ABSTRACT
All land plants must cope with phytopathogens. Algae face pathogens, too, and it is reasonable to assume that some of the strategies for dealing with pathogens evolved prior to the origin of embryophytes – plant terrestrialization simply changed the nature of the plant-pathogen interactions. Here we highlight that many potential components of the angiosperm defense toolkit are i) found in streptophyte algae and non-flowering embryophytes and ii) might be used in non-flowering plant defense as inferred from published experimental data. Nonetheless, the common signaling networks governing these defense responses appear to have become more intricate during embryophyte evolution. This includes the evolution of the antagonistic signaling pathways of jasmonic and salicylic acid, multiple independent expansions of resistance genes, and the evolution of resistance gene-regulating microRNAs. Future comparative studies will illuminate which modules of the streptophyte defense signaling network constitute the core and which constitute lineage- and/or environment-specific (peripheral) signaling circuits.
KEYWORDS: Plant evolution, molecular plant–microbe interaction, charophytes, streptophyte algae, plant defense, phytopathology
Introduction
Macroscopic algae and plants are bathed in microorganisms. Whatever their natural habitat, they are forced to interact with their microbial companions in some manner. Such interactions are diverse in nature. For example, various algae are known to depend on vitamin B12 provided by bacteria in their environment [1]. Another famous example is the “regulation” of algal blooms of the haptophyte Emiliania huxleyi by bacteria [2]. Interactions with microbes – both positive and negative – are thus part of every photosynthetic eukaryote’s life. This article will focus on the evolution of the framework that underlies molecular phytopathology in modern-day plants and algae. We review what is known about the recurrent evolution of plant defense signaling networks across streptophyte evolution (Figure 1). In so doing, we span the trajectory from streptophyte algae (the closest extant relatives to land plants [3-5]), mosses, gymnosperms, and angiosperms. Since most data have been gathered for angiosperms, we will use them primarily for comparative purposes.
Figure 1.
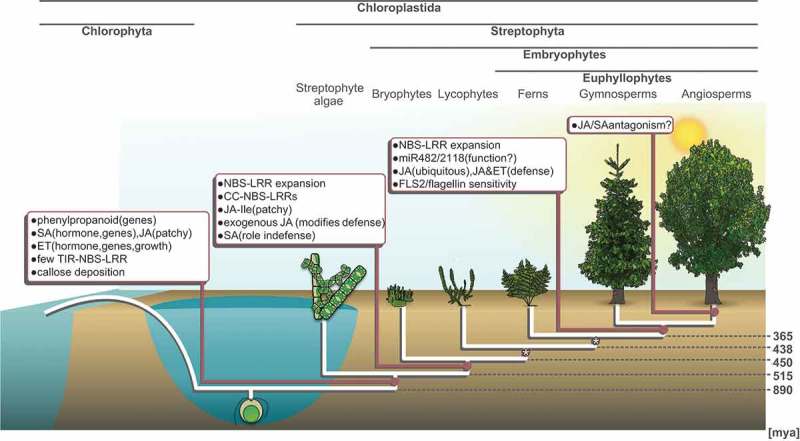
Key phytopathogen interaction factors across the trajectory of streptophyte evolution. Schematic cladogram (white lines) of the Chloroplastida depicts the deep split of the green lineage into Chlorophyta and Streptophyta about 900 million years ago (divergence times based on Morris et al. [45]). The Streptophyta encompass the paraphyletic streptophyte algae and the monophyletic land plants (Embryophyta). Land plants are likely >500 million years old and consist of the non-vascular bryophytes and the ~430 million year old clade of vascular plants, encompassing lycophytes and euphyllophytes. The euphyllophytes are the clade of ferns, gymnosperms and angiosperms (the latter two are the seed plants). Boxes highlight when – along this trajectory – signaling factors in plant defense are thought to have evolved; brackets further specify the type of data and/or functional significance of a given factor. The asterisks indicate nodes for which data are limited. JA, jasmonic acid; JA-Ile, jasmonic acid-isoleucine; SA, salicylic acid; ET, ethylene; NBS-LRR, nucleotide binding site-leucine-rich repeat; TIR-NBS-LRR, Toll-interleukin 1 receptor-nucleotide binding site-leucine-rich repeat; CC-NBS-LRR, coiled coil-nucleotide binding site-leucine-rich repeat; FLS2, FLAGELLIN SENSITIVE2.
Evolutionary phytopathology: The nuts-and-bolts of plant-microbe interactions
Common themes in the evolution of plant defense signaling networks become apparent when diverse species from different lineages are compared. Across angiosperm lineages, plant defense signaling is based on core sets of phytohormones (e.g., jasmonic acid; JA) and proteins (e.g., receptors that sense microbial proteins, such as Flagellin sensitive 2; FLS2). Genetic diversity is further shaped by co-evolution driven by arms race dynamics between plants and microbes – affecting, for example, both resistance genes [6,7] and the factors that regulate them, e.g. miRNAs [8,9]. Studying these factors in an evolutionary context has been summarized as the “coming of age” for the study of evolutionary molecular plant-microbe interactions (coined EvoMPMI) by Upson and colleagues [10]. Upson and colleagues [10] emphasized the need for evolutionarily informed studies that focus on a broad scale covering entire land plant diversity as well as on fine-scale variation within or between closely related species.
While the vast body of literature on how land plants deal with phytopathogens is focused primarily on angiosperms, research on gymnosperms and bryophytes is catching up [11-14] – yet, as highlighted by Upson et al. [10], ferns and lycophytes have yet to follow suit. Further, at the present time, little is known about the interactions between streptophyte algae and their phytopathogens. As outlined above, understanding commonalities in streptophyte algae and non-flowering plants is important to pinpoint how the defense signaling networks of plants arose. Because of the dynamics in plant-pathogen interactions, however, a plethora of different strategies in plant defense have come about.
Therefore, plant defense mechanisms are composed of common defense strategies as well as lineage-specific ones. A straightforward example of a lineage-specific defense strategy in gymnosperms is the flow of resin in wounded conifers, which depends upon resin ducts. Some resin ducts are formed during plant growth and flooded with resin in response to stress, while other resin ducts are only induced upon infection and wounding through the action of phytohormones [15-17]. By exploring the commonalities and differences, we will highlight both the evolutionary trajectories and underlying principles of land plant signaling upon phytopathogen attack – including the potential for this signaling in streptophyte algae. We will first consider an example from basal-branching embryophytes and their interactions with substrate-dwelling fungi.
Fungal symbioses exemplify ancient plant-microbe interactions
Symbioses with Glomeromycota-like fungi are hypothesized to have occurred during an early phase of land plant terrestrialization and to have contributed significantly to the global colonization of land [[5,18-20], see also [21]]. Motivated not least by these observations, there is a growing body of literature on bryophyte–fungus interactions.
Various fungal interactions have been observed in liverworts. For example, a recent study by Nelson and colleagues [22] describes several growth-promoting endophytes associated with the liverwort Marchantia polymorpha, providing fertile ground for future Evo-MPMI research (see [23]). Other Marchantia species were shown to engage in mutualistic interactions with Glomeromycota [24,25]. And other liverwort genera, such as Cephalozia bicuspidata [26], have also been shown to engage in mutualistic interactions with mycorrhizal fungi.
Several bryophytes form mycorrhizae by interacting with fungi [24,27,28], but the picture for bryophytes as a whole is patchy [29]. While many liverworts (outlined above) and hornworts [30,31] exhibit interactions with mycorrhizal fungi, mosses generally do not form mycorrhizae [32,33]; for a recent and comprehensive overview see [29]. That mosses do not form mycorrhizae is further corroborated by Wang and colleagues [34], who showed that moss arbuscular mycorrhizal symbiosis genes show high sequence divergence as compared to their homologous counterparts in all other land plants. Yet, in light of the recently supported monophyly of the bryophytes [35], the phenomenon that mosses do not form mycorrhizae likely represents a case of secondary loss [29]. On balance, symbiosis with arbuscular mycorrhizal fungi appears to be an ancestral feature of all land plants [36]. Indeed, molecular data presented by Wang et al. [34] indicate that genes associated with interactions with mycorrhizal fungi were likely present in the last common ancestor of land plants, which was corroborated by Delaux et al. [18]. But what about the algal progenitors of land plants?
Ancient land plant-microbe interactions and evidence from molecular data in streptophyte algae
Streptophyte algae are known to associate with various kinds of microorganisms. Knack and colleagues [37] performed a metagenomic study of aquatic streptophyte alga- and liverwort-associated microbes, including epiphytic microorganisms (e.g. those growing in the mucilage of streptophyte algae) as well as those colonizing the tissue. Their analyses of three higher-branching streptophyte algae (Coleochaete pulvinate, Chaetosphaeridium globosum and Nitella tenuissima) identified potentially beneficial microbes, for example nitrogen-fixing or cobalamin-producing bacteria, but also potentially harmful ones, such as bacteria associated with cellulose degradation [37]. Interestingly, Knack and colleagues [37] also detected some fungi in metagenomic data, an observation that warrants further investigation. Among the streptophyte algae investigated, the detected signals included sequences stemming from fungi belonging to the Cryptomycota and Chytridiomycota [37]. For the investigated liverwort Conocephalum conicum an association with glomalean fungi was demonstrated [37].
