Abstract
Reaction of [NBu4][LCuIIOH] with excess ROOH (R = cumyl or tBu) yielded [NBu4][LCuIIOOR], the reversible one-electron oxidation of which generated novel species with [CuOOR]2+ cores (formally CuIIIOOR), identified by spectroscopy and theory for the case R = cumyl. This species reacts with weak O−H bonds in TEMPO-H and 4-dimethylaminophenol (NMe2PhOH), the latter yielding LCu(OPhNMe2), which was also prepared independently. With the identification of [CuOOR]2+ complexes, the first precedent for this core in enzymes is provided, with implications for copper monooxygenase mechanisms.
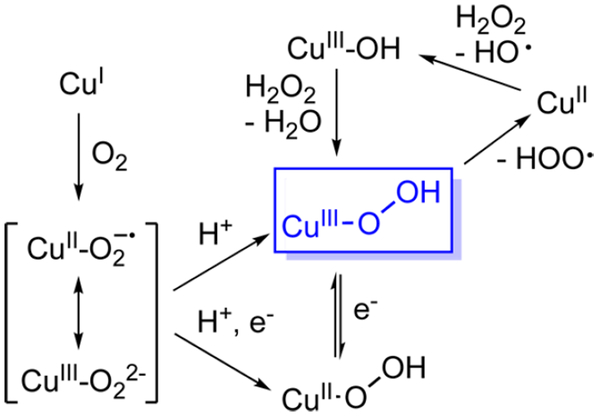
Understanding the mechanism(s) of monooxygenase reactions promoted by monocopper sites in enzymes1 and other catalysts2 is an important research goal. A generalized mechanistic scheme that highlights several key oxidants and processes under active discussion for many of these systems is shown in Figure 1. The first step in monooxygenases is reaction of the CuI form with O2 to yield a 1:1 Cu/O2 adduct typically formulated as a CuII-superoxide,1,3−5 but which in some model compounds can have CuIII-peroxide character.6 This intermediate may undergo proton-coupled electron transfer (PCET or “H+/e−”) via attack at a substrate C−H bond or delivery of a proton and an electron in another way to yield a CuII−OOH species (also written as [CuOOH]+), which may also attack the substrate or undergo PCET. Alternatively, protonation of the CuII-superoxide to yield a [CuOOH]2+ species (formally CuIIIOOH) that then attacks the substrate has been suggested on the basis of theory for peptidylglycine α-hydroxylating monooxygenase.7 A [CuOOH]2+ species also is implicated in a proposed mechanism for catalytic alkane oxidations, where reaction of a putative [CuIII−OH]2+ species with H2O2 to yield a hydroperoxyl radical and CuII presumably involves initial formation of [CuOOH]2+.8
Figure 1.
Generalized mechanistic scheme for O2 activation by monocopper sites with formal oxidation states indicated.
The characterization and study of the reactivity of discrete complexes comprising these and other copper−oxygen intermediates is an important strategy for determining their viability in catalytic systems.9 Examples of 1:1 Cu/O2 adducts,6,10,11 [CuOH]2+,12 and [CuOOR]+ (R = alkyl) complexes9a,13 have been characterized, but to our knowledge there are no reports of compounds with a [CuOOR]2+ unit.14 Herein, we report the isolation of two novel distorted square planar [CuIIOOR]+ (R = cumyl, tBu) complexes and their reversible 1-electron oxidation at low temperature to yield reactive species that we formulate as [CuOOR]2+complexes on the basis of spectroscopy and supporting theory. With this work, we experimentally demonstrate the feasibility of the [CuOOH]2+ unit, enabling new mechanistic insights for copper monooxygenases and other catalytic systems.
