Abstract
Non–small cell lung cancer (NSCLC) with activating EGFR mutations in exon 19 and 21 typically responds to EGFR tyrosine kinase inhibitors (TKI); however, for some patients, responses last only a few months. The underlying mechanisms of such short responses have not been fully elucidated. Here, we sequenced the genomes of 16 short-term responders (SR) that had progression-free survival (PFS) of less than 6 months on the first-generation EGFR TKI and compared them to 12 long-term responders (LR) that had more than 24 months of PFS. All patients were diagnosed with advanced lung adenocarcinoma and harbored EGFR 19del or L858R mutations before treatment. Paired tumor samples collected before treatment and after relapse (or at the last follow-up) were subjected to targeted next-generation sequencing of 416 cancer-related genes. SR patients were significantly younger than LR patients (P < .001). Collectively, 88% of SR patients had TP53 variations compared to 13% of LR patients (P < .001). Additionally, 37.5% of SR patients carried EGFR amplifications compared to 8% of LR patients. Other potential primary resistance factors were also identified in the pretreatment samples of 12 SR patients (75%), including PTEN loss; BIM deletion polymorphism; and amplifications of EGFR, ERBB2, MET, HRAS, and AKT2. Comparatively, only three LR patients (25%) were detected with EGFR or AKT1 amplifications that could possibly exert resistance. The diverse preexisting resistance mechanisms in SR patients revealed the complexity of defining treatment strategies even for EGFR-sensitive mutations.
Introduction
EGFR activating mutations, which are widely known for their prominent role in tumorigenesis and progression, are found in ~20% of patients with non–small cell lung cancer (NSCLC) in Western countries and ~50% in Asia [1]. The two most common mutations, the EGFR exon 19 deletion and the exon 21 L858R substitution, account for >90% of known EGFR activating mutations [2], [3] and are typically treated with first-generation (1st-gen) EGFR tyrosine kinase inhibitors (TKI), such as gefitinib, erlotinib. and icotinib [4], [5]. The overall response rate to these small molecular drugs is ~75%, and the median progression-free survival time (PFS) is ~10 months [6], [7], [8]. However, in clinical practice, patients who have the indications for 1st-gen TKI treatment demonstrated significant heterogeneity in treatment responses with a PFS ranging from a few months to several years [9], [10]. Recent studies have identified multiple factors that are associated with poor responses or resistance to 1st-gen TKIs, including the primary existence or acquisition of the EGFR T790 M mutation, BIM polymorphisms, inactivation of TP53, the activation of alternative signaling pathways (e.g., MET, ERBB2, FLT4), and downstream effectors (e.g., AKT amplifications, PTEN loss, PIK3CA activation) [11], [12], [13]. Tumor cells can also experience histology transformation, such as epithelial-mesenchymal transition (EMT) or the transformation to small cell lung cancer, which reduces treatment sensitivity [12].
Given the complexity of tumor heterogeneity, using EGFR-sensitizing mutations as the sole indicator for TKI treatment decision-making without considering the concurrent mutations in the tumor may be insufficient to yield optimal outcomes. To address this issue and identify biomarkers for more precise prognoses, we selected two groups of NSCLC patients who carried EGFR 19del or L858R mutations and demonstrated extremely short (≤6 months) or long (≥24 months) response periods following 1st-gen TKI treatments. Tumor samples prior and posttreatment were collected for tumor genomic profiling by pan-cancer gene next-generation sequencing. It was observed that TP53 inactivation mutations significantly accumulated in the short-term response group. Additionally, almost all patients in the short-term response group harbored multiple resistance factors that may have compromised TKI efficiency.
Material and Methods
Patient Enrollment and Sample Collection
Twenty-eight patients were enrolled from Zhejiang Cancer Hospital between 2011 and 2017, and the last follow-up was conducted in December 2017. Each patient provided written consent to contribute their clinical information and genetic test results for research and publication. The study was approved by the Ethics Committee of Zhejiang Cancer Hospital.
For each patient, formalin-fixed paraffin-embedded (FFPE) tissue sections or slides that were collected before TKI treatment were retrieved from the archive. Following TKI treatment, FFPE tissues or peripheral blood was collected again from patients, if clinical conditions permitted. All FFPE specimens were histologically examined by pathologists to ensure the tumor content was above 10% before proceeding to DNA extraction and sequencing.
