Abstract
Eucalyptusdimers A−C, three dimeric phellandrene-derived meroterpenoids featuring an unprecedented, fused skeleton between two phellandrene and two acylphloroglucinol subunits, along with one biogenetically related intermediate, (±)-eucalyprobusone A, were isolated from the fruits of Eucalyptus robusta. Their structures and absolute configurations were elucidated using spectroscopic data, X-ray crystallography, and electronic circular dichroism analysis. The isolated meroterpenoids were evaluated for their anti-inflammatory, acetylcholinesterase inhibitory, and protein tyrosine phosphatase 1B inhibitory effects.
The genus Eucalyptus (Myrtaceae family), comprising over 600 species distributed globally in tropical and sub-tropical areas and with 100 species found in South China,1 has afforded a large number of structurally interesting meroterpenoids.2–8 More importantly, these molecules exhibit diverse and significant bioactivities, including antimicrobial,2 anti-cancer,5 HGF/c-Met axis inhibitory,6 and PTP1B (protein tyrosine phosphatase 1B) inhibitory7 effects. The leaves of Eucalyptus robusta are generally used in Chinese folk medicine to treat dysentery, malaria, and other bacterial diseases.8 Our previous chemical studies on the plants of Psidium guajava and Callistemon salignus led to the isolation of a series of phloroglucinol derivatives with antibacterial and antitumor activities.9 As part of our continued effort to search for bioactive phloroglucinol derivatives, an investigation on the fruits of E. robusta resulted in the isolation of three phellandrene−phloroglucinol meroterpenoids (Figure 1, 1−3) and one biogenetically related intermediate, eucalyprobusone A (4). Their anti-inflammatory, AChE (acetylcholinesterase) inhibitory, and PTP1B inhibitory effects were also evaluated. Herein, the isolation, structural elucidation, and biological evaluation of these isolates are described in detail.
Figure 1.
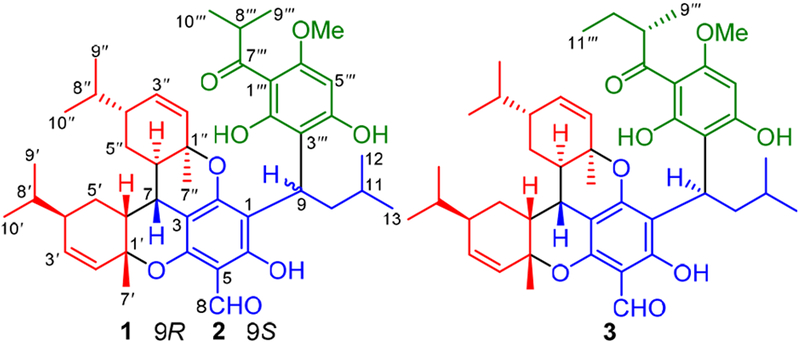
Structures of 1−3.
Eucalyptusdimer A (1) had a molecular formula of C44H58O8 as determined by the HRESIMS ion at m/z 737.4028 [M + Na]+ (calcd for C44H58O8Na, 737.4029), suggesting 16 degrees of unsaturation. The 1H NMR data (Table 1) showed two singlet methyls (δH 1.32 and 1.64), eight doublet methyls (δH 0.87, 0.89, 0.91, 0.93, 0.98, 1.00, 1.14, and1.16), a methoxy group (δH 3.80), five olefinic protons (δH 5.47, 5.58, 5.80, 5.89, and 5.97), an aldehydic proton (δH 9.94), as well as three phenolic hydroxy protons (δH 8.97, 13.82, and 14.69). With the aid of DEPT and HSQC spectra, analysis of the 13C NMR data (Table 1) disclosed 44 carbon resonances, including one ketone carbonyl carbon (δC 210.2), an aldehyde carbonyl carbon (δC 191.7), two benzene rings (six oxygenated carbon resonances), two disubstituted double bonds, two quaternary carbons, 10 methines, and three methylenes. The aforementioned functionalities accounted for 12 of the 16 degrees of unsaturation, thus requiring 1 to be tetracyclic.
Table 1.
