Abstract
Objective
To identify genetic loci through genome-wide association studies that associate with differences in fibroid size and number in a population of African American (AA) and European American (EA) women.
Design
Cross-sectional study
Setting
Not applicable.
Patient(s)
Using BioVU, a clinical population from the Vanderbilt University Medical Center, and the Coronary Artery Risk Development in Young Adults (CARDIA) cohort, a prospective cohort, we identified 1,520 women (609 AA and 911 EA) with documented fibroid characteristics.
Intervention(s)
None
Main Outcome Measure(s)
Outcome measurements include volume of largest fibroid, largest fibroid dimension, and number of fibroids (single vs. multiple).
Result(s)
In race-stratified analyses we achieved genome-wide significance at a variant located between MAT2B and TENM2 (rs57542984, β = 0.13; 95% CI: 0.09, 0.17; p = 2.36×10−9) for analyses of largest fibroid dimension in AAs. The strongest signal for transethnic analyses was at a variant on 1q31.1 located between PLA2G4A and BRINP3 (rs6605005, β = 0.24; 95% confidence interval: 0.15, 0.33; p = 7.68×10−8) for fibroid volume. Results from MetaXcan identified an association between predicted expression of the gene ER degradation enhancing alpha-mannosidase like protein 2 (EDEM2) in the thyroid and number of fibroids (Z-score = −4.51; p = 6.34×10−6).
Conclusion(s)
This study identified many novel associations between genetic loci and fibroid size and number in both race-stratified and transethnic analyses. Future studies are necessary to further validate our study findings and to better understand the mechanisms underlying these associations.
Keywords: Fibroids, leiomyomata, genome-wide association study, transethnic meta-analysis
INTRODUCTION
The majority of US women develop at least one leiomyoma, or uterine fibroid, by the age of menopause, (1) which results in around 5.9 to 34.4 billion dollars annually from treatments and costs associated with loss of work and healthcare (2). Fibroids are a heterogeneous disease that vary in size and number between women, likely leading to a range of symptoms including: pressure of the abdomen, chronic pelvic pain, and heavy or painful periods, (3).
Heritability estimates for fibroid number and size remain unknown while twin studies estimate that between 26% and 69% of fibroid risk is heritable (4, 5). Additional support for genetic etiology for fibroids comes from racial differences in fibroid risk (1, 6, 7), as well as well as the racial differences in fibroid size and number between African American (AA) and European American (EA) women (1, 8). For example, the incidence rate for fibroids is two to three times larger for AA women than for EA women (7) and AA women are more likely to have a hysterectomy because of fibroids than EA women (9). Additionally, AA women have larger and more numerous fibroids than EA women (1, 8). In an admixture mapping study on fibroid number and size, our lab group observed that AA women with more African ancestry were more likely to have multiple fibroids using genetic data from BioVU and Coronary Artery Risk Development in Young Adults (CARDIA) (10).
There have been no prior genome-wide association studies (GWAS) of fibroid characteristics, however, a few studies have shown a direct relationship between increasing fibroid size and gene variants (11, 12). Edwards et al. (2013) observed associations between increasing fibroid size in EAs with gene variants in trinucleotide repeat containing 6B (TNRC6B) and Bet1 golgi vesicular membrane trafficking protein like (BET1L) (11) that were originally found in a GWAS of fibroid risk (13). Aissani et al. (2015) showed associations between fibroid risk and largest fibroid dimension when evaluating a set of candidate gene variants (12). There have been two previous GWAS on fibroid risk (13–15). In the first GWAS examining fibroid risk using a population of Japanese women, Cha et al. (2011) found three chromosomal regions that were associated with increased fibroid risk, 10q24.33, 22q13.1 and 11p15.5 (13). The second GWAS on fibroid risk was performed in AA populations (15). In the study, the authors observed a genome-wide significant association between rs739187 in cytohesin 4 (CYTH4) in 22q13.1 and fibroid risk (15).
Fibroids are heterogeneous, and it is possible that there are distinct genetic variants that associate with fibroid size and number. Additionally, there may be genetic loci affecting fibroid size and number that are race-specific, as well as others that are common across racial/ancestral groups. The objective of this study is to identify genetic loci that associate with differences in fibroid size and number in a population of AA and EA individuals.
MATERIALS AND METHODS
Study Population
Coronary Artery Risk Development in Young Adults (CARDIA)
The CARDIA cohort was initiated in between 1985 and 1986 with the goal of measuring risk factors for coronary heart disease in a cohort of AA and EA individuals (16). The cohort consists of 5,115 AA and EA participants between 18 and 30 years of age who were selected based on approximately equal proportions of 18 to 24 and 25 to 30 year olds, sex, race (black and white), and education status with respect to high school graduation. Cohort recruitment took place at four places in the US: Birmingham, AL, Chicago IL, Minneapolis, MN, and Oakland, CA (16).
CARDIA Women’s Study (CWS) is an ancillary study of CARDIA that conducted pelvic ultrasounds among women in the CARDIA cohort at 16 years following enrollment. The goal of CWS was to evaluate the association between risk factors of polycystic ovary syndrome and cardiovascular disease. Largest fibroid dimensions, fibroid number, and relevant demographic data to our project was collected and recorded by trained CWS research staff (17). A transvaginal ultrasound was performed by sonographers who were certified by the American Registry of Diagnostic Medical Sonographers (ARMDS) and who had performed at least 50 prior transvaginal ultrasound examinations. The sonographers used a 5–7.5 MHz transvaginal probe. The dimensions of the largest fibroid were measured and number of fibroids was noted (17).
Our analyses used lifestyle and sociodemographic information that was collected via self and interviewer administered questionnaires (17). Measurements for height and weight were collected using a standardized protocol described previously (18).