As mentioned earlier, Glomeromycota-like fungi feature in discussions revolving around the beneficial symbioses that the earliest land plants engaged in [19,20]. Delaux et al. [18] found that streptophyte algae have most of the genes that land plants put to use during symbiosis signaling. These authors also performed functional complementation experiments in which the capacity to engage in symbiosis with arbuscular mycorrhizal fungi was rescued by heterologously expressing streptophyte algal CCaMK in Medicago ccamk mutants (which are deficient in interacting with mycorrhizal fungi). This underscores the functional conservation of symbiosis signaling across long evolutionary timescales.
The fossil record also provides insight into streptophyte-fungal symbioses. 400-plus million-year-old Horneophyton land plant fossils have been shown to harbor glomeromycotean- and mucoromycotean-resembling structures [38]. Together with the aforementioned molecular data, this information makes a strong case for the idea that the interaction with mycorrhizal fungi is ancient. The genes underlying these beneficial interactions likely predate the origin of the terrestrial flora.
Microorganisms are not only beneficial to plants and algae – they can exploit their hosts as (facultative) phytopathogens [39]. For example, several bacterial and fungal genera or species complexes include mutualistic, pathogenic and endophytic species [39]. Such microbe-host relationships can in fact switch between neutral, beneficial and detrimental in response to, for example, environmental factors [40,41]. It is noteworthy that some of the components necessary for a successful arbuscular mycorrhizal symbiosis, and which are present in streptophyte algae or bryophytes, can also be used for defense signaling [42]. For example, mutations in several symbiosis-associated LysM-RLKs (Lysin Motif Receptor-like Kinases) have been reported to impair defense signaling [42]. In contrast, in the case of Arabidopsis thaliana, which does not engage in symbioses with arbuscular mycorrhizal fungi, the oomycete pathogen Hyaloperonospora arabidopsidis seems to require components of the arbuscular mycorrhizal symbiosis-associated molecular machinery to successfully complete its life cycle [43]. Hence, “symbiosis genes” might not only tell a tale of ancient mutualism, but also ancient interactions with phytopathogens.
The terrestrial habitat was teeming with microbes before the dawn of land plants [reviewed by [44]]. Hence, during terrestrialization >500 million years ago [see [45] for the latest dating] one can imagine that the earliest land plants would have encountered a very different set of microbes than those in the freshwater environments from which they were emerging. Yet, a fluent passage scenario seems equally reasonable if one considers, e.g., a freshwater environment (microbe load A) that routinely dried out (microbe load B). It should further be noted that microbe load A and B may also have overlapped given that, for example, many oomycetes and fungi grow equally well in liquid or on solid medium in the laboratory – it goes without saying that this is a mere proxy for what might happen in nature and will require further studies. No matter the scenario, fossils have interesting stories to tell in this case, too.
Taylor et al. [46] reported the existence of a parasitic fungus in a likely more than 400-million-year-old fossil of Paleonitella, which appears to be related to extant charophyceaen streptophyte algae (such as Nitella). But the algal progenitors of land plants would have encountered (terrestrial and/or non-terrestrial) microbes even before this time. Berbee and colleagues [47] recently argued that the occurrence of pectinases (enzymes used for the degradation of pectin in plant cell walls) in even the earliest-diverging fungi [see [48]] argues for the antiquity of the fungal ability to exploit plant material. How so? Pectin is a cell wall component characteristic of land plants and streptophyte algae (reviewed in [49]). Berbee et al. [47] argue that since pectinase-harboring fungal lineages are older than the land plant clade, these fungi used their pectinases for the degradation of streptophyte algal cell walls. This is corroborated by the fact that i) phytoplankton are readily attacked by chytrid fungi [50] and ii) chytrid fungi have been found associated with streptophyte algal microbiomes [37].
In summary, land plants and their closest relatives are, and always have been, associated with both symbiotic and pathogenic microorganisms – their interactions with microbes are truly ancient. Because it is important for hosts to be able to distinguish between a pathogen or a symbiont – and to react accordingly – defense signaling mechanisms must presumably also be present in the algae that are most closely related to land plants. The question that remains is: how similar are these mechanisms in plants and algae? The answer will shed light on the plant-microbe interaction tool kit that was present in the earliest land plants.
PTI and ETI in non-flowering land plants and maybe streptophyte algae
Most of what we know about the plant immune system derives from studying angiosperms. The pathogen recognition system is based upon two components: pattern triggered immunity (PTI) and effector triggered immunity (ETI) [6]. The latter is more specific towards the infecting pathogen because plant resistance genes (R genes) recognize effector proteins secreted by, and specific to, a certain pathogen [51]. PTI causes, for example, stomata closure and cell wall reinforcements at the site of pathogen attack (e.g., through callose deposition, formation of papillae [deposits consisting of callose, phenolic compounds and polysaccharides], and lignification) [52-55]. PTI can also result in cell death caused by the release of reactive oxygen species (ROS) [56]. Additionally, ROS production and thus the initiation of the hypersensitive response (HR) is a classical hallmark of R gene-based immunity [51].
Not surprisingly, the potential for PTI can be found in early-branching land plant lineages as well as streptophyte algae (for a comprehensive discussion of genetic potential in streptophyte algae, see [5]). Streptophyte algae such as Coleochaete and Nitella have been found to contain lignin-like components [49,57-59], potentially used for cell wall reinforcement during pathogen attack. Moreover, the basal-branching streptophyte algae Klebsormidium spp. deposit callose in response to abiotic (desiccation) stress [60]. It is further noteworthy that even though Herburger and Holzinger [60] found that Zygnema spp. did not deposit callose in response to desiccation stress, callose was nonetheless present in these species. In the moss Physcomitrella patens, papillae formation is readily observed close to unsuccessful infection attempts by different Phytophthora pathogens [61]. Oomycete and fungal pathogens also induce ROS [62,63] and inoculations with oomycetes resulted in the accumulation of toxic phenolic compounds in P. patens [61,62]. Similarly, other mosses, including Funaria hygrometrica, also form papillae around fungal penetration sites to prohibit their entry [64,65]. Callose deposition was also observed in the interaction between the liverwort M. polymorpha and the oomycete Phytophthora palmivora [66].
PTI responses require receptors. One of the best-explored PTI-associated pattern recognition receptors (PRRs) in angiosperms is FLS2. FLS2 recognizes the microbe-associated molecular pattern (MAMP) flg22, a peptide component of the bacterial flagellin [56,67]. Orthologs of FLS2 were not found in the moss P. patens [68], although a homolog with appreciable sequence conservation was found [69]. Yet, P. patens is flg22-insensitive [70]. Likewise, the receptor for the bacterial translation elongation factor Tu (Ef-Tu), EFR [71,72], seems to be missing outside of the Brassicaceae [68,71] and Ef-Tu does not induce a PTI-like response in P. patens [70]. However, the moss does recognize bacteria and mounts a defense response accordingly [73]. This suggests that either more ancient or lineage-specific receptors are used in P. patens to recognize bacteria, and that FLS2 and EFR are more recent acquisitions. Indeed, in support of the presence of a more ancient type of receptor in mosses, P. patens is known to respond to bacterial peptidoglycan, which is recognized by the ortholog of the A. thaliana receptor CERK1 [70]. Moreover, CERK1 of P. patens recognizes chitin from fungi and triggers downstream signaling responses [70]. This might hint that CERK1 was present and functioning in peptidoglycan recognition in the last common ancestor of land plants – but this needs further clarification by investigating CERK1 function across a broader diversity of land plants. In contrast to P. patens, protoplasts of the conifer Pinus thunbergii produce ROS in response to flagellin treatment [74] and FLS2 is hypothesized to be present in gymnosperms [75]. This suggests that a diversification of PTI-associated PRRs occurred during the evolution of land plants, perhaps associated with the refinement of MAMP-triggered responses.
Components of the heterotrimeric G-protein complex, a signaling switch that consists of an α-, β- and several γ-subunits [76], are involved in land plant defense responses (e.g. [77],); the role of β- and γ-subunits in defense is also implicated to be mediated by FLS2, EF-Tu and CERK1 in A. thaliana [78]. Homologs of all three subunits are present in land plants and streptophyte algae [79,80]. Moreover, in the interaction of P. abies with the fungal pathogens Heterobasidion annosum, Heterobasidion parviporum and the saprotroph Phlebiopsis gigantea, genes for several subunits of the heterotrimeric G-protein complex were shown to be up-regulated [81]. It was further suggested that this response may be triggered by conserved molecular patterns of the fungi [81], hence possibly associated with PTI. Whether heterotrimeric G-proteins also play a role in defense responses of earlier-diverging land plants and streptophyte algae remains to be investigated.
