Abstract
Epigenetic changes have likely contributed to the large size and enhanced cognitive abilities of the human brain which evolved within the last 2 million years after the human–chimpanzee split. Using reduced representation bisulfite sequencing, we have compared the methylomes of neuronal and non-neuronal cells from 3 human and 3 chimpanzee cortices. Differentially methylated regions (DMRs) with genome-wide significance were enriched in specific genomic regions. Intraspecific methylation differences between neuronal and non-neuronal cells were approximately 3 times more abundant than interspecific methylation differences between human and chimpanzee cell types. The vast majority (>90%) of human intraspecific DMRs (including DMRs in retrotransposons) were hypomethylated in neurons, compared with glia. Intraspecific DMRs were enriched in genes associated with different neuropsychiatric disorders. Interspecific DMRs were enriched in genes showing human-specific brain histone modifications. Human–chimpanzee methylation differences were much more frequent in non-neuronal cells (n. DMRs = 666) than in neurons (n. DMRs = 96). More than 95% of interspecific DMRs in glia were hypermethylated in humans. Although without an outgroup we cannot assign whether a change in methylation occurred in the human or chimpanzee lineage, our results are consistent with a wave of methylation affecting several hundred non-neuronal genes during human brain evolution.
Keywords: cortex methylome, human brain evolution, human–chimpanzee epigenetic divergence, neuronal vs. non-neuronal cells, RRBS
Introduction
Compared to all other mammals and non-human primates, the human brain is characterized by its remarkable size, energy consumption, and cognitive abilities. Fossil records indicate that the encephalization quotient (EQ), which is defined as the ratio between actual brain mass and predicted brain mass for an animal of a given size, remained rather constant in the Australopithecines from 3.5 to 2 million years ago. Encephalization started to accelerate in early forms of Homo approximately 2 million years ago (Zollikofer et al. 2005; Hublin et al. 2015; Falk 2016). The molecular mechanisms underlying hominine brain evolution predating modern humans remain to be elucidated. Enhanced cognitive abilities, most importantly language traits that are attributed exclusively to humans may have been both a driving force and an outcome of enlarged brain size (Jerison 1973; Tomasello 2008). Because of lacking remains (i.e., brain tissue) that can be analyzed directly, studies on human brain evolution highly depend on comparative and deductive approaches, inferring the ancestral states. Endocasts provide information on brain size but also on structural innovations relevant to behavioral evolution (i.e., stone tool manufacture, sociality, and foraging) (Holloway et al. 2018). From a molecular perspective, genes which have been associated with intellectual disability (ID), language impairment, and/or neurodevelopmental disorders (such as autism and schizophrenia) in humans may also have played a role in evolution of the human brain (Fitch 2011; Schneider, Jensen et al. 2012).
The chimpanzee, our closest extant relative, diverged from humans 5–8 million years ago (Benton and Donoghue 2007; https://www.nature.com/scitable/knowledge/library/hominin-taxonomy-and-phylogeny-what-s-in-142102877). Human and chimpanzee exhibit approximately 1% fixed single nucleotide changes and approximately 3–5% DNA sequence divergence including indels (Chimpanzee Sequencing and Analysis Consortium 2005). It is unlikely that these relatively small genetic differences account for the dramatic phenotypic differences, in particular in EQ and cognitive abilities. Instead, changes in gene expression and regulation may have played a major role in human brain evolution (Khaitovich et al. 2004; Gilad et al. 2006; Blekhman et al. 2008). An individual organism is composed of many different tissues and cell types, each of which has a defined developmental trajectory during ontogenesis from fertilization to death of the organism. Because all these trajectories are blueprinted by the same genome, phenotypic development and diversity may be largely controlled by epigenetic mechanism (Smith and Meissner 2013; Schneider et al. 2016). Similarly, phenotypic differences between species, notably between human and non-human primates may be attributed to epigenetic changes (Hernando-Herraez et al. 2013; Gokhman et al. 2014).
Epigenetic mechanisms, in particular DNA methylation, which mainly occurs at position 5 of the pyrimidine ring in the context of CpG-dinucleotides, control temporal, spatial, and species-specific gene expression patterns. The human genome contains more than 28 million of such CpG sites, 7% of which are clustered within CpG islands in cis-regulatory regions of most mammalian genes (Rollins et al. 2006). Methylation of promoter regions or transcription start sites during development, differentiation, or disease processes generally leads to a chromatin structure which is no longer accessible to the transcriptional machinery and, consequently, gene silencing (Weber et al. 2007; Vaissière et al. 2008). In contrast, gene body methylation is usually associated with active genes and may play a role in exon definition and alternative splicing (Jones 2012). In addition, methylated CpGs are heavily enriched in repetitive DNA, including short (SINE) and long interspersed repetitive elements (LINEs). This is generally thought to suppress their retrotransposition activity (Yoder et al. 1997). Studies on RNA polymerase III occupancy suggest that methylation of histone H3 rather than DNA may prevent SINE transcription (Varshney et al. 2015).
Several studies have compared the methylation patterns of individual genes (Farcas et al. 2009; Schneider, Jensen et al. 2012; Schneider, Mayer et al. 2012; Schneider et al. 2014) or the entire methylome (Wang et al. 2012; Zeng et al. 2012; Chopra et al. 2014; Mendizabal et al. 2016) between human and chimpanzee brains. Collectively, the results suggest that considerable epigenetic changes have occurred during human brain evolution and that differentially methylated regions (DMRs) are associated with neurodevelopmental and neurodegenerative disorders. However, since all these studies were performed on bulk tissue (mainly frontal cortex), the observed species differences may at least partially be due to changes in cell compositions. Neuron densities in frontal cortex area BA10 of humans are substantially lower compared with chimpanzees and all other great apes (Semendeferi et al. 2001, 2011). Methylation arrays and genome-wide bisulfite sequencing revealed wide-spread methylation differences between neuronal and non-neuronal human cells (Guintivano et al. 2013; Lister et al. 2013; Kozlenkov et al. 2014; Kessler et al. 2016). To avoid artifacts due to differences in tissue composition, we have compared the methylomes of neuronal and glia cells between humans and chimpanzees.
Materials and Methods
Samples Preparation
Fresh-frozen frontal cortex samples (Brodmann area BA10) of 3 human females, who died at 67–77 years of cardiovascular disease, were obtained from the Munich Brain Bank at the Center for Neuropathology and Prion Research, LMU. Samples from BA10 of 3 female chimpanzees, who died at 12 (adolescent), 40 and 43 years (old age; Goodall 1983; Bronikowski et al. 2016) of accidents/euthanasia, were obtained from the Biomedical Primate Research Centre, Rijswijk, Netherlands with the support and through the European Primate Network EUPRIM-Net. Postmortem times were 10 h, 32 h, and 46 h in humans and 5–6 h in chimpanzees. Use of the human and chimpanzee brain samples was approved by the Ethics Committee of the Landesärztekammer Rheinland-Pfalz (no. 837.103.04_4261) and the Julius Maximilians Universität Würzburg (no. 262/14). Cortex tissue was fluorescence-activated cell sorted into neuronal and non-neuronal cells using a NeuN-specific antibody, as described previously (Wagner et al. 2015). Genomic DNA from sorted cell fractions was isolated with the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany).
Library Preparation and Sequencing
Libraries for reduced representation bisulfite sequencing (RRBS) were generated according to established protocols (Boyle et al. 2012; Hahn et al. 2015). Briefly, genomic DNA was digested with the methylation sensitive restriction enzyme MspI (New England BioLabs, Frankfurt a. M., Germany). End repair of the resulting 3’ overhangs and A-tailing were performed to enable adapter ligation. After purification with AMPure XP beads (Beckmann Coulter, Krefeld, Germany) and ligation of the methylated universal adapters (Illumina, San Diego, USA), an additional bead wash step was conducted. Bisulfite conversion was performed with the EpiTect Bisulfite Kit (Qiagen), followed by a wash step using the MinElute PCR Purification Kit (Qiagen). After barcoding of the bisulfite treated samples two additional rounds of bead clean-up were performed. The final libraries were sequenced (single end 76 bp reads) with the Illumina Genome Analyzer II (Illumina) according to the manufacturer’s instructions.
RRBS Data Analysis
The entire RRBS dataset consisting of 12 libraries (2 cell types × 2 species × 3 individuals) was deposited in NCBI GEO (accession number GSE109559). Adapter and low quality sequences were trimmed from RRBS reads by trim_galore_v0.4.0 (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/), a wrapper tool powered by Cutadapt 1.8.3 (Martin 2011) and FastQC v0.11.3 (Krueger and Andrews 2011). Trim galore parameters were set at --rrbs --stringency 1 -e 0.2 -a GATCGGAAGAGCA. The human genome hg19 (http://hgdownload.soe.ucsc.edu/goldenPath/hg19) and the chimpanzee genome panTro4 (http://hgdownload.soe.ucsc.edu/goldenPath/panTro4/) were used as reference for mapping human and chimpanzee reads, respectively. The curated reads were mapped in a directional manner using bismark_v0.14.5 (Krueger and Andrews 2011) with default parameters. Only uniquely aligning reads were retained and CpG methylation levels were extracted. For further analyses, the CpG positions in the chimpanzee genome were transferred onto the human genome via liftover (http://hgdownload.soe.ucsc.edu/admin/exe/linux.x86_64/liftOver; http://hgdownload.cse.ucsc.edu/goldenPath/panTro4/liftOver/panTro4ToHg19.over.chain.gz) (Kuhn et al. 2013). All methylation loci over all libraries were extracted for the principal component analysis using the rda function of the R package vegan (https://cran.r-project.org/web/packages/vegan/index.html) (Oksanen et al. 2017). The two axes with the most information contents were used for plotting.
The test for differential methylation was performed at the single base level via the DSS package (Feng et al. 2014; Park and Wu 2016). For each of the 4 (interspecific neuronal, interspecific non-neuronal, intraspecific human, and intraspecific chimpanzee) DMR classes, only CpGs covered by at least 3 reads in at least 2 of the 3 analyzed samples per group were considered. The function DML test was used to estimate mean methylation level and dispersion at each CpG site and to conduct a Wald test. The P values were corrected for multiple testing (Hochberg and Benjamini 1990). Detection of DMRs was based on the DML test results, by calling the callDMR function of the DSS package (Feng et al. 2014; Park and Wu 2016). Regions were classified as DMRs, if they had a minimum length of 100 bp, contained at least 2 CpG sites, and at least 50% statistically significant differentially methylated CpGs. Nearby DMRs with a distance less than 100 base pairs were merged into longer ones. According to the DSS package, DMRs were ranked by the sum of the test statics of the contained CpG sites.
Gene annotations were extracted from the human genome hg19 via BioMart (Kinsella et al. 2011). Promoter regions started 1500 bp upstream and ended 500 bp downstream of the transcription start site. Overlapping, preceding and following genes as well as promoter annotations were determined for each DMR using the R package GenomicRanges (Lawrence et al. 2013).