DNA extraction, library preparation, and targeted enrichment
FFPE samples were deparaffinized with xylene, and genomic DNA was extracted using the QIAamp DNA FFPE Tissue Kit (Qiagen) following manufacturer's instructions. For some patients, cell-free DNA (cfDNA) was extracted from plasma using the QIAmp Circulating Nucleic Acid Kit (Qiagen). Genomic DNA of whole blood was extracted using the DNeasy Tissue and Blood Kit (Qiagen) and used as the normal control to remove germline variations. DNA was quantified on a Qubit 3.0 Fluorometer with the Qubit dsDNA HS Assay kit (Thermo Fisher), and the quality was evaluated by a Nanodrop 2000 (Thermo Fisher). The size distribution of cfDNA was analyzed using a Bioanalyzer 2100 with High Sensitivity DNA kit (Agilent Technologies).
Library construction was performed as previously described [14]. Briefly, 1-2 μg of genomic DNA was sheared into ~350 bp fragments using a Covaris M220 instrument. For cfDNA, 2-200 ng was used as the input for library preparation, without DNA shearing. End repair, A-tailing, and adaptor ligation of fragmented DNA were performed using the KAPA Hyper DNA Library Prep kit (Roche Diagnostics), followed by size selection with Agencourt AMPure XP beads (Beckman Coulter). The ligated products were PCR amplified and purified by Agencourt AMPure XP beads.
Different DNA libraries with unique indexes were pooled and subjected to targeted enrichment using customized xGen lockdown probes (Integrated DNA Technologies) that were designed to capture 416 cancer-related genes and 16 frequently rearranged genes. Human cot-1 DNA (Life Technologies) and xGen Universal Blocking Oligos (Integrated DNA Technologies) were added as blocking reagents. The capture reaction was performed with Dynabeads M-270 (Life Technologies) and the xGen Lockdown Hybridization and Wash kit (Integrated DNA Technologies), according to manufacturers' protocols. Captured libraries were subjected to PCR amplification with KAPA HiFi HotStart ReadyMix (KAPA Biosystems). The purified library was quantified using the KAPA Library Quantification kit (KAPA Biosystems), and its fragment size distribution was analyzed using a Bioanalyzer 2100.
Sequencing and Bioinformatics Analysis
DNA libraries were sequenced on an Illumina Hiseq4000 platform (Illumina). Sequencing data were processed and genomic alterations were analyzed according to a previous report with minor modifications [14]. Briefly, Trimmomatic was used for FASTQ file quality control (QC) to remove leading/trailing low quality (quality reading below 15) or N bases before mapping to the reference genome. Only qualified reads were mapped to the reference human genome, hg19, using the Burrows-Wheller Aligner (BWA-mem, v0.7.12) with default parameters. Single nucleotide variants (SNVs) and indels were detected by VarScan. Single nucleotide polymorphisms (SNPs) with mutation allele frequencies (MAFs) >30% were filtered using the 1000 Genomes Project or 65,000 exomes project (ExAC) and were removed from the final reports if present in >1% population frequency in the databases. ADTEx (http://adtex.sourceforge.net) was used to identify copy number variants (CNVs) using a normal human HapMap DNA sample, NA18535, as the reference. The relative depth ratios were smoothed by discrete wavelet transformation techniques prior to applying the Hidden Markov Model to estimate polyploidy, normal contamination ratio, and absolute CNVs. Tumor mutation burden (TMB) was defined as the number of somatic mutations with synonymous somatic mutations included. Allele-specific copy number alterations and loss of heterozygosity (LOH) were analyzed by FACETS algorithms according to the instructions [15].
Drug Resistance Mechanism Analysis
Thirteen SR patients had tumor tissue or liquid biopsy samples sequenced following resistance to 1st-gen TKIs. Acquired mutations were defined as newly observed alterations following the development of resistance. For the analysis of relative MAFs for epigenetic modifiers, TP53 mutations or EGFR-activating mutations (only used in TP53 mutant-free samples) were considered clonal and used for MAF correction for other mutations [16]. The method was based on the theory that drug resistance clones in tumors demonstrate a growth advantage under selective pressure.
Results
Clinical Characteristics of the Study Cohort
In this study, 28 patients with advanced lung adenocarcinoma were retrospectively included based on their responses to 1st-gen EGFR TKI treatments. Twenty-six patients were at stage IV disease with either distant or in-chest metastasis, while the remaining two patients were stage III disease (Supplementary Table 1). All patients harbored either EGFR exon19 deletions (19del, including two exon19 insertion/deletion alterations) or the L858R mutation, and had received the 1st-gen EGFR TKI as the first- or second-line treatment (Supplementary Table 1). Gefitinib was administered in 11 patients, while icotinib was administered in 16 patients (Table 1).
Table 1.