The BioVU DNA Repository
The BioVU DNA Repository (2007–present) is a de-identified database of electronic health records (EHRs) that is linked to DNA. BioVU consists of stored de-identified demographic and clinical information for each patient who visits the Vanderbilt University Medical Center (19). A detailed description of BioVU has been previously described (19, 20). The Office of Human Research Protections and the Institutional Review Boards (21) deemed the BioVU DNA repository as non-human subjects research (20).
A validated phenotyping algorithm with a positive predictive value of 96% was used to identify fibroid cases (22). We included AA and EA women who were at least 18 years old, who had at least one documented fibroid and one pelvic imaging or surgery to treat fibroids, as indicated by the international classification of diseases, ninth revision, (ICD-9) or current procedural terminology (CPT) codes. Once fibroid cases were identified, we manually confirmed fibroid presence in patient EHRs by verifying that fibroids were visualized in pelvic imaging or surgery. We then manually abstracted fibroid measurements for the largest fibroid, as well as total number of reported fibroids, indication for imaging, subsequent treatment for fibroids, mode for fibroid confirmation (ultrasound, computed tomography [CT] scans, and magnetic resonance imaging [MRI] or from surgical reports comprising of hysterectomies and myomectomies), and pertinent demographic information. Our priority of recording patient information was from the first image report. If no image reports were available, we recorded patient information from surgical reports, which consisted of hysterectomies and myomectomies. If patient demographic information such as body mass index (BMI) was not listed in the image or surgical reports, we obtained the information from the nearest corresponding date.
Outcome measurements for analyses include: largest dimension of all fibroid measurements, volume of largest fibroid, and number of fibroids (single vs. multiple). To obtain an accurate estimate on fibroid volume, we used the following equation to calculate the volume of an ellipsoid for both CARDIA and BioVU samples: (Length × Width × Height × 0.523). The product of the three dimensions was multiplied by 0.523 to estimate volume assuming an ellipsoid shape. The total volume measurement and largest dimension were log10 transformed to create a normally distributed outcome for regression analyses. Some BioVU individuals with volume measurements (~33%) originally had only two measurements for their largest fibroid, but we imputed the third measurement by taking the average of the first two measurements. Many EHRs noted the presence of multiple fibroids but gave no specific number. In order to increase sample size and power, we coded fibroids as one versus multiple fibroids. Comparing subclasses of cases to controls would not necessarily tell us whether there are within case subphenotype (fibroid characteristics) differences, as evidence of an association from a case2 control analysis may still be a result of fibroid risk and not risk specific to a fibroid subphenotype. Because of this, the individuals in this study are limited to fibroid cases only. This study has been approved by the Vanderbilt University Medical Center Institutional Review Board (IRB).
Genotyping
Individuals from BioVU were genotyped on the on the Affymetrix Axiom Biobank array (Affymetrix, Inc., Santa Clara, CA), and BioVU AAs were further genotyped on the Axiom World Array 3 platform (Affymetrix, Inc., Santa Clara, CA) (Supplemental Table 1). Purification and quantification of DNA for BioVU was performed by PicoGreen (Invitrogen, Inc., Grand Island, NY). Individuals from CARDIA were genotyped at the Broad Institute of MIT and Harvard (Cambridge, MA) on the Affymetrix 6.0 array (Affymetrix, Santa Clara, CA).
GWAS Quality Control (QC)
The QC protocol was completed independently on race-stratified BioVU and CARDIA individuals using PLINK1.07 software (23) and the reference genome build GRCh37.p13. Briefly, the QC protocol included sample and SNP QC. Sample QC steps included: removing individuals with inconsistent reported versus genetic sex, removing related subjects (one subject from a pair with an identity by descent (IBD) probability between 0.20 and 0.95 and both subjects within a pair with an IBD probability > 0.95), and removing individuals with a low genotyping efficiency (≤0.95%). The SNP QC included: removing SNPs without a chromosome location, removing SNPs with a less than 1% minor allele frequency (MAF), removing low genotyping efficiency SNPs (≤0.95), and removing SNPs out of Hardy-Weinberg Equilibrium (p-value ≤ 1.0×10−6. Lastly, all SNPs were aligned to the 1000 Genomes (build 37, 2013) + strand.
Statistical Analyses
All covariate and demographic data were compiled using Stata/SE (College Station, Texas). Principal components were generated using EIGENSTRAT4.2 to adjust for potential confounding due to admixed ancestry (Supplemental Figures 1–4) (24). Principal components were also compared to the 1000 Genomes reference populations to help account for population stratification within each racial group of each study population. We phased genotyped data using SHAPEIT2 (25) and used the 1000 Genomes Cosmopolitan reference panel phase 1 version 3 to impute ungenotyped SNPs using IMPUTE2.3.0 software (26). Single SNP association analyses were performed on each outcome using a MAF ≥ 0.05, info score ≥ 0.4, and a HWE p-value ≥ 1.0×10−6 using SNPTESTv2.4.1 software (27) with an additive model adjusting for five PCs to control for population stratification, age, and BMI. The final number of SNPs available post-QC and post-imputation are shown in Supplemental Table 1. We performed a fixed effects inverse-variance weighted meta-analyses from single SNP association analyses using METAL software (28). We performed a transethnic meta-analysis using all individuals as well as race-specific meta-analyses. Quantile-quantile (QQ) plots of each meta-analysis for each outcome were created (Supplemental Figures 5–13). A p-value of 5×10−8 was used as the significance threshold for all meta-analyses resulting from GWAS. Suggestive signals included SNPs with a p-value of 1×10−6 for all meta-analyses resulting from GWAS. We listed the most significant index SNP using an r2 threshold of 0.6 to denote linkage between similar SNPs. In addition, we only listed characterized flanking genes in tables and excluded open reading frames and pseudogenes for results from meta-analyses resulting from GWAS. The regional plots were created by LocusZoom Standalone (http://csg.sph.umich.edu.locuszoom/) (29). The linkage disequilibrium (LD) between the index SNP and all other markers were estimated using the 1000 genomes build 37 using either African or European individuals.