Enrichment analyses
The methylation loci background model was created by extracting all CpG sites from the human and chimpanzee genome sequences. The DMR background models of each contrast for the enrichment analyses encompassed all potential DMRs based on the tested methylation loci. Each background was calculated with the same DSS parameters as the observed DMRs. Thus, any biases of methylation loci distribution over the genome, e.g., CpG distribution, technical biases towards MspI sites or parameter sets were accounted for.
For regional enrichment analyses, coordinates for the centromeric, telomeric, and subtelomeric regions were extracted for the human and the chimpanzee genome (Surrallés et al. 1999; Ding et al. 2014; Choudhury et al. 2016). To identify potential enrichment or depletion of measured methylation loci in centromeric, telomeric or subtelomeric regions, the number of measured methylation loci and methylation loci in the background model were counted and Fisher exact tests (Rödel 1971) applied. For regional DMR enrichment analyses, the overlap of DMRs and the 3 different regions were calculated for each inter- and intraspecific contrast. Fisher exact tests were performed with the help of the DMR background model. P values < 0.05 were considered significant. DMRs were enriched for odds ratios < 1 and depleted for odds ratio > 1.
To test enrichment for neuropsychiatric diseases and human-specific brain regulation, genes and promoters (1.5 kb upstream) overlapping the observed DMRs were extracted for the 4 inter- and intraspecific contrasts. Similarly, genes and promoters overlapping the background model were extracted. These gene sets were intersected with gene lists for neuropsychiatric diseases, including 138 genes that have been associated with intellectual disability (Supplementary Table 1), 203 associated with autism (Autism database AutDB 2017; http://autism.mindspec.org/autdb) (Basu et al. 2009), 408 with bipolar disorders (Genetic Database for Bipolar Disorder BDgene 2016; http://bdgene.psych.ac.cn), 351 with schizophrenia (Schizophrenia Working Group of the Psychiatric Genomics Consortium 2014), and 129 with Alzheimer disease (AlzGene database 2017; http://www.alzgene.org) (Bertram et al. 2007), as well as gene lists for human-specific brain expression patterns (Liu et al. 2012) and human-specific histone methylation signatures (Shulha et al. 2012). The intersections were used to perform Fisher exact tests (Rödel 1971) to identify enrichment or depletion in the overlap of DMRs and the gene lists. After correction for multiple testing (Hochberg and Benjamini 1990), P values < 0.05 were considered significant.
To compare human-specific brain expression values (Liu et al. 2012) with the identified interspecific DMRs, we downloaded and re-analyzed the corresponding microarray data from GEO (GSE22570). For the analysis all 25 human and 12 chimpanzee samples from the superior frontal gyrus were selected. The microarray datasets were mapped on the human genome (GRCh37) and tested for the differential expression between human and chimpanzee via limma (Ritchie et al. 2015), including the different age of the samples in a linear model. The age was normalized by grouping the samples into 5 age categories of 20% quantiles of the species lifespan. In this context, it is important to note that genes were classified as having human-specific expression patterns (Liu et al. 2012) when expression changes during ontogenesis differed significantly between humans and chimpanzees as well as between humans and macaques, but not between chimpanzees and macaques. Expression differences between species that were constant across lifespan were not considered.
Genes/promoters overlapping DMRs and the background model, respectively, were used for functional (gene ontology) enrichment analyses with the R package RDAVIDWebService (Fresno and Fernández 2013). The analyses were executed on the gene ontology annotations of GOTERM_BP_FAT, GOTERM_CC_FAT, and GOTERM_MF_FAT with the gene sets originating from the observed DMRs as foreground and gene sets originating from the background model as the background.
Bisulfite Pyrosequencing
PCR and sequencing primers for CAMTA1, PROSER2, RBFOX3, and RTN4R were designed with PyroMark Assay Design 2.0 software (Qiagen) (Supplementary Table 2). With exception of the forward primer for PROSER2, the same primers were used for human and chimpanzee. The 25 μL PCR reactions consisted 2.5 μL 10× PCR buffer, 20 mM MgCl2, 0.5 μL dNTP mix (10 mM), 1 μL of each forward and reverse primer (10 μM), 0.2 μL of FastStart Polymerase (5 U/μL), 1 μL bisulfite converted template DNA, and 18.8 μL PCR-grade water. Amplification was performed with an initial denaturation step at 95 °C for 5 min, 35 cycles of 95 °C for 30 s, primer-specific annealing temperature (57 °C for RTN4R and RBFOX3, 58 °C for human PROSER2, 59 °C for CAMTA1 and chimpanzee PROSER2) for 30 s, 72 °C for 45 s, and a final extension step at 72 °C for 5 min. Bisulfite pyrosequencing was performed on a PyroMark Q96 MD Pyrosequencing System using the PyroMark Gold Q96 CDT Reagent Kit (Qiagen) and 0.5 μL of sequencing primers (10 mM). Data analysis was done with the Pyro Q-CpG software (Qiagen). Mann–Whitney U test (Mann and Whitney 1947) was performed to identify methylation differences in the bisulfite pyrosequencing and the corresponding RRBS data.
Results
Reduced Representation Bisulfite Sequencing
RRBS was performed on neuronal (N+) and non-neuronal (N−) cells of 3 female human (Homo sapiens, HSA) and 3 female chimpanzee (Pan troglodytes, PTR) cortices. The entire RRBS dataset (NCBI GEO accession number GSE109559) comprised of 12 libraries yielding more than 149.5 million reads (between 5.9 and 19.6 million reads per library). 6.5 million reads (4.3%) were shorter than 16 bp and excluded from further analyses. The rates of uniquely mapped reads on the human and the chimpanzee genome, respectively, were between 48.4% and 78.1%, resulting in 90.4 million reads for methylation analyses (Supplementary Fig. 1, left diagrams). The uniquely mapped reads covered between 2.2 and 2.9 million methylation loci per human library and between 2.0 and 2.4 million per chimpanzee library. After removing loci which were not covered by at least 3 reads in at least 2 libraries per group, 1.3 million human methylation loci (49.1%) and 1.1 million chimpanzee methylation loci (49.5%) were tested for differential methylation in intraspecific and interspecific contrasts (Supplementary Fig. 1, right diagrams).
Principal component analysis revealed the highest methylation difference (axis 1, 28%) between neuronal versus non-neuronal cells, followed by the difference between humans and chimpanzees (axis 2, 19%) (Supplementary Fig. 2). According to anthropological age classifications (Goodall 1983; Bronikowski et al. 2016), 3 humans (67–77 years) and 2 chimpanzees (40 and 43 years) belonged to the old age class, whereas the 12-year old chimpanzee was adolescent. A global decline in DNA methylation occurs during ageing, although predominantly in domains with repetitive sequences (Kouzarides 2007). Age-associated methylation changes in genes are usually of much smaller effect size (in our experience in the order of several percentage points) than the observed methylation changes (in the order of 30–80%) described in our study (Supplementary Table S1). In addition, the postmortem intervals in humans (10–46 h) were considerably longer than in chimpanzees (5–6 h), which could affect DNA quality and DNA methylation variation (Rhein et al. 2015).
Therefore, we performed a redundancy analysis on the 539 023 methylation sites that were covered in all 12 libraries, including a linear model of the factors species, cell type, age, and postmortem interval. An ANOVA like permutation test (10 000 permutations) was applied to examine how well each factor explains the observed methylation levels. This test resulted in significant P values for the explanatory variables cell type (P = 0.0001) and species (P = 0.012) but not for the variables age (P = 0.13) and postmortem interval (P = 0.57). Thus, age and postmortem interval cannot significantly explain the response methylation level.
The majority of the tested methylation loci were located in CpG islands (70.7% for human and 75.7% for chimpanzee); CpG shores comprised 11.1% of the tested human and 11.2% of the chimpanzee loci, respectively. The distribution of methylation loci in the genomic landscape and gene model regions was consistent in both species (Supplementary Fig. 3). With 13.0% for human and 19.0% for chimpanzee, promoter regions (1.1% of the human and 0.9% of the chimpanzee genome) showed the highest density of tested loci. With 26.3% for human and 16.0% for chimpanzee libraries, 5’ UTRs (5.9% of the human and 2.5% of the chimpanzee genome) showed the second highest density.
For the intraspecific (neuronal vs. non-neuronal) contrasts, testing for differential methylation resulted in 34 182 significant loci in human and 20 393 significant loci in chimpanzee after multiple testing correction. The interspecific (human vs. chimpanzee) contrasts yielded 4778 significant loci in neuronal cells and 22 264 in non-neuronal cells after multiple testing correction.