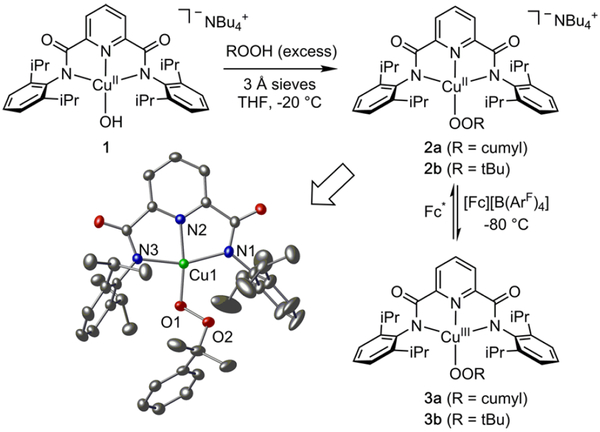
Reaction of blue solutions of [NBu4][LCuIIOH] ([NBu4]-[1], Figure 2)12 in THF with excess ROOH (R = cumyl, 80% v/v in cumene, or R = tBu, 87% in nonane) at −20 °C resulted in a color change to crimson. The complexes [NBu4]-[LCuIIOOR] ([NBu4][2a] and [NBu4][2b]) were isolated from these solutions in 68% (R = cumyl) or 81% (R = tBu) yields and characterized by UV−vis and EPR spectroscopy, and high resolution electrospray ionization mass spectrometry (Figure S2). In addition, we obtained X-ray crystal structures of congeners prepared using NEt4+ as a counterion ([NEt4]-[2a], Figure 2; [NEt4][2b], Figure S3). The complexes exhibit slightly rhombically distorted EPR signals typical for CuII complexes of L,12 but with g and A parameters suffciently different from those for [NBu4][1] to clearly indicate replacement of the hydroxide ligand by the alkylperoxides (Figures S4−S5, Table S1). In the X-ray structures, the O−O distances of 1.468(19) Å and 1.416(58) Å are similar to those measured for the only two [CuOOR]+ (R = H, alkyl) complexes reported previously (both of which are coincidentally 1.460(6) Å).13d,e DFT geometry optimizations of [2a]− and [2b]− showed good correlation to the XRD structures (Table S2), and spin density plots (Figure S6) were similar to those reported previously for [NBu4][1].12c Both [NBu4][2a] and [NBu4][2b] were found to be stable for weeks in solution at temperatures of −20 °C or lower but readily decomposed to compound [NBu4][1] at room temperature (UV−vis and EPR spectroscopy; Figures S7−S8).
Figure 2.
Syntheses of complexes and representation of the anionic portion of the X-ray crystal structure of 2a (NEt 4+ counterion), with formal copper oxidation states noted. Selected interatomic distances (Å): Cu1−O1, 1.841(14); Cu1−N1, 2.026(15); Cu1−N2, 1.934(14); Cu1−N3, 2.019(15); O1−O2, 1.468(19).
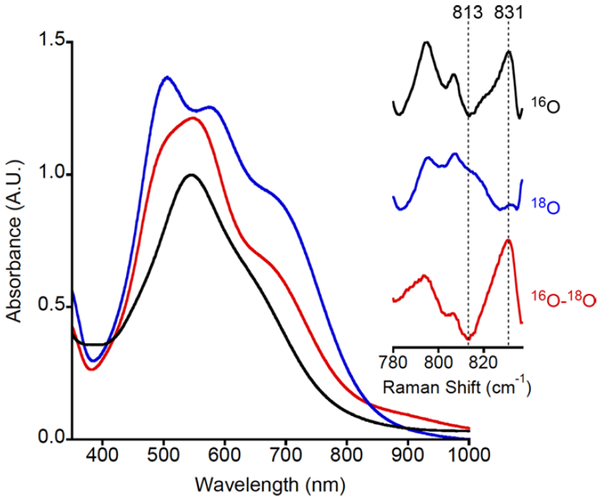
Cyclic voltammetry measurements of [NBu4][2a] and [NBu4][2b] (0.3 M Bu4NPF6 in THF, Figure S9) revealed quasireversible processes (ipa/ipc = 2.9 and 2.3 for 2a and b respectively, scan rate = 500 mV/s) with E1/2 values of −154 mV and −205 mV vs Fc+/Fc, respectively. Chemical oxidations of [NBu4][2a] and [NBu4][2b] in THF at −80 °C using ferrocenium tetrakis[3,5-bis(trifluoromethyl)phenyl]borate ([Fc][B(ArF)4]) resulted in an immediate color change to deep purple associated with intense absorptions in the 500− 700 nm region of UV−vis spectra (Figure 3). The absorptions are similar to those reported previously for LCuIIIOH (black line, Figure 3) and analogs, which were assigned by TD-DFT as ligand aryl π → Cu dx2-y2 transitions (see below).12c The oxidized