Abstract
Background
The interleukin (IL)-4/IL-13/signal transducer and activator of transcription (STAT) 6 signaling pathway and the SOCS3 gene, one of its main regulators, constitute an important link between the inflammation process in the epithelial cells and inflammatory-related tumorigenesis. The present study is the first to evaluate IL-4, IL-13, STAT6, and SOCS3 mRNA expression in non-small cell lung carcinoma (NSCLC) histopathological subtypes.
Methods
Gene expression levels were assessed using TaqMan® probes by quantitative reverse transcription PCR (qRT-PCR) in lung tumor samples and unchanged lung tissue samples.
Results
Increased expression of IL-4, IL-13, and STAT6 was observed in all histopathological NSCLC subtypes (squamous cell carcinoma [SCC], adenocarcinoma [AC], and large cell carcinoma [LCC]). Significantly higher expression of IL-13 and STAT6 (p = 0.019 and p = 0.008, respectively) was found in SCC than in LCC. No statistically significant differences were found for IL-4. Significantly higher SOCS3 expression was found in LCC than in AC (p = 0.027). A negative correlation (rho = –0.519) was observed for the STAT6 and SOCS3 genes in SCC (p = 0.005). No associations were found between gene expression and tumor staging (post-operative Tumor Node Metastasis [pTNM], American Joint Committee on Cancer [AJCC]), patients’ age, sex, or history of smoking.
Conclusions
As the number of LCC cases in our study was quite low, the statistically significant results obtained should be confirmed in a larger group of patients, particularly as the relationships identified between increased IL-4, IL-13, and STAT6 mRNA expression and decreased SOCS3 expression suggest that these genes may serve as potential diagnostic markers for differentiating between NSCLC histopathological subtypes.
Key Points
| The analysis of IL-13/STAT6/SOCS3 expression could be useful as a support tool in non-small cell lung carcinoma (NSCLC) histopathological examination. |
| The presence of a negative correlation between STAT6 and SOCS3 confirms that suppressors of cytokine signaling (SOCS) act as a negative regulator of signal transducer and activator of transcription (STAT). |
| An accurate subclassification based on molecular analysis could, in the future, offer more personalized therapies for NSCLC patients. |
Introduction
Interleukin (IL)-4, IL-13, and signal transducer and activator of transcription (STAT) 6 signaling pathways are known to exhibit a range of immunomodulatory functions, particularly IL-4 and IL-13 [1]. Both ILs are recognized as crucial cytokines that regulate the immune reactions taking place in the lung; these are characterized by the recruitment of inflammatory cells and lymphocytes and may be involved in lung remodeling [2]. Moreover, IL-4 and IL-13 are believed to play a key role in encouraging allergic airway inflammation by acting as the main stimulators of IgE production in B lymphocytes and T helper (Th) 2-type differentiation in T cells [3–5]. STAT6 seems to be the main target for IL-4 and IL-13 [6] and appears to be essential for Th1/Th2 balance during the autoimmunity response [7]. The results of animal studies suggest that IL-4, IL-13, or STAT6 deficiency may influence IgE synthesis and Th2-type reactions [8]. Additionally, it has been documented that STAT6 overexpression and secretion of IL-4 may significantly increase the level of cell apoptosis during cancer development [9]. In vitro studies have also demonstrated that IL-4 may directly inhibit the growth of transformed cells in human lung cancer, among other cancers [10, 11].
The negative regulation of Janus kinase (JAK)/STAT signaling is highly dependent on the action of suppressors of the cytokine signaling (SOCS) proteins SOCS1–SOCS7 and cytokine-inducible SH2 protein (CIS) [12, 13]. Overexpression of SOCS3 has been observed in human chronic inflammatory diseases [14, 15]; however, the deficiency of this protein has been recorded in some types of cancer, including lung cancer [16, 17]. Moreover, it has been confirmed that the main biochemical target for SOCS3, except STAT3, appears to also be STAT6 (via IL-4/IL-13/STAT6 signaling) [18]. STAT6 is an important protein known to be regulated by SOCS3, as two possible STAT-response elements (SREs) able to bind STAT6 have been identified within the SOCS3 gene promoter [19, 20]. Hence, SOCS3 deregulation in cancer development has been attributed to cross-talk between STAT6 and SOCS3 genes on both the protein and mRNA levels; however, this mechanism remains unclear.
Therefore, it is possible that molecular changes in the IL-4/IL-13/STAT6 signaling pathway on the transcriptional or translational level may have a significant influence on the development of human inflammatory diseases, as well as in the development of the inflammatory-related tumors, including lung cancer [8, 9, 21–23].
The present study evaluates the relationship between the expression of STAT6 and SOCS3 and their mRNA level in non-small cell lung carcinoma (NSCLC) histopathological subtypes to identify potential diagnostic molecular markers.
Materials and Methods
Clinical Characterization of the Studied Patients
Seventy-one patients with a confirmed diagnosis of NSCLC (25 women, mean age 63 ± 8.717 years; 46 men, mean age 65 ± 8.234 years) were enrolled into the study.
The smoking history was available for 71 patients: five patients were non-smokers and 66 were smokers or former smokers. They were divided into groups according to duration of tobacco addiction and number of cigarettes smoked; the latter was presented as pack years (PYs), and was calculated according to the NCI Dictionary of Cancer Terms [24] (1 PY is equal to 20 cigarettes smoked per day for 1 year) (see Table 1).
Table 1.
Characterization of the studied non-small cell lung cancer patients (n = 71) regarding their tobacco addiction and consumption (number of cigarettes smoked per day and pack years)
| Tobacco addiction and consumption | n (%) |
|---|---|
| Smoking period | |
| Smokers | 66 (93) |
| < 40 years | 37 (52) |
| ≥ 40 years | 29 (41) |
| Non-smokers | 5 (7) |
| Number of cigarettes smoked per day | |
| 10–15 | 6 (8) |
| 20 (1 pack) | 43 (61) |
| 30–40 (1.5–2 packs) | 17 (24) |
| PYs | |
| < 40 | 30 (42) |
| ≥ 40 | 36 (51) |
PYs pack years
Characterization of the Non-Small Cell Lung Carcinoma Tissue Samples
Lung tissue samples (100–150 mg) were received from patients with preoperatively diagnosed lung cancer who had undergone pulmonectomy or lobectomy at the Department of Thoracic Surgery, General and Oncologic Surgery, Medical University of Lodz, Lodz, Poland between July 2010 and June 2012. Immediately after resection, the tissue samples were collected in RNAlater® buffer (ThermoFisher Scientific, Waltham, MA, USA) and frozen at –80 °C. For each patient, two kinds of tissue samples were collected: a lung tissue sample from the primary lesion and a sample of adjacent non-cancerous macroscopically unchanged lung tissue (10 cm distant from the primary lesion) to serve as a control.