Summary of Clinical Characteristics of the SR and LR Groups
| SR (n = 16) |
LR (n = 12) |
P Value | ||
|---|---|---|---|---|
| Age | Median (range) | 52 (34-71) | 66 (53–75) | P = .001 |
| Sex | Female | 9 (56.3%) | 10 (83.3%) | NS |
| Male | 7 (43.7%) | 2 (16.7%) | ||
| Smoking | Yes | 6 (37.5%) | 2 (16.7%) | NS |
| No | 10 (62.5%) | 10 (83.3%) | ||
| Lines of 1st-gen TKI treatment | 1st line | 10 (62.5%) | 7 (58.3%) | NS |
| 2nd line | 6 (37.5%) | 5 (41.7%) | ||
| 1st-gen TKI | Gefitinib | 7 (43.75%) | 4 (33.3%) | NS |
| Icotinib | 8 (50%) | 8 (66.7%) | ||
| Erlotinib | 1 (6.26%) | 0 | ||
| Metastasis status | No | 1 (6.3%) | 1 (8.3%) | NS |
| Yes | 15 (93.7%) | 11 (91.7%) | ||
| EGFR mutations | 19del | 7 (43.8%) | 6 (50%) | NS |
| L858R | 9 (56.2%) | 6 (50%) |
NS, not significant.
Patients were divided into two groups: the short-term responders (SR, n = 16) with a progression-free survival (PFS) of less than 6 months on 1st-gen TKI (median: 3 months, range: 1-6 months) and the long-term responders (LR, n = 12) with extremely long PFS of over 24 months (median: 36.5 months, range: 24-60 months) (Supplementary Table 1).
Demographic and clinical characteristics were compared between the SR and LR groups and revealed that SR patients (median: 52 years old, range 34-71) were significantly younger than LR patients (median: 66 years old, range 53-75) (P = .001) (Table 1). Other features, such as sex, smoking history, the treatment lines of TKI, the administrated TKI, metastasis, and EGFR mutation status, were not significantly different between the two groups.
High Prevalence of TP53 Alterations and EGFR Amplification in the SR Group Prior to TKI Treatment
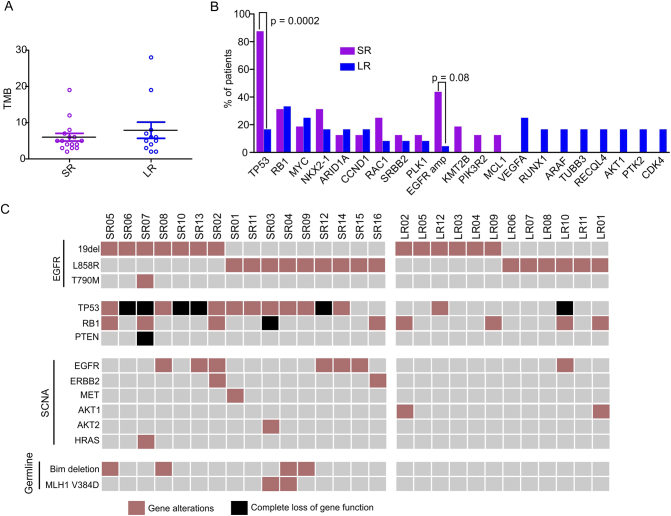
To explore the innate genomic alterations that might compromise 1st-gen TKI treatments, the tumor genomic backgrounds of all patients prior to TKI treatment were profiled using targeted next-generation sequencing (NGS) covering >400 cancer-related genes. The prevalence of each mutation is displayed in Supplementary Figure 1. Despite the substantial difference in PFS between the two groups, the tumor mutation burden (TMB) in the pretreatment samples of the SR and LR patients were not significantly different (Figure 1A, P = .67). However, TP53 was mutated in 87.5% (14/16) of SR patients, which was significantly higher than 16.7% (2/12) in LR patients (Figure 1B) (P = .0002). The TP53 mutations in SR patients were dispersed throughout exons 4-8, 10, and 11, with exon 5 mutations accounting for 33%. Mutations in LR patients were in exons 6-8 only (Supplementary Table 2).
Figure 1.
Genetic profiling of tumor samples from SR and LR groups. (A) Tumor mutation burden of each patient in SR and LR groups. Each circle represents one patient. The Mann-Whitney test was used to assess statistical differences between the two groups. (B) Comparison of the most frequently mutated genes in SR and LR groups. The chi-squared test was used to compare the proportions of each group that contained the mutations listed. (C) Mutation plot of potential resistance mechanisms in each patient (each column represents one patient). Complete loss of gene function (black block) was due to the double strikes on two gene alleles.