species has a t1/2 of 4.0 h at −80 °C in THF solution and is stable only for ~5 s at room temperature. Titration experiments with [NBu4][2a] showed that the absorption features generated upon oxidation reached maximum intensity upon addition of 1 equiv of [Fc][B(ArF)4], consistent with a one-electron process (Figure S10). In further experiments with the purple product of oxidation of [NBu4][2a], treatment of the solution with 1 equiv of decamethylferrocene (Fc*) at −80 °C resulted in immediate bleaching of the UV−vis spectrum, and subsequent addition of 1 equiv of [Fc][B(ArF)4] reformed the purple color and the associated UV−vis features (this process was repeated twice, albeit with some loss of intensity due to decomposition, Figure S11). Additionally, the reduction of 3a gives an EPR signal nearly identical to that of the starting [NBu4][2a] complex (Figure S12). Together, the data support a reversible one-electron redox reaction that interconverts [NBu4][2a] and 3a.
Figure 3.
UV−vis spectra (blue = 3a, red = 3b, and black = LCuIIIOH; all samples 0.1 mM in THF at −80 °C) and (inset) resonance Raman spectra for 3a, for samples prepared from R16O16OH (black, top) or R18O18OH (blue, middle), with difference spectrum shown in red (bottom). Conditions are noted in the text.
Additional data support the formulations of 3a and b as [CuOOR]2+ complexes. They exhibit no signal in perpendicular mode EPR spectra (Figure S13), consistent with S = 0 CuIII but not ruling out an antiferromagnetically coupled (S = 0) CuII- ligand radical or a ferromagnetically coupled (S = 1) CuII-ligand radical species (see theory results below). Prediction of the zero field splitting (E/D) and the J term (magnetic coupling) values requires high level multireference calculations (beyond the scope of this work), and even then the predicted values are generally unreliable.15 To confirm the presence of the −OOR ligand, we obtained resonance Raman spectra for frozen solutions of 3a in THF (λex = 561 nm) with samples prepared from natural abundance R16O16OH or labeled R18O18OH (R = cumyl, ~60% incorporation of 18O by ESI-MS; Figure S14). A peak at 831 cm−1 in the 16O sample shifts to 813 cm−1 upon 18O-labeling (Figure 3); it is the only feature that is sensitive to isotopic labeling (full spectra shown in Figure S15). The Δ16O−18O = 18 cm −1 is smaller than that calculated from Hooke’s law for ν(O−O) (48 cm−1), but close to values reported previously for CuIIOOR complexes (Δ16O−18O in the range 24−89 cm−1 with most between 24 and 30 cm−1).16 To rationalize these smaller values, mixing of the ν(O−O), ν(C−O), and ν(C−C) modes was proposed, and we surmise that similar mixing occurs for 3a.