We also predicted gene expression for specific tissues and performed association analyses between gene expression within the transethnic meta-analyses using MetaXcan software (30), a version of PrediXcan software that uses GWAS summary statistics (31). PrediXcan predicts gene expression from SNPs using the Genotype-Tissue Expression (GTEx) database, which aims to characterize gene expression of each tissue type with specific genotypes (32), and performs association analyses comparing tissue-specific gene expression to the outcome (31). MetaXcan predicts gene expression from GWAS summary statistics and performs association analyses comparing tissue-specific gene expression to the outcome (30). MetaXcan can predict tissue-specific gene expression for 44 different tissues. We limited to tissues that might interact with the uterus directly such as whole blood or indirectly and hormonally such as the adrenal gland. Selected tissues that were included in MetaXcan analyses included whole blood, vagina, uterus, thyroid, pituitary, ovary, cells transformed fibroblasts, breast mammary, adrenal gland, visceral omentum adipose, and subcutaneous adipose tissues We focused on these tissues due to their potential roles in hormonal pathways relating to uterine health or due to their physiologic similarity to fibroids. The GTEx database primarily genotyped DNA from blood samples, and RNA sequencing was performed on the individual tissues (33). Uterine tissue that was RNA sequenced mostly consisted of two pieces from varying places within the uterus (URL: https://www.gtexportal.org/home/histologyPage). Fibroid tumors were not included within the GTEx database. Significance thresholds for analyses involving MetaXcan were determined to be 9.91×10−7 for fibroid number, largest dimension, and volume by Bonferroni correction for multiple testing, and the suggestive threshold was 1.00×10−6.
Lastly, we used the cross phenotype association (CPASSOC) software (34, 35) to examine if there are loci that affect both fibroid number and size (fibroid number, volume, and max dimension) within the transethnic meta-analyses. We removed SNPs with an absolute Z-score value of 1.96 or greater when estimating the correlation matrix. There are two outputs for CPASSOC, SHom (which has more power when the summary statistics between traits are similar) and SHet (which has more power when the summary statistics between traits are different). We decided to focus on SHom a priori because racial differences in fibroid characteristics suggest that the genetic etiology of fibroid size and number are in the same direction (i.e., AA women get larger and more numerous fibroids than EA women) (1, 8). In addition, we only listed characterized flanking genes in tables and excluded open reading frames and pseudogenes for results from CPASSOC. The significance threshold for CPASSOC analyses was 5.00×10−8, and the corresponding suggestive threshold was 1.00×10−6.
RESULTS
Demographic Data
There were a total of 1,520 AA and EA women with genetic data (BioVU AAs = 438; BioVU EAs = 748; CARDIA AAs = 171; CARDIA EAs = 163) (Table 1). The mean age for AA women (BioVU = 41.5±11; CARDIA = 41.3±4) were lower than the mean age for EA women (BioVU = 47.7±12; CARDIA = 43.2±3). The mean BMI for AA women (BioVU = 32.9±8; CARDIA = 33.4±8) was higher than the mean BMI for EA women (BioVU = 28.9±8; CARDIA = 27.5±7). Additionally, there were more obese AA women (BioVU = 60%; CARDIA = 64%) than obese EA women (BioVU = 34%; CARDIA = 26%). AA women from BioVU had the largest fibroid volume (median: 19.6 cm3; interquartile range [IQR]: 5.3–76.0 cm3) and largest fibroid dimension (median: 3.5 cm; IQR: 2.2–5.7 cm) while EA women from CARDIA had the smallest fibroid volume (median: 2.8 cm3; IQR: 0.9–9.1 cm3) and fibroid dimension (median: 2.0 cm; IQR: 1.4–2.9 cm). AA women from CARDIA were most likely to have multiple fibroids (71%), and EA women from BioVU were least likely to have multiple fibroids (50%).
Table 1.
Demographics of BioVU and CARDIA AA and EA women
| Demographic Characteristics | N | BioVU AA (N=438) |
BioVU EA (N=748) |
CARDIA AA (N=171) |
CARDIA EA (N=163) |
|---|---|---|---|---|---|
| Age (mean±SD) | 1,520 | 41.5±11 | 47.7±12 | 41.3±4 | 43.2±3 |
| BMI (kg/m2) (mean±SD) | 1,515 | 32.9±8 | 28.9±8 | 33.4±8 | 27.5±7 |
| Underweight (<18.5) (%) | 17 | 1 | 1 | 1 | 1 |
| Normal weight (18.5–24.9) (%) | 401 | 16 | 33 | 11 | 42 |
| Overweight (25–29.9) (%) | 430 | 23 | 32 | 24 | 31 |
| Obese (≥30) (%) | 667 | 60 | 34 | 64 | 26 |
| Fibroid Volume (cm3) median (IQR) | 1,048 | 19.6 (5.3–76.0) | 9.7 (2.4–40.7) | 5.6 (2.1–19.3) | 2.8 (0.9–9.1) |
| Largest Fibroid Dimension (cm) (IQR) | 1,301 | 3.5 (2.2–5.7) | 2.7 (1.6–4.5) | 2.5 (1.8–3.7) | 2.0 (1.4–2.9) |
| Fibroid Number | 1,458 | ||||
| 1 (%) | 636 | 42 | 50 | 29 | 35 |
| >1 (%) | 822 | 58 | 50 | 71 | 65 |
BMI-body mass index; kg/m2-kilograms per meters squared; cm3-cubic centimeters; SD-standard deviation; IQR-interquartile range
Race Stratified Meta-analyses
There was one SNP from race-stratified analyses that reached GWAS threshold of significance (Table 2; Supplemental Table 2; Supplemental Figures 14–16). In the meta-analysis between BioVU and CARDIA AAs for fibroid max dimension, we observed that each effect allele of rs57542984 in 5q34 was associated with an increase in largest fibroid dimension (adjusted Beta: 0.13; 95% CI: 0.09, 0.17, p-value: 2.36×10−9) with flanking genes methionine adenosyltransferase 2B (MAT2B) and teneurin transmembrane protein 2 (TENM2) (Table 2). The most significant loci resulting from meta-analyses between BioVU and CARDIA EAs was rs141745233 where each effect allele was associated with a decrease in fibroid volume (adjusted Beta: −0.46; 95% CI: −0.64, −0.29; p-value: 3.97×10−7) (Supplemental Tables 3, 4; Supplemental Figures 17–19). The SNP, rs141745233, has flanking genes contactin associated protein like 2 (CNTNAP2) and cullin 1 (CUL1) (Supplemental Table 3).
