM Fmol-Cl and 8 mM DMAP for 2 hours (35). Diacylglycerols were characterized by derivatization with N,N-dimethylglycine (36). Non-esterized fatty acid species were measured by incubating with N-[4-(aminomethyl) phenyl] pyridinium (37). Internal standards were added before the extraction for normalization. For ESI direct infusion analysis, lipid extract was further diluted to a final concentration of approximately 500 fmol/µL by CHCl3/MeOH/isopropanol (1/2/4, v/v/v) with or without 0.02% (v/v) LiOH-saturated MeOH solution, and the mass spectrometric analysis was performed on a QqQ mass spectrometer (TSQ VANTAGE, Thermo, San Jose, California) or a Q-Exactive mass spectrometer (Thermo) equipped with an automated nanospray device (TriVersa NanoMate, Advion Bioscience Ltd., Ithaca, New York) and operated with Xcalibur software. Data processing of MS analyses including ion peak selection, data transferring, baseline correction, deisotoping, peak intensity comparison, and quantification is conducted by self-programmed MicroSoft Excel macros. Data from mouse soleus muscle samples were normalized to protein content.
Metabolomics
Acylcarnitines. A panel of acylcarnitines was quantitated by LC/MS/MS (Agilent 1290 HPLC/6490 triple quadrupole mass spectrometer). Calibration standards and mouse muscle homogenates were prepared in 50% acetonitrile (ACN)/0.3% formic acid spiked with stable isotope-labeled internal standards. Acylcarnitines were extracted from homogenates with methanol followed by vortexing and centrifugation. An aliquot of methanol was dried down, and acylcarnitines were derivatized with O-benzylhydroxylamine. After derivatization, acylcarnitines were separated and quantitated by LC/MS/MS. Organic Acids. Lactate, pyruvate, succinate, fumarate, malate, α-ketoglutarate, citrate, and 3-hydroxybutyrate were quantitated by LC/MS/MS with electrospray ionization (Thermo Scientific Ultimate 3000 UHPLC/Quantiva triple quadrupole mass spectrometer). Calibration standards and mouse muscle homogenates were prepared in 50% ACN/0.3% formic acid. Then, the calibration solutions and muscle homogenates were spiked with stable isotope-labeled internal standards and derivatized with O-benzylhydroxylamine followed by extraction with ethylacetate. The ethylacetate layer was dried and reconstituted in HPLC solvents prior to quantitation of organic acids by multiple reaction monitoring using LC/MS/MS. Amino Acids. Quantitation of amino acids was achieved using LC/MS/MS (Agilent 1290 HPLC/6490 triple quadrupole mass spectrometer). Calibration standards and mouse muscle homogenates were prepared in 50% ACN/0.3% formic acid spiked with stable isotope-labeled internal standards. Amino acids were extracted from homogenates with methanol followed by vortexing and centrifugation. An aliquot of methanol was dried down, and acylcarnitines were derivatized with O-benzylhydroxylamine. After derivatization, amino acids were separated and quantitated by LC/MS/MS.
Statistical Analysis
All data are represented as the mean ± SEM. All the statistical analyses were performed by GraphPad Prism. The differences between groups were conducted using analysis of variance (ANOVA) or t-test (paired and unpaired) approaches with Tukey’s/Bonferroni Multiple Comparison Test. When appropriate p <0.05, it was considered significant. The normalized (z-score) mean of each row was visualized in each heatmap, created with the “heatmap.2” function in the “gplots” package within the statistical program R (http://www.r-project.org/)
Results
Old Mice Have Impaired Early Recovery of Soleus Mass Following Unloading-Induced Muscle Atrophy
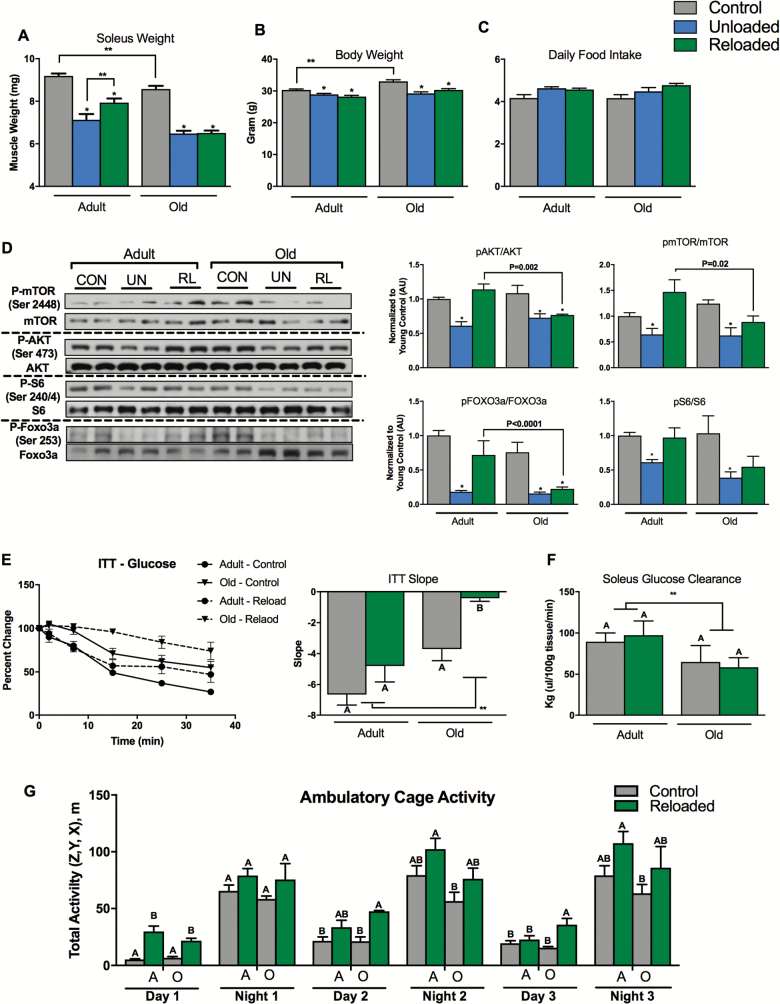
The overarching goal of the following experiments was to determine whether alterations in fuel metabolism and mitochondrial energetics are linked to poor early recovery of soleus muscle mass in old mice (22–24 months), compared to adult mice (6 months). To assess differences in the atrophy and recovery response, we used a modified version of the tail suspension hind limb unloading model to induce hind limb muscle atrophy over a 10-day period (28), followed by 3 days of reloading and voluntary cage ambulation. Although it is clear from previous reports that a full recovery of muscle mass and strength may take up to 40 days after unloading (6), we examined the response following a reloading period of 3 days, a time that others have shown to be important for induction of anabolic signal transduction and gene transcription networks (Akt, p70S6K, 4E-BP1, STAT3) involved in hypertrophy and recovery of muscle mass (38,39). Our goal was to understand the differences in response between adult and old mice at this key early period during reloading. The soleus of the old mice weighed less compared to young mice consistent with a sarcopenic phenotype and unloading-induced significant atrophy of the soleus (Figure 1A). A similar sarcopenic phenotype and response was seen for gastrocnemius and quadriceps muscle groups in old animals (data not shown), with soleus showing the greatest response to unloading. The soleus is a postural muscle consisting predominantly of type IIA and I oxidative myofibers making it particularly susceptible to unloading-induced atrophy and as such has been studied as a model of muscle atrophy and recovery by others. For this reason, we focused our analysis predominantly on the soleus. The magnitude of soleus muscle loss was similar between adult and old mice. However, following 3 days of reloading, significant recovery of soleus mass occurred in the adult, but not the old mice (Figure 1A). Similar responses were observed for the gastrocnemius and quadriceps (data not shown). In addition, body weight was greater in the old mice and decreased slightly during unloading and reloading in adult and old mice (Figure 1B), and there was no significant change in daily food intake (Figure 1C).