Consistent with this hypothesis, DFT numerical frequency calculations yielded Δ16O−18O = 26 cm 2−1 for a highly delocalized vibration at 856 cm−1 for the 16O isotopomer (video in Supporting Information). Both the singlet and triplet states were optimized for the one-electron oxidized complexes of 3a and b, where both spin density and molecular orbital plots (Figures S16−S18) showed significant delocalization onto the −OOR unit.17 No significant difference in the O−O stretching frequencies is predicted between singlet and triplet electronic states (within 2 cm−1, Figure S19). In prior work on the [CuOH]2+ moiety, we showed this core was best described as a singlet CuIII species.12a,c For the [CuOOR]2+ complexes, the electronic energies of the singlet and triplet states were tracked running relaxed surface scans about the Namide−Cu−O−O torsion angle (mPWPW/Def2-TZVP, Figures S20−S21). The data show that the torsion angle changes the energy of the singlet−triplet gap (ES−T) such that the singlet state is favored at 0° but the ES−T gap becomes much smaller at 90°. To better estimate the ES−T gap, broken-symmetry (BS) calculations starting at the singlet state optimized geometries (torsion angles of 4° (3a) or 3° (3b)) favored the singlet state by >10 kcal mol−1 after spin purification (using eq 4; Table S4). However, BS calculations starting at the triplet state optimized geometries (torsion angles of 80° (3a) or 63° (3b)) favored the singlet state by ~4 kcal mol−1 when using GGA or meta-GGA functionals, while hybrid-GGA, meta-hybrid-GGA, and double hybrid functionals predominantly favored the triplet ground state by ~2 kcal mol−1 (Table S3). Comparison of TD-DFT spectra calculated for both states (PBE0/Def2-TZVP) to the experimental UV−vis spectra for 3a and b indicated marginally better agreement for the singlet, but do not rule out the triplet (Figures S22−S24, Table S5). We conclude at this juncture that more extensive work is needed to unambiguously determine the ground state and note that because the accuracy of broken-symmetry predictions is notoriously diffcult for transition metals,18 future studies will involve reevaluation of the ES−T gap using multireference calculations.
Preliminary investigation of the reactivity of 3a revealed notable differences relative to LCuIIIOH.12 The rate of decay of solutions of 3a was not enhanced, and no new spectral features were observed upon addition of excess (200 equiv) 9,10-dihydroanthracene, 1,4-cyclohexadiene, 2,4,6-tri-tert-butyl phenol, 2,4-di-tert-butyl phenol, or 4-tert-butyl phenol, substrates with relatively weak C−H or O−H bonds that react rapidly with LCuIIIOH via PCET mechanisms. On the other hand, 3a (THF, −80 °C) decays rapidly in the presence of 1-hydroxy-2,2,6,6-tetramethylpiperidine (TEMPO-H, 25−200 equiv). Monitoring the decay by UV−vis spectroscopy showed the reaction to be first-order in TEMPO-H with an overall second-order rate constant of ~2.4 M−1 s−1 (Figures S25−S26). EPR spin quantification of reaction products was consistent with an ~70% yield of TEMPO· detected from the reaction of 3a with TEMPO-H (Figure S27).
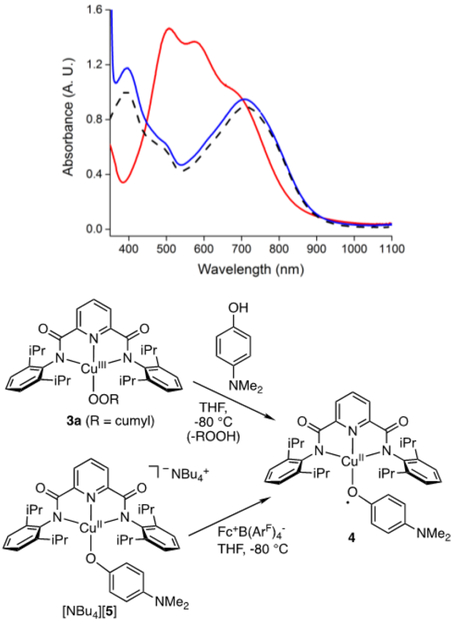
In addition, while various tert-butyl-substituted phenols were unreactive, treatment of 3a (THF, −80 °C) with 4-dimethylamino-phenol (NMe2PhOH) resulted in instantaneous loss of the UV−vis features associated with 3a and generation of a new feature at ~740 nm (Figure 4). This spectrum is identical to that reported previously12d to form upon reaction of LCuIIIOH with NMe2PhOH and tentatively assigned as the CuII-phenoxyl radical complex 4.19 To buttress this assignment via an independent synthetic route, we prepared the CuII- phenolate complex [NBu4][5] and characterized it by high resolution electrospray mass spectrometry, UV−vis spectroscopy, and cyclic voltammetry (Figures 4 and S28−S30). Treatment of [NBu4][5] (THF, −80 °C) with [Fc][B(ArF)4] yielded the same UV−vis spectrum as that seen for the reaction of 3a with NMe2PhOH,12d consistent with formation of 4 (Figure S31). The conversion of 3a to 4 involves a net proton transfer followed by a ligand exchange, but multiple possible mechanisms may be envisaged, including simple proton transfer, PCET, PT/ET, or ET/PT pathways. We note that NMe2PhOH is the least acidic phenol (pKa = 19.8 in DMSO)20 and contains the lowest O−H bond dissociation free energy (BDFE)19 of all the phenolic substrates that we have screened. Thus, at this juncture we view simple proton transfer to be unlikely and a PCET or ET/PT pathway to be more appropriate (further mechanistic work is underway).