The diagnosis of lung cancer was made according to the 2015 World Health Organization (WHO) Classification of Lung Tumours diagnostic criteria [25]. Squamous cell carcinoma (SCC) was diagnosed when keratinization, pearl formation, and/or intercellular bridges were observed. Furthermore, in case of high-grade SCC, immunohistochemical staining for cytokeratin (CK) 5/6 and CK 34 beta E12 was performed. Diagnosis of large cell carcinoma (LCC) was made after ruling out the presence of a component of SCC, adenocarcinoma (AC), or small-cell carcinoma. In case of AC diagnosis, glandular differentiation and/or mucin production were confirmed. Additionally, for unambiguous verification of AC, thyroid transcription factor-1 (TTF-1) and CK 7, immunohistochemical expression was assessed.
The resected specimens were subjected to post-operative histopathological evaluation and classified according to the American Joint Committee on Cancer (AJCC) staging [26] and post-operative Tumor Node Metastasis (pTNM) classification.
The results of the histopathological verification of tumor specimens, based on pathomorphological reports, are summarized in Table 2.
Table 2.
Histopathological verifications of non-small cell lung cancer samples
| Histopathological type of NSCLC | n (%) |
|---|---|
| Squamous cell carcinoma | 41 (58) |
| Non-squamous cell carcinoma | 30 (42) |
| Adenocarcinoma | 23 (32) |
| Large cell carcinoma | 7 (10) |
| AJCC | |
| AJCC IA | 14 (20) |
| AJCC IB | 11 (16) |
| AJCC IIA | 13 (18) |
| AJCC IIB | 10 (14) |
| AJCC IIIA/IIIB | 23 (32) |
| pTNM | |
| T1 | 19 (27) |
| T2 | 33 (47) |
| T3–4 | 19 (27) |
AJCC American Joint Committee on Cancer Staging according to the IASLC Staging Project [26], NSCLC non-small cell lung cancer, pTNM post-operative Tumor Node Metastasis classification according to the World Health Organization Histological Typing of Lung Tumor
RNA Extraction and Expression Analysis of IL-4, IL-13, STAT6, and SOCS3
RNA was extracted from tissue samples using a Universal RNA Purification Kit (Eurix, Gdańsk, Poland) according to the manufacturer’s recommendations. Complementary DNA (cDNA) was transcribed from 100 ng of total RNA using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA), in a total volume of 20 µL. Reverse transcription (RT) master mix contained the following: 10 × RT buffer, 25 × dNTP (deoxynucleotide) mix (100 mM), 10 × RT random primers, MultiScribe™ reverse transcriptase, RNase inhibitor, and nuclease-free water. The RT reaction was performed in a Personal Thermocycler (Eppendorf, Hamburg, Germany) in the following conditions: 10 min at 25 °C, followed by 120 min at 37 °C, then the samples were heated to 85 °C for 5 s, and hold at 4 °C.
The relative expression (RQ) of IL-4, IL-13, STAT6, and SOCS3 was assessed using TaqMan® probes (Applied Biosystems) for the studied genes (Hs00932431_m1, Hs00174379_m1, Hs00598625_m1, and Hs02330328_m1, respectively), with ACTB (Hs99999903_m1) as the reference gene. The procedure was performed in an Applied Biosystems 7900HT Fast Real-Time PCR System, for 39 cycles. The PCR mixture was as follows: cDNA (1–100 ng), 20 × TaqMan® Gene Expression Assay, 2 × TaqMan® Gene Expression Master Mix, and RNase-free water, in a total volume of 20 µL. The RQ of each sample was assessed using the Comparative ΔΔCT method adjusted to ACTB (endogenous control) expression and presented as the RQ value.
The following formula was used to determine the ΔΔCT value: ΔΔCT = ΔCT test sample − ΔCT calibrator sample. Higher ΔΔCT values indicated lower expression of IL-4, IL-13, STAT6, or SOCS3 in the specimen [27].
In relation to the expression level of the calibrator (macroscopically unchanged lung tissue), for which RQ = 1, the obtained results were compared between NSCLC patients in regard to histopathological NSCLC subtype, tumor staging (TNM, AJCC), patients’ age, sex, and smoking history. For the studied samples, an increased expression value was recognized when the RQ value was > 1 and decreased expression was when the RQ value was < 1.
Statistical Analysis
The analysis of variance (ANOVA) Kruskal-Wallis test was used to compare the RQ of IL-4, IL-13, STAT6, and SOCS3 between NSCLC subtypes (SCC, AC, and LCC). Neuman-Keuls’ multiple comparison test was used to identify possible significant differences in RQ values between the individual NSCLC subtypes.
Spearman’s rank correlation coefficient, the Mann-Whitney test, and the ANOVA Kruskal-Wallis test were performed to evaluate the relationships between the expression of the studied genes and the other examined parameters [i.e., patient age, sex, and tumor staging (pTNM, AJCC)]. Statistical significance was regarded as p < 0.05. Additionally, the statistical significance level for multiple analyses was adjusted to a p-value of 0.032 according to Bonferroni’s correction. Statistica™ for Windows 10.0 (TIBCO, Palo Alto, CA, USA) was used for calculations. The RQ values for the studied genes are presented as means ± standard error of the mean and means ± standard deviation.
Results
The results of the expression analysis were calculated using the ΔΔCT method adjusted to ACTB expression (endogenous control) and in relation to the expression level of calibrator (macroscopically unchanged lung tissue), for which RQ = 1.