Intraspecific and Interspecific DMRs
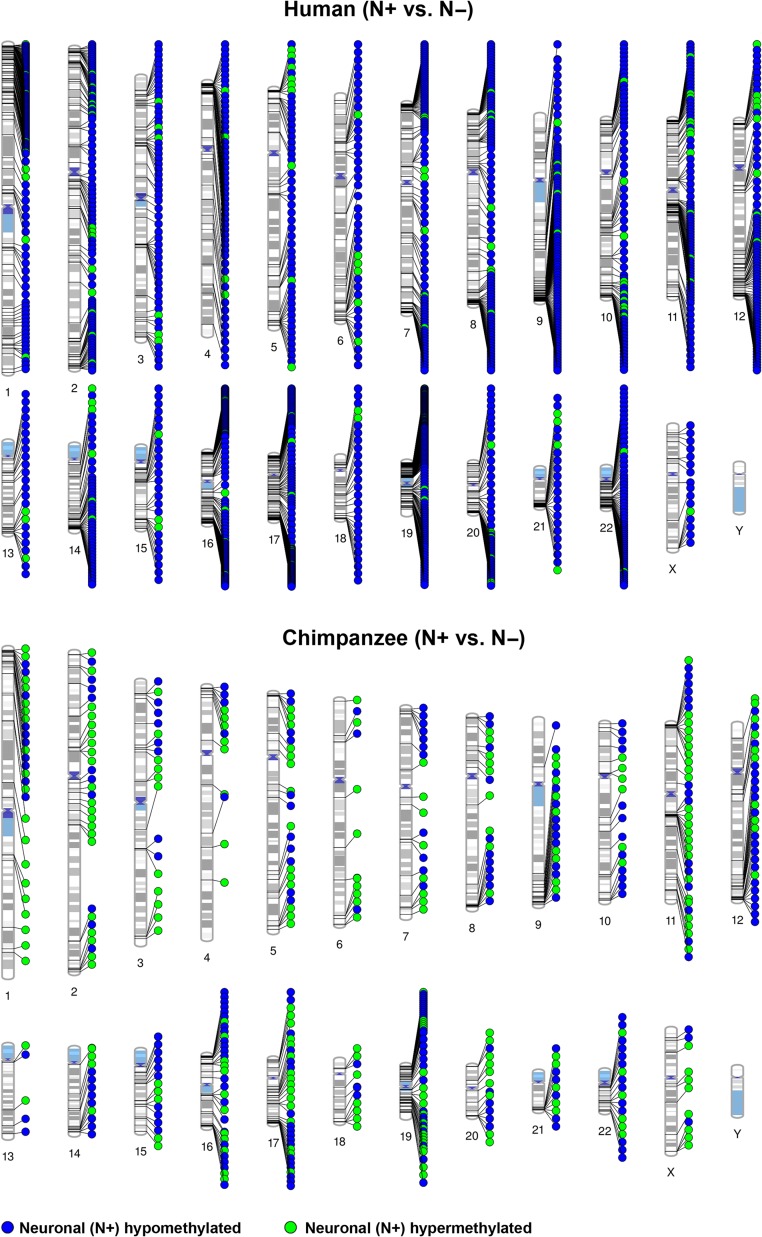
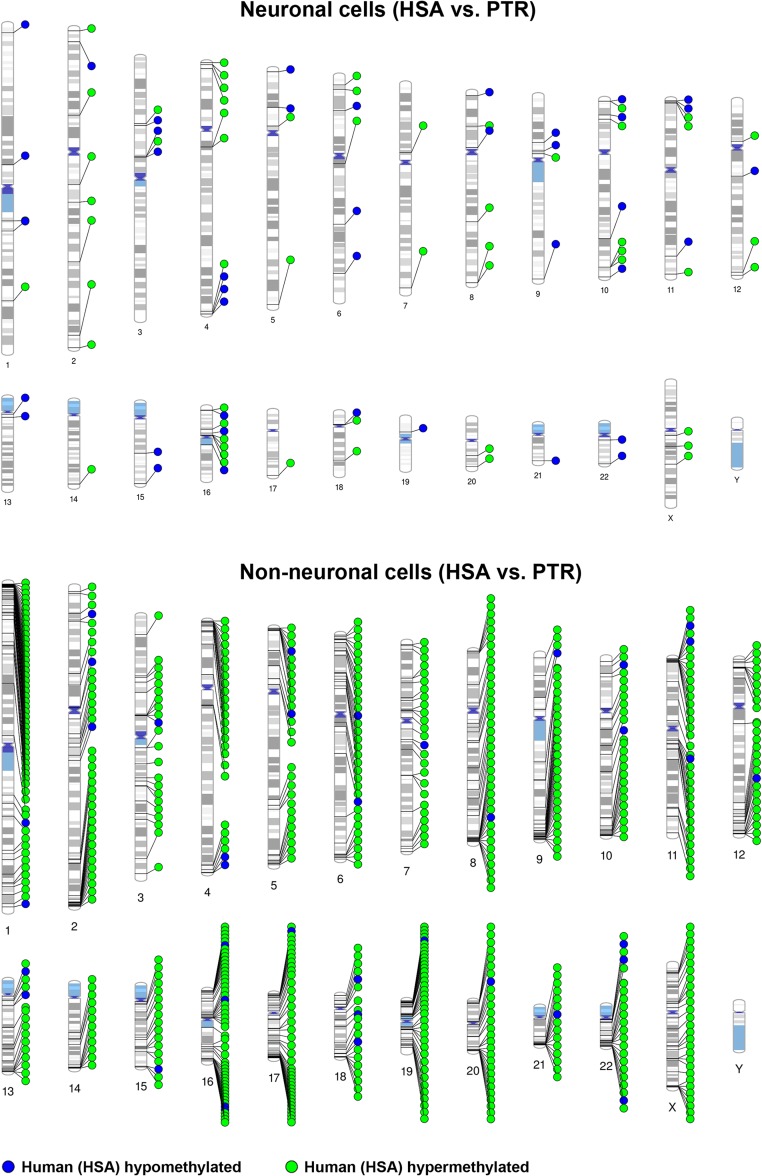
Figures 1 and 2 display the chromosomal distribution of intraspecific DMRs between N+ and N− cells in both species and of interspecific DMRs between HSA and PTR in both cell types. DMRs are present on all chromosomes (except Y, because only female samples were analyzed), with an obvious clustering in some chromosomal regions. In this context, it is important to mention that the CpG sites targeted by RRBS are also non-randomly distributed in the human genome (Supplementary Fig. 4). However, regional analysis considering the non-random distributions of targeted sites still revealed significant enrichment (Fisher test; P = 3.8E–46, factor 3.0 for human and P = 1.3E–125, factor 4.9 for chimpanzee) of differentially methylated CpGs in specific, i.e., subtelomeric chromosomal regions.
Figure 1.
Chromosomal distribution of intraspecific DMRs between neuronal (N+) and non-neuronal (N−) cells. The upper diagram shows human and the bottom diagram chimpanzee DMRs (mapped on the human karyotype). Blue dots represent DMRs that are hypomethylated, green dots DMRs that are hypermethylated in neuronal cells.
Figure 2.
Chromosomal distribution of interspecific DMRs between human (HSA) and chimpanzee (PTR). The upper diagram shows neuronal and the bottom non-neuronal cells. Blue dots represent DMRs that are hypomethylated, green dots DMRs that are hypermethylated in human cells.
Humans exhibited approximately 3 times more differences between N+ and N− cells than chimpanzees. Of 1636 human intraspecific DMRs, 128 were overlapping with promoters and 446 with genes. In chimpanzee, 50 and 358 of 496 DMRs were in promoters and genes, respectively. It is striking that in humans the vast majority (>90%) of intraspecific DMRs were hypomethylated in neurons, whereas the chimpanzee displayed comparable numbers of hypomethylated and hypermethylated neuronal DMRs (Fig. 1). A recent methylation array screen (Kozlenkov et al. 2014) identified 476 regions, which were differentially methylated between human N+ and N− cells, 166 of which overlapped with genes showing human intraspecific DMRs in our study. The vast majority (148 of 166; 89%) showed methylation changes in the same direction by both RRBS and microarrays. In contrast to our study, there was no bias towards hypomethylated neuronal DMRs detected by arrays. We also compared our list of 1636 intraspecific human DMRs with a list of 5163 intraspecific mouse DMRs (Kessler et al. 2016), which was based on re-analysis of a whole genome bisulfite sequencing dataset (Lister et al. 2013). One hundred twenty genes with intraspecific DMRs were hypomethylated and 2 hypermethylated in neurons of both species. The intraspecific DMRs in AP2A2, BAI1, BAIAP2, FBXL16, GABRB3, GRIN2A, KCNQ2, KDM4B, MYT1L, NGEF, NEU4, PLEC, PRKAG2, SHANK2, SYNPO, and TOLL1P have been replicated in independent studies in humans and conserved in humans, chimpanzee, and mouse (Supplementary Table 3).
The main focus of our study were differences between human and chimpanzee cell types. The number of such interspecific DMRs was considerably smaller in neuronal than in non-neuronal cells (Fig. 2). Of 96 interspecific neuronal DMRs, 36 were located in promoters and 64 in gene regions. Of 666 non-neuronal DMRs, 104 were in promoters and 446 in genes. Forty interspecific neuronal DMRs were hypomethylated and 56 hypermethylated in humans. In contrast, 95% of the interspecific non-neuronal DMRs were hypermethylated in humans. Table 1 shows 15 interspecific promoter DMRs in neuronal and 53 in non-neuronal cells which have been associated with bonafide genes. Notably, more than half of the identified interspecific promoter DMRs were associated with pseudogenes or transcripts of unknown significance. Cell-type specific methylation changes between human and chimpanzee cells ranged from >10% to 85%. Eight promoters exhibited almost identical evolutionary methylation changes in both cell types: ADM2, DUX4L9, and ZNF717 were hypomethylated, whereas FAM199X, FANK1/DHX32, GCNT2, KCNQ1OT1, and ZNF595 were hypermethylated in humans, compared with chimpanzee (Table 1). Collectively, these data suggest that human brain evolution was mainly associated with hypermethylation of human non-neuronal genes, resulting in a large number of human intraspecific DMRs with neuronal hypomethylation.
Table 1.