Complete loss of TP53 function was observed in five SR patients (31.3%) due to the loss-of-function mutation on one allele and LOH of the other wild-type allele. Only one LR patient (8.3%) exhibited such a phenomenon (Figure 1C). In addition, EGFR copy number gain was more common in SR patients (6/16, 37.5%) than in LR patients (1/12, 8%) (P = .08, Figure 1, B and C). Allele-specific amplification of EGFR was analyzed by FACETS [15], which confirmed that seven EGFR-amplified samples (6 SR and 1 LR patients) exhibited imbalanced amplification of the mutated EGFR allele (Supplementary Figure 2). As a result, the MAFs of the EGFR L858R or 19del mutations in those samples were all above 50% (Supplementary Figure 2A). One SR patient (#SR07) carried the EGFR T790 M mutation before TKI treatment, which caused intrinsic resistance to icotinib (Figure 1C).
Other Potential Factors Associated with the Short-Term Response to TKI Treatment
We summarized all known and putative factors that could shorten the response to TKIs, including the functional loss of TP53 and RB1, the EGFR T790 M mutation, alternative pathway activation via other tyrosine kinase receptors (e.g., ERBB2, MET, FLT4), downstream factors (e.g., PI3K/AKT and MEK/ERK), and germline variations (e.g., BIM deletion polymorphism and MLH1 V384D) [12], [17]. It was observed that 93.8% (15/16) of SR patients harbored at least one intrinsic factor that could decrease TKI efficacy, while 75% (12/16) of patients had more than one factor (Figure 1C). One patient (#SR15) had no resistance factors identified prior to treatment, except for EGFR amplification, but she acquired the EGFR T790 M mutation after 6 months of gefitinib treatment. Two other patients also acquired the EGFR T790 M mutation (patient #SR13, SR14), while one patient obtained EGFR amplification (#SR01) after resistance developed (Supplementary Table 3).
Aside from TP53 and EGFR variations, other recurrent resistance factors in SR patients included RB1 loss (31.3%, n = 5), germline BIM deletion (25%, n = 4), and the MLH V384D variant (12.5%, n = 2). Other genes that recurrently mutated in SR patients but not LR patients included KMT2B (20%, n = 3), PIK3R2 (13%, n = 2), and MCL1 (13%, n = 2) (Figure 1B) mutations, but the significance of those mutations requires further investigation in a larger patient cohort. Somatic copy number alterations (SCNA) were also more common in the SR group than in LR patients. In addition to EGFR amplification, five SR patients harbored SCNA for ERBB2, MET, AKT2, or HRAS (Figure 1C).
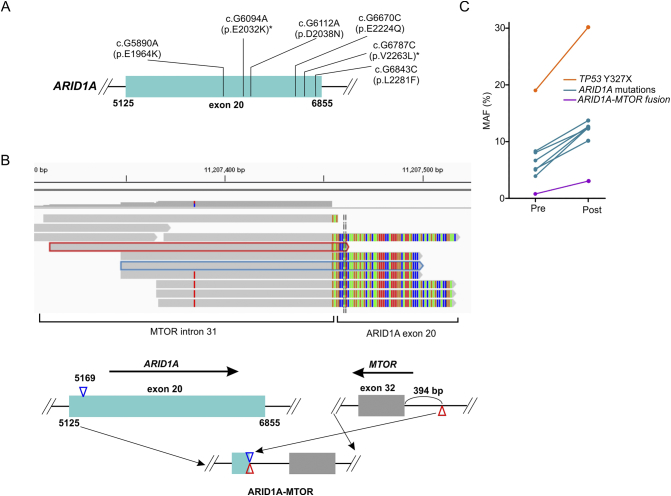
The mutation status of epigenetic regulators was also assessed since their abnormal activities are putative resistance mechanisms to TKIs [18]. Five SR patients and two LR patients had at least one genetic alteration in genes related to epigenetic regulation [19] (Table 2). In patient SR01, six ARID1A missense mutations were identified, as well as a fusion between ARID1A exon 20 and MTOR intron 31 that removed the glucocorticoid receptor–binding domain of ARID1A and impaired the chromatin-remodeling function of the SWI/SNF complex [20] (Figure 2, A and B). Following 3 months of gefitinib treatment, resequencing of the resistant tumor revealed that all ARID1A mutations were relatively enriched (the calculation method is described in Methods) during the course of treatment (Table 2, Figure 2C), suggesting that the functional loss of ARID1A might be associated with the TKI resistance. Moreover, patient #SR04 experienced TET2 V1064 fs and DAXX P540fs enrichment following disease progression, implicating those factors in TKI resistance as well (Supplementary Table 3).