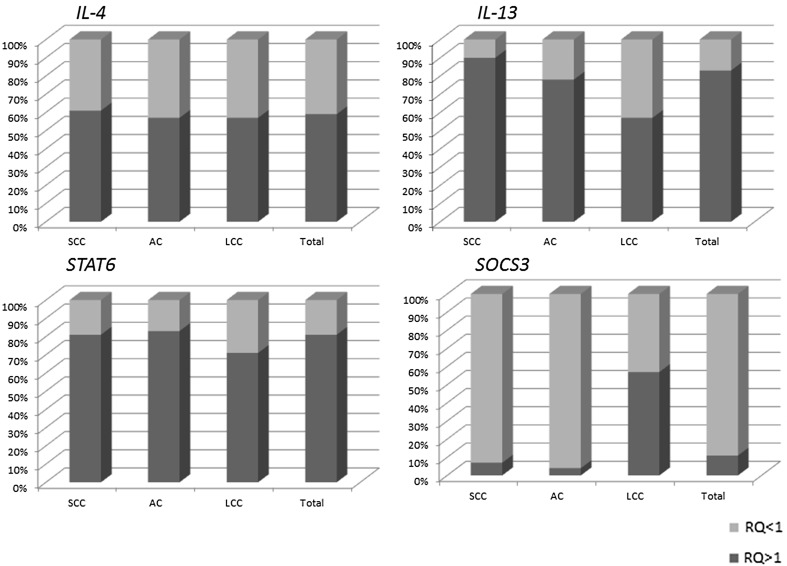
IL-4, IL-13, and STAT6 demonstrated increased expression (RQ value > 1) in all histopathological NSCLC subtypes, being seen in 57–90% of samples, depending on histotype and gene. For all studied genes, the highest level of expression was observed in the SCC subtype, followed by AC, with the lowest level being observed for LCC. SOCS3 was decreased (RQ value < 1) in 93–96% of SCC and AC samples, but increased (RQ value > 1) in 57% of LCC samples. The results are presented in Fig. 1.
Fig. 1.
Percentage of samples with increased (RQ > 1) and decreased (RQ < 1) expression of the examined genes in individual histopathological NSCLC subtypes. NSCLC non-small cell lung cancer, RQ relative expression
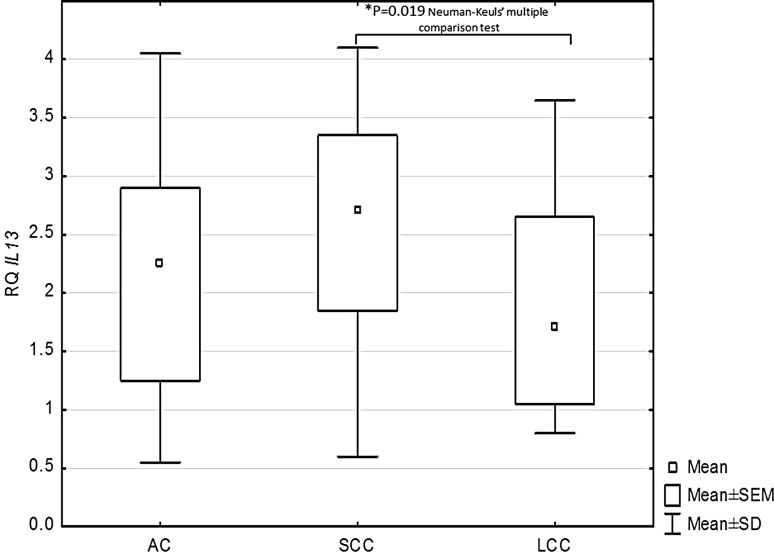
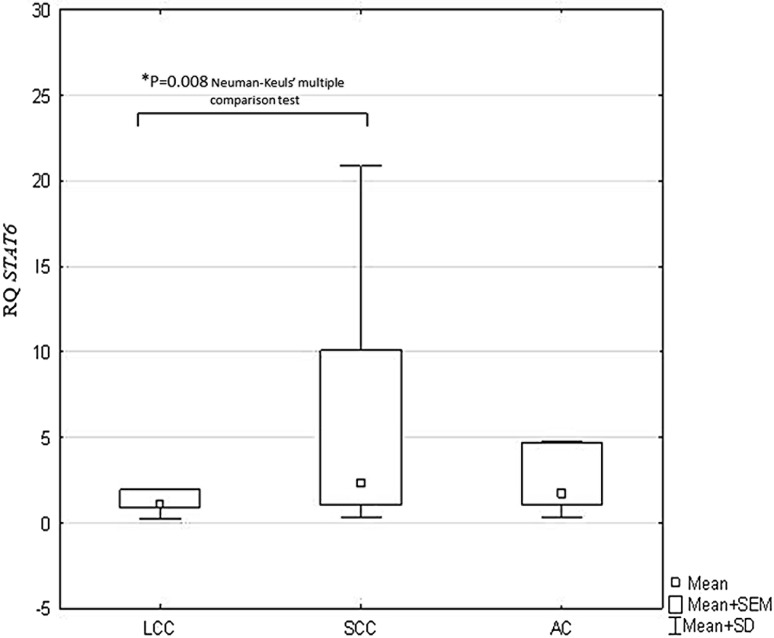
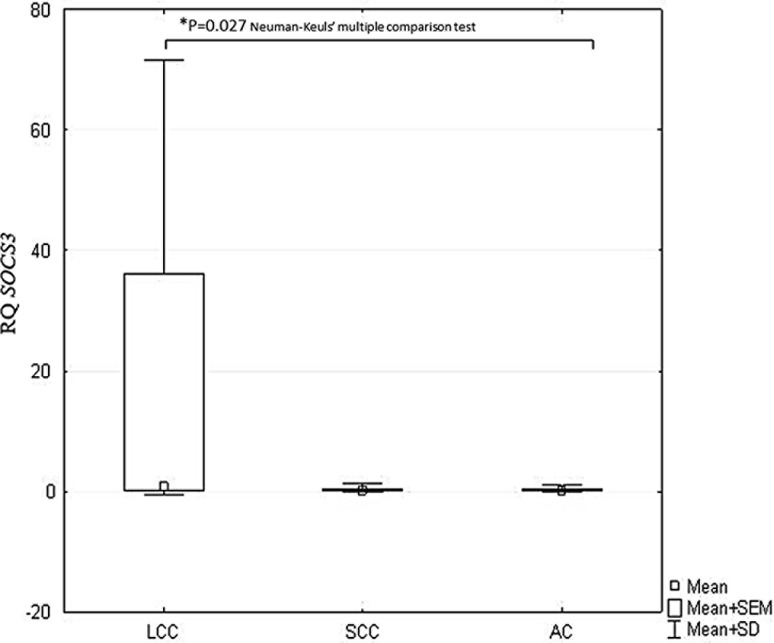
Statistically significant differences in IL-13, STAT6, and SOCS3 expression were observed between the histopathological NSCLC subtypes (p < 0.05, ANOVA Kruskal–Wallis test). The Neuman-Keuls’ multiple comparison test found significant differences between the SCC and LCC histotypes with regard to IL-13 (p = 0.019) and STAT6 expression (p = 0.008), as given in Figs. 2 and 3. Significantly lower SOCS3 expression was found in AC than in LCC (p = 0.027; Neuman-Keuls’ multiple comparison test) (Fig. 4).