1H (500 MHz) and 13C (125 MHz) NMR Data of 1 and 2 in CDCl3
| 1 | 2 | |||
|---|---|---|---|---|
| no. | δc | δH (mult, J (Hz)) | δc | δH (mult, J (Hz)) |
| 1 | 108.1 | 108.1 | ||
| 2 | 154.5 | 154.5 | ||
| 3 | 97.3 | 97.2 | ||
| 4 | 161.3 | 161.3 | ||
| 5 | 104.1 | 104.1 | ||
| 6 | 160.2 | 160.2 | ||
| 7 | 25.8 | 2.90 dd (11.5, 5.4) | 25.8 | 2.97 dd (11.6, 5.4) |
| 8 | 191.7 | 9.94 s | 191.7 | 9.95 s |
| 9 | 26.4 | 5.34 t (8.3) | 26.4 | 5.28 t (8.3) |
| 10a | 40.9 | 2.07 ddd (13.8, 8.3, 6.7) | 41.2 | 2.10 ddd (14.2, 8.3, 6.1) |
| 10b | 1.80 dt (13.8, 6.7) | 1.73 dt (14.2, 6.1) | ||
| 11 | 26.4 | 1.46 m | 26.2 | 1.46 m |
| 12 | 22.5 | 0.87 d (6.6) | 22.4 | 0.90 d (6.8) |
| 13 | 22.9 | 0.93 d (6.6) | 23.1 | 0.92 d (6.8) |
| 1′ | 75.2 | 75.1 | ||
| 2′ | 131.2 | 5.80 d (9.4) | 131.2 | 5.80 d (10.0) |
| 3′ | 134.6 | 5.97 dd (10.0, 4.5) | 134.5 | 5.96 dd (10.0, 4.5) |
| 4′ | 40.6 | 1.98 m | 40.5 | 1.94 m |
| 5′a | 20.2 | 1.62 m | 20.2 | 1.53 brd (13.7) |
| 5′b | 1.38 q (6.9) | 1.30 ddd (20.7, 9.0, 6.9) | ||
| 6′ | 32.0 | 1.86 m | 32.0 | 1.86 m |
| 7′ | 25.6 | 1.32 s | 25.7 | 1.38 s |
| 8′ | 31.6 | 1.67 m | 31.8 | 1.66 m |
| 9′ | 21.3 | 0.98 d (6.8) | 21.3 | 0.98 d (6.7) |
| 10′ | 21.0 | 1.00 d (6.8) | 20.9 | 0.96 d (6.7) |
| 1″ | 79.8 | 80.1 | ||
| 2″ | 132.8 | 5.58 d (10.3) | 132.9 | 5.81 d (10.3) |
| 3″ | 129.5 | 5.47 br d (10.3) | 129.7 | 5.59 br d (10.3) |
| 4″ | 37.5 | 1.95 m | 37.6 | 1.99 m |
| 5″a | 23.3 | 1.85 m | 23.3 | 1.84 m |
| 5″b | 1.69 m | 1.66 m | ||
| 6″ | 35.9 | 1.92 m | 35.8 | 1.83 m |
| 7″ | 29.6 | 1.64 s | 29.0 | 1.48 s |
| 8″ | 31.9 | 1.69 m | 31.6 | 1.68 m |
| 9″ | 19.3 | 0.91 d (6.8) | 19.2 | 0.92 d (6.8) |
| 10″ | 18.7 | 0.89 d (6.8) | 18.7 | 0.90 d (6.8) |
| 1‴ | 104.2 | 104.2 | ||
| 2‴ | 166.1 | 166.1 | ||
| 3‴ | 110.7 | 110.7 | ||
| 4‴ | 162.7 | 162.7 | ||
| 5‴ | 91.8 | 5.89 s | 91.8 | 5.89 s |
| 6‴ | 160.9 | 160.9 | ||
| 7‴ | 210.2 | 210.1 | ||
| 8‴ | 39.3 | 3.79 hept (6.8) | 39.3 | 3.77 hept (6.8) |
| 9‴ | 19.4 | 1.16 d (6.8) | 19.4 | 1.15 d (6.8) |
| 10‴ | 19.5 | 1.14 d (6.8) | 19.5 | 1.10 d (6.8) |
| OH-6 | 13.82 s | 13.76 s | ||