Figure 1.
Old mice do not recover muscle mass or insulin sensitivity following 10 days of hind limb unloading and 3 days of recovery. (A) Soleus weight following unloading, unloading/reloading, and control in adult and old mice, n = 11 per group. (B) Body weight for the adult and old mice following unloading, unloading/reloading, and control periods, n = 11 per group. (C) Average daily food intake for old and adult mice during unloading, unloading/reloading, and control periods, n = 11 per group. (D) Protein synthesis/breakdown signaling: pAKT/AKT, pmTOR/mTOR, pS6/S6, and pFOXO3a/FOXO3a ratios, n = 5 per group. (E) Blood glucose reductions (percent change from baseline) following intraperitoneal insulin injection. The slope of the fall in glucose levels from t = 0 to 15 minutes was used as an index of whole-body insulin action, n = 5 per group. (F) Soleus insulin-stimulated glucose clearance rates in control or unloaded/reloaded adult and old mice, n = 5 per group. (G) Day and night cage ambulatory activity (x, y, z) in control or unloaded/reloaded adult and old mice, n = 5 per group. Data are presented as mean ± SEM. The letters A and B denote significant differences between group/time points (p < 0.05, ANOVA/Tukey Correction). **p <0.05 ANOVA/Bonferroni correction. *p <0.05 versus control ANOVA/Bonferroni Correction. ANOVA = analysis of variance; CON = control; mTOR = mammalian target of rapamycin; RL = reloading recovery; FOXO3a = Forkhead box O3; UN = unloading.
Maintenance of skeletal muscle mass is a finely controlled balance between protein synthesis and degradation. We next examined the basal phosphorylation status of key signal transduction proteins that mediate protein synthesis and degradation in the soleus (Figure 1D). The AKT/mTOR pathway was inhibited following unloading in both adult and old mice as evidenced by reduced phosphorylation of AKT (Ser473), mTOR (Ser2448), and S6 (Ser240/244). However, the inhibition of the AKT/mTOR pathway was rescued by reloading in the adult mice, but not in the old mice. Forkhead box O3 (FOXO3a) is a transcription factor that regulates the muscle atrophy program. Under catabolic conditions, inhibition of AKT results in dephosphorylation and nuclear translocation of FOXO3a, which activates the expression of atrophy genes. Consistent with this role, we found that FOXO3a was dephosphorylated (Ser253) during unloading in both adult and old mouse soleus (Figure 1D). However during reloading, FOXO3a remained dephosphorylated in the old mice compared to the adults. Taken together, these data show that the key regulators of protein synthesis and degradation respond as expected during unloading in both adult and old mice. However, following 3 days of reloading, there is persistent activation of protein degradation and inhibition of protein synthesis mediators in old mice.
Aging and periods of disuse can induce whole-body and skeletal muscle insulin resistance. We next examined whether differences in metabolic phenotype may be related to the poor early recovery of muscle observed in old mice (Figure 1E and F). We found that whole-body insulin action following reloading was lower in old mice, when compared to control. This was not the case for adult mice (Figure 1E). We found that soleus glucose clearance was lower in the old mice (ANOVA main effect for age, Figure 1F) and that glucose clearance was similar between control and reloading in both adult and old mice.
Muscle protein synthesis and insulin sensitivity are both very sensitive to muscle contractile activity. To determine whether differences in ambulatory cage activity explained differences in muscle recovery and insulin sensitivity between adult and old mice, we examined cage activity over 3 days of the recovery period (Figure 2G). Generally, both the adult and old mice similarly recovered night and day total ambulatory cage activity, albeit in some cases (Day 1, for example) the reloaded animals were more active compared to controls.
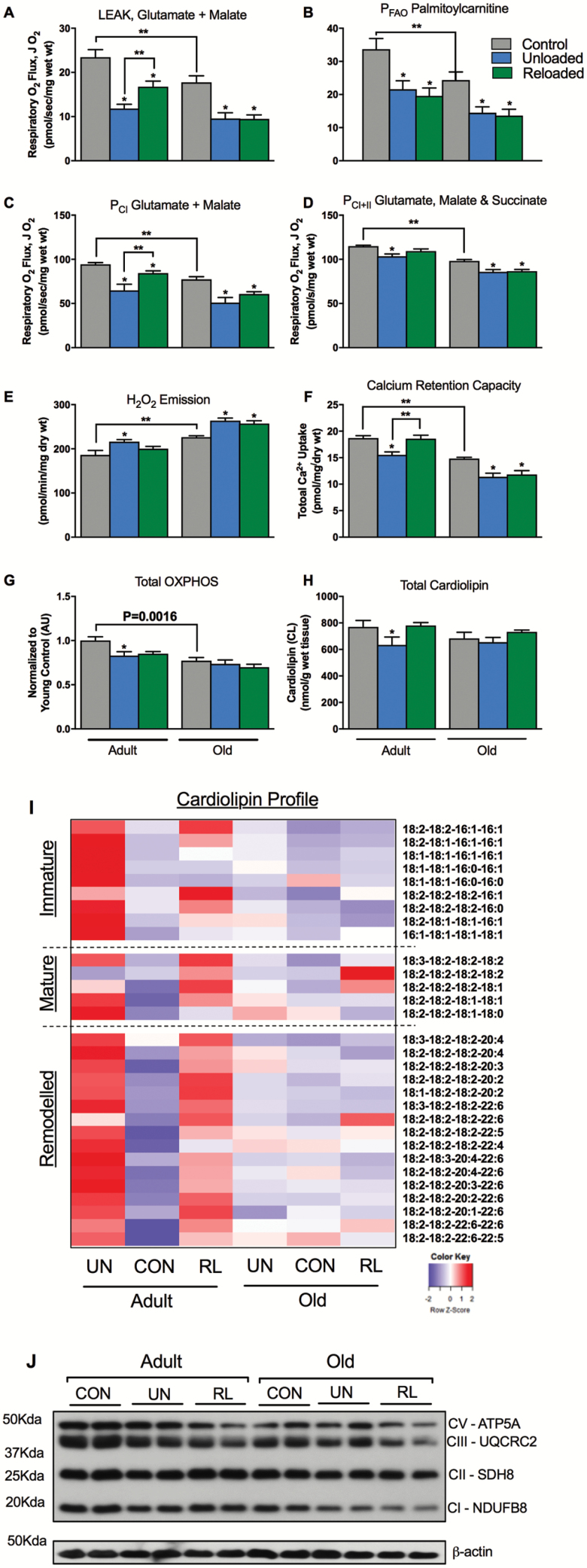
Figure 2.