ETI requires the presence of R proteins to detect pathogen secreted effector proteins either through direct binding or by monitoring whether other host proteins are altered by the actions of effectors [51]; such alterations can include changes in protein conformation and/or phosphorylation status [82,83]. Once R proteins detect an effector protein of a pathogen, they induce pathogen-specific immune responses [84-86]. Nucleotide-binding site leucine-rich repeats (NBS-LRRs) are one of the major classes of R proteins [87]. They are combined with various N-terminal domains, for example the coiled-coil (CC-NBS-LRR) or Toll-interleukin 1 receptor domain (TIR-NBS-LRR) [87].
Potential NBS-LRR-encoding genes have been found from streptophyte algae to angiosperms, but there is pronounced variation in the number of NBS-LRR genes present in any given genome. Conifers have undergone a dramatic expansion of their suite of NBS-LRR genes: while 69 putative NBS-LRR genes are predicted for P. patens and 16 for the lycophyte Selaginella moellendorffii, P. abies and Pinus taeda have been predicted to possess 562 and 677 putative NBS-LRR genes, respectively [88,89]. It is noteworthy that gymnosperms tend to have large genomes (often more than 10 Gbp in size [90]), which could suggest that the expansion of NBS-LRRs in plants is related to genome size of the respective plant. Yet, the large genomes of gymnosperms appear to be the result of an expansion of intron size because of repeated insertion of transposable elements and the total number of genes is in fact similar to that observed in A. thaliana [90]. Additionally, numbers of NBS-LRRs reported in Zhang et al. [88] seem to not necessarily be related to genome size. For example Medicago truncatula has “only” a 370 Mbp genome [91], but a similar number of NBS-LRRs as P. abies [88]. Likewise, the monocot Triticum aestivum has a 17 Gbp genome, similar in size to some gymnosperms [92], but has roughly double the number of NBS-LRRs than P. abies [88].
Species-specific expansions and reductions of NBS-LRRs have been observed throughout the Embryophyta [88] – including lineages with differentially expanded NBS-LRR subsets. For example, TIR-NBS-LRR-encoding genes are absent from the grasses (Poaceae; [e.g. [88]]). NBS-LRR genes also appear to be encoded in the genome of streptophyte algae, but whether they are required for streptophyte algal immunity is currently not known. Yue et al. [69] found three NBS-encoding sequences within the Coleochaetales (higher-branching streptophyte algae), two with sequence similarity to TIR-NBS-LRRs from angiosperms. Furthermore, Urbach and Ausubel ([93]; see supplementary appendix) reported the detection of two TIR-NBS-LRR genes in the genome of the early-branching streptophyte alga Klebsormidium nitens (whose whole genome sequence was reported by [94]). In agreement with this, Gao and colleagues [89] reported three TIR-NBS-LRRs in K. nitens as well as one NBS-LRR with an additional N-terminal domain (non-TIR-NBS-LRRs). Several non-TIR-NBS-LRRs were found in transcriptomes of six streptophyte algae [89]. Yet, CC-NBS-LRRs (a class of non-TIR-NBS-LRRs) have thus far only been found among land plants, including the moss P. patens [88]. It hence appears that one of the most prominent NBS-LRR combinations – the CC-NBS-LRRs – evolved on land.
Given that the recognition of effector proteins by NBS-LRRs results in the initiation of plant cell death, tight regulatory control is essential. Indeed, these proteins are regulated in many ways, including multiple posttranslational mechanisms, such as ubiquitination and oligomerization with different partners [95]. At the level of expression, they can be regulated by transcriptional as well as post-transcriptional means [95]. The latter is mediated by microRNAs (miRNAs) in angiosperms [96-99]. Several NBS-LRR-targeting miRNA families exist, but due to the broad distribution of the miR482/2118 family, this family has received more attention than others.
Members of the miR482/2118 family show low sequence conservation even between closely related species [8]. The family first emerged in gymnosperms [96,100], which seems to coincide with an expansion of NBS-LRRs during this time period [88]. The coniferous plant P. abies has one of the largest expansions of miR482/2118 [8,100] and the genes likely originated through inverted duplication of NBS-LRR genes [100]. miR482/2118 is a direct regulator of resistance to a diverse range of pathogens in dicots [9,98,101-103]. In monocots miR482/2118 is expressed in reproductive tissue and may function in its development [104]. Given the expression patterns of miR482/2118 in P. abies, with some members of this family solely expressed in reproductive organs [100], a broader function in the regulation of both reproductive organ development and disease resistance seems to be the more ancient mechanism.
Evolution of phytohormone defense networks
The plant immune system and phytohormone signaling are interwoven [105]. While almost all major phytohormones have been linked to plant immunity at some level [106], jasmonic acid (JA), salicylic acid (SA) and ethylene (ET) are key regulators [105]. In angiosperms, SA and JA act primarily antagonistically [107]. SA triggers immunity towards biotrophic pathogens, i.e., those requiring a living host [107]. SA regulates ROS levels by induction as well as scavenging [108]. Furthermore, SA is involved in the induction of HR, resulting in plant cell death [109]. On the other hand, JA is produced in response to herbivores, which induce wounding [110]. In concert with ET, JA also regulates responses towards several necrotrophic pathogens, i.e. those pathogens that actively induce host cell death [107].
It is likely that defense networks similar to those in land plants exist in streptophyte algae. A series of recent studies have revealed the presence of homologs of plant hormone biosynthesis and signaling pathway genes and/or the presence of various phytohormones in streptophyte algae [e.g. [94,111-114]]; yet we are only beginning to understand the function of these phytohormones in streptophyte algae [115-117]. All three canonical plant defense phytohormones, JA, SA and ET, have been detected in at least some species of streptophyte algae [94,112,115,118]. They also have been explored with regard to pathogen defense in non-flowering land plants. SA has been measured in mosses and gymnosperms – indeed, as in angiosperms, SA has been shown to accumulate in response to elicitors or pathogen attack [63,119,120], supporting its function in defense across land plant diversity.
In streptophyte algae, Ju et al. [112] detected Isochorismate Synthase 1 (ICS1) homologs; ICS1 catalyzes the first step in SA biosynthesis. Furthermore, an ortholog of phenylalanine ammonia lyase (PAL), the enzyme catalyzing the first step in the phenylpropanoid (PP) pathway, was detected in the genome of K. nitens [121]. The PP pathway is also a source for SA biosynthesis [122]. Moreover, potential homologs for the SA receptor Nonexpressor of PR genes 1 (NPR1) [123], were reported for all land plants [113] and the putative NPR1 homolog of P. patens can partially complement defense signaling-associated phenotypes of the Arabidopsis npr1 mutant [124]. As for ET, recent studies showed that streptophyte algae produce, sense and respond to ET [112,115], but these studies did not dissect the role of ET as a hormone involved in defense.
The existence and distribution of JA in early-diverging land plants and streptophyte algae is complex. The canonical pathway genes for JA biosynthesis (13-LOX, 13-Lipoxygenase; AOS, 13-Allene Oxide Synthase; AOC, Allene Oxide Cyclase, OPR3, OPDA Reductase 3 and JAR1, Jasmonate Resistant 1) are present in all land plant lineages [125], and some of its components were also detected in several streptophyte algae [94,112,125]. However, actual (mainly mass spectrometry-based) measurements of JA levels are suggestive of a patchier distribution among land plants and streptophyte algae. For example, while JA is reported to be produced in small quantities in the streptophyte algae K. nitens [94] and Chara australis [118], Hackenberg and Pandey [126] did not detect JA in Chara braunii. Furthermore, Gachet et al. [127] did not detect JA in Chara vulgaris and Klebsormidium elegans, while Koeduka et al. [128] found only minimal levels of JA in Klebsormidium flaccidum. Thus, within the genera Chara and Klebsormidium, the detection of JA is variable.
Like in K. flaccidum (a streptophyte alga) only non-existent or only minimal amounts of JA and its active derivative JA-Ile were detected for M. polymorpha (a liverwort) [128,129]. Furthermore, tissue wounding did not increase their amounts [128]. In bryophytes, like in streptophyte algae, JA seems to be produced in a species-specific manner [127,130]. However, JA appears to be absent from the model moss P. patens [131]. Yet, when the moss P. patens was infected with two species of Pythium, an increase in the production of endogenous JA was detected over time and compared to control plants [62] – although the levels of JA were minimal both before and after infection. In contrast, exposure to the fungal pathogen Botrytis cinerea did not result in an increase in JA, but instead an increase in SA and the JA-precursor 12-oxo-phytodienoic acid (OPDA [63];). OPDA also increased after wounding in M. polymorpha [129]. These patterns suggest that in bryophytes the function of JA in defense may in fact be conferred by OPDA. Indeed, the signaling pathway of JA in angiosperms is fully functional in M. polymorpha [132]. Yet, in contrast to Arabidopsis, a derivative of OPDA, 2,3-dinor-OPDA (dn-OPDA), is the functional ligand of the JA receptor ortholog in M. polymorpha, Coronatine insensitive 1 (COI1) [132].