| OH-2‴ | 14.69 s | 14.62 s | ||
| OH-4‴ | 8.97 s | 8.85 s | ||
| OMe-6‴ | 55.2 | 3.80 s | 55.2 | 3.80 s |
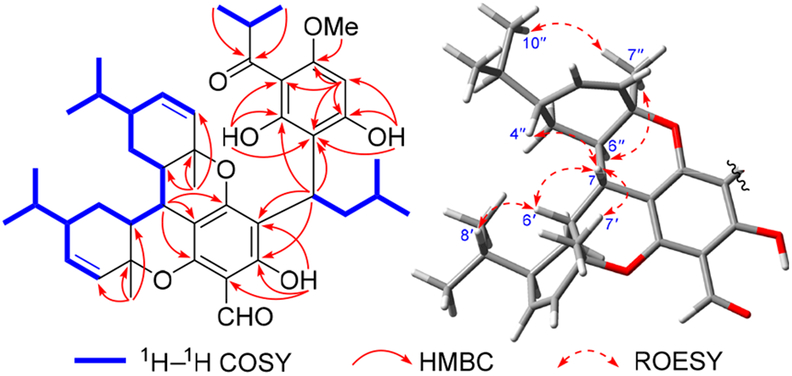
Comparison of 1H and 13C NMR data of 1 with those of phellandrene in the natural product viminadione A10 indicated the presence of two phellandrene moieties, which was further verified by the 1H−1H COSY and HMBC spectra (Figure 2). The 1H−1H COSY data revealed the presence of three spincoupling systems. Furthermore, the HMBC correlations from Me-7′ to C-1′/C-2′/C-6′ and from Me-7″ to C-1″/C-2″/C-6″ revealed the presence of two phellandrene moieties. Likewise, the HMBC correlations from Me-9′″/Me-10′″ to C-7′″, from H-5′″ to C-1′″/C-3′″/C-4′″/C-6′″, from δH 14.69 (OH-2′″) to C-1′″/C-2′″/C-3′″, from δH 3.80 (OMe- 6′″) to C-6′″, and from δH 8.97 (OH-4′″) to C-3′″/C-4′″/C-5′″ allowed the establishment of the acylphloroglucinol unit. Similarly, the HMBC correlations from H-9 to C-1/C-6/C-3′″/C-4′″ and from H-8 to C-4/C-5/C-6 not only indicated the existence of an isopentylacylphloroglucinal moiety but also proved that the isobutyrylphloroglucinal unit was connected to the former via a C-1−C-9−C-3′″ bond. Finally, two phellandrenes were linked via C-7 of the isopentylacylphlor oglucinal to furnish two additional dihydrobenzopyran rings11 by the key HMBC correlations from H-7 to C-2/C-3/C-4/C-6′/C-6″, which also satisfied the requirements for the remaining two degrees of unsaturation and MS information. The planar structure of 1 was thus defined as an unprecedented, dimeric phellandrene-derived meroterpenoid.
Figure 2.
Key 2D NMR correlations of 1 and 2.
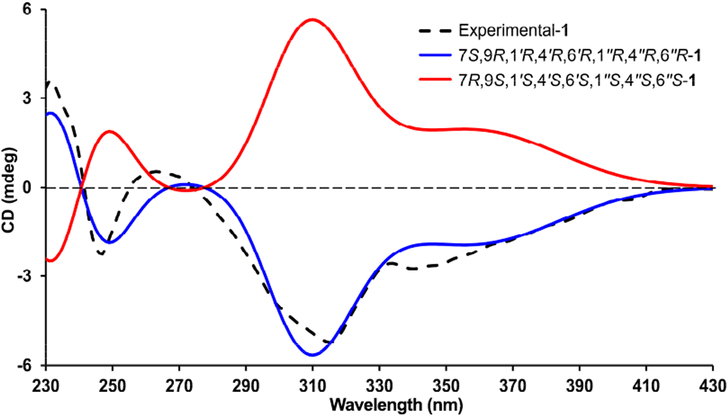
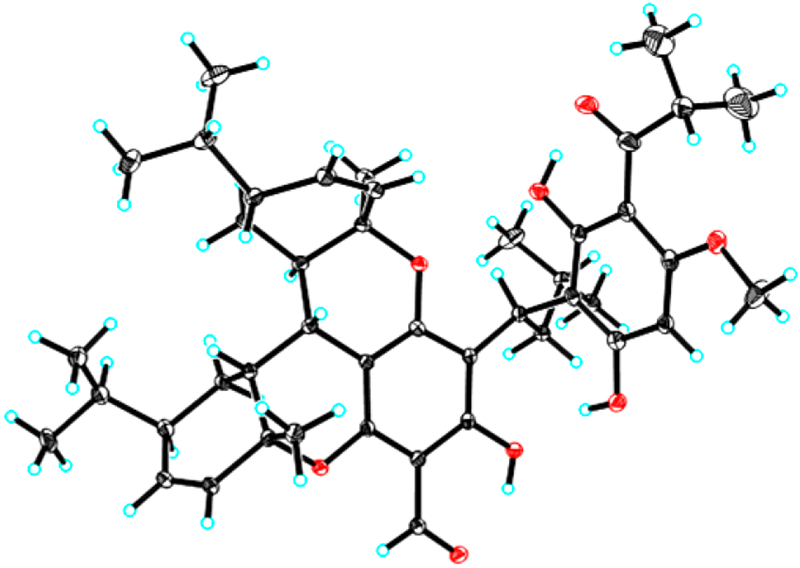
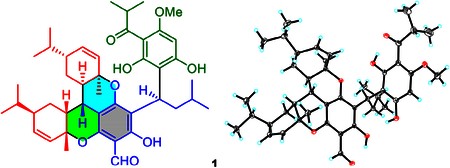
The partial relative configuration of 1 was determined by detailed interpretation of the ROESY NMR spectrum (Figure 2). The ROESY correlations of H-7 with H-6′/Me-7′/H-4″ and of H-6′ with H-8′ indicated that these protons were on the same side and were randomly assigned to be β-oriented. The ROESY correlations of Me-7″ with Me-10′′/H-6″ thus suggested that these protons were α-oriented. The configuration of C-9 was left unassigned due to the lack of the reliable NOESY correlations. The absolute configuration of 1 was determined to be 7S,9R,1′R,4′R,6′R,1″R,4″R,6″R by electronic circular dichroism (ECD) calculations (Figure 3). Fortunately, quality crystals of 1 were obtained, and further X-ray crystallography (Figure 4) unambiguously confirmed the proposed structure and also determined its absolute stereo-chemistry.
Figure 3.
Experimental and calculated ECD spectra of 1.
Figure 4.
X-ray crystallographic structure of 1.
Eucalyptusdimer B (2) displayed the same molecular formula of C44H58O8 as 1 based on the HRESIMS data. Apart from their similar 1H and 13C NMR data (Table 1), both the planar structure and relative configuration (except C-9) of 2 were identical to those of 1 (Figure 2). However, the absolute configuration of 2 was ambiguous. In order to confirm the absolute configuration of 2, ECD calculations were conducted. The calculated ECD curve of 7S,9S,1′R,4′R,6′R,1″R,4″R,6″R was in good agreement with the experimental spectrum obtained for 2 (Figure S1), which confirmed its absolute configuration.
Eucalyptusdimer C (3) gave a molecular formula of C45H60O8, as deduced from its HRESIMS data, which was 14 mass units more than those of 1 and 2. Comparison of its 1D NMR data (Table 2) with those of 1 indicated the replacement of an isobutyryl group at C-1′″ in 1 by a sec isovaleryl group in 3, as supported by HMBC correlations from Me-9′″ to C-7′″/C-8′″/C-10′″ and from Me-11′″ to C-8′″ and C-10′″. The configuration of C-8′ was assigned as S based on a related acylphloroglucinal derivative (callisalignone A)9b with an S absolute configuration that was previously obtained from Callistemon salignus. Finally, the absolute configuration of 7S,9R,1′R,4′R,6′R,1″R,4″R,6″R,8′″S for 3 was established by comparison of its experimental ECD spectrum with calculated ECD curves for two epimers of 7S,9R,1′R,4′R,6′R,1″R,4″R,−6″R,8′″S and 7S,9R,1′’R,4′R,6′R,1″R,4″R,6″R,8′″R (Figure S1).
Table 2.