Table 2.
Alterations in Epigenetic Modifiers Identified in Pre/Posttreatment Samples
| Patient ID | Gene | Alteration | Pretreatment MAF | Posttreatment MAF Fold Change | Alteration Type |
|---|---|---|---|---|---|
| SR01 | ARID1A | p.E2032K (c.G6094A) | 6.68% | 1.19 | Missense |
| ARID1A | p.E2224Q (c.G6670C) | 8.33% | 1.04 | Missense | |
| ARID1A | p.V2263 L (c.G6787C) | 5.04% | 1.56 | Missense | |
| ARID1A | p.D2038N (c.G6112A) | 8.09% | 0.96 | Missense | |
| ARID1A | p.E1964K (c.G5890A) | 5.20% | 1.23 | Missense | |
| ARID1A | p.L2281F (c.G6843C) | 3.91% | 2.01 | Missense | |
| ARID1A | ARID1A-MTOR | 1.00% | 2.08 | Fusion | |
| SR02 | KMT2B | p.L2335I (c.C7003A) | 44.40% | 0.72 | Missense |
| KMT2B | p.S2135X (c.C6404G) | 40.06% | 0.72 | Stop gained | |
| SR03 | KMT2B | KMT2B:exon2-ZNF254&LINC00662 | 5.63% | 0.28 | Fusion |
| SR04 | TET2 | p.V1064 fs (c.3191_3197delTTTTGAC) | 4.31% | 1.19 | Frameshift |
| DAXX | p.P540fs (c.1618_1637delCCCTCCAGCATAGATGCTGA) | 2.43% | 1.47 | Frameshift | |
| DAXX | p.S564F (c.C1691T) | 4.88% | 1.22 | Missense | |
| ARID1A | p.Q1519X (c.C4555T) | 13.25% | 0.77 | Stop gained | |
| SETD2 | p.E2528K (c.G7582A) | 7.37% | 0.69 | Missense | |
| SR08 | EP300 | p.M2278 V (c.A6832G) | 5.59% | N/A | Missense |
| KMT2B | p.G2286 V (c.G6857 T) | 7.09% | N/A | Missense | |
| LR08 | BRD4 | p.D650N (c.G1948A) | 6.63% | N/A | Missense |
| SETD2 | p.L483R (c.T1448G) | 22.01% | N/A | Missense | |
| KMT2A | c.G503-1A | 7.11% | N/A | Splicing variant | |
| CREBBP | p.L151 V (c.C451G) | 2.53% | N/A | Missense | |
| ARID1A | p.Y311N (c.T931A) | 5.24% | N/A | Missense | |
| LR09 | ARID1A | p.312_322del (c.936_965delCGGGGGCGACTACAGTGGCGGGCCCCAGGA) | 4.72% | N/A | In frame deletion |
Figure 2.
Gene alterations of ARID1A in patient #SR01. (A) A schematic map indicating multiple missense mutations identified in exon20 of the ARID1A gene in patient #SR01. Asterisks (*) indicate mutations present in the COSMIC database. (B) An integrative genomic viewer (IGV) visualization of the sequencing reads of the ARID1A-MTOR fusion gene (top panel). The gray portion indicates sequencing reads of MTOR intron 31, while the colored portion represents the sequencing reads of ARID1A exon 20. The schematic map (bottom panel) shows the structure of the ARID1A-MTOR fusion locus. Exons 1-20 of ARID1A (light green) were fused to intron 31 of MTOR (gray). Blue and red triangles indicate the broken positions of two genes. (C) Mutation allele frequency changes for genes before and after TKI treatment. Pre, pretreatment; Post, posttreatment.
Discussion
Previous reports indicated that 50%-60% of advanced EGFR-mutant lung cancers harbor TP53 mutations [10], [21], [22]. The concomitant loss of heterozygosity of TP53 also frequently occurs, resulting in a gain of function (GOF) of TP53 and more aggressive tumorigenesis [23], [24]. Among SR patients in this study, 88% (14 out of 16) carried loss of function alterations in TP53, with five having concurrent LOH. The LOH frequency might be underestimated in targeted NGS since the SNPs in a chromosomal segment that can be used by FACETS for heterozygosity analyses are limited. Therefore, we suspect that the TP53 GOF might be a more common phenomenon in advanced EGFR-sensitizing NSCLC patients that demonstrate poor responses to TKI treatments.