Table 2.
Summary of the most significant SNPs from the meta-analyses between BioVU and CARDIA AAs for each fibroid outcome
| Number | |||||||||
|
| |||||||||
| SNP | Region | BP | Genes | A2/A1 | EAF | OR [95% CI] | P-value | Het. P | Dir. |
|
| |||||||||
| rs35322806 | 4q32.1 | 161,236,141 | RAPGEF2||FSTL5 | T/A | 0.21 | 0.44 [0.32, 0.60] | 2.44×10−7 | 0.237 | −− |
| rs735658 | 17q25.3 | 75,858,706 | SEPT9||TNRC6C | T/C | 0.09 | 0.29 [0.18, 0.47] | 6.13×10−7 | 0.018 | −− |
| rs7625080 | 3p26.1 | 7,962,060 | GRM7||LMCD1 | G/A | 0.34 | 0.53 [0.41, 0.68] | 8.58×10−7 | 0.791 | −− |
| rs62203867 | 20p12.3 | 5,725,672 | GPCPD1||CHGB | G/C | 0.24 | 0.46 [0.34, 0.63] | 9.83×10−7 | 0.734 | −− |
|
| |||||||||
| Max Dimension | |||||||||
|
| |||||||||
| SNP | Region | BP | Genes | A2/A1 | EAF | Beta [95% CI] | P-value | Het. P | Dir. |
|
| |||||||||
| rs57542984 | 5q34 | 163,864,278 | MAT2B||TENM2 | G/GA | 0.81 | 0.13 [0.09, 0.17] | 2.36×10−9 | 0.697 | ++ |
| rs62484733 | 7q22.3 | 105,077,185 | SRPK2||PUS7 | A/G | 0.10 | 0.14 [0.09, 0.19] | 3.89×10−7 | 0.275 | ++ |
| rs13183849 | 5q14.3 | 85,058,124 | EDIL3||COX7C | C/T | 0.29 | −0.09 [−0.12, −0.05] | 8.11×10−7 | 0.359 | −− |
|
| |||||||||
| Volume | |||||||||
|
| |||||||||
| SNP | Region | BP | Genes | A2/A1 | EAF | Beta [95% CI] | P-value | Het. P | Dir. |
|
| |||||||||
| rs6938199 | 6q22.33 | 128,434,932 | PTPRK | G/A | 0.10 | −0.41 [−0.56, −0.25] | 3.90×10−7 | 0.378 | −− |
SNP-single nucleotide polymorphism; BP-base pairs; A2-effect allele; A1-reference allele; OR-odds ratio; CI-confidence interval; Het. P-heterogeneity p-value; Dir.-direction.
Transethnic Meta-analyses
We observed no associations from the transethnic meta-analyses from outcomes fibroid number, largest dimension, and volume that reached the GWAS threshold of significance (Table 3, Supplemental Figures 20–22). The most significant SNP from the transethnic meta-analyses was rs6605005 (adjusted Beta: 0.24; 95% CI: 0.15, 0.33; p-value: 7.68×10−8) from fibroid volume in 1q31.1 with flanking genes phospholipase A2 group IVA (PLA2G4A) and BMP/retinoic acid inducible neural specific 3 (BRINP3) (Table 3). The effect allele frequency was more common in EAs (BioVU = 0.91; CARDIA = 0.87) than in AAs (BioVU = 0.75; CARDIA = 0.74) (Supplemental Table 5). The second most significant SNP from the transethnic meta-analyses was rs10024805 (adjusted Beta: 0.19; 95% CI: 0.12, 0.26; p-value: 9.30×10−8) from fibroid volume in 4q35.1 in the intron of gene sorbin and SH3 domain containing 2 (SORBS2) (Table 3). The effect allele frequency was higher among AAs (BioVU = 0.36; CARDIA = 0.39) than EAs (BioVU = 0.29; CARDIA = 0.26) (Supplemental Table 5).
Table 3.