1H (600 MHz) and 13C (155 MHz) NMR Data of 3 in CDCl3
| no. | δc | δH (mult, J (Hz)) | no. | δc | δH (mult, J (Hz)) |
|---|---|---|---|---|---|
| 1 | 108.1 | 1″ | 79.8 | ||
| 2 | 154.6 | 2″ | 132.9 | 5.54 d (10.2) | |
| 3 | 97.3 | 3″ | 129.4 | 5.44 br d (10.2) | |
| 4 | 161.3 | 4″ | 37.5 | 1.95 m | |
| 5 | 104.1 | 5″a | 23.3 | 1.85 m | |
| 6 | 160.2 | 5″b | 1.66 br dd (12.8, 6.2) | ||
| 7 | 25.8 | 2.90 dd (11.5, 5.4) | 6″ | 35.9 | 1.91 m |
| 8 | 191.7 | 9.95 s | 7″ | 29.6 | 1.63 s |
| 9 | 26.4 | 5.34 t (8.2) | 8″ | 31.9 | 1.67 m |
| 10a | 40.9 | 2.07 m | 9″ | 19.4 | 0.90 d (6.8) |
| 10b | 1.79 m | 10″ | 18.3 | 0.89 d (6.8) | |
| 11 | 26.4 | 1.47 m | 1‴ | 104.8 | |
| 12 | 22.6 | 0.87 d (6.6) | 2‴ | 166.1 | |
| 13 | 22.9 | 0.93 d (6.6) | 3‴ | 110.7 | |
| 1′ | 75.2 | 4‴ | 162.7 | ||
| 2′ | 131.2 | 5.80 d (10.2) | 5‴ | 91.8 | 5.88 s |
| 3′ | 134.6 | 5.97 dd (10.2, 4.5) | 6‴ | 160.9 | |
| 4′ | 40.6 | 1.98 m | 7‴ | 210.0 | |
| 5′a | 20.3 | 1.61 m | 8‴ | 46.0 | 3.64 sext (6.6) |
| 5′b | 1.38 ddd (20.9, 14.8, 6.6) | 9‴ | 16.7 | 1.16 d (6.7) | |
| 6′ | 32.0 | 1.85 m | 10‴a | 27.3 | 1.82 m |
| 7′ | 25.6 | 1.32 s | 10‴b | 1.40 m | |
| 8′ | 31.6 | 1.66 m | 11‴ | 12.0 | 0.86 t (7.2) |
| 9′ | 21.3 | 0.98 d (6.8) | 2‴–OH | 14.73 s | |
| 10′ | 21.0 | 1.00 d (6.8) | 4‴-OH | 8.97 s | |
| 6-OH | 13.82 s | 6‴-OMe | 55.3 | 3.80 s |
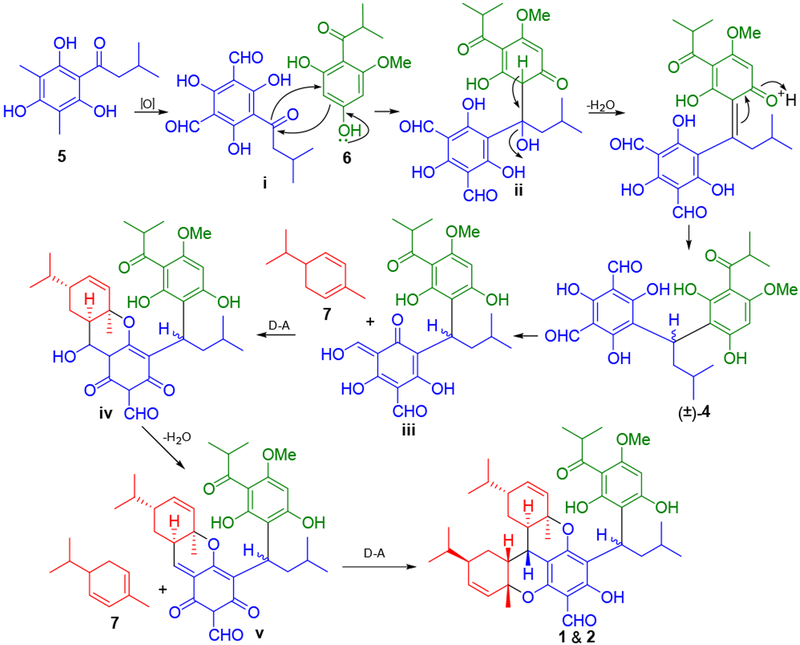
Additionally, one biogenetically related intermediate of 1−3, eucalyprobusone A (Scheme 1, 4), was isolated from the title species. Coincidentally, 4 was found to be a racemic mixture as revealed by a chiral HPLC analysis; subsequent chiral separation and ECD calculations established absolute configurations as 9S for (+)-4 and 9R for (−)-4 (Supporting Information).
Scheme 1.