On the other hand, whether EGFR amplification can confer TKI resistance remains unknown. Shan et al. found that the concurrence of EGFR amplification and TKI-sensitive EGFR mutations in lung cancer patients was correlated with a longer PFS of TKI treatment [25]. However, another study found that EGFR amplification was more prevalent in patients with innate resistance to the third-generation TKI rociletinib than patients with acquired resistance, suggesting that EGFR amplification might be a resistance mechanism to TKIs. A third study by Shigenari stated that amplification of EGFR wild-type alleles conveyed resistance to EGFR TKIs [26]; however, in our study, allele-specific analysis of EGFR in six patients (6 SR and 1 LR patients) found uneven amplification of the mutant allele rather than the wild-type allele. Thus, it is more likely that EGFR amplification—particularly on the mutant allele—is a resistance mechanism in EGFR-sensitizing TKI treatments.
The resistance-conveying role of epigenetic modifiers in treatment is garnering attention due to their dynamic features compared to genetic heterogeneity, as well as the potential for reversing resistance or extending the treatment benefits by targeting such modifiers [18]. ARID1A is one of the most frequently mutated genes in human cancers and is present in 7% of lung adenocarcinomas [27]. Previous studies observed that loss of ARID1A was correlated with resistance to the ERBB2-targeting antibody trastuzumab by activating the AKT pathway [28]. In this study, we observed the outbreak of multiple missense mutations and one function-impairing fusion in ARID1A in patient #SR01. All mutation clones were enriched postprogression on gefitinib, indicating a possible correlation to resistance.
In patient #SR8, a frame-shift mutation of the DAXX gene, which encodes a chromatin-remodeling factor that regulates gene transcription, was also dramatically enriched following treatment. It has been suggested that the Daxx protein is involved in the suppression of the EMT of tumor cells and its functional loss facilitates tumor invasion [29]. Since EMT is another resistance mechanism in TKI treatment, the functional damage of Daxx might also accelerate resistance.
By investigating the genetic profiles of two groups of patients with different response periods to 1st-gen TKI treatments (SR vs LR), we observed a significant accumulation of TP53 alterations and more frequent EGFR amplifications in patients with short-term responses (SR), thus indicating their possible roles in delivering the poor outcomes. Additionally, mutations in epigenetic modifiers such as ARID1A and DAXX were more common in short-term responders, and their enrichment in the resistance samples suggests their possible involvement in TKI resistance. The validation of those findings in a large cohort will be useful in identifying molecular biomarkers for patient selection prior to TKI treatment.
The following are the supplementary data related to this article.
Mutation plot of SR and LR patients. Only genes that have at least one mutation in all patients are shown. Each column represents a patient with patient IDs shown at the bottom. The percentage for each row indicates the mutation frequency for each gene. The bar graph at the top represents the mutation counts for each patient.
FACETS analysis of copy number changes and allele imbalances. (A) The embedded table lists the MAF of EGFR in pretreatment samples. (B) Allele-specific copy number analysis of six samples that had imbalanced EGFR amplification using FACETS. For each sample, the top panel shows the total copy number log-ratio of different chromosomal segments. The middle panel shows the allele-specific log-odds-ratio, which indicates the copy number differences between two alleles of the EGFR segment. The blue arrows indicate the copy number increases of EGFR in chromosome 7, while the green arrows indicate the allele imbalance of two alleles. The third panel displays the integer copy number calls (black line: total; red line: minor). The color bar at the bottom indicates the predicted tumor cellular fraction.
FACETS analysis of copy number changes and allele imbalances in patient #SR07. The tumor genome reveals triploidy, and the TP53 gene on chromosome 17 demonstrates the loss of heterozygosity (black arrow).
Demographic and Clinical Characteristics of All Enrolled Patients
TP53 Mutations in SR and LR Patients
Acquired Mutations Posttreatment
Footnotes
Conflict of Interest: Y. W. Shao, X. Wu, and X. Tong are shareholders or employees of Geneseeq Technology Inc. J. Yan is an employee of Nanjing Geneseeq Technology Inc.
This work was supported by Natural Scientific Foundation of Zhejiang Province (LGF18H160017). The funders had no role in data collection, decision to publish, or preparation of the manuscript.