Figure 4.
(top) Overlay of UV−vis spectra (THF, −80 °C) of 3a (red), the product of the reaction of 3a with NMe2PhOH (blue), and the product of the reaction of [NBu4][5] with [Fc][B(ArF)4] (black, dashed). (bottom) Reactions yielding 4, with formal copper oxidation states noted.
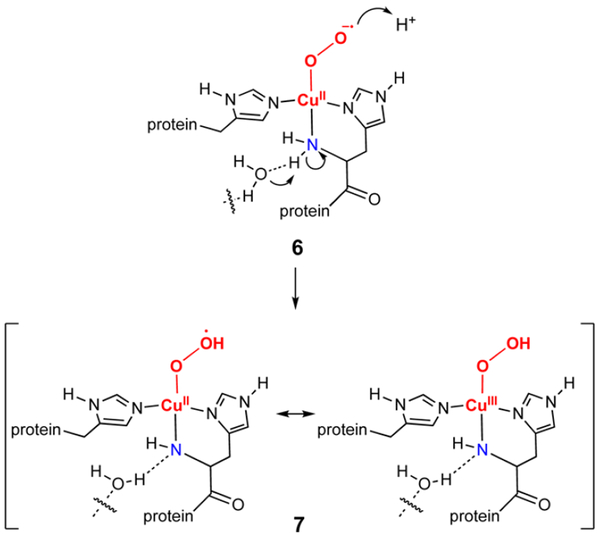
In conclusion, we have prepared two new, anionic [CuOOR]+ complexes ([NBu4][2a] and [NBu4][2b]) and have obtained evidence from spectroscopy and theory that one-electron oxidation at low temperature yields novel [CuOOR]2+ species. This discovery provides initial precedent for the possible existence of such species in oxidations catalyzed by copper enzymes. For example, one could envision the involvement of a related [CuOOH]2+ species in lytic polysaccharide monooxygenase, an intriguing enzyme due to its utility in lignocellulosic degradation for biofuel generation (Figure 5).21 According to this hypothetical scheme, deprotonation of the amino group of the histidine brace enhances the basicity of the initial Cu(II)-superoxide intermediate 6 (the formation of which may also be promoted by such deprotonation). Such deprotonation of the amino group has been suggested on the basis of X-ray crystallography.22 The enhanced basicity of 6 would facilitate protonation to yield a [CuOOH]2+ species (7) analogous to the species we have prepared and similarly stabilized by an anionic supporting ligand (for 7, a trans amide donor). Whether 7 is suffciently reactive to be a viable oxidizing intermediate is an open question.23 Preliminary studies indicate the significantly decreased reactivity of 3a with C−H and O−H bonds compared to LCuIIIOH, consistent with the low oxidation potential and expected low pKa for the ROO− ligand of the former. Still, further work is needed to understand the reactivity of the [CuOOR]2+ core, key issues being the relative propensity for Cu−O vs O−O bond scission.
Figure 5.
Proposed intermediates in LPMO.
Supplementary Material
ACKNOWLEDGMENTS
X-ray diffraction experiments were performed using a diffractometer acquired through NSF-MRI Award CHE-1229400. We thank the NIH (R37GM47365) for financial support and Prof. Christopher J. Cramer for helpful discussions. The authors acknowledge the Minnesota Supercomputing Institute (MSI) at the University of Minnesota for providing resources that contributed to the research results reported within this paper.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.7b05754.