Mitochondrial function is impaired during unloading and improves following 3 days of recovery only in adult mice. (A) Complex I (Glutamate and Malate) supported LEAK (CIL or State 4) respiration. (B) Palmitoylcarnitine supported OXPHOS (State 3) respiration. (C) Complex I supported OXPHOS respiration. (D) Complex I & II supported OXPHOS respiration. (E) Mitochondrial maximal H2O2 emission as determined with Amplex Red, during oligomycin-induced state 4 respiration. (F) Mitochondrial calcium retention capacity. n = 8–10 per group for permeabilized fiber bundle experiments. (G) Total OXPHOS protein content by immunoblot, n = 5 per group. (H) Total cardiolipin content by tandem mass spectrometry. (I) Heatmap depicting change in individual immature, mature, and remodeled cardiolipin species. The normalized (z-score) mean of each row (cardiolipin species) was visualized in the heatmap, n = 7 per group. (J) Representative immunoblot for OXPHOS proteins. Data are presented as mean ± SEM. **p <0.05 ANOVA/Bonferroni correction. *p <0.05 versus control ANOVA/Bonferroni correction. ANOVA = analysis of variance.
Mitochondrial Function Does Not Improve During Early Recovery in Old Mice
Our central hypothesis is that alterations in mitochondrial energetics and fuel metabolism play a key role in the poor recovery of muscle mass in older animals. Functional assays of soleus mitochondria were conducted using saponin-permeabilized myofiber bundles, an approach that preserves the native reticular structure of the mitochondria (40). When compared to adult, soleus myofibers isolated from old control mice displayed lower respiration, OXPHOS content, and CRC, an index of apoptotic susceptibility, while H2O2 emission was elevated (Figure 2A–G). Following unloading, LEAK, complex I, I & II and FAO-supported OXPHOS respiration, and CRC in soleus were all reduced in both the adult and old mice. In addition, mitochondrial H2O2 emission was elevated in both young and aged animals (Figure 2A–F). Total cardiolipin and OXPHOS protein (measures of mitochondrial content) were both reduced with unloading in the adult mice, but remained unchanged in the older animals indicating that changes in function were independent of changes in mitochondrial content during unloading in the old animals (Figure 4G,H, and J).
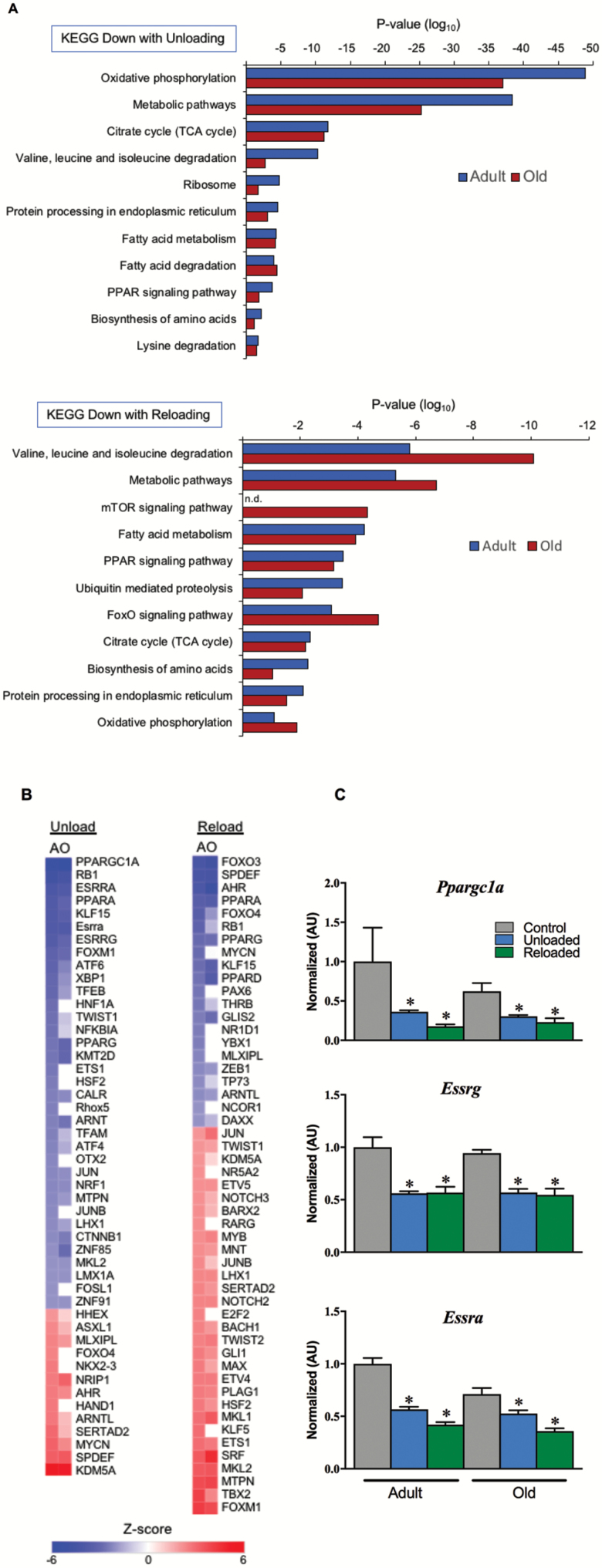
Figure 4.
Gene regulatory networks for oxidative metabolism are suppressed during unloading and do not recovery following 3 days of reloading. (A) KEGG pathway analysis of RNA-sequencing data set representing significantly downregulated pathways relating to mitochondria, metabolism, and protein metabolism during unloading and after 3 days of reloading. (B) Heatmap depicting IPA Upstream Regulator analysis to identify upstream transcriptional regulators responsible for the transcriptomic profile seen with unloading and reloading, plotted as a centralized Z-score for each transcription factor. The RNA-sequencing data set used was filtered with reads per kilobase per million mapped reads (RPKM) >1, p <0.05. (C) RT-qPCR assay for expression of key nuclear transcription factors that drive mitochondrial energy metabolism. n = 4–9 per group. Data are presented as mean ± SEM. *p <0.05 versus control ANOVA/Bonferroni correction. FOXO = Forkhead box O; KEGG = Kyoto Encyclopedia of Genes and Genomes; mTOR = mammalian target of rapamycin; RT-qPCR = reverse transcription-quantitative polymerase chain reaction; TCA = tricarboxylic acid.
Following 3 days of recovery, complex I & II supported mitochondrial respiration, CRC, and H2O2 emission improved in the young mice. Interestingly, FAO-supported respiration did not recover in the young animals. Old mice did not recover any aspect of mitochondrial function as evidenced by sustained elevated H2O2 emission and suppressed CRC and respiration. Taken together, these data indicate that mitochondrial function does not improve during the early recovery phase in old mice.