Unlike P. patens, the model lycophyte S. moellendorffii produces JA and is able to sense the phytohormone [133]. However, other lycophytes including another species from the genus Selaginella did not produce measurable levels of JA [127], supporting the notion of a high species-specificity in JA biosynthesis. Similarly, some species of ferns show pronounced JA responses, while others do not [134-136]; likewise, the production of JA was shown to be species-specific in ferns [127]. This distribution of JA biosynthesis becomes less patchy in gymnosperms and angiosperms, as shown in the dataset by Gachet and colleagues [127], where only one species in each of these two lineages was identified that did not produce a detectable amount of JA.
How do non-flowering land plants mount their defense responses? In P. patens, infection by oomycete and fungal pathogens leads to up-regulation of the usual suspects of angiosperm defense signaling: PAL, Dirigent (DIR), Chalcone synthase (CHS)andPathogen related (PR) genes, as well as genes involved in JA and JA-precursor biosynthesis, such as LOX, AOS and OPR [62,63,137]. This is, however, not surprising, since infections with B. cinerea or two Pythium pathogens lead to OPDA production [62,63]. In the spruce P. abies, the pathogens H. parviporum and H. annosum induce the expression of, among other genes, the JA biosynthesis and signaling genes LOX and Jasmonate Zim Domain (JAZ), as well as genes for the biosynthesis of ET (ACO, 1-aminocyclopropane-1-carboxylic acid [ACC]-oxidase; ACS, ACC-synthase), and PAL, DIR2/32 and PR1 [138-140]. JA and ET act in concert to induce defense responses against necrotrophic pathogens in angiosperms [107]. Hence, the activation of both JA and ET biosynthesis genes in response to necrotrophic fungal pathogens in P. abies suggests a similar interaction between the two phytohormones. In agreement with this, in the two conifers Pseudotsuga menziesii and Sequoiadendron giganteum the application of MeJA and wounding induce ET biosynthesis, as measured by the activation of ACO [16]. In that study, ET was (at least partially) required for the plants’ defense responses induced by MeJA and wounding [16]. This suggests that both mosses and gymnosperms not only induce similar defense pathways during infection with necrotrophic pathogens, but also that non-flowering land plants produce immune reactions similar to those observed in angiosperms.
Despite the apparent similarities in immune responses in non-flowering land plants and angiosperms, some differences have been discovered. As mentioned earlier, in the moss P. patens, the necrotrophic pathogen B. cinerea induced SA production in addition to the biosynthesis of the JA-precursor OPDA [63]. In agreement with this, expression of moss PpPAL is induced by SA, JA, MeJA, and OPDA [62,63], suggesting that exogenous JA and SA at least partially activate similar pathways. Indeed, Thaler et al. [141] and Han [125] suggested that the JA/SA-antagonism arose at the earliest in seed plants. Along these lines, it was hypothesized that in the fern Azolla some JA-orchestrated signaling responses may be initiated via SA instead of JA because MeSA application induced the expression of Plant Defensin 1.4 (AfPDF1.4 [136]); in Arabidopsis, PDFs are JA-responsive [142]. These results, together with the data from mosses, speak in favor of a reduced antagonism – or perhaps complete lack thereof – between JA and SA in mosses and ferns. In contrast to the hypothesis of Thaler et al. [141], a lack of a canonical antagonism between JA and SA was also suggested for P. abies [143]. Both MeJA and MeSA induce marker genes of SA signaling (PR1 and Late up-regulated in response to Hyaloperonospora parasitica 1 [LURP1]) [143]. These genes are also up-regulated in response to the fungal pathogen H. parviporum, and upon inhibition of JA signaling, PR1 expression is significantly reduced after fungal attack [143]. Furthermore, Kozlowski et al. [119] showed that exogenous MeJA can increase SA levels in P. abies. In Ginkgo biloba an elicitor from Phytophthora boehmeriae causes an increase in both endogenous JA and SA [120]. Moreover, both JA and SA were required to produce a defense-associated metabolite in response to the elicitor treatment in G. biloba [120]. Yet this study also found that artificially reduced SA led to an increase in JA levels, complementing the loss of SA-derived production of the defense metabolite. This points to some negative regulatory effects of SA on JA, although the downstream signaling pathways of both hormones do not seem to be antagonistic.
Overall, it seems that JA synthesis was either lost or highly reduced several times throughout the evolution of land plants. Therefore, the requirement for JA in defense responses may be lineage specific. JA precursors, on the other hand, such as OPDA and other oxylipins, are involved in immune signaling in early-branching land plants [144]. As the production of JA became more consistent in gymnosperms and angiosperms, and levels of JA increased compared to earlier-branching lineages, its use in defense signaling was cemented. Long before that, however, at the base of the vascular plants, COI1 acquired a mutation leading to a broader binding pocket, which enabled binding to JA-Ile, the active JA-derivative [132]. After the establishment of JA as another regulator of defense responses, JA and SA signaling evolved into a highly specific antagonistic network.
There are, however, many complexities with regard to the antagonism of JA and SA in A. thaliana [145]. Liu et al. [145] showed that SA promotes the synthesis of JA and the activation of its signaling during ETI. However, a recent study by Betsuyaku et al. [146] showed that SA and JA act antagonistically during ETI on a narrow spatial scale. So far, spatial information on JA responses in non-flowering plants is only available for conifers, where MeJA treatment results in cell type-specific PAL activation [17]. Moreover, cell type-specific transcriptomes of Picea glauca showed strong cell-specific modulation of gene expression by MeJA treatment, including PP pathway-associated genes, such as PAL [147]. Nevertheless, these studies did not dissect the JA/SA antagonism on spatial scales. Moreover, we cannot exclude the possibility that JA/SA antagonism (or in organisms lacking JA, dn-OPDA/SA antagonism) is lineage-specific in non-angiosperms. However, for the time being, the evidence points to the evolution of JA/SA antagonism with regard to the regulation of defense responses after the split of gymnosperms and angiosperms.
Phenylpropanoids and their derivatives in streptophyte defense responses
Many of the defense- and JA/SA-regulated genes described above encode enzymes in the PP pathway or those downstream of it. PPs and PP-derived compounds, such as lignins, lignans, flavonoids and stilbenes, are defense metabolites, because they i) can be toxic for pathogens and/or ii) reinforce cell wall structures, thereby reducing the possibility of penetration by pathogens [122,148]. PAL encodes the first enzyme in the PP pathway [122]. It shows a strong responsiveness to pathogens or exogenously applied JA in gymnosperms and JA and SA in mosses [62,63,73,143,149,150]. Therefore, it is not surprising that the defense response of P. patens following the inoculation with oomycete and fungal pathogens includes the production of phenolic compounds [61-63]. Cell wall reinforcements in P. patens by lignification after B. cinerea infection was also suggested because of the enhanced expression of the Dirigent-like gene, PpDIR [63]; DIR and DIR-like enzymes function both in lignan and lignin formation [151]. Moreover, in a study focused on gene expression of nearly all enzymes required for lignin production in P. abies, Koutaniemi and colleagues [138] found that PAL and at least one representative of the nine tested gene families were up-regulated in response to H. annosum – a pathogen inducing JA biosynthesis and signaling genes in its host [139]. This points to enhanced lignification as a pathogen defense response in conifers. Indeed, enhanced lignification in cell walls was observed for conifer species from the Cupressaceae and Podocarpaceae after MeJA application [17]. Furthermore, in conifers from different families, the application of MeJA increased the amount of PAL in polyphenolic and ray parenchyma cells [17]. These cell types also accumulated phenolic compounds after the treatment with MeJA in several of the species tested [17].
While it was previously thought that the PP pathway was limited to land plants, de Vries and colleagues [121] showed that streptophyte algae likely possess genes (orthologous to their respective, well-characterized land plant counterparts) for the production of PPs and lignins. As discussed above, a PAL-encoding orthologous gene was detected in the genome of K. nitens [121], suggesting that this early-branching streptophyte alga is capable of producing PPs. This is in agreement with the aforementioned detection of lignin-like compounds in streptophyte algae [see 49, 57, 58, 59], which are also derived from the PP pathway. While this suggests that both mechanisms are ancient, we do not know whether PPs and their derivatives are used by streptophyte algae for pathogen and parasite defense.