References
- 1.Midha A, Dearden S, McCormack R. EGFR mutation incidence in non–small-cell lung cancer of adenocarcinoma histology: a systematic review and global map by ethnicity (mutMapII) Am J Cancer Res. 2015;5:2892–2911. [PMC free article] [PubMed] [Google Scholar]
- 2.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non–small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 3.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 4.Cabanero M, Sangha R, Sheffield BS, Sukhai M, Pakkal M, Kamel-Reid S, Karsan A, Ionescu D, Juergens RA, Butts C. Management of EGFR-mutated non–small-cell lung cancer: practical implications from a clinical and pathology perspective. Curr Oncol. 2017;24:111–119. doi: 10.3747/co.24.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen KS, Neal JW. First-line treatment of EGFR-mutant non–small-cell lung cancer: the role of erlotinib and other tyrosine kinase inhibitors. Biologics. 2012;6:337–345. doi: 10.2147/BTT.S26558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 7.Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, Palmero R, Garcia-Gomez R, Pallares C, Sanchez JM. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non–small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 8.Song Z, Yu X, Cai J, Shao L, Lin B, He C, Zhang B, Zhang Y. Efficacy of icotinib for advanced non–small cell lung cancer patients with EGFR status identified. Zhongguo Fei Ai Za Zhi. 2013;16:138–143. doi: 10.3779/j.issn.1009-3419.2013.03.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong S, Gao F, Fu S, Wang Y, Fang W, Huang Y, Zhang L. Concomitant genetic alterations with response to treatment and epidermal growth factor receptor tyrosine kinase inhibitors in patients with EGFR-mutant advanced non–small cell lung cancer. JAMA Oncol. 2018;4(5):739–742. doi: 10.1001/jamaoncol.2018.0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu H, Suzawa K, Jordan EJ, Zehir A, Ni A, Kim HR, Kris MG, Hellmann MD, Li BT, Somwar R. Concurrent alterations in EGFR-mutant lung cancers associated with resistance to EGFR kinase inhibitors and characterization of MTOR as a mediator of resistance. Clin Cancer Res. 2018;24(13):3108–3118. doi: 10.1158/1078-0432.CCR-17-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Canale M, Petracci E, Delmonte A, Chiadini E, Dazzi C, Papi M, Capelli L, Casanova C, De Luigi N, Mariotti M. Impact of TP53 mutations on outcome in EGFR-mutated patients treated with first-line tyrosine kinase inhibitors. Clin Cancer Res. 2017;23:2195–2202. doi: 10.1158/1078-0432.CCR-16-0966. [DOI] [PubMed] [Google Scholar]
- 12.Stewart EL, Tan SZ, Liu G, Tsao MS. Known and putative mechanisms of resistance to EGFR targeted therapies in NSCLC patients with EGFR mutations—a review. Transl Lung Cancer Res. 2015;4:67–81. doi: 10.3978/j.issn.2218-6751.2014.11.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.VanderLaan PA, Rangachari D, Mockus SM, Spotlow V, Reddi HV, Malcolm J, Huberman MS, Joseph LJ, Kobayashi SS, Costa DB. Mutations in TP53, PIK3CA, PTEN and other genes in EGFR mutated lung cancers: correlation with clinical outcomes. Lung Cancer. 2017;106:17–21. doi: 10.1016/j.lungcan.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shu Y, Wu X, Tong X, Wang X, Chang Z, Mao Y, Chen X, Sun J, Wang Z, Hong Z. Circulating tumor DNA mutation profiling by targeted next generation sequencing provides guidance for personalized treatments in multiple cancer types. Sci Rep. 2017;7:583–594. doi: 10.1038/s41598-017-00520-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016;44:e131. doi: 10.1093/nar/gkw520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chabon JJ, Simmons AD, Lovejoy AF, Esfahani MS, Newman AM, Haringsma HJ, Kurtz DM, Stehr H, Scherer F, Karlovich CA. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun. 2016;7 doi: 10.1038/ncomms11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiu CH, Ho HL, Doong H, Yeh YC, Chen MY, Chou TY, Tsai CM. MLH1 V384D polymorphism associates with poor response to EGFR tyrosine kinase inhibitors in patients with EGFR L858R-positive lung adenocarcinoma. Oncotarget. 2015;6:8407–8417. doi: 10.18632/oncotarget.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L, Dong X, Ren Y, Luo J, Liu P, Su D, Yang X. Targeting EHMT2 reverses EGFR-TKI resistance in NSCLC by epigenetically regulating the PTEN/AKT signaling pathway. Cell Death Dis. 2018;9:129–142. doi: 10.1038/s41419-017-0120-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfister SX, Ashworth A. Marked for death: targeting epigenetic changes in cancer. Nat Rev Drug Discov. 2017;16:241–263. doi: 10.1038/nrd.2016.256. [DOI] [PubMed] [Google Scholar]