Fig. 2.
Box-and-whisker plots representing mean IL-13 expression in the studied NSCLC subtypes (p = 0.019; ANOVA Kruskal-Wallis test). *Indicates a statistically significant difference between SCC and LCC (p = 0.019; Neuman-Keuls’ multiple comparison test). ANOVA analysis of variance, LCC large cell carcinoma, NSCLC non-small cell lung cancer, SCC squamous cell carcinoma
Fig. 3.
Box-and-whisker plots representing mean STAT6 expression in the studied NSCLC subtypes (p = 0.037; ANOVA Kruskal-Wallis test). *Indicates a statistically significant difference between LCC and SCC (p = 0.008; Neuman-Keuls’ multiple comparison test). ANOVA analysis of variance, LCC large cell carcinoma, NSCLC non-small cell lung cancer, SCC squamous cell carcinoma
Fig. 4.
Box-and-whisker plots representing mean SOCS3 expression in the studied NSCLC subtypes (p = 0.029; ANOVA Kruskal-Wallis test). *Indicates a statistically significant difference between LCC and AC (p = 0.027; Neuman-Keuls’ multiple comparison test). AC adenocarcinoma, ANOVA analysis of variance, LCC large cell carcinoma, NSCLC non-small cell lung cancer
No significant differences were found between any of the NSCLC subtypes with regard to IL-4 expression (p = 0.079; ANOVA Kruskal–Wallis test).
Spearman’s rank correlation revealed a statistically significant negative correlation (rho = –0.519) between the RQ values of the STAT6 and SOCS3 genes in the SCC subtype (p = 0.005). While a negative correlation (rho = –0.538) was also observed between the expression of STAT6 and SOCS3 in the AC subtype, this relationship was not significant (p = 0.08; Spearman’s rank correlation). A positive correlation (rho = 0.671) between STAT6 and SOCS3 was found in the LCC subtype, but, again, this was insignificant (p = 0.56; Spearman’s rank correlation).
No statistically significant correlations were found between the RQ values of the studied genes and the clinical features of NSCLC patients, i.e., patient age, sex, or history of smoking assessed as PYs (p > 0.05; ANOVA Kruskal-Wallis test, Mann-Whitney test, followed by Spearman’s rank correlation coefficient). Similarly, no associations were found with tumor staging according to the pTNM and AJCC classifications (p > 0.05; ANOVA Kruskal-Wallis test, Mann-Whitney test).
No statistically significant mutual correlations were found between IL-4, IL-13, STAT6, and SOCS3 gene expression and clinical features, i.e., patient age, sex, history of smoking, or tumor staging according to the pTNM or AJCC classifications.
Study Limitations
The main limitation of our study is that it is based on only a small number of patients with diagnosed LCC, which can be attributed to the fact that it was a single-center study. The research material was obtained from one hospital, in which only a small number of cases were reported during the study period. However, it is important to bear in mind that genetic and environmental factors have a strong influence on lung cancer incidence and the contribution of its histopathological subtypes, especially in polluted areas such as large urban areas.
However, we believe that the results obtained indicating significant differences between NSCLC histopathological subtypes, including the LCC group, are nevertheless valuable and encourage further research aimed at supporting lung cancer diagnostics (NSCLC subtyping) with molecular methods.
Discussion
There is a growing need for molecular studies of the etiology of NSCLC, as lung cancer constitutes a leading cause of mortality worldwide. Hence, to identify informative molecular diagnostic markers in NSCLC the present study evaluates the mRNA expression of IL-4, IL-13, STAT6, and SOCS3, these being important factors in signal transduction and transcription regulation. Due to the high heterogeneity of NSCLC cells, histological subtyping should be supplemented by molecular analysis of cancer cells. Therefore, the study analyzes the differences in the expressions of the studied genes between three different NSCLC histopathological subtypes: SCC, AC, and LCC.
The present study is the first to compare the expression of IL-4 on the mRNA level between different subtypes of NSCLC. Higher expression was observed in SCC than LCC; however, the difference was not statistically significant. Although our results should be interpreted with a degree of caution due to the small number of LCC cases, they confirm those of Huang et al. [28], who also report the expression of IL-4 to be highest in an SCC line, but not significantly so. The same authors confirmed higher immunoexpression of IL-4 in SCC than AC at the protein level [28]. Our results indicate that IL-4 mRNA expression varies depending on the NSCLC subtype; this is an important observation, considering that IL-4 mediates pleiotropic effects in the NSCLC microenvironment and demonstrates both stimulatory and inhibitory effects on antitumor immune responses and tumor proliferation [29–31]. Interestingly, it has been documented that IL-4 may decrease the proliferation of some solid tumor cell lines [32], including lung cancers [31], and may inhibit tumor vascularization [33]. On the other hand, IL-4 influences cancer-associated fibroblasts, which are recognized as key factors in cancer growth and progression [34]. Moreover, patients with advanced cancer often have reduced cell immunity associated with a ‘switching’ from Th1 to Th2 [35]. Tumor cells often demonstrate inhibition of the Thl cytokine response combined with an elevated type 2 (Th2) cytokine response (including IL-2) [36].
To better understand the biological role of IL-4 in cancer cells, the study compared IL-4 mRNA expression between various stages of lung tumor development (TNM staging, AJCC classification). It should be emphasized that our work is the first study to address this issue. However, no statistically significant differences were found in IL-4 expression with regard to the TNM/AJCC classifications. On the other hand, a growing body of evidence suggests that overexpression of IL-4 receptor (IL-4R) may be observed in lung cancer cells as a tissue-specific molecular biomarker [37–39]. Based on this observation, IL-4 signaling through IL-4R has been recognized as a cytotoxic antitumor cytokine, regulating tumor cell survival via the activation of antiapoptotic proteins [31, 39–42]. Moreover, at the protein level, both IL-4R and IL-4 are considered prognostic biomarkers for various types of human epithelial cancer, including lung cancer [41–44].