Genes with interspecific promoter DMRs in neuronal and non-neuronal cells
| Gene promoter | Chromosomal localization (bp)a | DMR size (bp) | Number of CpGs | Methylation (%) in HSA | Methylation (%) in PTR |
|---|---|---|---|---|---|
| Interspecific (HSA vs. PTR) promoter DMRs in neuronal cells | |||||
| SNORA26 | chr2:10 231 774–10 231 895 | 122 | 10 | 16↓ | 93↑ |
| HYAL2 | chr3:50 359 59–50 359 977 | 119 | 4 | 46↑ | 17↓ |
| ZNF717 | chr3:75 834 642–75 834 760 | 119 | 16 | 1↓ | 51↑ |
| ZNF595 | chr4:53 199–53 650 | 452 | 46 | 41↑ | 1↓ |
| DUX4L9 | chr4:190 943 271–190 943 995 | 725 | 12 | 6↓ | 70↑ |
| GCNT2 | chr6:10 491 902–10 492 049 | 148 | 10 | 89↑ | 6↓ |
| HCG11 | chr6:26 522 175–26 522 438 | 264 | 36 | 9↓ | 56↑ |
| FAM166B | chr9:35 562 915–35 563 884 | 970 | 3 | 31↓ | 46↑ |
| FANK1; DHX32 | chr10:127 584 458–127 584 670 | 213 | 27 | 42↑ | 5↓ |
| chr10:127 584 810–127 585 087 | 278 | 38 | 44↑ | 1↓ | |
| chr10:127 585 282–127 585 428 | 147 | 14 | 37↑ | 1↓ | |
| KCNQ1OT1 | chr11:2 720 727–2 720 965 | 239 | 8 | 35↑ | 24↓ |
| FOXR1 | chr11:118 842 511–118 842 623 | 113 | 8 | 12↓ | 57↑ |
| ADM2 | chr22:50 919 427–50 919 550 | 124 | 9 | 17↓ | 49↑ |
| FAM199X | chrX: 103 411 195–103 411 368 | 174 | 24 | 59↑ | 14↓ |
| Interspecific (HSA vs. PTR) promoter DMRs in non-neuronal cells | |||||
| MSH4 | chr1:76 262 740–76 262 858 | 119 | 9 | 38↑ | 1↓ |
| NDUFS2 | chr1:161 166 064–161 166 168 | 105 | 3 | 64↑ | 24↓ |
| EDARADD | chr1:236 511 571–236 511 701 | 131 | 8 | 81↑ | 6↓ |
| FAM132B | chr2:239 066 937–239 067 044 | 108 | 8 | 59↑ | 17↓ |
| ZNF717 | chr3:75 834 604–75 834 754 | 151 | 18 | 7↓ | 33↑ |
| ZNF595 | chr4:53 199–53 650 | 452 | 51 | 42↑ | 1↓ |
| DUX4L9 | chr4:190 943 271–190 943 980 | 710 | 11 | 5↓ | 71↑ |
| LRRC14B | chr5:192 104–192 228 | 125 | 16 | 63↑ | 16↓ |
| HUS1B | chr6:656 770–656 921 | 152 | 14 | 83↑ | 31↓ |
| C6orf201 | chr6:4 078 806–4 079 160 | 355 | 9 | 63↑ | 19↓ |
| GCNT2 | chr6:10 491 902–10 492 049 | 148 | 10 | 87↑ | 4↓ |
| ZFP57 | chr6:29 648 624–29 648 848 | 225 | 11 | 49↑ | 8↓ |
| LY6G5C | chr6:31 651 249–31 651 416 | 168 | 10 | 70↑ | 12↓ |
| LFNG | chr7:2 551 610–2 556 071 | 4462 | 4 | 88↑ | 29↓ |
| MYL10 | chr7:101 273 888–101 274 019 | 132 | 3 | 67↑ | 16↓ |
| GBX1 | chr7:150 871 359–150 872 590 | 1232 | 30 | 68↑ | 28↓ |
| LCN9 | chr9:138 553 850–138 559 100 | 5251 | 3 | 78↑ | 16↓ |
| FANK1;DHX32 | chr10:127 584 458–127 585 087 | 630 | 88 | 46↑ | 1↓ |
| chr10:127 585 267–127 585 428 | 162 | 15 | 40↑ | 1↓ | |
| KCNQ1OT1 | chr11:2 720 727–2 720 942 | 216 | 6 | 72↑ | 22↓ |
| PAX6 | chr11:31 840 149–31 840 575 | 427 | 4 | 70↑ | 26↓ |
| DAGLA | chr11:61 448 209–61 448 360 | 152 | 6 | 13↓ | 41↑ |
| RNF17 | chr13:25 338 594–25 338 721 | 128 | 6 | 95↑ | 45↓ |
| ADPRHL1 | chr13:114 107 515–114 107 633 | 119 | 11 | 84↑ | 3↓ |
| CCDC177 | chr14:70 039 335–70 039 894 | 560 | 13 | 79↑ | 23↓ |
| MAGEL2 | chr15:23 892 617–23 892 778 | 162 | 9 | 44↑ | 8↓ |
| KBTBD13; RASL12 | chr15:65 369 267–65 369 492 | 226 | 40 | 57↑ | 5↓ |
| SKOR1 | chr15:68 112 105–68 112 229 | 125 | 3 | 66↑ | 22↓ |
| DDX11L9 | chr15:102 520 460–102 520 753 | 294 | 11 | 76↑ | 1↓ |
| MSLN | chr16:810 668–811 141 | 474 | 5 | 62↑ | 15↓ |
| NDUFB10; RPL3L | chr16:2 004 823–2 008 441 | 3619 | 4 | 59↑ | 44↓ |
| QPRT | chr16:29 674 999–29 675 137 | 139 | 4 | 47↑ | 13↓ |
| NRN1L | chr16:67 918 866–67 919 928 | 1063 | 5 | 67↑ | 5↓ |
| MIR5189 | chr16:88 535 740–88 539 534 | 3795 | 5 | 79↑ | 36↓ |
| CCDC144NL | chr17:20 799 775–20 799 923 | 149 | 13 | 68↑ | 15↓ |
| HSA-MIR-6080 | chr17:62 776 922–62 777 639 | 718 | 18 | 69↑ | 15↓ |
| CTXN1 | chr19:7 991 826–7 992 062 | 237 | 14 | 54↑ | 5↓ |
| LINC00028 | chr20:30 072 017–30 072 157 | 141 | 24 | 33↑ | 2↓ |
| MIR4326 | chr20:61 915 520–61 919 256 | 3737 | 4 | 68↑ | 8↓ |
| LINC00319 | chr21:44 865 876–44 869 682 | 3807 | 3 | 72↑ | 20↓ |
| SNOZ6 | chr21:45 855 261–45 857 894 | 2634 | 4 | 78↑ | 29↓ |
| GAB4 | chr22:17 488 852–17 488 993 | 142 | 14 | 62↓ | 91↑ |
| MPST | chr22:37 414 515–37 414 724 | 210 | 25 | 44↑ | 8↓ |
| ADM2 | chr22:50 919 424–50 919 551 | 128 | 7 | 17↓ | 50↑ |
| CCT8L2 | chr22:17 073 105–17 077 894 | 4790 | 5 | 87↑ | 2↓ |
| SMS | chrX: 21 958 673–21 961 090 | 2418 | 7 | 45↑ | 15↓ |
| PORCN | chrX: 48 367 203–48 367 406 | 204 | 37 | 53↑ | 9↓ |
| PCSK1N | chrX: 48 693 805–48 693 951 | 147 | 29 | 54↑ | 17↓ |
| GPRASP2 | chrX: 101 967 560–101 967 714 | 155 | 9 | 33↑ | 2↓ |
| FAM199X | chrX: 103 411 119–103 411 368 | 250 | 33 | 60↑ | 15↓ |
| DOCK11 | chrX: 117 630 252–117 630 392 | 141 | 16 | 67↑ | 16↓ |
| DDX26B-AS1 | chrX: 134 655 175–134 655 343 | 169 | 14 | 41↑ | 2↓ |
| GDI1 | chrX: 153 665 659–153 665 766 | 108 | 19 | 55↑ | 10↓ |
aGenomic coordinates are based on Ensembl release 75.
DMRs and Neuropsychiatric Diseases
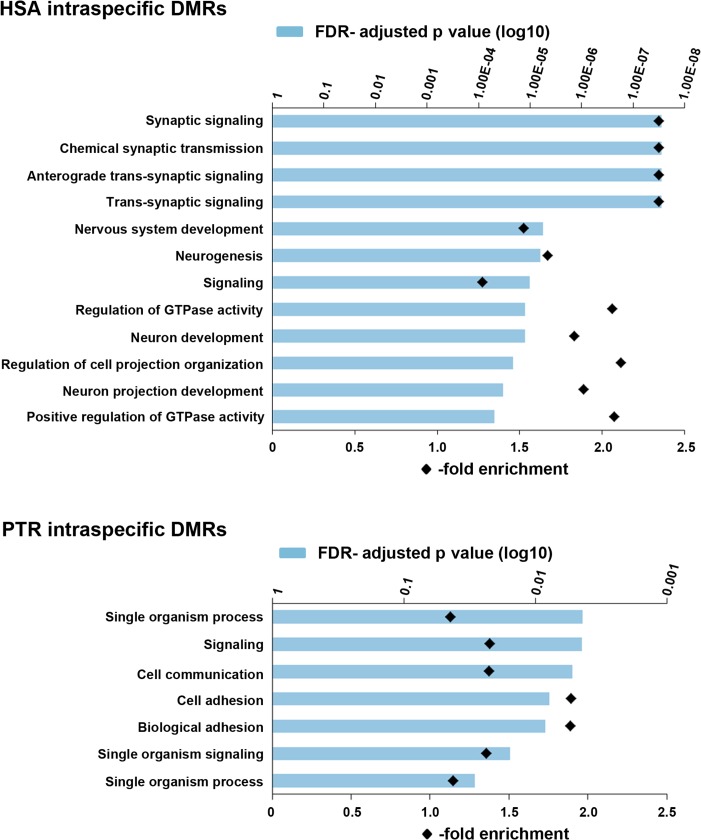
To gain further insights into the functional and biological role of the identified DMRs, genes and promoter regions overlapping the 2313 intraspecific and 762 interspecific DMRs were used to run an Gene ontology (GO) enrichment analysis. Human intraspecific DMRs showed significant enrichment for 52 biological processes, many of which were associated with neurogenesis and synaptic signaling. Figure 3 presents the top 12 GO terms with FDR-adjusted P < 1.00E–04. Chimpanzee intraspecific DMRs showed significant enrichment for 7 GO terms with P values between 0.05 and 0.005. Genes involved in neurogenesis, including CELSR1 (in N− cells), CDH4 (N+, N−), GDI1 (N−) ILRAPL1 (N−), OPCML (N+, N−), OPHN1 (N+, N−), and SHANK3 (N−) were approximately 2 times enriched with interspecific neuronal and/or non-neuronal DMRs (Table 2). These genes are primary candidates for epigenetic regulators underlying accelerated human brain evolution.
Figure 3.
Enrichment of GO terms for biological processes in genes with intraspecific DMRs (N+ vs. N− cells). The blue bars indicate the FDR-adjusted P value for enrichment of a given term (on the Y-axis). Black diamonds indicate the degree of enrichment (fold overrepresentation). The upper diagram shows the top 12 (of 52) terms in human intraspecific DMRs, the bottom the 7 significant terms in chimpanzee intraspecific DMRs.
Table 2.