- 20.Muratcioglu S, Presman DM, Pooley JR, Grontved L, Hager GL, Nussinov R, Keskin O, Gursoy A. Structural modeling of GR interactions with the SWI/SNF chromatin remodeling complex and C/EBP. Biophys J. 2015;109:1227–1239. doi: 10.1016/j.bpj.2015.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blakely CM, Watkins TBK, Wu W, Gini B, Chabon JJ, McCoach CE, McGranahan N, Wilson GA, Birkbak NJ, Olivas VR. Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat Genet. 2017;49:1693–1704. doi: 10.1038/ng.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Labbe C, Cabanero M, Korpanty GJ, Tomasini P, Doherty MK, Mascaux C, Jao K, Pitcher B, Wang R, Pintilie M. Prognostic and predictive effects of TP53 co-mutation in patients with EGFR-mutated non–small cell lung cancer (NSCLC) Lung Cancer. 2017;111:23–29. doi: 10.1016/j.lungcan.2017.06.014. [DOI] [PubMed] [Google Scholar]
- 23.Hanel W, Marchenko N, Xu S, Yu X, Weng W, Moll U. Two hot spot mutant p53 mouse models display differential gain of function in tumorigenesis. Cell Death Differ. 2013;20:898–909. doi: 10.1038/cdd.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson HH, Wilkojmen M, Marsit CJ, Kelsey KT. TP53 mutation, allelism and survival in non–small cell lung cancer. Carcinogenesis. 2005;26:1770–1773. doi: 10.1093/carcin/bgi125. [DOI] [PubMed] [Google Scholar]
- 25.Shan L, Wang Z, Guo L, Sun H, Qiu T, Ling Y, Li W, Li L, Liu X, Zheng B. Concurrence of EGFR amplification and sensitizing mutations indicate a better survival benefit from EGFR-TKI therapy in lung adenocarcinoma patients. Lung Cancer. 2015;89:337–342. doi: 10.1016/j.lungcan.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 26.Nukaga S, Yasuda H, Tsuchihara K, Hamamoto J, Masuzawa K, Kawada I, Naoki K, Matsumoto S, Mimaki S, Ikemura S. Amplification of EGFR wild-type alleles in non–small cell lung cancer cells confers acquired resistance to mutation-selective EGFR tyrosine kinase inhibitors. Cancer Res. 2017;77:2078–2089. doi: 10.1158/0008-5472.CAN-16-2359. [DOI] [PubMed] [Google Scholar]
- 27.N. Cancer Genome Atlas Research Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berns K, Sonnenblick A, Gennissen A, Brohee S, Hijmans EM, Evers B, Fumagalli D, Desmedt C, Loibl S, Denkert C. Loss of ARID1A activates ANXA1, which serves as a predictive biomarker for trastuzumab resistance. Clin Cancer Res. 2016;22:5238–5248. doi: 10.1158/1078-0432.CCR-15-2996. [DOI] [PubMed] [Google Scholar]
- 29.Lin CW, Wang LK, Wang SP, Chang YL, Wu YY, Chen HY, Hsiao TH, Lai WY, Lu HH, Chang YH. Daxx inhibits hypoxia-induced lung cancer cell metastasis by suppressing the HIF-1alpha/HDAC1/Slug axis. Nat Commun. 2016;7 doi: 10.1038/ncomms13867. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mutation plot of SR and LR patients. Only genes that have at least one mutation in all patients are shown. Each column represents a patient with patient IDs shown at the bottom. The percentage for each row indicates the mutation frequency for each gene. The bar graph at the top represents the mutation counts for each patient.
FACETS analysis of copy number changes and allele imbalances. (A) The embedded table lists the MAF of EGFR in pretreatment samples. (B) Allele-specific copy number analysis of six samples that had imbalanced EGFR amplification using FACETS. For each sample, the top panel shows the total copy number log-ratio of different chromosomal segments. The middle panel shows the allele-specific log-odds-ratio, which indicates the copy number differences between two alleles of the EGFR segment. The blue arrows indicate the copy number increases of EGFR in chromosome 7, while the green arrows indicate the allele imbalance of two alleles. The third panel displays the integer copy number calls (black line: total; red line: minor). The color bar at the bottom indicates the predicted tumor cellular fraction.
FACETS analysis of copy number changes and allele imbalances in patient #SR07. The tumor genome reveals triploidy, and the TP53 gene on chromosome 17 demonstrates the loss of heterozygosity (black arrow).
Demographic and Clinical Characteristics of All Enrolled Patients
TP53 Mutations in SR and LR Patients
Acquired Mutations Posttreatment