We used shotgun lipidomics to quantify the cardiolipin pool (41). Cardiolipin is a phospholipid specific to the inner mitochondrial membrane and is critical for ETC integrity and mitochondrial function (42). Cardiolipin species can be broadly categorized based on acyl chain length and degree of saturation, into: immature, mature, and remodeled species. Immature cardiolipins are those that have heterogeneous acyl chain length and degree of saturation and typically contain one or more 16:1 acyl chains. Mature cardiolipins have more uniform acyl chain length and degree of saturation, for example, tetralinolyol cardiolipin (18:2–18:2–18:2–18:2). Remodeled cardiolipin species contain greater PUFA (typically 22:6) acyl chains and may have been remodeled due to oxidative insult (43). The soleus of older animals had slightly lower content of many cardiolipin species, again likely due to reduced mitochondrial content with aging (Figure 2I). Unloading elicited a striking reduction in many immature, mature, and remodeled species of cardiolipin in adult mice with a partial recovery following reloading. The same dynamic response of the cardiolipin pool was not evident in the older mice.
Divergent Metabolomic Response Between Adult and Old Mice During Unloading and Recovery
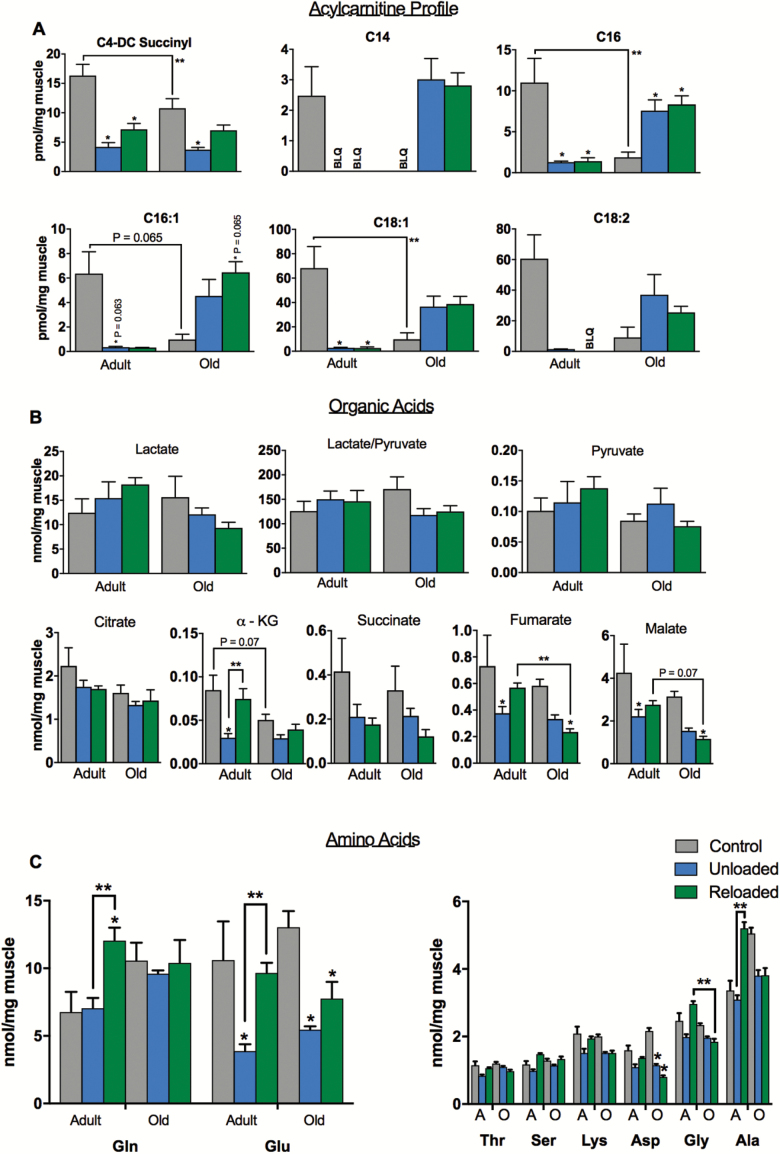
Targeted metabolomic profiling was conducted on soleus samples to quantify acylcarnitines, organic acids, and amino acids in order to assess perturbations in mitochondrial fuel metabolism that may occur with unloading and reloading. We show for the first time a striking decrease in long-chain acylcarnitines during unloading in the adult animals (Figure 3A), in-line with the observations of others who have shown that contractile inactivity induces a switch to carbohydrate fuel source and reductions in fatty acid oxidation (44). Interestingly, following reloading, acylcarnitines remained suppressed in the adult mice, consistent with lower FAO-supported respiration. However, certain tricarboxylic acid (TCA) cycle intermediates (fumarate, malate, and α-ketoglutarate) and amino acids (Gln, Glu, Ala, and Citrulline), increased during reloading in the adult soleus, consistent with recovery in TCA/CI&II supported respiration (Figure 3B and C). These data suggest that FAO remains suppressed during the early reloading phase in adult muscle and that recovery of mitochondrial respiration is supported by glycolysis and TCA cycle anaplerosis via amino acids. The increase in C4 and C5 acylcarnitines during reloading are also consistent with increased amino acid catabolism and anaplerosis.
Figure 3.
Acylcarnitine, organic acid, and amino acid profiles during unloading and reloading. (A) Acylcarnitine profile in soleus of adult and old mice during control, unloading, and reloading conditions. Acyl chain length and degree of saturation are denoted by standard nomenclature (C:D, where C is the number of carbon atoms and D is the number of double bonds). (B) Metabolite profiles showing lactate and pyruvate and organic acid intermediates of the tricarboxylic acid (TCA) cycle. (C) Amino acids profile in soleus during control, unloading, and reloading conditions. n = 6 per group. Data are presented as mean ± SEM. **p <0.05 ANOVA/Bonferroni correction. *p <0.05 versus control ANOVA/Bonferroni correction. ANOVA = analysis of variance; BLQ = below limit of quantification.
In old mice, the metabolite profile in response to unloading and reloading was distinctly different from that of the adult animals. The majority of long-chain acylcarnitines increased during unloading and reloading in the old mice (Figure 3A). This, in concert with reduced TCA cycle intermediates and reduced capacity for respiration, suggests a bottleneck in FAO resulting in accumulation of acylcarnitine species. Interestingly, many of the amino acids that were increased on reloading in the adult animals did not rise in the older animals (Figure 3C).