Genes for neurogenesis with interspecific DMRs
| Gene | Chromosomal localization (bp)a | DMR size (bp) | Number of CpGs | Methylation (%) in HSA | Methylation (%) in PTR |
|---|---|---|---|---|---|
| Interspecific (HSA vs. PTR) DMRs for neurogenesis in neuronal cells | |||||
| NOTCH1 | Chr9: 139 389 718–139 393 758 | 4041 | 6 | 36↓ | 85↑ |
| KNDC1 | Chr10: 135 010 300–135 010 673 | 374 | 6 | 7↓ | 37↑ |
| OPCML | Chr11: 132 662 857–132 663 105 | 249 | 7 | 84↑ | 10↓ |
| LRP1 | Chr12: 57 591 001–57 591 112 | 112 | 5 | 17↓ | 70↑ |
| MAP1S | Chr19: 17 838 761–17 838 874 | 114 | 5 | 6↓ | 55↑ |
| CDH4 | Chr20: 60 511 870–60 516 006 | 4137 | 5 | 69↑ | 37↓ |
| OPHN1 | ChrX: 67 352 685–67 352 815 | 131 | 11 | 70↑ | 29↓ |
| Interspecific (HSA vs. PTR) DMRs for neurogenesis in non-neuronal cells | |||||
| PRDM16 | Chr1: 3 011 508–3 011 608 | 101 | 8 | 60↑ | 23↓ |
| Chr1: 3 028 919–3 029 108 | 190 | 12 | 87↑ | 4↓ | |
| Chr1: 3 038 277–3 038 394 | 118 | 9 | 65↑ | 14↓ | |
| Chr1: 3 148 336–3 148 455 | 120 | 5 | 91↑ | 49↓ | |
| Chr1: 3 103 003–3 103 272 | 270 | 11 | 73↑ | 38↓ | |
| CHD5 | Chr1: 6 170 013–61 70 141 | 129 | 12 | 61↑ | 11↓ |
| CASZ1 | Chr1: 10 764 500–10 764 665 | 166 | 12 | 64↑ | 26↓ |
| Chr1: 10 811 734–10 813 837 | 2104 | 10 | 77↑ | 32↓ | |
| PTCHD2 | Chr1: 11 561 458–11 562 143 | 686 | 26 | 53↑ | 13↓ |
| MINOS;NBL1 | Chr1: 19 983 375–19 983 497 | 123 | 7 | 55↑ | 9↓ |
| EPHA8 | Chr1: 22 918 367–22 918 520 | 154 | 3 | 87↑ | 28↓ |
| ASTN1 | Chr1: 177 001 756–177 001 904 | 149 | 7 | 89↑ | 5↓ |
| SLC45A3 | Chr1: 205 631 992–205 632 102 | 111 | 9 | 87↑ | 29↓ |
| SDCCAG8 | Chr1: 243 637 718–243 637 844 | 127 | 10 | 89↑ | 42↓ |
| KIF26B | Chr1: 245 851 577–245 851 741 | 165 | 13 | 12↓ | 60↑ |
| GLI2 | Chr2: 121 532 650–121 532 965 | 316 | 2 | 95↑ | 56↓ |
| NRP2 | Chr2: 206 572 534–206 572 645 | 112 | 5 | 79↑ | 21↓ |
| PLXND1 | Chr3: 129 317 949–129 318 104 | 156 | 12 | 54↑ | 10↓ |
| EIF2B5 | Chr3: 184 377 554–184 377 665 | 112 | 5 | 67↑ | 24↓ |
| GFRA3 | Chr5: 137 593 424–137 593 714 | 291 | 18 | 67↑ | 18↓ |
| LHFPL5 | Chr6: 35 778 755–35 778 892 | 138 | 4 | 83↑ | 49↓ |
| GLI3 | Chr7: 42 004 666–42 004 966 | 301 | 14 | 69↑ | 24↓ |
| CAMK2B | Chr7: 44 279 728–44 279 853 | 126 | 3 | 81↑ | 26↓ |
| GBX1 | Chr7: 150 871 359–150 872 590 | 1232 | 30 | 68↑ | 28↓ |
| MNX1 | Chr7: 156 800 542–156 800 664 | 123 | 3 | 42↑ | 2↓ |
| ARHGEF10 | Chr8: 1 788 181–1 788 341 | 161 | 3 | 95↑ | 18↓ |
| Chr8: 1 877 950–1 879 119 | 1170 | 9 | 72↑ | 15↓ | |
| Chr8: 1 882 222–1 885 011 | 2790 | 3 | 89↑ | 11↓ | |
| ARFGEF1 | Chr8: 68 113 651–68 113 763 | 113 | 4 | 93↑ | 25↓ |
| TRAPPC9 | Chr8: 141 454 885–141 455 166 | 282 | 4 | 75↑ | 27↓ |
| ABL1 | Chr9: 133 748 333–133 748 450 | 118 | 5 | 73↑ | 29↓ |
| CDH23 | Chr10: 73 222 997–73 223 132 | 136 | 3 | 39↑ | 1↓ |
| PAX6 | Chr11: 31 840 149–31 840 575 | 427 | 4 | 70↑ | 26↓ |
| DAGLA | Chr11: 61 448 209–61 448 360 | 152 | 6 | 13↓ | 41↑ |
| MYRF | Chr11: 61 545 421–61 546 428 | 1008 | 5 | 56↑ | 17↓ |
| MYO7A | Chr11: 76 848 977–76 849 143 | 167 | 15 | 66↑ | 31↓ |
| DRD2 | Chr11: 113 283 246–113 283 362 | 117 | 6 | 59↑ | 4↓ |
| OPCML | Chr11: 132 662 753–132 663 105 | 353 | 17 | 75↑ | 4↓ |
| FMN1 | Chr15: 33 360 195–33 360 385 | 191 | 11 | 65↑ | 7↓ |
| SKOR1 | Chr15: 68 112 105–68 112 229 | 125 | 3 | 66↑ | 22↓ |
| NRN1L | Chr16: 67 918 866–67 919 928 | 1063 | 5 | 67↑ | 5↓ |
| RTN4RL1 | Chr17: 1 918 769–1 918 869 | 101 | 5 | 93↑ | 45↓ |
| GFAP | Chr17: 42 989 090–42 989 653 | 564 | 4 | 76↑ | 32↓ |
| SDK2 | Chr17: 71 391 468–71 393 610 | 2143 | 7 | 86↑ | 60↓ |
| BAIAP2 | Chr17: 79 022 690–79 023 511 | 822 | 5 | 69↑ | 19↓ |
| Chr17: 79 045 375–79 045 513 | 139 | 8 | 69↑ | 6↓ | |
| MAG | Chr19: 35 800 743–35 801 231 | 489 | 12 | 85↑ | 34↓ |
| SPTBN4 | Chr19: 41 025 452–41 025 556 | 105 | 14 | 68↑ | 20↓ |
| CDH4 | Chr20: 60 277 050–60 277 173 | 124 | 5 | 61↑ | 14↓ |
| RTN4R | Chr22: 20 229 564–20 229 670 | 107 | 7 | 8↓ | 53↑ |
| TSPO | Chr22: 43 553 694–43 555 961 | 2268 | 4 | 75↑ | 21↓ |
| CELSR1 | Chr22: 46 847 905–46 848 027 | 123 | 6 | 89↑ | 45↓ |
| WNT7B | Chr22: 46 315 247–46 318 508 | 3262 | 4 | 84↑ | 27↓ |
| SHANK3 | Chr22: 51 169 861–51 172 813 | 2953 | 4 | 83↑ | 21↓ |
| IL1RAPL1 | ChrX: 28 632 961–28 633 099 | 139 | 5 | 71↑ | 20↓ |
| OPHN1 | ChrX: 67 352 685–67 352 923 | 239 | 26 | 68↑ | 12↓ |
| PLXNB3 | ChrX: 153 043 548–153 044 190 | 643 | 9 | 62↑ | 13↓ |
| GDI1 | ChrX: 153 665 659–153 665 766 | 108 | 19 | 55↑ | 10↓ |
| ChrX: 153 673 108–153 673 230 | 123 | 7 | 60↑ | 11↓ | |
aGenomic coordinates are based on Ensembl release 75.
To test whether genes/promoters overlapping DMRs are enriched with genes for neuropsychiatric disorders, we used published gene lists (Supplementary Table 1). With exception of the GDI1 promoter, all ID genes with interspecific DMRs were hypermethylated in the human gene body, which is usually associated with gene activation (Jones 2012). The X-linked OPHN1 gene was hypermethylated in both human N+ and N− cells, compared with the chimpanzee. Two other X-linked ID genes, IL1RAPL1 and GDI1, were hypermethylated in human non-neuronal cells. MYT1L was hypermethylated in a CTCF binding region (Chr2:1 817 284–1 818 177) of human versus chimpanzee neurons. The remaining gene body was hypomethylated in N+ versus N− cells in both species. Intraspecific human DMRs were significantly enriched (Fisher exact test) in candidate genes for ID, autism, bipolar disorders and Alzheimer disease, intraspecific chimpanzee DMRs in genes for autism and bipolar disorders (Table 3; Supplementary Table 1). In contrast, interspecific DMRs did not show any significant enrichment for genes associated with neuropsychiatric disorders.
Table 3.
P values and odds ratios for an enrichment of DMRs in gene lists for neuropsychiatric diseases
| Intraspecific (N+ vs. N−) DMRs | Interspecific (HSA vs. PTR) DMRs | |||
|---|---|---|---|---|
| HSA | PTR | N+ | N− | |
| Intellectual disability | 0.002* [OR 2.73] (n = 17) | 0.46 [OR 1.68] (n = 4) | 0.15 [OR 4.89] (n = 2) | 0.08 [OR 2.42] (n = 7) |
| Autism | 6.1E–05* [OR 2.95] (n = 27) | 0.002* [OR 3.52] (n = 12) | 0.61 [OR 1.57] (n = 1) | 0.11 [OR 2.09] (n = 9) |
| Bipolar disorders | 0.001* [OR 2.24] (n = 39) | 0.005* [OR 2.49] (n = 16) | 1.0 [OR 0.84] (n = 1) | 0.61 [OR 1.21] (n = 10) |
| Schizophrenia | 0.61 [OR 1.19] (n = 20) | 0.34 [OR 1.49] (n = 9) | 1.0 [OR 0.93] (n = 1) | 1.0 [OR 0.91] (n = 7) |
| Alzheimer disease | 0.007* [OR 2.73] (n = 13) | 1.0 [OR 0.50] (n = 1) | 1.0 [OR 0] (n = 0) | 0.32 [OR 0] (n = 0) |
Candidate gene lists consisted of 138 genes for ID, 203 genes for autism, 408 genes for bipolar disorders, 351 genes for schizophrenia, and 129 genes for Alzheimer disease.
n indicates the number of candidate genes with DMRs.
Significant enrichments are indicated by asterisks.
DMRs and Human-specific Changes in Brain Gene Regulation during Hominine Brain Evolution
Next we compared genes/promoters overlapping DMRs with 540 genes showing known human-specific expression patterns (Liu et al. 2012) and 441 genes close to human-specific histone methylation signatures (Shulha et al. 2012) in frontal cortex. Intraspecific DMRs between human N+ and N− cells were significantly enriched in genes with human-specific expression (P = 6E–05, OR = 2.1) and histone modifications (P = 1E–04, OR = 2.4). In total, 87 genes with known human-specific regulation were endowed with intraspecific, often multiple DMRs in humans and 23 in chimpanzee (Supplementary Table 4).
Interspecific DMRs were significantly enriched (P = 0.007, OR = 5.7 in neuronal and P = 0.001, OR = 2.8 in non-neuronal cells) in genes with human-specific histone signatures. Altogether, 5 interspecific DMRs in neuronal cells and 24 in non-neuronal cells were associated with genes showing human-specific regulation in the brain (Supplementary Table 4). Since the expression values of genes with human-specific brain expression (Liu et al. 2012) were not publicly available, we re-analyzed the corresponding microarray dataset (GSE22570) of 25 human and 12 chimpanzee frontal cortex samples. Consistent with an inverse relationship between DNA methylation and gene expression, 7 of 9 interspecific DMRs (OPCML in N+, ARHGEF10, CAMK2B, OPCML, PLXND1, SYNJ2, and TCERG1L in N−) showed higher methylation and lower gene expression in humans. GSTO2 displayed hypomethylation and underexpression in human neuronal, GFAP hypermethylation and overexpression in human non-neuronal cells.
Three genes with human-specific regulation showed species-specific methylation changes in both neuronal and non-neuronal cells. OPCML and TNFRSF11A were hypermethylated in both human cell types. The transcription factor ZNF717 displayed complex species- and cell-type-specific methylation. The promoter region was hypomethylated in human neuronal (chr3:75 834 642–75 834 760) and non-neuronal cells (75 834 604–75 834 754), compared with chimpanzee. The gene region following the promoter (75 827 859–75 829 467) was hypermethylated in human versus chimpanzee non-neuronal cells, but not in neuronal cells. The gene body (75 827 263–75 827 404, 75 827 718–75 827 965 in HSA; 75 826 348–75 827 894, 75 829 292–75 829 467 in PTR) was hypomethylated in human and hypermethylated in chimpanzee neurons, compared with non-neuronal cells.