The IL-13 mRNA level was increased in all histopathological NSCLC subtypes and was significantly higher in SCC than LCC. Although SCC is thought to be associated with smoking behavior [45], this was not indicated by our findings. As only a small number of LCC samples could be included in the present study, our findings should be confirmed in a larger group of patients diagnosed with this NSCLC subtype. LCC displays an aggressive phenotype, as it tends to grow rapidly and spreads more quickly than other NSCLC subtypes [46, 47]. Moreover, LCC frequently presents problems in terms of preoperative diagnosis and often requires additional histological examination. From this point of view, the results obtained seem to be encouraging for further research, in terms of the suitability of IL-13 as a differentiating biomarker for NSCLC.
Similarly, increased IL-13 protein and mRNA expression has been confirmed in NSCLC cell lines by Huang et al. [28]. Little is known about the importance of IL-13 in lung tumors, but it is claimed that IL-4 and IL-13 are important immunological antitumor response factors in lung cancer cells. In contrast to other human epithelial cancers, where the association with invasion, metastasis and poor prognosis has been confirmed [48, 49], the present study is the first to evaluate the significance of IL-4 and IL-13 expression as molecular markers in lung cancer (in relation to TNM/AJCC classification). Unfortunately, as no relationship was found between IL-13 or IL-4 expression and TNM/AJCC classification, it was not possible to confirm whether their mRNA expression could be used for NSCLC prognosis.
The study also found STAT6 upregulation in all studied histopathological NSCLC subtypes (SCC, AC, LCC). These findings are similar to those of previous studies, which indicate the indirect significance of STAT6 in apoptosis resistance in NSCLC cells lines via cyclo-oxygenase (COX)-2 upregulation. Moreover, constitutive activation of STATs and their overexpression has been observed in vivo in many human tumors, including lung cancer [50–57]. It has been observed that cells harboring upregulated STAT6 displayed expression profiles supporting Th2 cytokine secretion, cell cycle promotion, anti-apoptosis, and pro-metastasis processes [51]. Our findings also confirm increased expression of the active form of STAT6 protein in lung cancer patients [58].
It should be pointed out that molecular studies on STAT6 in lung cancer are rare and have received limited attention. This is surprising, given that among many STAT proteins, STAT6 acts as a positive regulator of cancer cell proliferation and tumor metastasis capacity [50, 51]. It is well-recognized that lung cancer, apart from genetic factors, which include JAK/STAT signaling, may be induced by smoking [59–61]. However, no published reports could be found on the relationship between STAT6 expression and patient smoking behavior, and our findings do not reveal any such association between smoking history and STAT6 expression. The reason may be the small number of patients studied. However, it is worth continuing this investigation of the relationship between smoking and changes in STAT6 expression in view of raising awareness of the part played by STAT6 in allergic airway chronic inflammation and the promotion of inflammatory-related cancer.
Similar to IL-13, the STAT6 expression level was significantly higher in SCC than LCC samples, and these results should also be interpreted with caution because of the small number of LCC patients. Still, we hope that our observation might be beneficial for clinical practice because some authors have demonstrated a survival advantage for patients with the SCC subtype [45]. However, a more recent study has reported better prognosis for AC stage I than SCC stage I, and the opposite in stage II [62], while other studies indicate significantly more frequent metastasis occurrence in AC than SCC [46]. So, as the prognostic significance of different histological subtypes remains controversial [47, 63], there is a real need to search for the molecular differences between histological subtypes and the differentiating molecular markers.
The expression of SOCS3 mRNA, a cytokine suppressor, was decreased in both the AC and SCC subtypes, with significant differences being observed between AC and LCC. Although, as mentioned earlier, our LCC group was restricted in size, the differences observed in SOCS3, IL-13, and STAT6 expression between NSCLC subtypes can provide valuable information in the search for additional diagnostic markers in lung cancer. In future, an accurate subclassification based on molecular analysis could act as the basis for targeted therapies in NSCLC patients.
Reduced SOCS3 expression in NSCLC has been reported in a number of previous studies [16, 17, 58, 64]. Significant downregulation of the SOCS3 protein or loss of gene expression in lung tumors or NSCLC cell lines due to promoter hypermethylation was revealed [16]. In the present study, more than 80% of the tested NSCLC samples, with the exception of the LCC subtype, had decreased SOCS3 gene expression, confirming its role as a negative regulator of STAT. However, as SOCS3 may be down- or upregulated by STATs, depending on tumor type [64–68], it is difficult to determine the diagnostic or prognostic value of SOCS3 gene expression. As observed in the present study, the combined downregulation of SOCS3 and upregulation of IL-4, IL-13, and STAT6 indicate the presence of a functional relationship between SOCS3 and the IL-4/IL-13/STAT6 signaling pathway, which has been confirmed elsewhere [20, 52]. The recognized interaction of STAT and SOCS3 genes by binding to SRE (STAT-response elements) in SOCS3 promoter sequences seems to be responsible for IL-4/IL-13/STAT6 stimulation [20, 52].
The present study attempted to check whether the studied genes may be regarded as molecular diagnostic markers in NSCLC, based on an assessment of the relationship between gene expression and TNM/AJCC staging. The most interesting findings seem to be related to the STAT6 and SOCS3 genes, largely due to the paucity of reports focused on similar correlations. The present study is the first to examine the correlation of STAT6 expression with TNM/AJCC staging in lung cancer, and hence may be compared only with results from studies on gliomas and colon or prostate tumors, which indicate an association between STAT6 expression and higher histological grades and larger tumor size, apoptosis resistance, metastasis, and shorter survival [20, 51, 57]. However, these ambiguous observations concerning NSCLC should not be surprising. It has been documented that depending on the cancer cell type, STAT6 can act as either a pro- or anti-apoptotic factor, and, by interacting with other transcription factors, plays a dual role as a STAT [69]. In addition, regarding the association between SOCS3 mRNA expression and tumor progression, no statistically significant differences were found between TNM/AJCC stages. As in the case of STAT6, no comparable studies examining lung cancer can be found. However, Zhang et al. [17] report that restored expression of the SOCS3 protein in lung cancer cells increased apoptosis and decreased cell invasiveness, while significantly downregulated SOCS3 protein expression was correlated with more aggressive phenotypes and poor prognosis in other tumor types, such as breast and prostate cancers [64, 68, 69].
Interestingly, an inverse correlation was found between the STAT6/SOCS3 relationship (as regulators of mutual expression) and tumor progression stage (TNM/AJCC) and NSCLC subtypes. It is therefore reasonable to assume that SOCS3 may be a negative regulator of STAT6, especially in the SCC histopathological subtype.