Transcriptomic Response to Unloading and Reloading Is Similar in Adult and Old Mice
Gene-expression profiling was conducted on soleus samples from the six groups using RNA sequencing. Our goal was to determine whether the divergent mitochondrial energetic and fuel metabolic responses to unloading and particularly reloading in old mice were underpinned by a distinct transcriptomic response. The DAVID v6.8 was used to identify significantly regulated KEGG pathways (45) during unloading and reloading. Given the mitochondrial respiration and metabolomic data, we focused our attention on pathways related to mitochondrial function and fuel metabolism. Many of these pathways were downregulated during unloading in both adult and old mice including oxidative phosphorylation and the TCA cycle (Figure 4A). Remarkably, several of these pathways were further downregulated following 3 days of reloading, again consistent with the lack of recovery of fatty acid supported respiration or acylcarnitines (Figure 4A). We next used the IPA Upstream Regulator to identify upstream transcriptional regulators responsible for the transcriptomic profile seen with unloading and reloading (Figure 4B). Many known transcriptional regulators of mitochondrial biogenesis and function such as PGC-1α (PPARGC1A), ERRα (ESRRA), and PPARα (PPARA) were all predicted to be inhibited during unloading. mRNA levels of PGC-1α (Ppargc1a), ERRα (Esrra), and ERRγ (Esrrg) were all decreased during unloading suggesting that inhibition of these factors is at least in part due to decreased expression (Figure 4C). The expression of these factors did not recover during the reloading phase.
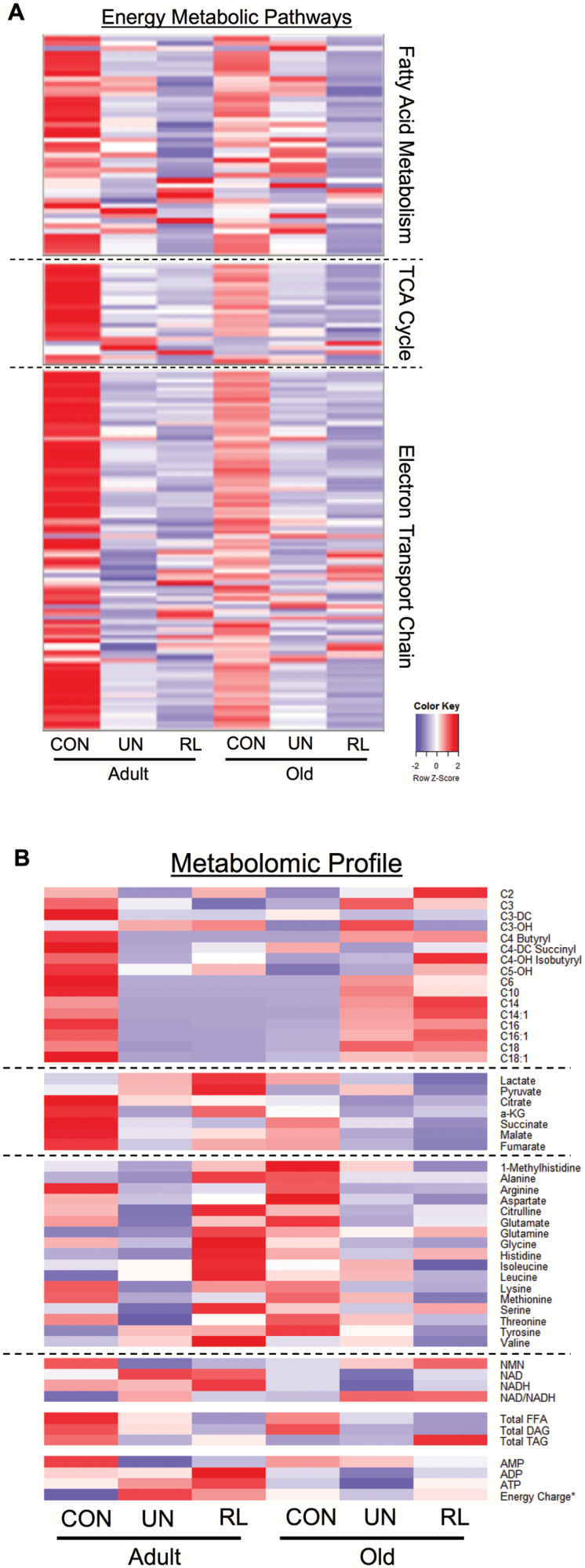
Interestingly, in contrast to the metabolomic results, the transcriptomic changes in energy metabolic pathways were similar between the adult and old mice. Therefore, we compared the collective results of the transcriptomic and metabolomic profiling. This comparison highlights striking differences in the metabolomic and transcriptomic responses following 3 days of reloading for adult and old mice. The relative expression level of transcripts of genes encoding mitochondrial proteins involved in fatty acid metabolism, the TCA cycle, and oxidative phosphorylation is shown in the heatmap in Figure 5A. The relative expression level of these genes is lower in control old versus adult mice. However, in both groups, there is a marked decrease in expression during unloading that further slightly decreases with reloading (Figure 5A). This is in stark contrast to the pattern observed with changes in metabolite levels. As shown in Figure 5B, in adult mice, the recovery of mitochondrial respiratory capacity occurs along with partial normalization of metabolites, particularly amino acids and organic acids, but not acylcarnitines. However, in old mice, the metabolite response during recovery is distinctly different. Namely, acylcarnitines are elevated while organic acids and amino acids remain low. Taken together, these results indicate that mitochondrial energetics gene and metabolite profiles are repressed in concert with unloading and muscle atrophy. However, metabolite levels clearly distinguish the early muscle hypertrophy response in adult animals versus the lack of recovery in old animals upon reloading and to a far greater degree than mitochondrial energetic gene-expression profile.
Figure 5.
Comparative analysis of transcriptomic and metabolomics profiles during unloading and reloading. (A) Heatmap containing RNA-sequencing data set representing the level of expression of genes with reads per kilobase per million mapped reads (RPKM) >1, plotted as a centralized Z-score for each gene in fatty acid metabolism, tricarboxylic acid (TCA) cycle, and electron transport chain/oxidative phosphorylation pathways defined by Ingenuity Pathway Analysis. (B) Heatmap containing the levels of acylcarnitines, organic acids, amino acids, pyridine, and adenine nucleotides and total level of free fatty acids (FFA), diacylglycerol (DAG), and triacylglycerol (TAG). For all heatmaps, relative downregulation is in blue and upregulation in red. The intensity of color indicates the magnitude of the change. *The energy charge ratio is defined as ([ATP] + 1/2[ADP])/([AMP] + [ADP] + [ATP]). CON = control; RL = reloading recovery; UN = unloading.
Discussion
Poor recovery of muscle mass following periods of disuse due to illness or immobilization leads to increased risk of frailty and mobility disability, and thus represents a significant health concern for older adults. The mechanisms responsible for the lack of muscle mass recovery in older adults following disuse have not been fully elucidated but are likely complex and multifactorial. Here, we report that 22- to 24-month-old mice with low muscle mass, display poor early recovery of muscle mass following 10 days of hind limb unloading. Importantly, this was linked to worsened whole-body insulin resistance. We profiled changes in energy metabolic gene expression and metabolite pools and show for the first time that mitochondrial dysfunction and a dysregulated metabolic response occur concomitantly with poor early recovery of soleus mass in old mice. Interestingly, while metabolite profile distinguished the phenotype of poor muscle recovery in old mice, transcriptomic profiles for energy metabolic pathways were similar between old and adult mice during unloading and reloading. Our results demonstrate the power of using unbiased molecular profiling to define signatures of complex physiological states combined with deep phenotyping of mitochondrial energetics.