DMR Validation by Bisulfite Pyrosequencing
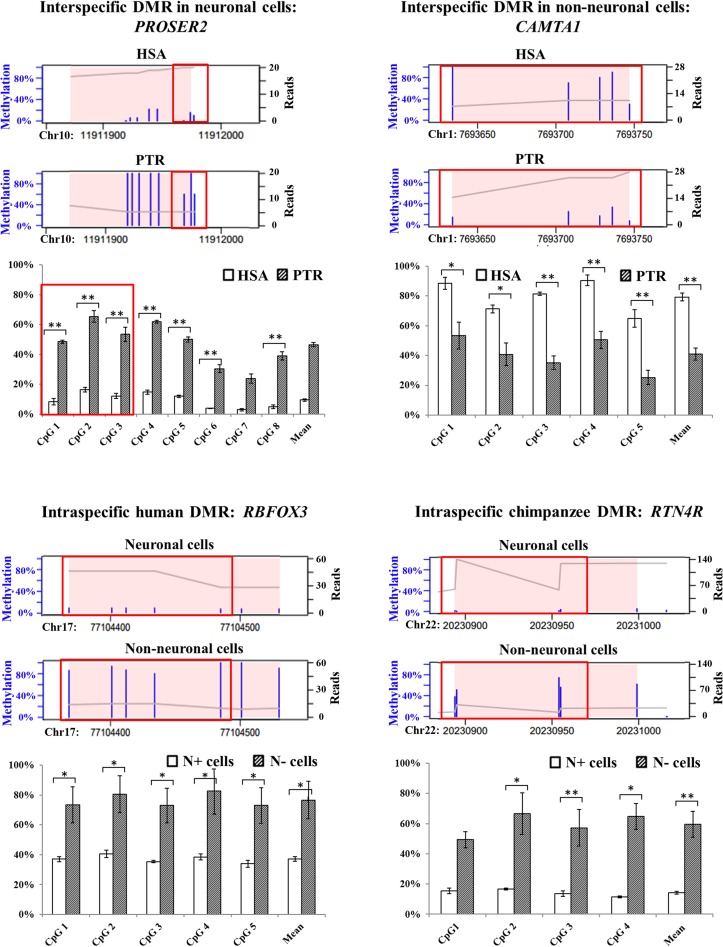
To test the quality of our RRBS data, we selected one DMR from each contrast for validation: PROSER2 as an interspecific neuronal DMR (HSA 25%, PTR 89%), CAMTA1 as an interspecific non-neuronal DMR (77% in HSA, 21% in PTR), RBFOX3 as an intraspecific human DMR (N+ 4%, N− 65%), and RTN4R as an intraspecific chimpanzee DMR (N+ 2%, N− 45%). Bisulfite pyrosequencing was used to quantify methylation of 8 RRBS CpGs in PROSER2, 5 RRBS CpGs in CAMTA1, 5 RRBS CpGs in RBFOX3, and 4 RRBS CpGs in RTN4R (Fig. 4). With exception of CpG1 in RTN4R, all target RRBS CpGs exhibited significant methylation differences in the same direction by pyrosequencing. The mean methylation difference of the targeted CpGs in a given region was −38% (P = 0.0002) for PROSER2 (−66%, P = 0.0009 by RRBS), +43% (P = 0.008) for CAMTA1 (+56%, P = 0.008 by RRBS), −39% (P = 0.008) for RBFOX3 (−62%, P = 0.012 by RRBS), and −45% (P = 0.029) for RTN4R (−41%, P = 0.029 by RRBS).
Figure 4.
Comparison of RRBS and bisulfite pyrosequencing results for PROSER2 (interspecific neuronal DMR), CAMTA1 (interspecific non-neuronal DMR), RBFOX3 (intraspecific human DMR), and RTN4R (intraspecific chimpanzee DMR). Thin blue bars in the RRBS plots indicate the methylation levels (0–100%) of single CpGs. The X-axis indicates the genomic position of the identified DMR, which is highlighted by a pink background. The gray line represents the sequence coverage (number of reads for each nucleotide). The bar diagrams below the RRBS plots indicate the single CpG methylation levels of a given region measured by bisulfite pyrosequencing. Significance levels for an inter- or intraspecific methylation difference are indicated by one star (P < 0.05) and two stars (P < 0.01), respectively. Identical CpGs in the RRBS and bisulfite pyrosequencing data are framed in red.
DMRs in Repetitive Elements
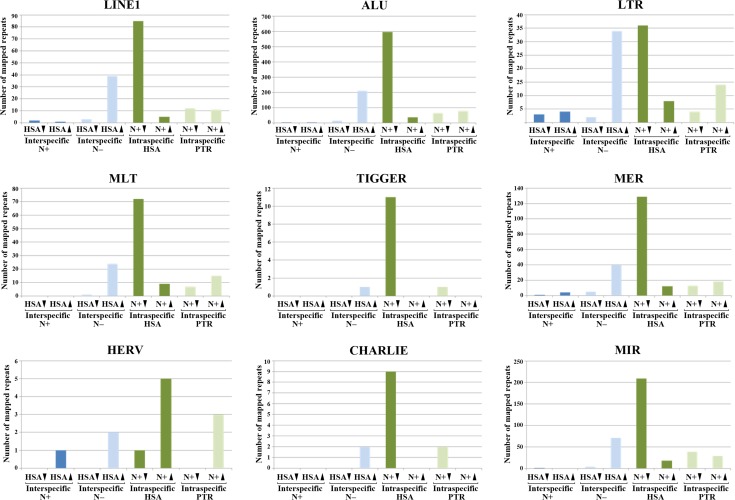
A small proportion of DMRs can be mapped to specific genomic positions of repetitive elements because of unique sequence associated with the repeat in the respective reads. For example, we identified DMRs in approximately 150 LINE1 and 1000 ALU repeats. Although these mappable DMRs represent only approximately 0.1% of >100 000 LINE1 and >1 000 000 ALU elements in the human genome (Treangen and Salzberg 2011), it is striking that most DMRs in LINE1, ALU, and other repetitive elements (Fig. 5) were hypomethylated in human neurons, compared with non-neuronal cells. The second most frequent class of repeat DMRs were hypermethylated in human versus chimpanzee glia cells. The majority (>70%) of repeat DMRs were located in genes, many of which are linked to brain function and/or neurological diseases. One interesting example is ARHGEF10, a gene with human-specific brain expression and 3 interspecific non-neuronal DMRs (Supplementary Table 4). The largest of these DMRs (chr8:1 882 222–1 885 011) contains two LINE1 elements (chr8:1 882 726–1 883 281 and 1 883 277–1 883 653) with 89% methylation in humans and 11% in chimpanzees. It is also noteworthy that 25 (16%) of 156 identified LINE1 DMRs are associated with regulatory microRNAs. Similar to all DMRs (Figs 1 and 2), repeat DMRs are clustered in particular chromosome regions (Supplementary Fig. 5).
Figure 5.
Interspecific (blue bars) and intraspecific DMRs (green bars) in different repeat classes. The number of repeat DMRs which could be uniquely mapped on the reference genome is indicated on the Y-axis. Different repeat classes appear to be undermethylated in human neurons, compared with non-neuronal cells. In addition, many repeats exhibit higher methylation in human than in chimpanzee glia.
Discussion
Neuronal Hypomethylation of Human Intraspecific DMRs
RRBS identified intraspecific DMRs between neuronal and non-neuronal cells as well as interspecific DMRs between human and chimpanzee cell fractions. In humans, most intraspecific DMRs displayed neuronal hypomethylation. A subset of these intraspecific DMRs have been conserved in human (Kozlenkov et al. 2014; this study), chimpanzee (this study), and mouse (Kessler et al. 2016), showing the same cell-type specific methylation in rodents and primates. Intraspecific DMRs were enriched in candidate genes for ID, autism, bipolar disorders, and Alzheimer disease. GO analysis revealed enrichments with numerous biological processes which are essential for brain development and function. This is consistent with the view that intraspecific DMRs play an essential role in regulating the highly complex (temporal and spatial) cooperation between neuronal and glia cells in the brain.
Neuronal hypomethylation also affects a number of human intraspecific DMRs containing repetitive elements. Because retrotransposition activity of LINE1 and other transposable elements is associated with genomic instability, their expression and mobilization is usually suppressed in most cell types. DNA methylation and alterations in chromatin structure/condensation play an important role in this process (Slotkin and Martienssen 2007). In the developing human brain, LINE1 promoters become transcriptionally activated during differentiation of neural precursor cells into neurons and glia (Muotri et al. 2005; Coufal et al. 2009). Retrotransposition events are more common in neurons than in other neural cell types, generating neuronal somatic mosaicism (Evrony et al. 2012; Bundo et al. 2014). This increased retrotransposition activity could be consequence or cause of neuronal hypomethylation during human brain evolution.
Interspecific DMRs
This is the first study systematically exploring methylation differences between human versus chimpanzee neurons and glia, respectively. Only a limited number of 96 interspecific DMRs was identified in neurons, compared with 666 interspecific DMRs in non-neuronal cells. Because of its large size, the human brain has an extraordinary energy demand. In humans the developing newborn brain requires 60–80% and the adult brain 20–25% of the resting metabolic rate (RMR). This is outstanding compared to an average RMR of <10% in non-human primates (Aiello and Wheeler 1995; Gibbons 1998). It is interesting to speculate that the much higher number of evolutionary DMRs in non-neuronal cells reflects adaptive changes in brain homeostasis to increase human brain metabolism and synaptic transmission. The evolution of higher cognitive functions in humans is associated with an increased utilization of glucose and oxygen. The trophic functions of glial cells are pivotal for metabolic maintenance of the nervous system and meeting the increased energy requirements of active neurons (Deitmer 2001). Accumulating evidence suggests that glial-neuronal interactions can also modulate neuronal transmission and synaptic plasticity (Ben Acour and Pascual 2010; Perea and Araque 2010). With only few exceptions, the non-neuronal interspecific DMRs were hypermethylated in the human brain.