Conclusions
The present study presents some evidence that STAT6/SOCS3 mRNA expression analysis could be useful as a support tool in NSCLC histopathological examination. However, our observations require further studies on larger numbers of patients diagnosed with non-small cell lung carcinoma.
Funding
This study was supported by the Medical University of Lodz. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
Dorota Pastuszak-Lewandoska, Daria Domańska-Senderowska, Adam Antczak, Jacek Kordiak, Paweł Górski, Karolina H. Czarnecka, Monika Migdalska-Sęk, Ewa Nawrot, Justyna M. Kiszałkiewicz, and Ewa Brzeziańska-Lasota declare that they have no conflict of interest.
Ethical Approval and Informed Consent
The study has been approved by the Ethical Committee of the Medical University of Lodz, Poland, no. RNN/64/11/KE.
References
- 1.Minty A, Chalon P, Derocq JM, Dumont X, Guillemot JC, Kaghad M, et al. Interleukin-13 is a new human lymphokine regulating inflammatory and immune responses. Nature. 1993;362:248–250. doi: 10.1038/362248a0. [DOI] [PubMed] [Google Scholar]
- 2.Tepper TI, Levinson DA, Stanger BZ, Campos-Torres J, Abbas AK, Leder P. IL-4 induces allergic-like inflammatory disease and alters T cell development in transgenic mice. Cell. 1990;62:457–467. doi: 10.1016/0092-8674(90)90011-3. [DOI] [PubMed] [Google Scholar]
- 3.Elias JA, Lee CG, Zheng T, Ma B, Homer RJ, Zhu Z. New insights into the pathogenesis of asthma. J Clin Invest. 2003;111:291–297. doi: 10.1172/JCI17748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grunig G. Requirement for IL-13 independently of IL-4 in experimental asthma. Science. 1998;282:2261–2263. doi: 10.1126/science.282.5397.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chatila TA. Interleukine-4 receptor signaling pathways in asthma pathogenesis. Trends Mol Med. 2004;10:493–499. doi: 10.1016/j.molmed.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 6.Takeda K, Kamanaka M, Tanaka T, Kishimoto T, Akira S. Impaired IL-13-mediated functions of macrophages in STAT6-deficient mice. J Immunol. 1996;157:3220–3222. [PubMed] [Google Scholar]
- 7.Tamachi T, Takatori H, Fujiwara M. STAT6 inhibits T-bet-independent Th1 cell differentiation. Biochem Biophys Res Commun. 2009;382:751–755. doi: 10.1016/j.bbrc.2009.03.101. [DOI] [PubMed] [Google Scholar]
- 8.McKenzie GJ, Fallon PG, Emson CL, Grencis RK, McKenzie AN. Simultaneous disruption of interleukin (IL)-4 and IL-13 defines individual roles in T helper cell type 2-mediated responses. J Exp Med. 1999;189:1565–1572. doi: 10.1084/jem.189.10.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gooch JL, Christy B, Yee D. STAT6 mediates interleukin-4 growth inhibition in human breast cancer cells. Neoplasia. 2002;4:324–331. doi: 10.1038/sj.neo.7900248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Topp MS, Papadimitriou CA, Eitelbach F, Koenigsmann M, Oelmann E, Koehler B, et al. Recombinant human interleukin 4 has antiproliferative activity on human tumor cell lines derived from epithelial and nonepithelial histologies. Cancer Res. 1995;55:2173–2176. [PubMed] [Google Scholar]
- 11.Vokes E, Figlin R, Hochster H, Lotze M, Rybak M. A phase II study of recombinant human interleukin-4 for advanced or recurrent non-small cell lung cancer. Cancer J. 1998;4:46–51. [PubMed] [Google Scholar]
- 12.Yamada S, Shiono S, Joo A, Yoshimura A. Control mechanism of JAK/STAT signal transduction pathway. FEBS Lett. 2003;534:190–196. doi: 10.1016/S0014-5793(02)03842-5. [DOI] [PubMed] [Google Scholar]
- 13.Krebs DL, Hilton DJ. SOCS proteins: negative regulators of cytokine signaling. Stem Cells. 2001;19:378–387. doi: 10.1634/stemcells.19-5-378. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki A, Hanada T, Mitsuyama K, Yoshida T, Kamizono S, Hoshino T. CIS3/SOCS3/SSI3 plays a negative regulatory role in STAT3 activation and intestinal inflammation. J Exp Med. 2001;193:471–481. doi: 10.1084/jem.193.4.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shouda T, Yoshida T, Hanada T, Wakioka T, Oishi M, Miyoshi K, et al. Induction of the cytokine signal regulator SOCS3/CIS3 as a therapeutic strategy for treating inflammatory arthritis. J Clin Invest. 2001;108:1781–1788. doi: 10.1172/JCI13568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He B, You L, Uematsu K. SOCS-3 is frequently silenced by hypermethylation and suppresses cell growth in human lung cancer. Proc Natl Acad Sci USA. 2003;100:14133–14138. doi: 10.1073/pnas.2232790100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang S, Wang W, Wang E, Qiu X. SOCS3 expression is inversely correlated with Pyk2 in non-small cell lung cancer and exogenous SOCS3 inhibits proliferation and invasion of A549 cells. Pathology. 2012;44:434–440. doi: 10.1097/PAT.0b013e328354ffdf. [DOI] [PubMed] [Google Scholar]
- 18.Albanesi C, Fairchild HR, Madonna S, Scarponi C, De Pità O, Leung DY, Howell MD. IL-4 and IL-13 negatively regulate TNF-alpha- and IFN-gamma-induced beta-defensin expression through STAT-6, suppressor of cytokine signaling (SOCS)-1, and SOCS-3. J Immunol. 2007;179:984–992. doi: 10.4049/jimmunol.179.2.984. [DOI] [PubMed] [Google Scholar]
- 19.Yu CR, Mahdi RM, Ebong S. Cell proliferation and STAT6 pathways are negatively regulated in T cells by STAT1 and suppressors of cytokine signaling. J Immunol. 2004;173:737–746. doi: 10.4049/jimmunol.173.2.737. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Deuring J, Peppelenbosch MP, Kuipers EJ, de Haar C, van der Woude CJ. STAT1, STAT6 and adenosine 3’,5’-cyclic monophosphate (cAMP) signaling drive SOCS3 expression in inactive ulcerative colitis. Mol Med. 2012;18:1412–1419. doi: 10.2119/molmed.2012.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 22.De Monte L, Reni M, Tassi E, Clavenna D, Papa I, Recalde H, et al. Intratumor T helper type 2 cell infiltrate correlates with cancer-associated fibroblast thymic stromal lymphopoietin production and reduced survival in pancreatic cancer. J Exp Med. 2011;208:469–478. doi: 10.1084/jem.20101876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Natoli A, Lüpertz R, Merz C, Müller WW, Köhler R, Krammer PH, Li-Weber M. Targeting the IL-4/IL-13 signaling pathway sensitizes Hodgkin lymphoma cells to chemotherapeutic drugs. Int J Cancer. 2013;133:1945–1954. doi: 10.1002/ijc.28189. [DOI] [PubMed] [Google Scholar]
- 24.National Cancer Institute. NCI dictionary of cancer terms. https://www.cancer.gov/publications/dictionaries/cancer-terms/def/pack-year.