Hypothetical Biosynthetic Pathways to 1, 2, and 4
Biogenetically, eucalyptusdimers A−C (1−3) are the first meroterpenoid dimers constructed by two phellandrene and two acylphloroglucinol subunits. Their plausible biogenetic pathways are proposed as shown in Scheme 1, which includes 3,5-dimethyl-2,4,6,-trihydroxyisovalerophenone phloroglucinol (5), isobutyrylphloroglucinol (6), and α-phellandrene (7). Initially, phloroglucinol 5 may undergo oxidation to generate i, which can then be attacked by nucleophilic phloroglucinol 6 to produce ii. Thereafter, intermediate ii could be readily transformed into (±)-4 and its dearomatized tautomer iii, followed by a hetero-Diels−Alder reaction with 7 to form adduct iv.12 Subsequent loss of H2O from iv can afford o-quinone methide (o-QM) v, which may undergo a second hetero-Diels−Alder cycloaddition with 7 to afford 1 and 2. A similar pathway may be envisioned to afford 3.
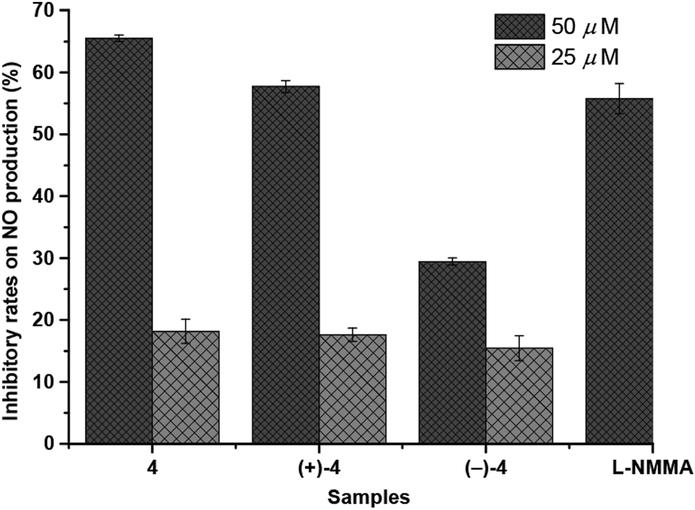
Compounds 1−4 were evaluated for their anti-inflammatory, AChE (acetylcholinesterase), and PTP1B (protein tyrosine phosphatase-1B) inhibitory activities. Under the concentration of 50 μM, only (±)-4 and (+)-4 showed remarkable anti-inflammatory inhibition rates of 65.51 and 57.72%, respectively (Figure 5). Moreover, 1, 4, (+)-4, and (−)-4 were found to exhibit AChE inhibitory activities with IC50 values of 17.71, 13.61, 18.55, and 22.61 μM, respectively. None of them displayed significant PTP1B inhibitory activity at 50 μM.
Figure 5.
Inhibitory effects on NO production of (±)-4.
In conclusion, eucalyptus dimers A−C (1−3) obtained from the fruits of E. robusta have been found to have an unprecedented skeleton fused with two phellandrene and two phloroglucinol moieties. These isolates could enrich the chemodiversity of phloroglucinol−terpenoid derivatives. Bio-genetic cycloaddition of two α-phellandrene units with a 3,5-diformylphloroglucinol and a isobutyrylphloroglucinol scaffold has been proposed to construct the two dihydrobenzopyran rings. Accordingly, 1, 4, (+)-4, and (−)-4 which possess AChE inhibitory properties are excellent scaffolds for further chemical synthesis and pharmacological investigations, which are currently in progress.
Supplementary Material
ACKNOWLEDGMENTS
This research work was funded by the National Natural Science Foundation of China (31570363, 31600283, and 31770391), National Institutes of Health (R35 GM-118173), Major Bio-Medical Project of Yunnan Province (2017AB001 and 2018ZF005), Natural Science Foundation of Yunnan Province (2017FB128), Key Research and Development Plan of Yunnan Province−Special Project of Science and Technology in Yunnan Province (2017IB007), Innovation Team of the Ministry of Education (IRT-17R49), and Foundation of State Key Laboratory of Phytochemistry and Plant Resources in West China (P2017-ZZ04), Kunming Institute of Botany, Chinese Academy of Sciences.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.orglett.8b02259.