Insulin signaling intersects with protein synthesis and degradation pathways via AKT/FOXO/mTOR. Hence, a logical supposition is that baseline insulin resistance worsens muscle atrophy response during disuse. Similarly, mitochondrial dysfunction and ROS emission are linked to activation of protein degradation pathways and may worsen muscle atrophy due to disuse (46). Indeed, older adults with type 2 diabetes, a condition defined by muscle insulin resistance and mitochondrial dysfunction, have an accelerated loss of muscle mass over time (47), indicating a propensity for catabolic metabolism. However, we found that old mice with mitochondrial dysfunction did not have an exacerbated muscle atrophy during disuse. The relative magnitude of muscle loss and alterations in the activity (phosphorylation state) of AKT/mTOR/FOXO3a, and the majority of endpoint measurements were similar in adult and old animals in response to unloading. These findings suggest that the presence of mitochondrial dysfunction in aging does not influence the muscle atrophy response due to disuse or unloading.
Physical inactivity results in attenuated mitochondria capacity along with a shift in fuel selection to carbohydrate and suppressed FAO (44). Here, in adult mice, we show a distinct remodeling of soleus mitochondrial energetics and fuel metabolism during unloading as evidenced by reduced respiration, elevated H2O2 emission, and reduced long-chain acylcarnitines and organic acids. This change in mitochondrial phenotype occurred along with striking reductions in many individual molecular species of cardiolipin. Indeed, synthesis and remodeling of cardiolipin is crucial for the structural and functional integrity of the mitochondria. Our findings expand upon the work of Ostojic and colleagues, others who reported decreased cardiolipin during muscle atrophy (48).
Although impaired recovery by aged muscle is reasonably well documented, the mechanisms responsible for the lack of growth are not clearly defined. Investigations to date have focused on attenuated protein synthesis possibly due to ER stress and impaired neuromuscular transmission (39). In separate lines of investigation, mitochondrial energetics and processes that govern quality (fusion/fission) have been linked to the regulation of muscle mass (14,49). We show that impaired early recovery of muscle mass occur concomitantly with insulin resistance and persistent mitochondrial dysfunction in old animals. In stark contrast to adult mice, there was no change in the cardiolipin species of old mice that likely contributed to persistent mitochondrial dysfunction, perhaps due to a lack of mitophagy and clearance of damaged mitochondria. Cardiolipin synthesis and remodeling is critical for correct cristae folding and functional integrity of the ETC complexes. The increase in acylcarnitines (long chain in particular) in concert with reductions in particular TCA cycle organic acids (fumarate and malate), and no change in pyruvate or lactate, suggest a bottleneck in carbon flux from ß-oxidation into the TCA cycle. The metabolite profile served to robustly distinguish the phenotype of poor muscle recovery in old mice and is similar to that observed with high fat feeding and obesity (50). This could reduce the capacity to generate the reducing equivalents needed to produce ATP and in turn contribute to poor anabolic response as the energetic cost of protein synthesis is quite substantial. It is estimated that the energetic cost of protein synthesis accounts for approximately 20% of basal metabolism (51,52). Protein breakdown is estimated to account for another 5% to 15% of basal metabolism (53).
Our findings are also in line with observations that insulin resistance, obesity, and elevated plasma free fatty acids, physiological contexts that also encompass impaired mitochondrial energetics, are linked with blunted amino acid stimulated muscle protein synthesis (54,55). Indeed, elevated mitochondrial ROS emission during reloading in the old mice along with inhibition of the AKT/mTOR pathway is consistent with previously reported links between mitochondrial mediated oxidative stress and insulin resistance (56), and represents a putative mechanism by which impaired mitochondrial energetics cause poor recovery of muscle mass. Further studies are needed to examine whether targeting mitochondria may impact early recovery of muscle in old mice, potentially via improved insulin sensitivity.
Interestingly, while metabolite profile distinguished the phenotype of poor muscle recovery in old mice, transcriptomic profiles for energy metabolic pathways were similar between old and adult mice during unloading and reloading. In addition, the recovery of mitochondrial respiratory capacity in adult mice occurred along with partial recovery of metabolites, while FAO, TCA, and ETC genes remain repressed. Indeed, FAO genes appeared to be further repressed during unloading in adult mice. These data suggest that the early recovery of mitochondrial function following reloading seems to be independent of FAO and mediated by cardiolipin remodeling and alterations in metabolite profile. Further studies using tracer methodologies to detect changes in lipid metabolism and metabolic flux would be advantageous to understand early changes in metabolic flux during recovery. In addition, posttranslational modification, such as lysine acetylation and succinylation have been recently shown to alter activity of ETC complexes, and may play a role in improved mitochondrial function independent of changes in expression of gene regulatory networks, although this remains to be tested. Taken together, these results indicate that mitochondrial energetics, gene, and metabolite profiles are repressed in concert with unloading and muscle atrophy.
There are a few minor limitations that should be considered when interpreting this work. We focused primarily on the soleus and the same responses may not be evident in locomotor muscle groups with a greater proportion of fast fiber types. In addition, there were variable sample numbers (n) per analysis, in part due to the amount of material required for the analysis, and the small size of the soleus (~9 mg).
In summary, our data suggest that dysregulated FAO and elevated H2O2 emission from mitochondria likely contribute to the lack of recovery of soleus muscle mass following unloading in sarcopenic mice. These findings suggest that pathways governing muscle fuel metabolism and mitochondrial energetics might harbor promising therapeutic targets for improving the recovery of muscle mass following periods of disuse in older patients.
Supplementary Material
Supplementary data is available at The Journals of Gerontology, Series A: Biological Sciences and Medical Sciences online.
Funding
This study was supported by funding from the National Institutes of Health | National Institute on Aging (K01 AG044437) awarded to PMC.
Conflict of Interest
None reported.
Acknowledgments
We are grateful for the excellent technical assistance of the staff at the Vivarium, the Cardiometabolic Phenotyping core, and the Metabolomics core of the Sanford Burnham Prebys Medical Discovery Institute at Lake Nona. We are also grateful to Feng Qi for bioinformatics support and Fanchao Yi for statistics support.