- 25.Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, et al. The 2015 World Health Organization Classification of Lung Tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10:1243–1260. doi: 10.1097/JTO.0000000000000630. [DOI] [PubMed] [Google Scholar]
- 26.Goldstraw P, Crowley J, Chansky K, Giroux DJ, Groome PA, et al. The IASLC lung cancer staging project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM classification of malignant tumours. J Thorac Oncol. 2007;2:706–14. doi: 10.1097/JTO.0b013e31812f3c1a. [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Huang M, Wang J, Lee P, Stiantila S, Mao J, Meissner H, et al. Human non-small cell lung cancer cells express a type 2 cytokine pattern. Cancer Res. 1995;55:3847–3853. [PubMed] [Google Scholar]
- 29.Paul WE. Interleukin-4: a prototypic immunoregulatory lymphokine. Blood. 1991;77:1859–1870. [PubMed] [Google Scholar]
- 30.Stremmel C, Greenfield EA, Howard E, Freeman GJ, Kuchroo VK. B7-2 expressed on EL4 lymphoma suppresses antitumor immunity by an interleukin 4–dependent mechanism. J Exp Med. 1999;189:919–930. doi: 10.1084/jem.189.6.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Topp MS, Koenigsmann M, Mire-Sluis A, Oberberg D, Eitelbach F, Marschall ZV, et al. Recombinant human intcrleukin-4 inhibits growth of some human lung tumor cell lines in vitro and in vivo. Blood. 1993;82:2837–2844. [PubMed] [Google Scholar]
- 32.Yannopoulos A, Nikiteas N, Chatzitheofylaktou A, Tsigris C. The (-590 C/T) polymorphism in the interleukin-4 gene is associated with increased risk for early stages of colorectal adenocarcinoma. In Vivo. 2007;21:1031–1035. [PubMed] [Google Scholar]
- 33.Lee IY, Kim J, Ko EM, Jeoung EJ, Kwon YG, Choe J. Interleukin-4 inhibits the vascular endothelial growth factor- and basic fibroblast growth factor-induced angiogenesis in vitro. Mol Cells. 2002;14:115–121. [PubMed] [Google Scholar]
- 34.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 35.Shurin MR, Lu L, Kalinski P, Stewart-Akers AM, Lotze MT. Th1/Th2 balance in cancer, transplantation and pregnancy. Springer Semin Immunopathol. 1999;21:339–359. doi: 10.1007/BF00812261. [DOI] [PubMed] [Google Scholar]
- 36.Kopf M, Le Gros G, Nachmann M, Lamers M, Bluethmann H, Köhler G. Disruption of the murine IL-4 gene blocks Th2 cytokine responses. Nature. 1993;2:245–248. doi: 10.1038/362245a0. [DOI] [PubMed] [Google Scholar]
- 37.Doucet C, Jasmin C, Azzarone B. Unusual interleukin-4 and -13 signaling in human normal and tumor lung fibroblasts. Oncogene. 2000;19:5898–5905. doi: 10.1038/sj.onc.1203933. [DOI] [PubMed] [Google Scholar]
- 38.Chi L, Na MH, Jung HK, Vadevoo SM, Kim CW, Padmanaban G, et al. Enhanced delivery of liposomes to lung tumor through targeting interleukin-4 receptor on both tumor cells and tumor endothelial cells. J Control Release. 2015;209:327–336. doi: 10.1016/j.jconrel.2015.05.260. [DOI] [PubMed] [Google Scholar]
- 39.Kawakami M, Kawakami K, Stepensky VA, Maki RA, Robin H, Muller W, et al. Interleukin 4 receptor on human lung cancer: a molecular target for cytotoxin therapy. Clin Cancer Res. 2002;8:3503–3511. [PubMed] [Google Scholar]
- 40.Prokopchuk O, Liu Y, Henne-Bruns D, Kornmann M. Interleukin-4 enhances proliferation of human pancreatic cancer cells: evidence for autocrine and paracrine actions. Br J Cancer. 2005;92:921–928. doi: 10.1038/sj.bjc.6602416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Todaro M, Alea MP, Di Stefano AB, Cammareri P, Vermeulen L, Iovino F, et al. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell. 2007;1:389–402. doi: 10.1016/j.stem.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 42.Todaro M, Lombardo Y, Francipane MG, Alea MP, Cammareri P, Iovino F, et al. Apoptosis resistance in epithelial tumors is mediated by tumor-cell-derived interleukin-4. Cell Death Differ. 2008;15:762–772. doi: 10.1038/sj.cdd.4402305. [DOI] [PubMed] [Google Scholar]
- 43.Joshi BH, Leland P, Lababidi S, Varrichio F, Puri RK. Interleukin-4 receptor alpha overexpression in human bladder cancer correlates with the pathological grade and stage of the disease. Cancer Med. 2014;3:1615–1628. doi: 10.1002/cam4.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kwon M, Kim JW, Roh JL, Park Y, Cho KJ, Choi SH, et al. Recurrence and cancer-specific survival according to the expression of IL-4Ralpha and IL-13Ralpha1 in patients with oral cavity cancer. Eur J Cancer. 2015;51:177–185. doi: 10.1016/j.ejca.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 45.Maeshima AM, Maeshima A, Asamura H, Matsuno Y. Histologic prognostic factors for small-sized squamous cell carcinomas of the peripheral lung. Lung Cancer. 2006;52:53–58. doi: 10.1016/j.lungcan.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 46.Charloux A, Hedelin G, Dietemann A, Ifoundza T, Roeslin N, et al. Prognostic value of histology in patients with non-small cell lung cancer. Lung Cancer. 1997;17:123–134. doi: 10.1016/S0169-5002(97)00655-7. [DOI] [PubMed] [Google Scholar]
- 47.Nesbitt JC, Putnam JB, Walsh GL, Roth JA, Mountain CF. Survival in early-stage non-small cell lung cancer. Ann Thorac Surg. 1995;60:466–472. doi: 10.1016/0003-4975(95)00169-L. [DOI] [PubMed] [Google Scholar]