The expression of flavonoid-associated genes is also triggered by pathogens: Pinaceae up-regulate genes from the flavonoid biosynthesis pathway during infection [149,152]. The expression of flavonoid biosynthesis genes was also correlated with an increase in the flavonoid (+)-catechin in P. abies 15 days after infection with H. annosum [149]. However, in this study, the increase was genotype dependent, with more susceptible genotypes showing no increase or less of the flavonoid. In P. patens flavonoids seem to also play a role in defense responses, as bacterial elicitors as well as oomycete and fungal pathogens induce CHS [61,62,73]. Furthermore, other genes of the flavonoid biosynthesis pathway are induced by bacterial elicitors [153]. In streptophyte algae, several homologs, but few orthologs of the genes required for flavonoid biosynthesis were detected [121]. That being said, Goiris et al. [154] reported the presence of flavonoids in algae from various lineages, including chlorophytes. A 1969 study by Markham and Porter [155] reported on the presence of flavonoids in the charophyceae Nitella, highlighting the need to further investigate streptophyte algae with regard to the presence of these metabolites. It is noteworthy that Van de Poel and colleagues [115] found ET-dependent regulation of a homolog of TRANSPARENT TESTA 8 (TT8) in the Zygnematophyceae Spirogyra pratensis; TT8 is a known regulator of flavonoid biosynthesis [156]. A TT8 ortholog is also present in the dataset for the Coleochaetophyceae Coleochaete scutata [114], where it is induced by high light stress.
Conclusion
Angiosperms have evolved complex and fine-tuned regulatory networks to mount their defense responses against microbial pathogens. Many molecular components of these networks can be found in the closest relatives of land plants, the streptophyte algae. We are, however, just beginning to understand whether these pathways are required for streptophyte algal defense responses – and hence likely to have served this purpose in the ancestor of land plants – or whether other pathways are more important in these lineages. We know that non-flowering land plants induce many of these pathways for defense against bacteria, fungi and oomycetes. Defense responses in non-flowering land plants utilize different regulatory modes than do angiosperms, as exemplified by the lack of the JA/SA antagonism in non-flowering land plants (Figure 1). Moreover, regulatory circuits have become seemingly more elaborate throughout land plant evolution, with the expansion of PTI-associated PRRs and NBS-LRRs and the occurrence of NBS-LRR-regulating miRNAs (Figure 1). In conclusion, it seems that many defense pathways of angiosperms existed in the last common land plant ancestor. The same pathways have, however, been reinvented and interwoven during subsequent land plant evolution, resulting in highly intertwined, specific and complex regulatory networks for plant defense.
Funding Statement
This work was supported by the Canadian Network for Research and Innovation in Machining Technology, Natural Sciences and Engineering Research Council of Canada [Discovery grant RGPIN/05754-2015]; Deutsche Forschungsgemeinschaft [VR 132/1-1]; Deutsche Forschungsgemeinschaft [Research Training Group GRK1525]; Killam Trusts [Izaak Walton Killam Postdoctoral Fellowship].
Acknowledgments
SdV thanks the Killam Trusts for the Izaak Walton Killam Postdoctoral Fellowship. JdV (Research Fellowship, VR132/1-1), JKvD and LER (Research Training Group, GRK1525) thank the German Research Foundation (Deutsche Forschungsgemeinschaft, DFG) for funding. CHS acknowledges funding by NSERC (Discovery grant RGPIN/05754-2015).
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Croft MT, Lawrence AD, Raux-Deery E, et al. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature. 2005;438:90–93. [DOI] [PubMed] [Google Scholar]
- [2].Segev E, Wyche TP, Kim KH, et al. Dynamic metabolic exchange governs a marine algal-bacterial interaction. Elife. 2016;18:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wickett NJ, Mirarab S, Nguyen N, et al. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc Natl Acad Sci USA. 2014;111:E4859–E4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Delwiche CF, Cooper ED.. The evolutionary origin of a terrestrial flora. Curr Biol. 2015;25:R899–R910. [DOI] [PubMed] [Google Scholar]
- [5].de Vries J, Archibald JM. Plant evolution: Landmarks on the path to terrestrial life. New Phytol. 2018;217:1428–1434. [DOI] [PubMed] [Google Scholar]
- [6].Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. [DOI] [PubMed] [Google Scholar]
- [7].Stukenbrock EH, McDonald BA. Population genetics of fungal and oomycete effectors involved in gene-for-gene interactions. Mol Plant Microbe In. 2009;22:371–380. [DOI] [PubMed] [Google Scholar]
- [8].de Vries S, Kloesges T, Rose LE. Evolutionarily dynamic, but robust, targeting of resistance genes by the miR482/2118 gene family in the Solanaceae. Genome Biol Evol. 2015;7:3307–3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].de Vries S, Kukuk A, von Dahlen JK, et al. Expression profiling across wild and cultivated tomatoes supports the relevance of early miR482/2118 suppression for Phytophthora resistance. Proc R Soc B. 2018;285:20172560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Upson JL, Zess EK, Białas A, et al. The coming of age of EvoMPMI: evolutionary molecular plant–microbe interactions across multiple timescales. Curr Opin Plant Biol. 2018;44:108–116. [DOI] [PubMed] [Google Scholar]
- [11].Kovalchuk A, Keriö S, Oghenekaro AO, et al. Antimicrobial defenses and resistance in forest trees: Challenges and perspectives in a genomic era. Annu Rev Phytopathol. 2013;51:221–244. [DOI] [PubMed] [Google Scholar]
- [12].Parent GJ, Raherison E, Sena J, et al. Chapter two – Forest Tree Genomics: Review of Progress In: Plomion C, Adam-Blondon A-Feditors. Land Plants – Trees: Advances in Botanical Research. London (UK), Oxford (UK), Waltham (MA), San Diego (CA): Academic Press; 2015. p. 39–92. [Google Scholar]
- [13].Ponce De León I, Montesano M. Adaptation mechanisms of moss defenses to microbes. Front Plant Sci. 2017;8:366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Carella P, Schornack S. Manipulation of bryophyte hosts by pathogenic and symbiotic microbes. Plant Cell Physiol. 2018;59:651–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Nagy NE, Franceschi VR, Solheim H, et al. Wound-induced traumatic resin duct development in stems of Norway spruce (Pinaceae): Anatomy and cytochemical traits. Am J Bot. 2000;87:302–313. [PubMed] [Google Scholar]
- [16].Hudgins JW, Franceschi VR. Methyl jasmonate-induced ethylene production is responsible for conifer phloem defense responses and reprogramming of stem cambial zone for traumatic resin duct formation. Plant Physiol. 2004;135:2134–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hudgins JW, Christiansen E, Franceschi VR. Induction of anatomically based defense responses in stems of diverse conifers by methyl jasmonate: a phylogenetic perspective. Tree Physiol. 2004;24:251–264. [DOI] [PubMed] [Google Scholar]
- [18].Delaux PM, Radhakrishnan GV, Jayaraman D, et al. Algal ancestor of land plants was preadapted for symbiosis. Proc Natl Acad Sci USA. 2015;112:13390–13395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Field KJ, Pressel S, Duckat JG, et al. Symbiotic options for the conquest of land. Trends Ecol Evol. 2015;30:477–486. [DOI] [PubMed] [Google Scholar]
- [20].Selosse MA, Strullu‐Derrien C, Martin FM, et al. Plants, fungi and oomycetes: a 400‐million year affair that shapes the biosphere. New Phytol. 2015;206:501–506. [DOI] [PubMed] [Google Scholar]
- [21].Edwards D, Kenrick P. The early evolution of land plants, from fossils to genomics: a commentary on Lang (1937) ‘On the plant‐remains from the Downtonian of England and Wales’. Phil Trans R Soc B. 2015;370:20140343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Nelson JM, Hauser DA, Hinson R, et al. A novel experimental system using the liverwort Marchantia polymorpha and its fungal endophytes reveals diverse and context-dependent effects. New Phytol. 2018. DOI: 10.1111/nph.15012. [DOI] [PubMed] [Google Scholar]
- [23].Rich M, Delaux P-M. Taking the step: from Evo-Devo to plant–microbe interaction evolution with the liverwort Marchantia. New Phytol. 2018;218:882–884. [DOI] [PubMed] [Google Scholar]
- [24].Humphreys CP, Franks PJ, Rees M, et al. Mutualistic mycorrhiza-like symbiosis in the most ancient group of land plants. Nat Commun. 2010;1:103. [DOI] [PubMed] [Google Scholar]
- [25].Field KJ, Cameron DD, Leake JR, et al. Contrasting arbuscular mycorrhizal responses of vascular and non-vascular plants to a simulated Paleozoic CO2 decline. Nat Commun. 2012;3:835. [DOI] [PubMed] [Google Scholar]