References
- 1. Cruz-Jentoft AJ, Baeyens JP, Bauer JM et al. ; European Working Group on Sarcopenia in Older People Sarcopenia: European consensus on definition and diagnosis: report of the European Working Group on Sarcopenia in Older People. Age Ageing. 2010;39: 412–423. doi:10.1093/ageing/afq034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rolland Y, Czerwinski S, Abellan Van Kan G et al. . Sarcopenia: its assessment, etiology, pathogenesis, consequences and future perspectives. J Nutr Health Aging. 2008;12: 433–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zarzhevsky N, Carmeli E, Fuchs D, Coleman R, Stein H, Reznick AZ. Recovery of muscles of old rats after hindlimb immobilisation by external fixation is impaired compared with those of young rats. Exp Gerontol. 2001;36: 125–140. [DOI] [PubMed] [Google Scholar]
- 4. Dardevet D, Sornet C, Taillandier D, Savary I, Attaix D, Grizard J. Sensitivity and protein turnover response to glucocorticoids are different in skeletal muscle from adult and old rats. Lack of regulation of the ubiquitin-proteasome proteolytic pathway in aging. J Clin Invest. 1995;96: 2113–2119. doi:10.1172/JCI118264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hao Y, Jackson JR, Wang Y, Edens N, Pereira SL, Alway SE. β-Hydroxy-β-methylbutyrate reduces myonuclear apoptosis during recovery from hind limb suspension-induced muscle fiber atrophy in aged rats. Am J Physiol Regul Integr Comp Physiol. 2011;301: R701–R715. doi:10.1152/ajpregu.00840.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Magne H, Savary-Auzeloux I, Vazeille E et al. . Lack of muscle recovery after immobilization in old rats does not result from a defect in normalization of the ubiquitin-proteasome and the caspase-dependent apoptotic pathways. J Physiol. 2011;589(Pt 3): 511–524. doi:10.1113/jphysiol.2010.201707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Degens H, Alway SE. Skeletal muscle function and hypertrophy are diminished in old age. Muscle Nerve. 2003;27: 339–347. doi:10.1002/mus.10314 [DOI] [PubMed] [Google Scholar]
- 8. Suetta C, Frandsen U, Mackey AL et al. . Ageing is associated with diminished muscle re-growth and myogenic precursor cell expansion early after immobility-induced atrophy in human skeletal muscle. J Physiol. 2013;591: 3789–3804. doi:10.1113/jphysiol.2013.257121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. English KL, Paddon-Jones D. Protecting muscle mass and function in older adults during bed rest. Curr Opin Clin Nutr Metab Care. 2010;13: 34–39. doi:10.1097/MCO.0b013e328333aa66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hoppeler H. Molecular networks in skeletal muscle plasticity. J Exp Biol. 2016;219(Pt 2): 205–213. doi:10.1242/jeb.128207 [DOI] [PubMed] [Google Scholar]
- 11. Bonaldo P, Sandri M. Cellular and molecular mechanisms of muscle atrophy. Dis Model Mech. 2013;6: 25–39. doi:10.1242/dmm.010389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Egerman MA, Glass DJ. Signaling pathways controlling skeletal muscle mass. Crit Rev Biochem Mol Biol. 2014;49: 59–68. doi:10.3109/10409238.2013.857291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Joseph AM, Adhihetty PJ, Leeuwenburgh C. Beneficial effects of exercise on age-related mitochondrial dysfunction and oxidative stress in skeletal muscle. J Physiol. 2016;594: 5105–5123. doi:10.1113/JP270659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Powers SK, Wiggs MP, Duarte JA, Zergeroglu AM, Demirel HA. Mitochondrial signaling contributes to disuse muscle atrophy. Am J Physiol Endocrinol Metab. 2012;303: E31–E39. doi:10.1152/ajpendo.00609.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Talbert EE, Smuder AJ, Min K, Kwon OS, Szeto HH, Powers SK. Immobilization-induced activation of key proteolytic systems in skeletal muscles is prevented by a mitochondria-targeted antioxidant. J Appl Physiol (1985). 2013;115: 529–538. doi:10.1152/japplphysiol.00471.2013 [DOI] [PubMed] [Google Scholar]
- 16. Min K, Smuder AJ, Kwon OS, Kavazis AN, Szeto HH, Powers SK. Mitochondrial-targeted antioxidants protect skeletal muscle against immobilization-induced muscle atrophy. J Appl Physiol (1985). 2011;111: 1459–1466. doi:10.1152/japplphysiol.00591.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jang YC, Lustgarten MS, Liu Y et al. . Increased superoxide in vivo accelerates age-associated muscle atrophy through mitochondrial dysfunction and neuromuscular junction degeneration. FASEB J. 2010;24: 1376–1390. doi:10.1096/fj.09-146308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aucello M, Dobrowolny G, Musarò A. Localized accumulation of oxidative stress causes muscle atrophy through activation of an autophagic pathway. Autophagy. 2009;5: 527–529. [DOI] [PubMed] [Google Scholar]
- 19. Li YP, Chen Y, Li AS, Reid MB. Hydrogen peroxide stimulates ubiquitin-conjugating activity and expression of genes for specific E2 and E3 proteins in skeletal muscle myotubes. Am J Physiol Cell Physiol. 2003;285: C806–C812. doi:10.1152/ajpcell.00129.2003 [DOI] [PubMed] [Google Scholar]
- 20. Grune T, Merker K, Sandig G, Davies KJ. Selective degradation of oxidatively modified protein substrates by the proteasome. Biochem Biophys Res Commun. 2003;305: 709–718. [DOI] [PubMed] [Google Scholar]
- 21. Smuder AJ, Kavazis AN, Hudson MB, Nelson WB, Powers SK. Oxidation enhances myofibrillar protein degradation via calpain and caspase-3. Free Radic Biol Med. 2010;49: 1152–1160. doi:10.1016/j.freeradbiomed.2010.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Romanello V, Guadagnin E, Gomes L et al. . Mitochondrial fission and remodelling contributes to muscle atrophy. EMBO J. 2010;29: 1774–1785. doi:10.1038/emboj.2010.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Greer EL, Oskoui PR, Banko MR et al. . The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem. 2007;282: 30107–30119. doi:10.1074/jbc.M705325200 [DOI] [PubMed] [Google Scholar]
- 24. Brault JJ, Jespersen JG, Goldberg AL. Peroxisome proliferator-activated receptor gamma coactivator 1alpha or 1beta overexpression inhibits muscle protein degradation, induction of ubiquitin ligases, and disuse atrophy. J Biol Chem. 2010;285: 19460–19471. doi:10.1074/jbc.M110.113092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sandri M, Lin J, Handschin C et al. . PGC-1alpha protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc Natl Acad Sci USA. 2006;103: 16260–16265. doi:10.1073/pnas.0607795103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cannavino J, Brocca L, Sandri M, Bottinelli R, Pellegrino MA. PGC1-α over-expression prevents metabolic alterations and soleus muscle atrophy in hindlimb unloaded mice. J Physiol. 2014;592: 4575–4589. doi:10.1113/jphysiol.2014.275545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Masiero E, Sandri M. Autophagy inhibition induces atrophy and myopathy in adult skeletal muscles. Autophagy. 2010;6: 307–309. [DOI] [PubMed] [Google Scholar]