Detailed experimental procedures and spectroscopic data (NMR, MS, and CD) for 1−4 and structure elucidation of 4 (PDF)
Accession Codes
CCDC 1853638 contains the supplementary crystallographic data for this paper. These data can be obtained free of charge via www.ccdc.cam.ac.uk/data_request/cif, or by e-mailing data_request@ccdc.cam.ac.uk, or by contacting The Cambridge Crystallographic Data Centre, 12 Union Road, Cambridge CB2 1EZ, U.K.; fax: + 44 1223 36033.
The authors declare no competing financial interest.
REFERENCES
- (1).Chen J; Craven LA In Flora of China; Science Press: Beijing, 2007; Vol. 13, pp 321–328. [Google Scholar]
- (2) (a).Osawa K; Yasuda H J. Nat. Prod. 1996, 59, 823–827. [DOI] [PubMed] [Google Scholar]; (b) Tian LW; Xu M; Li XC; Yang CR; Zhu HJ; Zhang YJ RSC Adv. 2014, 4, 21373–21378. [Google Scholar]; (c) Shang ZC; Yang MH; Liu RH; Wang XB; Kong LY Sci. Rep 2016, 6, 39815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Singh IP; Umehara K; Asai T; Etoh H; Takasaki M; Konoshima T Phytochemistry 1998, 47, 1157–1159. [DOI] [PubMed] [Google Scholar]
- (4).Pal Singh I; Bharate SB Nat. Prod. Rep 2006, 23, 558–591. [DOI] [PubMed] [Google Scholar]
- (5) (a).Yin S; Xue JJ; Fan CQ; Miao ZH; Ding J; Yue JM Org. Lett 2007, 9, 5549–5552. [DOI] [PubMed] [Google Scholar]; (b) Peng LY; He J; Xu G; Wu XD; Dong LB; Gao X; Cheng X; Su J; Li Y; Zhao QS Nat. Prod. Bioprospect 2011, 1, 101–103. [Google Scholar]; (c) Shang ZC; Yang MH; Jian KL; Wang XB; Kong LY Chem. - Eur. J 2016, 22, 11778– 11784. [DOI] [PubMed] [Google Scholar]
- (6).Yang SP; Zhang XW; Ai J; Gan LS; Xu JB; Wang Y; Su ZS; Wang L; Ding J; Geng MY; Yue JM J. Med. Chem 2012, 55, 8183–8187. [DOI] [PubMed] [Google Scholar]
- (7).Yu Y; Gan LS; Yang SP; Sheng L; Liu QF; Chen SN; Li J; Yue JM J. Nat. Prod 2016, 79, 1365–1372. [DOI] [PubMed] [Google Scholar]
- (8).Qin GW; Chen ZX; Wang HC; Qian MK Acta Chim.Sin 1981, 39, 83–89. [Google Scholar]
- (9) (a).Qin XJ; Yu Q; Yan H; Khan A; Feng MY; Li PP; Hao XJ; An LK; Liu HY J. Agric. Food Chem. 2017, 65, 4993– 4999. [DOI] [PubMed] [Google Scholar]; (b) Qin XJ; Liu H; Yu Q; Yan H; Tang JF; An LK; Khan A; Chen QR; Hao XJ; Liu HY Tetrahedron 2017, 73, 1803–1811. [Google Scholar]; (c) Qin XJ; Shu T; Yu Q; Yan H; Ni W; An LK; Li PP; Zhi YE; Khan A; Liu HY Nat. Prod. Bioprospect 2017, 7, 315–321. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Qin XJ; Yan H; Ni W; Yu MY; Khan A; Liu H; Zhang HX; He L; Hao XJ; Di YT; Liu HY Sci. Rep 2016, 6, 32748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Khambay BPS; Beddie DG; Hooper AM; Simmonds MSJ; Green PW C. J. Nat. Prod. 1999, 62, 1666–1667. [DOI] [PubMed] [Google Scholar]
- (11).Li CJ; Ma J; Sun H; Zhang D; Zhang DM Org. Lett. 2016, 18, 168–171. [DOI] [PubMed] [Google Scholar]
- (12) (a).Lawrence AL; Adlington RM; Baldwin JE; Lee V; Kershaw JA; Thompson AL Org. Lett 2010, 12, 1676–1679. [DOI] [PubMed] [Google Scholar]; (b) Guo Y; Zhang Y; Xiao M; Xie Z Org. Lett 2018, 20, 2509– 2512. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.