- [26].Kowal J, Pressel S, Ducket JG, et al. From rhizoid to roots? Experimental evidence of mutualism between liverworts and ascomycete fungi. Ann Bot. 2018;121:221–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhang Y, Guo LD. Arbuscular mycorrhizal structure and fungi associated with mosses. Mycorrhiza. 2007;17:319–325. [DOI] [PubMed] [Google Scholar]
- [28].Field KJ, Rimington WR, Bidartondo MI, et al. First evidence of mutualism between ancient plant lineages (Haplomitriopsida liverworts) and Mucoromycotina fungi and its response to simulated Palaeozoic changes in atmospheric CO2. New Phtyol. 2015;205:743–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Field KJ, Pressel S. Unity in diversity: structural and functional insights into the ancient partnerships between plants and fungi. New Phytol. 2018. DOI: 10.1111/nph.15158 [DOI] [PubMed] [Google Scholar]
- [30].Schüßler A. Glomus claroideum forms an arbuscular mycorrhiza-like symbiosis with the hornwort Anthoceros punctatus. Mycorrhiza. 2000;10:15–21. [Google Scholar]
- [31].Desirò A, Duckett JG, Pressel S, et al. Fungal symbioses in hornworts: a chequered history. Proc R Soc B. 2013;280:20130207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang B, Qiu YL. Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza. 2006;16:299–363. [DOI] [PubMed] [Google Scholar]
- [33].Pressel S, Bidartondo MI, Ligrone R, et al. Fungal symbioses in bryophytes: new insights in the Twenty First Century. Phytotaxa. 2010;9:238–253. [Google Scholar]
- [34].Wang B, Yeun LH, Xue J-Y, et al. Presence of three mycorrhizal genes in the common ancestor of land plants suggests a key role of mycorrhizas in the colonization of land by plants. New Phytol. 2010;186:514–525. [DOI] [PubMed] [Google Scholar]
- [35].Puttick MN, Morris JL, William TA, et al. The interrelationships of land plants and the nature of the ancestral embryophyte. Curr Biol. 2018;28:733–745. [DOI] [PubMed] [Google Scholar]
- [36].Bonfante P, Selosse M-A. A glimpse into the past of land plants and of their mycorrhizal affairs: from fossils to evo-devo. New Phytol. 2010;186:267–270. [DOI] [PubMed] [Google Scholar]
- [37].Knack JJ, Wilcox LW, Delaux P‐M, et al. Microbiomes of streptophyte algae and bryophytes suggest that a functional suite of microbiota fostered plant colonization of land. Int J Plant Sci. 2015;176:405–420. [Google Scholar]
- [38].Strullu‐Derrien C, Kenrick P, Pressel S, et al. Fungal associations in Horneophyton ligneri from the Rhynie Chert (c. 407 million year old) closely resemble those in extant lower land plants: novel insights into ancestral plant–fungus symbioses. New Phytol. 2014;203:964–979. [DOI] [PubMed] [Google Scholar]
- [39].Brader G, Compant S, Vescio K, et al. Ecology and genomic insights into plant-pathogenic and plant-nonpathogenic endophytes. Annu Rev Phytopathol. 2017;55:61–83. [DOI] [PubMed] [Google Scholar]
- [40].Schulz B, Boyle C. The endophytic continuum. Mycol Res. 2005;109:661–686. [DOI] [PubMed] [Google Scholar]
- [41].Junker C, Draeger S, Schulz B. A fine line — edophytes or pathogens in Arabidopsis thaliana. Fungal Ecol. 2012;5:657–662. [Google Scholar]
- [42].Rey T, Jacquet C. Symbiosis genes for immunity and vice versa. Curr Opin Plant Biol. 2018;44:64–71. [DOI] [PubMed] [Google Scholar]
- [43].Ried MK, Banhara A, Binder A, et al. Symbiosis-related genes sustain the development of a downy mildew pathogen on Arabidopsis thaliana. bioRxiv. 2018. DOI: 10.1101/286872. [DOI] [Google Scholar]
- [44].Wellman CH, Strother PK. The terrestrial biota prior to the origin of land plants (embryophytes): a review of the evidence. Paleontology. 2015;58:601–627. [Google Scholar]
- [45].Morris JL, Puttick MN, Clark JW, et al. The timescale of early land plant evolution. Proc Natl Acad Sci USA. 2018;115:E2274– E2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Taylor TN, Remy W, Hass H. Parasitism in a 400-mllion-year-old green alga. Nature. 1992;357:493–494. [Google Scholar]
- [47].Berbee ML, James TY, Strulle-Derrien C. Early diverging fungi: diversity and impact at the dawn of terrestrial life. Ann Rev Microbiol. 2017;71:41–60. [DOI] [PubMed] [Google Scholar]
- [48].Spatafora JW, Chang Y, Benny GL, et al. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia. 2016;108:1028–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Sørensen I, Pettolino FA, Bacic A, et al. The charophycean green algae provide insights into the early origins of plant cell walls. Plant J. 2011;68:201–211. [DOI] [PubMed] [Google Scholar]
- [50].Rasconi S, Niquil N, Sime-Ngando T. Phytoplankton chytridiomycosis: community structure and infectivity of fungal parasites in aquatic ecosystems. Environ Microbiol. 2012;14:2151–2170. [DOI] [PubMed] [Google Scholar]
- [51].Cui H, Tsuda K, Parker JE. Effector-triggered immunity: from pathogen perception to robust defense. Annu Rev Plant Biol. 2015;66:487–511. [DOI] [PubMed] [Google Scholar]
- [52].Melotto M, Underwood W, Koczan J, et al. Plant stomata function in innate immunity against bacterial invasion. Cell. 2006;126:969–980. [DOI] [PubMed] [Google Scholar]
- [53].Underwood W. The plant cell wall: a dynamic barrier against pathogen invasion. Front Plant Sci. 2012;3:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Lloyd SR, Schoonbeek H-J, Trick M, et al. Methods to studies PAMP-triggered immunity in Brassica species. Mol Plant Microbe Int. 2014;27:286–295. [DOI] [PubMed] [Google Scholar]
- [55].Mott GA, Guttman DS, Desveaux D. The study of pattern-triggered immunity in Arabidopsis. Can J Plant Pathol. 2017;39:275–281. [Google Scholar]
- [56].Felix G, Duran JD, Volko S, et al. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999;18:265–276. [DOI] [PubMed] [Google Scholar]
- [57].Delwiche CF, Graham LE, Thomson N. Lignin-like compounds and sporopollenin in Coleochaete, an algal model for land plant ancestry. Science. 1989;245:399–401. [DOI] [PubMed] [Google Scholar]
- [58].Kroken SB, Graham LE, Cook ME. Occurrence and evolutionary significance of resistant cell walls in charophytes and bryophytes. Am J Bot. 1996;83:1241–1254. [Google Scholar]
- [59].Ligrone R, Carafa A, Duckett JG, et al. Immunocytochemical detection of lignin-related epitopes in cell walls in bryophytes and the charalean alga Nitella. Plant Syst Evol. 2008;270:257–272. [Google Scholar]
- [60].Herburger K, Holzinger A. Localization and quantification of callose in the streptophyte green algae Zygnema and Klebsormidium: correlation with desiccation tolerance. Plant Cell Physiol. 2015;56:2259–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Overdijk EJR, de Keijzer J, de Groot D, et al. Interaction between the moss Physcomitrella patens and Phytophthora: a novel pathosystem for live-cell imaging of subcellular defence. J Microsc. 2016;263:171–180. [DOI] [PubMed] [Google Scholar]
- [62].Oliver JP, Castro A, Gaggero C, et al. Pythium infection activates conserved plant defense responses in mosses. Planta. 2009;230:569–579. [DOI] [PubMed] [Google Scholar]
- [63].Ponce de León I, Schmelz EA, Gaggero C, et al. Physcomitrella patens activates reinforcement of the cell wall, programmed cell death and accumulation of evolutionary conserved defence signals, such as salicylic acid and 12-oxo-phytodienoic acid, but not jasmonic acid, upon Botrytis cinerea infection. Mol Plant Pathol. 2012;13:960–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Davey ML, Tsuneda A, Currah RS. Pathogenesis of bryophyte hosts by the ascomycete Atradidymella muscivora. Am J Bot. 2009;96:1274–1280. [DOI] [PubMed] [Google Scholar]
- [65].Davey ML, Tsuneda A, Currah RS. Saprobic and parasitic interactions of Coniochaeta velutina with mosses. Botany. 2010;88:258–265. [Google Scholar]
- [66].Carella P, Gogleva A, Tomaselli M, et al. Phytophthora palmivora establishes tissue-specific intracellular infection structures in the earliest divergent land plant lineage. Proc Natl Acad Sci USA. 2018;115:E3846– E3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Gómez-Gómez L, Boller T. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5:1003–1011. [DOI] [PubMed] [Google Scholar]
- [68].Boller T, Felix GA. Renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. [DOI] [PubMed] [Google Scholar]
- [69].Yue J-X, Meyers BC, Chen J-Q, et al. Tracing the origin and evolutionary history of plant nucleotide-binding site-leucine-rich repeat (NBS-LRR) genes. New Phytol. 2012;193:1049–1063. [DOI] [PubMed] [Google Scholar]
- [70].Bressendorf S, Azevedo R, Kenchappa CS, et al. An innate immunity pathway in the moss Physcomitrella patens. Plant Cell. 2016;28:1328–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Kunze G, Zipfel C, Robatzek S, et al. The N terminus of bacterial Elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell. 2004;16:3496–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Zipfel C, Kunze G, Chinchilla D, et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. [DOI] [PubMed] [Google Scholar]