Summary of the most significant SNPs from the transethnic meta-analysis between all BioVU and CARDIA women for each
| Number | |||||||||
|
| |||||||||
| SNP | Region | BP | Genes | A2/A1 | EAF | OR [95% CI] | P-value | Dir. | Het. P |
|
| |||||||||
| rs55964111 | 15q15.3 | 44,323,171 | FRMD5 | GA/G | 0.70 | 1.79 [1.44, 2.23] | 2.00×10−7 | ++++ | 0.315 |
| rs1419784 | 7p14.3 | 34,493,947 | BMPER||NPSR1 | A/T | 0.73 | 1.54 [1.30, 1.83] | 7.60×10−7 | ++++ | 0.284 |
|
| |||||||||
| Max Dimension | |||||||||
|
| |||||||||
| SNP | Region | BP | Genes | A2/A1 | EAF | Beta [95% CI] | P-value | Dir. | Het. P |
|
| |||||||||
| rs7751006 | 6q27 | 167774927 | TTLL2||TCP10 | C/T | 0.73 | 0.07 [0.04, 0.10] | 2.80×10−7 | ++++ | 0.928 |
| rs111855550 | 4p16.1 | 11106351 | CLNK||MIR572 | G/T | 0.13 | −0.08 [−0.11, −0.05] | 8.70×10−7 | −−−− | 0.185 |
|
| |||||||||
| Volume | |||||||||
|
| |||||||||
| SNP | Region | BP | Genes | A2/A1 | EAF | Beta [95% CI] | P-value | Dir. | Het. P |
|
| |||||||||
| rs6605005 | 1q31.1 | 188,621,556 | PLA2G4A|| BRINP3 | T/C | 0.79 | 0.24 [0.15, 0.33] | 7.68×10−8 | ++++ | 0.081 |
| rs10024805 | 4q35.1 | 186,677,519 | SORBS2 | T/G | 0.33 | 0.19 [0.12, 0.26] | 9.30×10−8 | ++++ | 0.392 |
| rs7968890 | 12p12.1 | 24,990,707 | BCAT1 | G/A | 0.44 | 0.17 [0.11, 0.24] | 6.11×10−7 | ++++ | 0.311 |
SNP-single nucleotide polymorphism; BP-base pairs; A2-effect allele; A1-reference allele; EAF-effect allele frequency; OR-odds ratio; CI-confidence interval; Dir- direction; Het. P-heterogeneity p-value.
Genetically Predicted Gene Expression and Correlated Outcome (CPASSOC) Analyses
In addition to the GWAS meta-analyses, we performed subanalyses using MetaXcan on selected tissues potentially related to fibroids from the transethnic meta-analyses. No tissue-specific gene expression associations from MetaXcan reached multiple testing significance (p-value threshold = 9.91×10−7) (Supplemental Table 6; Supplemental Figures 23–31). The most significant tissue-specific gene expression association from the transethnic meta-analyses was for the gene ER degradation enhancing alpha-mannosidase like protein 2 (EDEM2) within thyroid tissue for fibroid number, where increasing EDEM2 expression within the thyroid was associated with single fibroids (Z-score = −4.51; p-value = 6.34 ×10−6) (Supplemental Figure 29). We also performed CPASSOC and found novel associations from SHom analyses most notably within SNP rs200348 in chromosomal region 14.32.2 (p-value 7.13×10−7) (fibroid number, max dimension, and volume Z-scores being −4.26 −3.12, and −1.69 respectively) (Supplemental Table 7).
DISCUSSION
This is the first GWAS and subsequent transethnic meta-analysis on fibroid size and number using both AAs and EAs. In this study we found many loci that were suggestively associated with fibroid characteristics from the transethnic meta-analyses including rs6605005 associating with fibroid volume in chromosomal region 1q31.1 (Figure 1). There was a single significant SNP, rs57542984, in region 5q34 that associated with max fibroid dimension in AA women only (Figure 1). MetaXcan analyses associated EDEM2 expression in thyroid tissue with fibroid number. Additionally, CPASSOC analyses of SHom revealed SNPs associating with fibroid number, max dimension, and volume that were not observed from that transethnic meta9 analyses.
Figure 1. Regional association plot of the most significant marker for the AA meta-analyses.
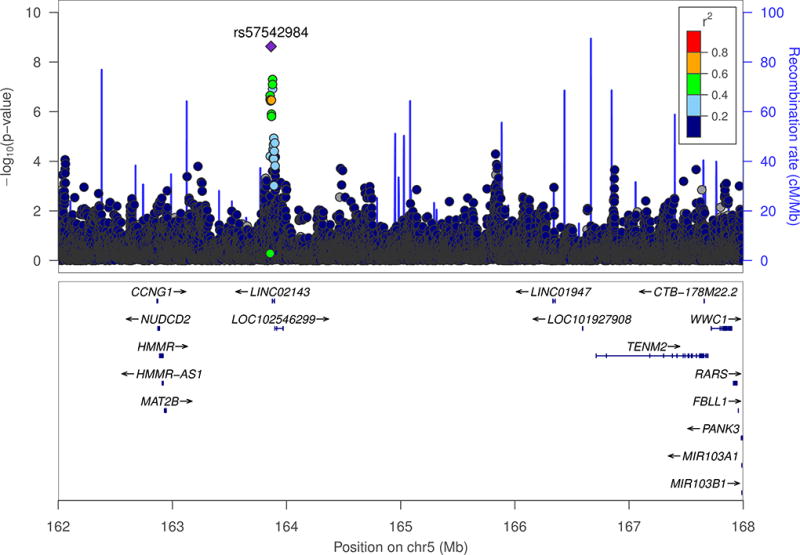
The index SNP from this regional plot is rs57542984 from the AA meta-analysis of largest fibroid dimension. African individuals from the 1000 genomes was used to estimate LD for this plot. The X-axis represents the genomic position along each chromosome in megabases (Mb). The Y-axis represents both the −log10 P-values for each SNP as well as the recombination rate. The color of each SNP represents the strength of the correlation (r2) to the index SNP (purple). Nearby genes are listed below the regional plots.