- 28. Morey-Holton ER, Globus RK. Hindlimb unloading rodent model: technical aspects. J Appl Physiol (1985). 2002;92: 1367–1377. doi:10.1152/japplphysiol.00969.2001 [DOI] [PubMed] [Google Scholar]
- 29. Ayala JE, Bracy DP, Julien BM, Rottman JN, Fueger PT, Wasserman DH. Chronic treatment with sildenafil improves energy balance and insulin action in high fat-fed conscious mice. Diabetes. 2007;56: 1025–1033. doi:10.2337/db06-0883 [DOI] [PubMed] [Google Scholar]
- 30. Coen PM, Jubrias SA, Distefano G et al. . Skeletal muscle mitochondrial energetics are associated with maximal aerobic capacity and walking speed in older adults. J Gerontol A Biol Sci Med Sci. 2013;68: 447–455. doi:10.1093/gerona/gls196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Coen PM, Menshikova EV, Distefano G et al. . Exercise and weight loss improve muscle mitochondrial respiration, lipid partitioning, and insulin sensitivity after gastric bypass surgery. Diabetes. 2015;64: 3737–3750. doi:10.2337/db15-0809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee S, Leone TC, Rogosa L et al. . Skeletal muscle PGC-1β signaling is sufficient to drive an endurance exercise phenotype and to counteract components of detraining in mice. Am J Physiol Endocrinol Metab. 2017;312: E394–E406. doi:10.1152/ajpendo.00380.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang M, Han X. Multidimensional mass spectrometry-based shotgun lipidomics. Methods Mol Biol. 2014;1198: 203–220. doi:10.1007/978- 1-4939-1258-2_13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang M, Fang H, Han X. Shotgun lipidomics analysis of 4-hydroxyalkenal species directly from lipid extracts after one-step in situ derivatization. Anal Chem. 2012;84: 4580–4586. doi:10.1021/ac300695p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Han X, Yang K, Cheng H, Fikes KN, Gross RW. Shotgun lipidomics of phosphoethanolamine-containing lipids in biological samples after one-step in situ derivatization. J Lipid Res. 2005;46: 1548–1560. doi:10.1194/jlr.D500007-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang M, Hayakawa J, Yang K, Han X. Characterization and quantification of diacylglycerol species in biological extracts after one-step derivatization: a shotgun lipidomics approach. Anal Chem. 2014;86: 2146–2155. doi:10.1021/ac403798q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang M, Han RH, Han X. Fatty acidomics: global analysis of lipid species containing a carboxyl group with a charge-remote fragmentation-assisted approach. Anal Chem. 2013;85: 9312–9320. doi:10.1021/ac402078p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Childs TE, Spangenburg EE, Vyas DR, Booth FW. Temporal alterations in protein signaling cascades during recovery from muscle atrophy. Am J Physiol Cell Physiol. 2003;285: C391–C398. doi:10.1152/ajpcell.00478.2002 [DOI] [PubMed] [Google Scholar]
- 39. Baehr LM, West DW, Marcotte G et al. . Age-related deficits in skeletal muscle recovery following disuse are associated with neuromuscular junction instability and ER stress, not impaired protein synthesis. Aging (Albany NY). 2016;8: 127–146. doi:10.18632/aging.100879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Picard M, Taivassalo T, Ritchie D et al. . Mitochondrial structure and function are disrupted by standard isolation methods. PLoS One. 2011;6: e18317. doi:10.1371/journal.pone.0018317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Han X, Yang K, Yang J, Cheng H, Gross RW. Shotgun lipidomics of cardiolipin molecular species in lipid extracts of biological samples. J Lipid Res. 2006;47: 864–879. doi:10.1194/jlr.D500044-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Paradies G, Paradies V, Ruggiero FM, Petrosillo G. Cardiolipin and mitochondrial function in health and disease. Antioxid Redox Signal. 2014;20: 1925–1953. doi:10.1089/ars.2013.5280 [DOI] [PubMed] [Google Scholar]
- 43. Chicco AJ, Sparagna GC. Role of cardiolipin alterations in mitochondrial dysfunction and disease. Am J Physiol Cell Physiol. 2007;292: C33–C44. doi:10.1152/ajpcell.00243.2006 [DOI] [PubMed] [Google Scholar]
- 44. Bergouignan A, Rudwill F, Simon C, Blanc S. Physical inactivity as the culprit of metabolic inflexibility: evidence from bed-rest studies. J Appl Physiol (1985). 2011;111: 1201–1210. doi:10.1152/japplphysiol. 00698.2011 [DOI] [PubMed] [Google Scholar]
- 45. Dennis G Jr, Sherman BT, Hosack DA et al. . DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4: P3. [PubMed] [Google Scholar]
- 46. Romanello V, Sandri M. Mitochondrial biogenesis and fragmentation as regulators of muscle protein degradation. Curr Hypertens Rep. 2010;12: 433–439. doi:10.1007/s11906-010-0157-8 [DOI] [PubMed] [Google Scholar]
- 47. Park SW, Goodpaster BH, Lee JS et al. ; Health, Aging, and Body Composition Study Excessive loss of skeletal muscle mass in older adults with type 2 diabetes. Diabetes Care. 2009;32: 1993–1997. doi:10.2337/dc09-0264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ostojic O, O’Leary MF, Singh K, Menzies KJ, Vainshtein A, Hood DA. The effects of chronic muscle use and disuse on cardiolipin metabolism. J Appl Physiol (1985). 2013;114: 444–452. doi:10.1152/japplphysiol.01312.2012 [DOI] [PubMed] [Google Scholar]
- 49. Romanello V, Sandri M. Mitochondrial quality control and muscle mass maintenance. Front Physiol. 2015;6: 422. doi:10.3389/fphys.2015.00422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Koves TR, Ussher JR, Noland RC et al. . Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. 2008;7: 45–56. doi:10.1016/j.cmet.2007.10.013 [DOI] [PubMed] [Google Scholar]
- 51. Waterlow JC. Protein turnover with special reference to man. Q J Exp Physiol. 1984;69: 409–438. [DOI] [PubMed] [Google Scholar]
- 52. Musci RV, Hamilton KL, Miller BF. Targeting mitochondrial function and proteostasis to mitigate dynapenia. Eur J Appl Physiol. 2018;118: 1–9. doi:10.1007/s00421-017-3730-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rolfe DF, Brown GC. Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiol Rev. 1997;77: 731–758. doi: 10.1152/physrev.1997.77.3.731 [DOI] [PubMed] [Google Scholar]
- 54. Murton AJ, Marimuthu K, Mallinson JE et al. . Obesity appears to be associated with altered muscle protein synthetic and breakdown responses to increased nutrient delivery in older men, but not reduced muscle mass or contractile function. Diabetes. 2015;64: 3160–3171. doi:10.2337/db15-0021 [DOI] [PubMed] [Google Scholar]
- 55. Stephens FB, Chee C, Wall BT et al. . Lipid-induced insulin resistance is associated with an impaired skeletal muscle protein synthetic response to amino acid ingestion in healthy young men. Diabetes. 2015;64: 1615–1620. doi:10.2337/db14-0961 [DOI] [PubMed] [Google Scholar]
- 56. Muoio DM, Neufer PD. Lipid-induced mitochondrial stress and insulin action in muscle. Cell Metab. 2012;15: 595–605. doi:10.1016/j.cmet.2012.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.