Experimental and computational details (PDF)
A vibrational mode (AVI)
Notes
The authors declare no competing financial interest.
REFERENCES
- (1).Solomon EI; Heppner DE; Johnston EM; Ginsbach JW; Cirera J; Qayyum M; Kieber-Emmons MT; Kjaergaard CH; Hadt RG; Tian L Chem. Rev 2014, 114, 3659–3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).(a) Allen SE; Walvoord RR; Padilla-Salinas R; Kozlowski MC Chem. Rev 2013, 113, 6234–458. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wendlandt AE; Suess AM; Stahl SS Angew. Chem., Int. Ed 2011, 50, 11062–11087. [DOI] [PubMed] [Google Scholar]
- (3).Klinman JP J. Biol. Chem. 2006, 281, 3013–3016. [DOI] [PubMed] [Google Scholar]
- (4).(a) Chen P; Solomon EI J. Am. Chem. Soc 2004, 126, 4991–5000. [DOI] [PubMed] [Google Scholar]; (b) Crespo A; Marti MA; Roitberg AE; Amzel LM; Estrin DA J. Am. Chem. Soc. 2006, 128, 12817–12828. [DOI] [PubMed] [Google Scholar]
- (5).Kjaergaard CH; Qayyum MF; Wong SD; Xu F; Hemsworth GR; Walton DJ; Young NA; Davies GJ; Walton PH; Johansen KS; Hodgson KO; Hedman B; Solomon EI Proc. Natl. Acad. Sci. U. S. A 2014, 111, 8797–8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Cramer CJ; Tolman WB Acc. Chem. Res 2007, 40, 601–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Abad E; Rommel JB; Kastner JJ Biol. Chem 2014, 289, 13726–13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Garcia-Bosch I; Siegler MA Angew. Chem., Int. Ed 2016, 55, 12873–12876. [DOI] [PubMed] [Google Scholar]
- (9).(a) Elwell CE; Gagnon NL; Neisen BD; Dhar D; Spaeth AD; Yee GM; Tolman WB Chem. Rev 2017, 117, 2059–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Liu JJ; Diaz DE; Quist DA; Karlin KD Isr. J. Chem 2016, 56, 9–10. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Itoh S Acc. Chem. Res 2015, 48, 2066–2074. [DOI] [PubMed] [Google Scholar]; (d) Mirica LM; Ottenwaelder X; Stack TDP Chem. Rev 2004, 104, 1013–1045. [DOI] [PubMed] [Google Scholar]; (e) Lewis EA; Tolman WB Chem. Rev 2004, 104, 1047–1076. [DOI] [PubMed] [Google Scholar]
- (10).Itoh S Curr. Opin. Chem. Biol 2006, 10, 115–122. [DOI] [PubMed] [Google Scholar]
- (11).Selected examples:Kunishita A; Kubo M; Sugimoto H; Ogura T; Sato K; Takui T; Itoh S J. Am. Chem. Soc 2009, 131, 2788–2789.Woertink JS; Tian L; Maiti D; Lucas HR; Himes RA; Karlin KD; Neese F; Würtele C; Holthausen MC;Bill E; Sundermeyer J. r.; Schindler S; Solomon EI Inorg. Chem 2010, 49, 9450–9459.Ginsbach JW; Peterson RL; Cowley RE; Karlin KD; Solomon EI Inorg. Chem 2013, 52, 12872–12874.Peterson RL; Ginsbach JW; Cowley RE; Qayyum MF; Himes RA; Siegler MA; Moore CD; Hedman B; Hodgson KO; Fukuzumi S; Solomon EI; Karlin KD J. Am. Chem. Soc 2013, 135, 16454–16467.