There was a single SNP, rs57542984, associating with fibroid max dimension which reached genome-wide significance in 5q34 in AAs from BioVU and CARDIA. The SNP was flanked by MAT2B and TENM2. MAT2B expression has been linked with cirrhosis, a fibroproliferative disease, and proliferate advantages in hepatoma cells (36). Interestingly, MAT2B is expressed in the uterus (>5 reads per kilobase of transcript per million mapped reads [RPKM]) (GTEx Analysis Release V7 [dbGaP Accession phs000424.v7.p2]) (32). In addition, TENM2 has been found to be expressed in cells transformed fibroblasts (>5 RPKM) (GTEx Analysis Release V7 [dbGaP Accession phs000424.v7.p2]). Interestingly, TENM2 has exhibited altered expression levels depending on the cancer subtype (37). For example, TENM2 mRNA was observed to be overexpressed in ovarian cancer cells, and decreased expression of TENM2 was documented in premalignant lesions such as breast hyperplastic enlarged lobular units, a precursor to breast cancer (37). It is possible that genetic variation around rs57542984 could lead to overexpression of MAT2B or susceptibility of TENM2 dysregulation in the uterus leading to an increase in fibroid largest dimension. The most significant transethnic meta-analysis SNP was rs6605005 associating with fibroid volume in chromosomal region 1q31.1 where each effect increased the volume of largest fibroid. The genes PLA2G4A and BRINP3 flanked rs6605005. PLA2G4A, or cytosolic phospholipase A2 α (cPLA2α), expression is increased by certain growth and pro-inflammatory factors (38). Inflammatory diseases implicated by PLA2G4A include pulmonary fibrosis, arthritis, and allergic reactions (38). Fibroids and inflammatory diseases are types of fibroproliferative diseases (39). It is possible that PLA2G4A could lead to an increased risk of developing more exacerbated forms of fibroproliferative diseases, which could include a larger fibroid volume. Evaluating PLA2G4A expression in GTEx revealed that PLA2G4A is expressed in the uterus (>5 RPKM) (GTEx Analysis Release V7 [dbGaP Accession phs000424.v7.p2]) (32). The gene BRINP3, however, is primarily expressed in neuronal cells (40). Interestingly, when BRINP3 was expressed in nonneuronal cells, cell cycle progression was halted (40). Lastly, rs10024805 is in the intron of SORBS2. SORBS2 is expressed in the uterus (>5 RPKM) (GTEx Analysis Release V7 [dbGaP Accession phs000424.v7.p2]) (32). Expression of SORBS2 in fibroid tissues has been shown to be down regulated in fibroids compared to myometrium tissue (41). It is possible that there is genetic variation near rs10024805 could lead to a decrease in SORBS2 expression resulting in larger fibroid volumes.
MetaXcan analyses highlighted an association between gene expression of EDEM2 in thyroid tissue and number of fibroids. EDEM2 has been shown to be localized in the endoplasmic reticulum and is involved with ER-associated degradation (42). It is possible that increased expression of EDEM2 could increase degradation of proteins in thyroid tissue. This could lead to altered hormone levels reducing the odds of an individual having multiple fibroids. MetaXcan was applied in a previous study GWAS on uterine fibroid risk using AAs by Hellwege et al. (2017) (15). The authors found that lower predicted CYTH4 expression was associated with fibroid risk in thyroid tissue. This result was not replicated in our MetaXcan race-stratified meta-analyses on fibroid characteristics for AAs (Supplemental Figures 23–25) or EAs (Supplemental Figures 26–28) or the transethnic meta-analyses (Supplemental Figures 29–31) on fibroid characteristics. This could be because fibroid risk and fibroid size and number could have different etiologies for tissue-specific gene expression levels.
We applied CPASSOC analyses to highlight SNPs that associate with fibroid size and number that might have been missed via conventional association analyses involving a single outcome. For example, Zhu et al. (2015) performed CPASSOC analyses on three blood pressure traits (systolic blood pressure, diastolic blood pressure, and hypertension status) using an African American population and identified four genetic loci (CHIC2, HOXA-EVX1, IGFBP1/IGFBP3, and CDH17) that reached genome-wide significance and an additional six loci which were suggestively associated (34). Each of these loci were missed in single trait GWAS from the original report (34). In our analyses, SHom discovered SNPs that were not previously found in the individual meta-analyses within this study. The most significant identified SNP was rs200348 in 14q32.2 being flanked by genes vaccinia related kinase 1 (VRK1) and B cell CLL/lymphoma 11B (BCL11B). In a study by Cai et al. (2017), the authors demonstrated that the mouse gene Bcl11b expression is needed to maintain mammary gland homeostasis and epithelial cell proliferation capability (43).
The previous GWAS on uterine fibroids by Cha et al. (2011) using a Japanese population and Hellwege et al. (2017) using population of AAs identified several loci associating with fibroid risk (13, 15); these loci did not overlap with our study findings. There were, however, notable differences between the previous studies (13, 15) and ours. First, the previous GWAS on uterine fibroids focused on the presence of fibroids, while our study focused on fibroid number and size of individuals who had a diagnosis of fibroids. Second, our study’s main emphasis examined AAs and EAs in a transethnic meta-analysis. The transethnic meta-analysis highlighted genetic variants that were present with a MAF >5% in both US AA and EA populations.
We acknowledge that variation in both the technology used to visualize fibroids and how fibroid measurements we documented in electronic health records is a potential limitation for this study. All women included from the CARDIA Women’s Study and most BioVU women had their fibroids accessed by ultrasounds. However, we included other imaging technology for fibroid measurements obtained from BioVU, including MRI and CT scans. Imaging methods such as MRI have a higher sensitivity for detecting fibroids than ultrasounds (44). Since most individuals had ultrasounds in our study, there is likely some degree of misclassification occurring where some women with multiple fibroids are being misclassified as having single fibroids. When examining fibroid size, there was less discrepancy differences between MRI and ultrasound accuracy meaning that fibroid size measurements might have lower misclassification rates than fibroid number (44).
This study was the first transethnic meta-analysis to assess the association between genetic markers throughout the genome and fibroid size and number using both AAs and EAs. We found many novel genetic loci associating with fibroid characteristics that were not previously associated with fibroid risk suggesting different genetic etiologies exist between fibroid risk and fibroid characteristics. Further studies are needed to explain the underlying mechanisms observed in these genetic associations.
Supplementary Material
Acknowledgments
We thank Ayush Giri for his expert technical assistance. This study was funded by the National Institutes of Health (NIH) grants (R01HD074711, R03HD078567, and R01HD093671) to Digna R. Velez Edwards and by the Human Genetic Training Grant (5T32GM080178) and the VICTR Training Grant (6TL1TR000447) to Michael J. Bray.