- [73].Ponce de León I, Oliver JP, Castro A, et al. Erwinia carotovora elicitors and Botrytis cinerea activate defense responses in Physcomitrella patens. BMC Plant Biol. 2007;7:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Xu Z, Yu J, Cui L, et al. Effects of Pseudomonas fluorescens flagellin on physiological and biochemical characteristics in the suspension cells of Pinus thunbergii. Eur J Plant Pathol. 2013;136:729–736. [Google Scholar]
- [75].Albert M, Jehle AK, Lipschis M, et al. Regulation of cell behaviour by plant receptor kinases: Pattern recognition receptors as prototypical models. Eur J Cell Biol. 2010;89:200–207. [DOI] [PubMed] [Google Scholar]
- [76].Urano D, Jones AM. Heterotrimeric G protein-coupled signaling in plants. Annu Rev Plant Biol. 2014;65:365–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Delgado-Cerezo M, Sánchez-Rodríguez C, Escudero V, et al. Arabidopsis heterotrimeric G-protein regulates cell wall defense and resistance to necrotrophic fungi. Mol Plant. 2012;5:98–114. [DOI] [PubMed] [Google Scholar]
- [78].Liu J, Ding P, Sun T, et al. Heterotrimeric G proteins serve as a converging point in plant defense signaling activated by multiple Receptor-like Kinases. Plant Physiol. 2013;161:2146–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Hackenberg D, Sakayama H, Nishiyama T, et al. Characterization of the heterotrimeric G-protein complex and its regulator from the green alga Chara braunii expands the evolutionary breadth of plant G-protein signaling. Plant Physiol. 2013;163:1510–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Urano D, Chen J-G, Botello JR, et al. Heterotrimeric G protein signalling in the plant kingdom. Open Biol. 2013;3:120186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].de Vries S, Nemesio-Gorriz M, Blair PB, et al. Heterotrimeric G-protein in Picea abies and their regulation in response to Heterobasidion annosum s.l. infection. BMC Plant Biol. 2015;15:287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Chung E-H, El-Kasmi F, Loehr A, et al. A plant phosphoswitch platform repeatedly targeted by type III effector proteins regulates the output of both tiers of plant immunity receptors. Cell Host Microbe. 2014;16:484–494. [DOI] [PubMed] [Google Scholar]
- [83].Li M, Ma X, Chiang Y-H, et al. Proline isomerization of the immune receptor-interacting protein RIN4 by a cyclophilin inhibits effector-triggered immunity in Arabidopsis. Cell Host Microbe. 2014;16:473–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Rehmany AP, Gordon A, Rose LE. Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 genes from two Arabidopsis lines. Plant Cell. 2005;17:1839–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Allen RL, Meitz JC, Baumber RE, et al. Natural variation reveals key amino acids for recognition specificity between downy mildew effector and an Arabidopsis resistance gene. Mol Plant Pathol. 2008;9:511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Krasileva KV, Dahlbeck D, Staskawicz BJ. Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell. 2010;22:2444–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Meyers BC, Kaushik S, Nandety RS. Evolving disease resistance genes. Curr Opin Plant Biol. 2005;8:129–134. [DOI] [PubMed] [Google Scholar]
- [88].Zhang Y, Xia R, Kuang H, et al. The diversification of plant NBS-LRR defense genes directs the evolution of microRNAs that target them. Mol Biol Evol. 2016;33:2692–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Gao Y, Wang W, Zhang T, et al. Out of water: The origin and early diversification of plant R-genes. Plant Phys. 2018;177:82–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Nystedt B, Street NR, Wetterbom A, et al. The Norway spruce genome sequence and conifer genome evolution. Nature. 2013;497:579–584. [DOI] [PubMed] [Google Scholar]
- [91].Young ND, Debellé F, Oldroyd GE, et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature. 2011;480:520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].International Wheat Genome Sequencing Consortium A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science. 2014;345:1251788. [DOI] [PubMed] [Google Scholar]
- [93].Urbach JM, Ausubel FM. The NBS-LRR architectures of plant R-proteins and metazoan NLRs evolved in independent events. Proc Natl Acad Sci USA. 2017;114:1063–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Hori K, Maruyama F, Fujisawa T, et al. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat Commun. 2014;5:3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Li X, Kapos P, Zhang Y. NLRs in plants. Curr Opin Immunol. 2015;32:114–121. [DOI] [PubMed] [Google Scholar]
- [96].Zhai J, Jeong D-H, De Paoli E, et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011;25:2540–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Li F, Pignatta D, Bendix C, et al. MicroRNA regulation of plant innate immune receptors. Proc Natl Acad Sci USA. 2012;109:1790–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Shivaprasad PV, Chen HM, Patel K, et al. A microRNA superfamily regulates nucleotide binding site-leucine rich repeats and other mRNAs. Plant Cell. 2012;24:859–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Fei Q, Xia R, Meyers BC. Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell. 2013;25:2400–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Xia R, Xu J, Arikit S, et al. Extensive families of miRNAs and PHAS loci in Norway spruce demonstrate the origins of complex phasiRNA networks in seed plants. Mol Biol Evol. 2015;32:2905–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Boccara M, Sarazin A, Thiébeauld O, et al. The Arabidopsis miR472-RDR6 silencing pathway modulates PAMP- and effector-triggered immunity through the post-transcriptional control of disease resistance genes. PLoS Pathog. 2014;10:e1003883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Ouyang S, Park G, Atamian HS, et al. MicroRNAs suppress NB domain genes in tomato that confer resistance to Fusarium oxysporum. PLoS Pathog. 2014;10:e1004464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Ji H-M, Zhao M, Gao Y, et al. FRG3, a target of slmiR482e-3p, provides resistance against the fungal pathogen Fusarium oxysporum in tomato. Front Plant Sci. 2018;9:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Zhai J, Zhang H, Arikit S, et al. Spatiotemporally dynamic, cell-type-dependent premeiotic and meiotic phasiRNAs in maize anthers. Proc Natl Acad Sci USA. 2015;112:3146–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Shigenaga AM, Argueso CT. No hormone to rule them all: Interactions of plant hormones during the responses of plants to pathogens. Semin Cell Dev Biol. 2016;56:174–189. [DOI] [PubMed] [Google Scholar]
- [106].Berens ML, Berry HM, Mine A, et al. Evolution of hormone signaling networks in plant defense. Annu Rev Phytopathol. 2017;55:401–425. [DOI] [PubMed] [Google Scholar]
- [107].Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol. 2005;43:205–227. [DOI] [PubMed] [Google Scholar]
- [108].Herrera-Vásquez A, Salinas P, Holuigue L. Salicylic acid and reactive oxygen species interplay in the transcriptional control of defense genes expression. Front Plant Sci. 2015;6:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Ishikawa A, Kimura Y, Yasuda M, et al. Salicylic acid-mediated cell death in the Arabidopsis len3 mutant. Biosci Biotechnol Biochem. 2006;70:1447–1453. [DOI] [PubMed] [Google Scholar]
- [110].McConn M, Creelman RA, Bell E, et al. Jasmonate is essential for insect defense in Arabidopsis. Proc Natl Acad Sci USA. 1997;94:5473–5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Delaux P-M, Xie X, Timme RE, et al. Origin of strigolactones in the green lineage. New Phytol. 2012;195:857–871. [DOI] [PubMed] [Google Scholar]
- [112].Ju C, Van De Poel B, Cooper ED, et al. Conservation of ethylene as a plant hormone over 450 million years of evolution. Nat Plants. 2015;1:14004. [DOI] [PubMed] [Google Scholar]