The observation that interspecific (both neuronal and non-neuronal) DMRs are enriched in genes showing human-specific brain regulation supports a role of interspecific DMRs in human brain evolution. One of the most interesting candidates is ZNF717, which displays promoter hypomethylation in the human brain (in neuronal and non-neuronal cells), followed by a hypermethylated region (in human non-neuronal cells). Moreover, the gene body contains an intraspecific DMR which is hypomethylated in human and hypermethylated in chimpanzee neurons. The function of the Kruppel-associated box (KRAB) zinc finger protein 717 as a transcriptional regulator is not well known. A microarray screen (Nowick et al. 2009) identified 90 transcription factors with expression differences between human and chimpanzee brain, forming a regulatory network. This network consists of two distinct modules, which are linked by ZNF717. Because of its central role in coordinating concerted changes in a gene expression network, changes in the epigenetic regulation of ZNF717 may have functioned as a trigger in human brain evolution. KCNQ1OT1 is slightly hypermethylated (11 percentage points) in human neurons, and extensively hypermethylated (50 percentage points) in human glia, compared with chimpanzee. The analyzed DMR lies in the Beckwith-Wiedemann imprinting control region 2, which regulates the imprinted expression of the antisense transcript KCNQ1OT1 (LIT1) and the growth inhibitor CDKN1C (Azzi et al. 2014). ARHGEF10 is endowed with 3 interspecific non-neuronal DMRs, which are hypermethylated (>50 percentage points) in humans versus chimpanzee. The Rho guanine nucleotide exchange factor 10 is highly expressed in the developing brain and thought to play a role in developmental myelination (Verhoeven et al. 2003). Arhgef10 knockout mice are a model for impaired social behaviors in autism (Lu et al. 2018). Moreover, ARHGEF10 has been associated with schizophrenia (Jungerius et al. 2008).
Limitations
Our study has several limitations that have to be taken into consideration for interpretation of the results. Since we were not able to include an outgroup in our cell-type specific methylome analysis, polarity of epigenetic characters was not unequivocally defined. Although it seems plausible to assume that the most dramatic changes occurred during human brain evolution, we cannot assign whether a specific change occurred in the human or the chimpanzee lineage.
Because of ethical and other issues, the number of cortex samples was relatively small and they were not perfectly matched. For example, the postmortem intervals differed between human (10–46 h) and chimpanzee brain samples (5–6 h). However, DNA methylation patterns are much more stable than RNA. In fact, bisulfite sequencing has even been used to study DNA methylation of archeological samples (Llamas et al. 2012). Candidate gene analyses in Alzheimer brains did not show changes in methylation profiles with postmortem delay up to 48 h (Barrachina and Ferrer 2009). Prolonged postmortem times leading to DNA degradation did not change mean methylation but methylation variance increased (Rhein et al. 2015).
Three humans and 2 chimpanzees were of old age, whereas one chimpanzee was adolescent. The increased inner-group variance due to higher age variability in chimpanzees than in humans may reduce the number of observed DMRs in the chimpanzee intraspecific (N+ vs. N−) contrast. However, it should have the same effect on the interspecific (HSA vs. PTR) contrasts. Thus it does not explain the much higher frequency of interspecific DMRs in non-neuronal, compared with neuronal cells.
A redundancy analysis showed only weak effects of age and postmortem interval on our methylation data, compared with cell type and species. Although differences in age and postmortem intervals may well increase technical noise and inner-group variance, they are highly unlikely to feign significant changes of large effect size (>50% methylation difference) between cell types and/or species.
Conclusions
The epigenetic differences between human neuronal and non-neuronal cells (1636 intraspecific human DMRs) are much more frequent than between human and chimpanzee (96 interspecific neuronal and 666 interspecific non-neuronal DMRs). Evolutionary DNA methylation changes in neuronal and more abundantly in non-neuronal cells may have contributed to the increased size and cognitive abilities of the human brain. We propose that adaptive changes in the human brain were accompanied by hypermethylation of several hundred genes in non-neuronal cells. From an epigenetic point of view, the contribution of the different non-neuronal cell populations (microglia, astrocytes, and oligodendrocytes) to human brain evolution has been underestimated. In fact, the metabolic capabilities of the enhanced human brain, which requires a much higher RMR than the chimpanzee, were pushed to the limit (Khaitovich et al. 2008). In addition, non-neuronal cells do not only provide physical and nutritional support for neurons but substantially impact neuronal functions. Neuron–glia interactions have important roles in the process of mental activities and neuropsychiatric disorders (Kato et al. 2013; Jäkel and Dimou 2017). Therefore, it is not unexpected that many of the identified intra- and interspecific DMRs have been associated with neuropsychiatric disorders.
Supplementary Material
Authors’ Contributions
J.B. and N.E.H. performed the molecular genetic experiments. J.B. and E.S. drafted an initial version of the manuscript. C.R., M.D., and T.M. carried out bioinformatic and statistical analyses. I.K., S.P., and R.B. provided chimpanzee samples. T.K. and A.G. provided human samples and carried out cell purifications. C.A. helped with next generation sequencing. E.S. and T.H. designed and supervised the study. T.H. coordinated the experiments and drafted the final version of the manuscript. All authors read and approved the manuscript.
Funding
This work was supported by a research grant from the German Research Foundation (SCHN 1367/1-1).
Footnotes
We would like to thank Virginie Guibourt for excellent technical support in brain tissue preparation and Sebastian Bultmann and Heinrich Leonhardt for support with nuclei sorting. Conflict of Interest: The authors declare that they have no competing interests.
References
- Aiello LC, Wheeler P. 1995. The expensive-tissue hypothesis: the brain and the digestive system in human and primate evolution. Curr Anthropol. 36:199–221. [Google Scholar]
- Azzi S, Abi Habib W, Netchine I. 2014. Beckwith-Wiedemann and Russell-Silver syndromes: from new molecular insights to the comprehension of imprinting regulation. Curr Opin Endocrinol Diabetes Obes. 21:30–38. [DOI] [PubMed] [Google Scholar]
- Barrachina M, Ferrer I. 2009. DNA methylation of Alzheimer disease and tauopathy-related genes in postmortem brain. J Neuropathol Exp Neurol. 68:880–891. [DOI] [PubMed] [Google Scholar]
- Basu SN, Kollu R, Banerjee-Basu S. 2009. AutDB: a gene reference resource for autism research. Nucleic Acids Res. 37:D832–D836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Achour S, Pascual O. 2010. Glia: the many ways to modulate synaptic plasticity. Neurochem Int. 57:440–445. [DOI] [PubMed] [Google Scholar]
- Benton MJ, Donoghue PC. 2007. Paleontological evidence to date the tree of life. Mol Biol Evol. 24:26–53. [DOI] [PubMed] [Google Scholar]
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. 2007. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 39:17–23. [DOI] [PubMed] [Google Scholar]
- Blekhman R, Oshlack A, Chabot AE, Smyth GK, Gilad Y. 2008. Gene regulation in primates evolves under tissue-specific selection pressures. PLoS Genet. 4:e1000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle P, Clement K, Gu H, Smith ZD, Ziller M, Fostel JL, Holmes L, Meldrim J, Kelley F, Gnirke A, et al. . 2012. Gel-free multiplexed reduced representation bisulfite sequencing for large-scale DNA methylation profiling. Genome Biol. 13:R92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronikowski AM, Cords M, Alberts SC, Altmann J, Brockman DK, Fedigan LM, Pusey A, Stoinski T, Strier KB, Morris WF. 2016. Female and male life tables for seven wild primate species. Sci Data. 3:160006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundo M, Toyoshima M, Okada Y, Akamatsu W, Ueda J, Nemoto-Miyauchi T, Sunaga F, Toritsuka M, Ikawa D, Kakita A, et al. . 2014. Increased l1 retrotransposition in the neuronal genome in schizophrenia. Neuron. 81:306–313. [DOI] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 437:69–87. [DOI] [PubMed] [Google Scholar]
- Chopra P, Papale LA, White AT, Hatch A, Brown RM, Garthwaite MA, Roseboom PH, Golos TG, Warren ST, Alisch RS. 2014. Array-based assay detects genome-wide 5-mC and 5-hmC in the brains of humans, non-human primates, and mice. BMC Genomics. 15:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury SR, Cui Y, Narayanan A, Gilley DP, Huda N, Lo CL, Zhou FC, Yernool D, Irudayaraj J. 2016. Optogenetic regulation of site-specific subtelomeric DNA methylation. Oncotarget. 7:50380–50391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O’Shea KS, Moran JV, Gage FH. 2009. L1 retrotransposition in human neural progenitor cells. Nature. 460:1127–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitmer JW. 2001. Strategies for metabolic exchange between glial cells and neurons. Respir Physiol. 129:71–81. [DOI] [PubMed] [Google Scholar]
- Ding Z, Mangino M, Aviv A, Spector T, Durbin R, UK10K Consortium . 2014. Estimating telomere length from whole genome sequence data. Nucleic Acids Res. 42:e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrony GD, Cai X, Lee E, Hills LB, Elhosary PC, Lehmann HS, Parker JJ, Atabay KD, Gilmore EC, Poduri A, et al. . 2012. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. 151:483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk D. 2016. Evolution of brain and culture: the neurological and cognitive journey from Australopithecus to Albert Einstein. J Anthropol Sci. 94:99–111. [DOI] [PubMed] [Google Scholar]
- Farcas R, Schneider E, Frauenknecht K, Kondova I, Bontrop R, Bohl J, Navarro B, Metzler M, Zischler H, Zechner U, et al. . 2009. Differences in DNA methylation patterns and expression of the CCRK gene in human and nonhuman primate cortices. Mol Biol Evol. 26:1379–1389. [DOI] [PubMed] [Google Scholar]
- Feng H, Conneely KN, Wu H. 2014. A Bayesian hierarchical model to detect differentially methylated loci from single nucleotide resolution sequencing data. Nucleic Acids Res. 42:e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch WT. 2011. Unity and diversity in human language. Philos Trans R Soc Lond B Biol Sci. 366:376–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fresno C, Fernández EA. 2013. RDAVIDWebService: a versatile R interface to DAVID. Bioinformatics. 29:2810–2811. [DOI] [PubMed] [Google Scholar]
- Gibbons A. 1998. Solving the brain’s energy crisis. Science. 280:1345–1347. [DOI] [PubMed] [Google Scholar]
- Gilad Y, Oshlack A, Smyth GK, Speed TP, White KP. 2006. Expression profiling in primates reveals a rapid evolution of human transcription factors. Nature. 440:242–245. [DOI] [PubMed] [Google Scholar]
- Gokhman D, Lavi E, Prüfer K, Fraga MF, Riancho JA, Kelso J, Pääbo S, Meshorer E, Carmel L. 2014. Reconstructing the DNA methylation maps of the Neandertal and the Denisovan. Science. 344:523–527. [DOI] [PubMed] [Google Scholar]
- Goodall J. 1983. Population dynamics during a 15 year period in one community of free-living chimpanzees in the Gombe national park, Tanzania. Z Tierpsychol. 61:1–60. [Google Scholar]