The publication described was supported by CTSA award No. UL1TR000445 from the National Center for Advancing Translational Sciences. Its contents are solely the responsibility of the authors and do not necessarily represent official views of the National Center for Advancing Translational Sciences or the National Institutes of Health.
The Coronary Artery Risk Development in Young Adults Study (CARDIA) is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with the University of Alabama at Birmingham (HHSN268201300025C & HHSN268201300026C), Northwestern University (HHSN268201300027C), University of Minnesota (HHSN268201300028C), Kaiser Foundation Research Institute (HHSN268201300029C), and Johns Hopkins University School of Medicine (HHSN268200900041C). CARDIA is also partially supported by the Intramural Research Program of the National Institute on Aging (NIA) and an intra-agency agreement between NIA and NHLBI (AG0005). This manuscript has been reviewed by CARDIA for scientific content. The CARDIA Women’s Study was supported by the NHLBI (R01-HL-065611). Genotyping was funded as part of the NHLBI Candidate-gene Association Resource (N01-HC-65226) and the NHGRI Gene Environment Association Studies (GENEVA) (U01-HG004729, U01-HG04424, and U01-HG004446).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST STATEMENT
The authors disclose that there is no conflict of interest.
References
- 1.Baird DD, Dunson DB, Hill MC, Cousins D, Schectman JM. High cumulative incidence of uterine leiomyoma in black and white women: ultrasound evidence. American journal of obstetrics and gynecology. 2003;188:100–7. doi: 10.1067/mob.2003.99. [DOI] [PubMed] [Google Scholar]
- 2.Cardozo ER, Clark AD, Banks NK, Henne MB, Stegmann BJ, Segars JH. The estimated annual cost of uterine leiomyomata in the United States. American journal of obstetrics and gynecology. 2012;206:211, e1–9. doi: 10.1016/j.ajog.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zimmermann A, Bernuit D, Gerlinger C, Schaefers M, Geppert K. Prevalence, symptoms and management of uterine fibroids: an international internet-based survey of 21,746 women. BMC women's health. 2012;12:6. doi: 10.1186/1472-6874-12-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snieder H, MacGregor AJ, Spector TD. Genes control the cessation of a woman's reproductive life: a twin study of hysterectomy and age at menopause. The Journal of clinical endocrinology and metabolism. 1998;83:1875–80. doi: 10.1210/jcem.83.6.4890. [DOI] [PubMed] [Google Scholar]
- 5.Luoto R, Kaprio J, Rutanen EM, Taipale P, Perola M, Koskenvuo M. Heritability and risk factors of uterine fibroids--the Finnish Twin Cohort study. Maturitas. 2000;37:15–26. doi: 10.1016/s0378-5122(00)00160-2. [DOI] [PubMed] [Google Scholar]
- 6.Walker CL, Stewart EA. Uterine fibroids: the elephant in the room. Science (New York, NY) 2005;308:1589–92. doi: 10.1126/science.1112063. [DOI] [PubMed] [Google Scholar]
- 7.Marshall LM, Spiegelman D, Barbieri RL, Goldman MB, Manson JE, Colditz GA, et al. Variation in the incidence of uterine leiomyoma among premenopausal women by age and race. Obstetrics and gynecology. 1997;90:967–73. doi: 10.1016/s0029-7844(97)00534-6. [DOI] [PubMed] [Google Scholar]
- 8.Moorman PG, Leppert P, Myers ER, Wang F. Comparison of characteristics of fibroids in African American and white women undergoing premenopausal hysterectomy. Fertility and sterility. 2013;99:768–76.e1. doi: 10.1016/j.fertnstert.2012.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilcox LS, Koonin LM, Pokras R, Strauss LT, Xia Z, Peterson HB. Hysterectomy in the United States, 1988–1990. Obstetrics and gynecology. 1994;83:549–55. doi: 10.1097/00006250-199404000-00011. [DOI] [PubMed] [Google Scholar]
- 10.Bray MJ, Edwards TL, Wellons MF, Jones SH, Hartmann KE, Velez Edwards DR. Admixture mapping of uterine fibroid size and number in African American women. Fertility and sterility. 2017;108:1034–42. e26. doi: 10.1016/j.fertnstert.2017.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edwards TL, Hartmann KE, Velez Edwards DR. Variants in BET1L and TNRC6B associate with increasing fibroid volume and fibroid type among European Americans. Human genetics. 2013;132:1361–9. doi: 10.1007/s00439-013-1340-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aissani B, Zhang K, Wiener H. Follow-up to genome-wide linkage and admixture mapping studies implicates components of the extracellular matrix in susceptibility to and size of uterine fibroids. Fertility and sterility. 2015;103:528–34. e13. doi: 10.1016/j.fertnstert.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cha PC, Takahashi A, Hosono N, Low SK, Kamatani N, Kubo M, et al. A genome-wide association study identifies three loci associated with susceptibility to uterine fibroids. Nature genetics. 2011;43:447–50. doi: 10.1038/ng.805. [DOI] [PubMed] [Google Scholar]
- 14.Eggert SL, Huyck KL, Somasundaram P, Kavalla R, Stewart EA, Lu AT, et al. Genome-wide linkage and association analyses implicate FASN in predisposition to Uterine Leiomyomata. American journal of human genetics. 2012;91:621–8. doi: 10.1016/j.ajhg.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hellwege JN, Jeff JM, Wise LA, Gallagher CS, Wellons M, Hartmann KE, et al. A multi-stage genome-wide association study of uterine fibroids in African Americans. Human genetics. 2017 doi: 10.1007/s00439-017-1836-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedman GD, Cutter GR, Donahue RP, Hughes GH, Hulley SB, Jacobs DR, Jr, et al. CARDIA: study design, recruitment, and some characteristics of the examined subjects. Journal of clinical epidemiology. 1988;41:1105–16. doi: 10.1016/0895-4356(88)90080-7. [DOI] [PubMed] [Google Scholar]