- [113].Wang C, Liu Y, Li -S-S, et al. Insights into the origin and evolution of the plant hormone signaling machinery. Plant Physiol. 2015;167:872–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].de Vries J, Curtis BA, Gould SB, et al. Embryophyte stress signaling evolved in the algal progenitors of land plants. Proc Natl Acad Sci USA. 2018;115:E3471– E3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Van de Poel B, Cooper ED, Van Der Straeten D, et al. Transcriptome profiling of the green alga Spirogyra pratensis (Charophyta) suggests an ancestral role for ethylene in cell wall metabolism, photosynthesis, and abiotic stress responses. Plant Physiol. 2016;172:533–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Ohtaka K, Hori K, Kanno Y, et al. Primitive auxin response without TIR1 and Aux/IAA in the charophyte alga Klebsormidium nitens. Plant Physiol. 2017;174:1621–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Mutte S, Kato H, Rothfels C, et al. Origin and evolution of the nuclear auxin response system. eLife. 2018;7:e33399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Beilby MJ, Turi CE, Baker TC, et al. Circadian changes in endogenous concentrations of indole-3-acetic acid, melatonin, serotonin, abscisic acid and jasmonic acid in Characeae (Chara australis Brown). Plant Signal Behav. 2015;10:e1082697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Kozlowski G, Métraux J-P. Infection of Norway spruce (Picea abies (L.) Karst.) seedlings with Pythium irregularre Buism. and Pythium ultimum Trow.: histological and biochemical responses. Eur J Plant Pathol. 1998;104:225–234. [Google Scholar]
- [120].Xu M, Dong J, Wang H, et al. Complementary action of jasmonic acid on salicylic acid in mediating fungal elicitor-induced flavonol glycoside accumulation of Gingko biloba cells. Plant Cell Environ. 2009;32:960–967. [DOI] [PubMed] [Google Scholar]
- [121].de Vries J, de Vries S, Slamovits CH, et al. How embryophytic is the biosynthesis of phenylpropanoids and their derivatives in streptophyte algae. Plant Cell Physiol. 2017;58:934–945. [DOI] [PubMed] [Google Scholar]
- [122].Dixon RA, Achnine L, Kota P, et al. The phenylpropanoid pathway and plant defence – genomics perspective. Mol Plant Pathol. 2002;3:371–390. [DOI] [PubMed] [Google Scholar]
- [123].Wu Y, Zhang D, Chu JY, et al. The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 2012;1:639–647. [DOI] [PubMed] [Google Scholar]
- [124].Peng Y, Sun T, Zhang Y. Perception of salicylic acid in Physcomitrella patens. Front Plant Sci. 2017;8:2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Han G-Z. Evolution of jasmonate biosynthesis and signaling mechanisms. J Exp Bot. 2017;68:1323–1331. [DOI] [PubMed] [Google Scholar]
- [126].Hackenberg D, Pandey S. Heterotrimeric G-proteins in green algae. Plant Signal Behav. 2014;9:e28457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Gachet MS, Schubert A, Calarco S, et al. Targeted metabolomics shows plasticity in the evolution of signaling lipids and uncovers old and new endocannabinoids in the plant kingdom. Sci Rep. 2017;7:41177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Koeduka T, Ishizaki K, Mugo Mwenda M, et al. Biochemical characterization of allene oxide synthases from the liverwort Marachantia polymorpha and green microalgae Klebsormidium flaccidum provides insight into the evolutionary divergence of the plant CYP74 family. Planta. 2015;242:1175–1186. [DOI] [PubMed] [Google Scholar]
- [129].Yamamoto Y, Ohshika J, Takahashi T, et al. Functional analysis of allene oxide cyclase, MpAOC, in the liverwort Marchantia polymorpha. Phytochemistry. 2015;116:48–56. [DOI] [PubMed] [Google Scholar]
- [130].Záveská Drábková L, Dobrev PI, Motyka V. Phytohormone profiling across the bryophytes. PLoS One. 2015;10:e0125411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Stumpe M, Göbel C, Faltin B, et al. The moss Physcomitrella patens contains cyclopentenones but no jasmonates: mutations in allene oxide cyclase lead to reduced fertility and altered sporophyte morphology. New Phytol. 2010;188:740–749. [DOI] [PubMed] [Google Scholar]
- [132].Monte I, Ishida S, Zamarreño AM, et al. Ligand-receptor co-evolution shaped the jasmonate pathway in land plants. Nat Chem Biol. 2018;14:480–488. [DOI] [PubMed] [Google Scholar]
- [133].Pratiwi P, Tanaka G, Takahashi T, et al. Identification of jasmonic acid and jasmonoyl-isoleucine, and characterization of AOS, AOC, OPR and JAR1 in the model lycophyte Selaginella moellendorffii. Plant Cell Physiol. 2017;58:789–801. [DOI] [PubMed] [Google Scholar]
- [134].Camloh M, Ravinkar M, Žel J. Jasmonic acid promotes division of fern protoplasts, elongation of rhizoids and early development of gametophytes. Physiol Plantarum. 1996;97:659–664. [Google Scholar]
- [135].Camloh M, Vilhar B, Žel J, et al. Jasmonic acid stimulates development of rhizoids and shoots in fern leaf culture. J Plant Physiol. 1999;155:798–801. [Google Scholar]
- [136].de Vries S, de Vries J, Teschke H, et al. Jasmonic and salicylic acid response in the fern Azolla filiculoides and its cyanobiont. Plant Cell Environ. 2018. DOI: 10.1111/pce.13131. [DOI] [PubMed] [Google Scholar]
- [137].Castro A, Vidal S, Ponce De León I. Moss pathogenesis-related-10 protein enhances resistance to Pythium irregulare in Physcomitrella patens and Arabidopsis thaliana. Front Plant Sci. 2016;7:580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Koutaniemi S, Warinowski T, Kärkönen A, et al. Expression profiling of the lignin biosynthetic pathway in Norway spruce using EST sequencing and real-time RT-PCR. Plant Mol Biol. 2007;65:311–328. [DOI] [PubMed] [Google Scholar]
- [139].Arnerup J, Lind M, Olson Å, et al. The pathogenic white-rot fungus Heterobasidion parviporum triggers non-specific defence response in the bark of Norway spruce. Tree Physiol. 2011;31:1262–1272. [DOI] [PubMed] [Google Scholar]
- [140].Oliva J, Rommel S, Fossdal CG, et al. Transcriptional responses of Norway spruce (Picea abies) inner sapwood against Heterobasidion parviporum. Tree Physiol. 2015;35:1007–1015. [DOI] [PubMed] [Google Scholar]
- [141].Thaler JS, Humphrey PT, Whiteman NK. Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 2012;17:260–270. [DOI] [PubMed] [Google Scholar]
- [142].Zimmerli L, Stein M, Lipka V, et al. Host and non-host pathogens elicit different jasmonate/ethylene responses in Arabidopsis. Plant J. 2004;40:633–646. [DOI] [PubMed] [Google Scholar]
- [143].Arnerup J, Nemesio-Gorriz M, Lunden K, et al. The primary module in Norway spruce defence signaling against H. annosum s.l. seems to be jasmonate-mediated signalling without antagonism of salicylate-mediated signalling. Planta. 2013;237:1037–1045. [DOI] [PubMed] [Google Scholar]
- [144].Ponce De León I, Hamberg M, Castresana C. Oxylipins in moss development and defense. Front Plant Sci. 2015;6:483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Liu L, Sonbol F-M, Huot B, et al. Salicylic acid receptors activate jasmonic acid signaling through a non-canonical pathway to promote effector-triggered immunity. Nat Commun. 2016;7:13099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Betsuyaku S, Katou S, Takebayashi Y, et al. Salicylic acid and jasmonic acid pathways are activated in spatially different domains around the infection site during effector-triggered immunity in Arabidopsis thaliana. Plant Cell Physiol. 2018;59:8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Celedon JM, Yuen MMS, Chiang A, et al. Cell-type- and tissue-specific transcriptomes of the white spruce (Picea glauca) bark unmask fine-scale spatial patterns of constitute and induced conifer defense. Plant J. 2017;92:710–726. [DOI] [PubMed] [Google Scholar]
- [148].Miedes E, Vanholme R, Boerjan W, et al. The role of the secondary cell wall in plant resistance to pathogens. Front Plant Sci. 2014;5:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Danielsson M, Lunden K, Elfstrand M, et al. Chemical and transcriptional responses of Norway spruce genotypes with different susceptibility to Heterobasidion spp. infection. BMC Plant Biol. 2011;11:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Deflorio G, Horgan G, Woodward S, et al. Gene expression profiles, phenolics and lignin of Sikta spruce bark and sapwood before and after wounding and inoculation with Heterobasidion annosum. Physiol Mol Plant P. 2011;75:180–187. [Google Scholar]
- [151].Paniagua C, Bilkova A, Jackson P, et al. Dirigent proteins in plants: modulating cell wall metabolism during abiotic and biotic stress exposure. J Exp Bot. 2017;68:3287–3301. [DOI] [PubMed] [Google Scholar]
- [152].Adomas A, Heller G, Li G, et al. Transcript profiling of a conifer pathosystem: response of Pinus sylvestris root tissue to pathogen (Heterobasidion annosum) invasion. Tree Physiol. 2007;27:1441–1458. [DOI] [PubMed] [Google Scholar]
- [153].Ponce de León I, Montesano M. Activation of defense mechanisms against pathogens in mosses and flowering plants. Int J Mol Sci. 2013;14:3178–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Goiris K, Muylaert K, Voorspoels S, et al. Detection of flavonoids in microalgae from different evolutionary lineages. J Phycol. 2014;50:483–492. [DOI] [PubMed] [Google Scholar]
- [155].Markham KR, Poster LJ. Flavonoids in the green algae (Chlorophyta). Phytochemistry. 1969;8:1777–1781. [Google Scholar]
- [156].Nesi N, Debeaujon I, Jond C, et al. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell. 2000;12:1863–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]