- (12).(a) Donoghue PJ; Tehranchi J; Cramer CJ; Sarangi R; Solomon EI; Tolman WB J. Am. Chem. Soc 2011, 133, 17602–17605. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Dhar D; Tolman WB J. Am. Chem. Soc 2015, 137, 1322–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Dhar D; Yee GM; Spaeth AD; Boyce DW; Zhang H; Dereli B; Cramer CJ; Tolman WB J. Am. Chem. Soc 2016, 138, 356–368. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Dhar D; Yee GM; Markle TF; Mayer JM; Tolman WB Chem. Sci 2017, 8, 1075–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Spaeth AD; Gagnon NL; Dhar D; Yee GM; Tolman WB J. Am. Chem. Soc 2017, 139, 4477–4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Selected examples:Kim S; Saracini C; Siegler MA; Drichko N; Karlin KD Inorg. Chem 2012, 51, 12603–12605.Choi YJ; Cho K-B; Kubo M; Ogura T; Karlin KD; Cho J; Nam W Dalton Trans 2011, 40, 2234–2241.Tano T; Ertem MZ; Yamaguchi S; Kunishita A; Sugimoto H; Fujieda N; Ogura T; Cramer CJ; Itoh S Dalton Trans 2011, 40, 10326.Yamaguchi S; Masuda H Sci. Technol. Adv. Mater 2005, 6, 34–47.Kitajima N; Katayama T; Fujisawa K; Iwata Y; Moro-oka Y J. Am. Chem. Soc 1993, 115, 7872–7873.
- (14).A complex with a [CuOOR]+ unit appended to an anilino radical has been reported:Paria S; Ohta T; Morimoto Y; Ogura T; Sugimoto H; Fujieda N; Goto K; Asano K; Suzuki T; Itoh S J. Am. Chem. Soc 2015, 137, 10870–10873.
- (15).(a) Neese F; Liakos DG; Ye S JBIC, J. Biol. Inorg. Chem 2011, 16, 821–829. [DOI] [PubMed] [Google Scholar]; (b) Güell M; Luis JM; Sola,` M.; Swart, M. J. Phys. Chem. A 2008, 112, 6384–6391. [DOI] [PubMed] [Google Scholar]
- (16).Chen P; Fujisawa K; Solomon EI J. Am. Chem. Soc 2000, 122, 10177–10193. [Google Scholar]
- (17).We considered the possibility that the complexes comprise a [CuOOR]+/ligand-centered radical pair (ref 14), but the calculations do not support significant radical character on the N-donor ligand.
- (18).(a) Cramer CJ; Truhlar DG Phys. Chem. Chem. Phys 2009, 11, 10757–10816. [DOI] [PubMed] [Google Scholar]; (b) Neese F Coord. Chem. Rev 2009, 253, 526–563. [Google Scholar]
- (19).An alternative CuIII-phenolate ground state structure is also possible. A more detailed evaluation of the ground state of LCu(phenolate) complexes will be reported separately.
- (20).Warren JJ; Tronic TA; Mayer JM Chem. Rev 2010, 110, 6961–7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).(a) Hemsworth GR; Johnston EM; Davies GJ; Walton PH Trends Biotechnol 2015, 33, 747–761. [DOI] [PubMed] [Google Scholar]; (b) Johansen KS Biochem. Soc. Trans 2016, 44, 143–149. [DOI] [PubMed] [Google Scholar]
- (22).(a) Frandsen KEH; Simmons TJ; Dupree P; Poulsen J-CN; Hemsworth GR; Ciano L; Johnston EM; Tovborg M; Johansen KS; von Freiesleben P; Marmuse L; Fort S; Cottaz S; Driguez H; Henrissat B; Lenfant N; Tuna F; Baldansuren A; Davies GJ; Lo Leggio L; Walton PH Nat. Chem. Biol 2016, 12, 298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Bacik JP; Mekasha S; Forsberg Z; Kovalevsky AY; Vaaje-Kolstad G; Eijsink VGH; Nix JC; Coates L; Cuneo MJ; Unkefer CJ; Chen JC Biochemistry 2017, 56, 2529–2532. [DOI] [PubMed] [Google Scholar]
- (23).According to theoretical calculations reported recently, hydrogen abstraction by complexes with an intact O−O bond such as 7 is not energetically feasible.Hedegård ED; Ryde U J. Biol. Inorg. Chem 2017, DOI: 10.1007/s00775-017-1480-1.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.