- 17.Wellons MF, Lewis CE, Schwartz SM, Gunderson EP, Schreiner PJ, Sternfeld B, et al. Racial differences in self-reported infertility and risk factors for infertility in a cohort of black and white women: the CARDIA Women's Study. Fertility and sterility. 2008;90:1640–8. doi: 10.1016/j.fertnstert.2007.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis CE, Smith DE, Wallace DD, Williams OD, Bild DE, Jacobs DR., Jr Seven-year trends in body weight and associations with lifestyle and behavioral characteristics in black and white young adults: the CARDIA study. American journal of public health. 1997;87:635–42. doi: 10.2105/ajph.87.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clinical pharmacology and therapeutics. 2008;84:362–9. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pulley J, Clayton E, Bernard GR, Roden DM, Masys DR. Principles of human subjects protections applied in an opt-out, de-identified biobank. Clinical and translational science. 2010;3:42–8. doi: 10.1111/j.1752-8062.2010.00175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirby KN, Gerlanc D. BootES: an R package for bootstrap confidence intervals on effect sizes. Behavior research methods. 2013;45:905–27. doi: 10.3758/s13428-013-0330-5. [DOI] [PubMed] [Google Scholar]
- 22.Feingold-Link L, Edwards TL, Jones S, Hartmann KE, Velez Edwards DR. Enhancing uterine fibroid research through utilization of biorepositories linked to electronic medical record data. Journal of women's health. 2014;23:1027–32. doi: 10.1089/jwh.2014.4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature genetics. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 25.Delaneau O, Howie B, Cox AJ, Zagury JF, Marchini J. Haplotype estimation using sequencing reads. American journal of human genetics. 2013;93:687–96. doi: 10.1016/j.ajhg.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS genetics. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nature genetics. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 28.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barbeira A, Dickinson SP, Torres JM, Torstenson ES, Zheng J, Wheeler HE, et al. Integrating tissue specific mechanisms into GWAS summary results. bioRxiv. 2016 [Google Scholar]
- 31.Gamazon ER, Wheeler HE, Shah KP, Mozaffari SV, Aquino-Michaels K, Carroll RJ, et al. A gene-based association method for mapping traits using reference transcriptome data. Nature genetics. 2015;47:1091–8. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science (New York, NY) 2015;348:648–60. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Consortium GT, Laboratory DA, Coordinating Center -Analysis Working G, Statistical Methods groups-Analysis Working G, Enhancing Gg, Fund NIHC et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–13. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu X, Feng T, Tayo BO, Liang J, Young JH, Franceschini N, et al. Meta-analysis of correlated traits via summary statistics from GWASs with an application in hypertension. American journal of human genetics. 2015;96:21–36. doi: 10.1016/j.ajhg.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kayima J, Liang J, Natanzon Y, Nankabirwa J, Ssinabulya I, Nakibuuka J, et al. Association of genetic variation with blood pressure traits among East Africans. Clinical genetics. 2017 doi: 10.1111/cge.12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez-Chantar ML, Garcia-Trevijano ER, Latasa MU, Martin-Duce A, Fortes P, Caballeria J, et al. Methionine adenosyltransferase II beta subunit gene expression provides a proliferative advantage in human hepatoma. Gastroenterology. 2003;124:940–8. doi: 10.1053/gast.2003.50151. [DOI] [PubMed] [Google Scholar]
- 37.Ziegler A, Corvalan A, Roa I, Branes JA, Wollscheid B. Teneurin protein family: an emerging role in human tumorigenesis and drug resistance. Cancer letters. 2012;326:1–7. doi: 10.1016/j.canlet.2012.07.021. [DOI] [PubMed] [Google Scholar]
- 38.Ghosh M, Tucker DE, Burchett SA, Leslie CC. Properties of the Group IV phospholipase A2 family. Progress in lipid research. 2006;45:487–510. doi: 10.1016/j.plipres.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 39.Russell SB, Smith JC, Huang M, Trupin JS, Williams SM. Pleiotropic Effects of Immune Responses Explain Variation in the Prevalence of Fibroproliferative Diseases. PLoS genetics. 2015;11:e1005568. doi: 10.1371/journal.pgen.1005568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawano H, Nakatani T, Mori T, Ueno S, Fukaya M, Abe A, et al. Identification and characterization of novel developmentally regulated neural-specific proteins, BRINP family. Brain research Molecular brain research. 2004;125:60–75. doi: 10.1016/j.molbrainres.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 41.Hever A, Roth RB, Hevezi PA, Lee J, Willhite D, White EC, et al. Molecular characterization of human adenomyosis. Molecular human reproduction. 2006;12:737–48. doi: 10.1093/molehr/gal076. [DOI] [PubMed] [Google Scholar]
- 42.Mast SW, Diekman K, Karaveg K, Davis A, Sifers RN, Moremen KW. Human EDEM2, a novel homolog of family 47 glycosidases, is involved in ER-associated degradation of glycoproteins. Glycobiology. 2005;15:421–36. doi: 10.1093/glycob/cwi014. [DOI] [PubMed] [Google Scholar]
- 43.Cai S, Kalisky T, Sahoo D, Dalerba P, Feng W, Lin Y, et al. A Quiescent Bcl11b High Stem Cell Population Is Required for Maintenance of the Mammary Gland. Cell stem cell. 2017;20:247–60. e5. doi: 10.1016/j.stem.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levens ED, Wesley R, Premkumar A, Blocker W, Nieman LK. Magnetic resonance imaging and transvaginal ultrasound for determining fibroid burden: implications for research and clinical care. American journal of obstetrics and gynecology. 2009;200:537, e1–7. doi: 10.1016/j.ajog.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