- Guintivano J, Aryee MJ, Kaminsky ZA. 2013. A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics. 8:290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MA, Li AX, Wu X, Pfeifer GP. 2015. Single base resolution analysis of 5-methylcytosine and 5-hydroxymethylcytosine by RRBS and TAB-RRBS. Methods Mol Biol. 1238:273–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernando-Herraez I, Prado-Martinez J, Garg P, Fernandez-Callejo M, Heyn H, Hvilsom C, Navarro A, Esteller M, Sharp AJ, Marques-Bonet T. 2013. Dynamics of DNA methylation in recent human and great ape evolution. PLoS Genet. 9:e1003763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg Y, Benjamini Y. 1990. More powerful procedures for multiple significance testing. Stat Med. 9:811–818. [DOI] [PubMed] [Google Scholar]
- Holloway RL, Hurst SD, Garvin HM, Schoenemann PT, Vanti WB, Berger LR, Hawks J. 2018. Endocast morphology of Homo naledi from the Dinaledi Chamber, South Africa. Proc Natl Acad Sci USA. 115:5738–5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hublin JJ, Neubauer S, Gunz P. 2015. Brain ontogeny and life history in Pleistocene hominins. Philos Trans R Soc Lond B Biol Sci. 370:20140062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerison HJ. 1973. Evolution of the brain and intelligence. New York: Academic Press. [Google Scholar]
- Jones PA. 2012. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 13:484–492. [DOI] [PubMed] [Google Scholar]
- Jungerius BJ, Hoogendoorn ML, Bakker SC, Van’t Slot R, Bardoel AF, Ophoff RA, Wijmenga C, Kahn RS, Sinke RJ. 2008. An association screen of myelin-related genes implicates the chromosome 22q11 PIK4CA gene in schizophrenia. Mol Psychiatry. 13:1060–1068. [DOI] [PubMed] [Google Scholar]
- Jäkel S, Dimou L. 2017. Glial cells and their function in the adult brain: a journey through the history of their ablation. Front Cell Neurosci. 11:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato TA, Watabe M, Kanba S. 2013. Neuron-glia interaction as a possible glue to translate the mind-brain gap: a novel multi-dimensional approach toward psychology and psychiatry. Front Psychiatry. 4:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler NJ, Van Baak TE, Baker MS, Laritsky E, Coarfa C, Waterland RA. 2016. CpG methylation differences between neurons and glia are highly conserved from mouse to human. Hum Mol Genet. 25:223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, Lockstone HE, Wayland MT, Tsang TM, Jayatilaka SD, Guo AJ, Zhou J, Somel M, Harris LW, Holmes E, et al. . 2008. Metabolic changes in schizophrenia and human brain evolution. Genome Biol. 9:R124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, Muetzel B, She X, Lachmann M, Hellmann I, Dietzsch J, Steigele S, Do HH, Weiss G, Enard W, et al. . 2004. Regional patterns of gene expression in human and chimpanzee brains. Genome Res. 14:1462–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsella RJ, Kähäri A, Haider S, Zamora J, Proctor G, Spudich G, Almeida-King J, Staines D, Derwent P, Kerhornou A, et al. . 2011. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database. 2011:bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. 2007. Chromatin modifications and their function. Cell. 128:693–705. [DOI] [PubMed] [Google Scholar]
- Kozlenkov A, Roussos P, Timashpolsky A, Barbu M, Rudchenko S, Bibikova M, Klotzle B, Byne W, Lyddon R, Di Narzo AF, et al. . 2014. Differences in DNA methylation between human neuronal and glial cells are concentrated in enhancers and non-CpG sites. Nucleic Acids Res. 42:109–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. 2011. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 27:1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RM, Haussler D, Kent WJ. 2013. The UCSC genome browser and associated tools. Brief Bioinform. 14:144–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. 2013. Software for computing and annotating genomic ranges. PLoS Comput Biol. 9:e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, et al. . 2013. Global epigenomic reconfiguration during mammalian brain development. Science. 341:1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Somel M, Tang L, Yan Z, Jiang X, Guo S, Yuan Y, He L, Oleksiak A, Zhang Y, et al. . 2012. Extension of cortical synaptic development distinguishes humans from chimpanzees and macaques. Genome Res. 22:611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llamas B, Holland ML, Chen K, Cropley JE, Cooper A, Suter CM. 2012. High-resolution analysis of cytosine methylation in ancient DNA. PLoS One. 7:e30226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu DH, Liao HM, Chen CH, Tu HJ, Liou HC, Gau SS, Fu WM. 2018. Impairment of social behaviors in Arhgef10 knockout mice. Mol Autism. 9:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann HB, Whitney DR. 1947. On a test of whether one of two random variables is stochastically larger than the other. Ann Math Statist. 18:50–60. [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17:10–12. [Google Scholar]
- Mendizabal I, Shi L, Keller TE, Konopka G, Preuss TM, Hsieh TF, Hu E, Zhang Z, Su B, Yi SV. 2016. Comparative methylome analyses identify epigenetic regulatory loci of human brain evolution. Mol Biol Evol. 33:2947–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. 2005. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 435:903–910. [DOI] [PubMed] [Google Scholar]
- Nowick K, Gernat T, Almaas E, Stubbs L. 2009. Differences in human and chimpanzee gene expression patterns define an evolving network of transcription factors in brain. Proc Natl Acad Sci USA. 106:22358–22363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinnd D, Minchin PR, O’Hara RB, Simpson GL, Solmos P, et al. . 2017. Vegan: Community Ecology Package. https://cran.r-project.org/web/packages/vegan/index.html
- Park Y, Wu H. 2016. Differential methylation analysis for BS-seq data under general experimental design. Bioinformatics. 32:1446–1453. [DOI] [PubMed] [Google Scholar]
- Perea G, Araque A. 2010. GLIA modulates synaptic transmission. Brain Res Rev. 63:93–102. [DOI] [PubMed] [Google Scholar]
- Rhein M, Hagemeier L, Klintschar M, Muschler M, Bleich S, Frieling H. 2015. DNA methylation results depend on DNA integrity-role of post mortem interval. Front Genet. 6:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins RA, Haghighi F, Edwards JR, Das R, Zhang MQ, Ju J, Bestor TH. 2006. Large-scale structure of genomic methylation patterns. Genome Res. 16:157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rödel R. 1971. Fisher, R.A.: Statistical methods for research workers, 14. Aufl., Oliver & Boyd, Edinburgh, London 1970. XIII, 362 S., 12 Abb., 74 Tab., 40 s. Biom Z. 1971:13429–13430. [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium 2014. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 511:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider E, Dittrich M, Böck J, Nanda I, Müller T, Seidmann L, Tralau T, Galetzka D, El Hajj N, Haaf T. 2016. CpG sites with continuously increasing or decreasing methylation from early to late human fetal brain development. Gene. 592:110–118. [DOI] [PubMed] [Google Scholar]
- Schneider E, El Hajj N, Richter S, Roche-Santiago J, Nanda I, Schempp W, Riederer P, Navarro B, Bontrop RE, Kondova I, et al. . 2014. Widespread differences in cortex DNA methylation of the “language gene” CNTNAP2 between humans and chimpanzees. Epigenetics. 9:533–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider E, Jensen LR, Farcas R, Kondova I, Bontrop RE, Navarro B, Fuchs E, Kuss AW, Haaf T. 2012. A high density of human communication-associated genes in chromosome 7q31-q36: differential expression in human and non-human primate cortices. Cytogenet Genome Res. 136:97–106. [DOI] [PubMed] [Google Scholar]
- Schneider E, Mayer S, El Hajj N, Jensen LR, Kuss AW, Zischler H, Kondova I, Bontrop RE, Navarro B, Fuchs E, et al. . 2012. Methylation and expression analyses of the 7q autism susceptibility locus genes MEST, COPG2, and TSGA14 in human and anthropoid primate cortices. Cytogenet Genome Res. 136:278–287. [DOI] [PubMed] [Google Scholar]
- Semendeferi K, Armstrong E, Schleicher A, Zilles K, Van Hoesen GW. 2001. Prefrontal cortex in humans and apes: a comparative study of area 10. Am J Phys Anthropol. 114:224–241. [DOI] [PubMed] [Google Scholar]
- Semendeferi K, Teffer K, Buxhoeveden DP, Park MS, Bludau S, Amunts K, Travis K, Buckwalter J. 2011. Spatial organization of neurons in the frontal pole sets humans apart from great apes. Cereb Cortex. 21:1485–1497. [DOI] [PubMed] [Google Scholar]
- Shulha HP, Crisci JL, Reshetov D, Tushir JS, Cheung I, Bharadwaj R, Chou HJ, Houston IB, Peter CJ, Mitchell AC, et al. . 2012. Human-specific histone methylation signatures at transcription start sites in prefrontal neurons. PLoS Biol. 10:e1001427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R. 2007. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 8:272–285. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. 2013. DNA methylation: roles in mammalian development. Nat Rev Genet. 14:204–220. [DOI] [PubMed] [Google Scholar]
- Surrallés J, Hande MP, Marcos R, Lansdorp PM. 1999. Accelerated telomere shortening in the human inactive X chromosome. Am J Hum Genet. 65:1617–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasello M. 2008. Origins of human communication. Cambridge (MA): The MIT Press. [Google Scholar]
- Treangen TJ, Salzberg SL. 2011. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet. 13:36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaissière T, Sawan C, Herceg Z. 2008. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat Res. 59:40–48. [DOI] [PubMed] [Google Scholar]
- Varshney D, Vavrova-Anderson J, Oler AJ, Cowling VH, Cairns BR, White RJ. 2015. SINE transcription by RNA polymerase III is suppressed by histone methylation but not by DNA methylation. Nat Commun. 6:6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven K, De Jonghe P, Van de Putte T, Nelis E, Zwijsen A, Verpoorten N, De Vriendt E, Jacobs A, Van Gerwen V, Francis A, et al. . 2003. Slowed conduction and thin myelination of peripheral nerves associated with mutant rho guanine-nucleotide exchange factor 10. Am J Hum Genet. 73:926–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M, Steinbacher J, Kraus TF, Michalakis S, Hackner B, Pfaffeneder T, Perera A, Müller M, Giese A, Kretzschmar HA, et al. . 2015. Age-dependent levels of 5-methyl-, 5-hydroxymethyl-, and 5-formylcytosine in human and mouse brain tissues. Angew Chem Int Ed Engl. 54:12511–12514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Cao X, Zhang Y, Su B. 2012. Genome-wide DNA methylation analyses in the brain reveal four differentially methylated regions between humans and non-human primates. BMC Evol Biol. 12:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M, Schübeler D. 2007. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 39:457–466. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13:335–340. [DOI] [PubMed] [Google Scholar]
- Zeng J, Konopka G, Hunt BG, Preuss TM, Geschwind D, Yi SV. 2012. Divergent whole-genome methylation maps of human and chimpanzee brains reveal epigenetic basis of human regulatory evolution. Am J Hum Genet. 91:455–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zollikofer CP, Ponce de León MS, Lieberman DE, Guy F, Pilbeam D, Likius A, Mackaye HT, Vignaud P, Brunet M. 2005. Virtual cranial reconstruction of Sahelanthropus tchadensis. Nature. 434:755–759. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.