- 48.Hallett MA, Venmar KT, Fingleton B. Cytokine stimulation of epithelial cancer cells: the similar and divergent functions of IL-4 and IL-13. Cancer Res. 2012;72:6338–6343. doi: 10.1158/0008-5472.CAN-12-3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barderas R, Bartolomé RA, Fernandez-Aceñero MJ, Torres S, Casal JI. High expression of IL-13 receptor α2 in colorectal cancer is associated with invasion, liver metastasis, and poor prognosis. Cancer Res. 2012;72:2780–2790. doi: 10.1158/0008-5472.CAN-11-4090. [DOI] [PubMed] [Google Scholar]
- 50.Ni Z, Lou W, Lee SO, Dhir R, DeMiguel F, et al. Selective activation of members of the signal transducers and activators of transcription family in prostate carcinoma. J Urol. 2002;167:1859–1862. doi: 10.1016/S0022-5347(05)65249-4. [DOI] [PubMed] [Google Scholar]
- 51.Li BH, Yang XZ, Li PD, Yuan Q, Liu XH, et al. IL-4/Stat6 activities correlate with apoptosis and metastasis in colon cancer cells. Biochem Biophys Res Comm. 2008;369:554–560. doi: 10.1016/j.bbrc.2008.02.052. [DOI] [PubMed] [Google Scholar]
- 52.Ehret GB, Reichenbach P, Schindler U, Horvath CM, Fritz S, et al. DNA binding specificity of different STAT proteins. Comparison of in vitro specificity with natural target sites. J Biol Chem. 2001;276:6675–6688. doi: 10.1074/jbc.M001748200. [DOI] [PubMed] [Google Scholar]
- 53.Magkou C, Giannopoulou I, Theohari I, Fytou A, Rafailidis P, et al. Prognostic significance of phosphorylated STAT-1 expression in premenopausal and postmenopausal patients with invasive breast cancer. Histopathology. 2012;60:1125–1132. doi: 10.1111/j.1365-2559.2011.04143.x. [DOI] [PubMed] [Google Scholar]
- 54.Grandis J, Drenning S, Zeng Q, et al. Constitutive activation of stat3 signaling abrogates apoptosis in squamous cell carcinogenesis in vivo. Proc Natl Acad Sci USA. 2000;97:4227. doi: 10.1073/pnas.97.8.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cui X, Zhang L, Luo J, Rajasekaran A, Hazra S, Cacalano N, Dubinett SM. Unphosphorylated STAT6 contributes to constitutive cyclooxygenase-2 expression in human non-small cell lung cancer. Oncogene. 2007;26:4253–4260. doi: 10.1038/sj.onc.1210222. [DOI] [PubMed] [Google Scholar]
- 56.Bruns HA, Kaplan MH. The role of constitutively active Stat6 in leukemia and lymphoma. Crit Rev Oncol Hematol. 2006;57:245–253. doi: 10.1016/j.critrevonc.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 57.Merk BC, Owens JL, Lopes MB, Silva CM, Hussaini IM. STAT6 expression in glioblastoma promotes invasive growth. BMC Cancer. 2011;11:184. doi: 10.1186/1471-2407-11-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pastuszak-Lewandoska D, Domańska-Senderowska D, Kordiak J, Antczak A, Czarnecka KH, Migdalska-Sęk M, et al. Immunoexpression analysis of selected JAK/STAT pathway molecules in patients with non- small-cell lung cancer. Pol Arch Intern Med. 2017;127:758–764. doi: 10.20452/pamw.4115. [DOI] [PubMed] [Google Scholar]
- 59.Chapoval SP, Dasgupta P, Smith EP, DeTolla LJ, Lipsky MM, et al. STAT6 expression in multiple cell types mediates the cooperative development of allergic airway disease. J Immunol. 2011;186:2571–2583. doi: 10.4049/jimmunol.1002567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pesch B, Kendzia B, Gustavsson P, Jöckel KH, Johnen G, et al. Cigarette smoking and lung cancer–relative risk estimates for the major histological types from a pooled analysis of case-control studies. Int J Cancer. 2012;131:1210–1219. doi: 10.1002/ijc.27339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karamouzis MV, Konstantinopoulos PA, Papavassiliou A. The role of STATs in lung carcinogenesis: an emerging target for novel therapeutics. J Mol Med. 2007;85:427–436. doi: 10.1007/s00109-006-0152-3. [DOI] [PubMed] [Google Scholar]
- 62.Okamoto T, Maruyama R, Suemitsu R, Aoki Y, Wataya H, Kojo M, et al. Prognostic value of the histological subtype in completely resected non-small cell lung cancer. Interact Cardiovasc Thorac Surg. 2006;5:362–366. doi: 10.1510/icvts.2005.125989. [DOI] [PubMed] [Google Scholar]
- 63.Lubin JH, Blot WJ. Assessment of lung cancer risk factors by histologic category. J Natl Cancer Inst. 1984;74:383–389. doi: 10.1093/jnci/73.2.383. [DOI] [PubMed] [Google Scholar]
- 64.Ying M, Li D, Yang L, Wang M, Wang N, et al. Loss of SOCS3 expression is associated with an increased risk of recurrent disease in breast carcinoma. J Cancer Res Clin Oncol. 2010;136:1617–1626. doi: 10.1007/s00432-010-0819-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Evans MK, Yu CR, Lohani A, Mahdi RM, Liu X, et al. Expression of SOCS1 and SOCS3 genes is differentially regulated in breast cancer cells in response to proinflammatory cytokine and growth factor signals. Oncogene. 2007;26:1941–1948. doi: 10.1038/sj.onc.1209993. [DOI] [PubMed] [Google Scholar]
- 66.Neuwirt H, Puhr M, Cavarretta IT, Mitterberger M, Hobisch A, et al. Suppressor of cytokine signalling-3 is up-regulated by androgen in prostate cancer cell lines and inhibits androgen-mediated proliferation and secretion. Endocr Relat Cancer. 2007;14:1007–1019. doi: 10.1677/ERC-07-0172. [DOI] [PubMed] [Google Scholar]
- 67.Rossa C, Jr, Sommer G, Spolidorio LC, Rosenzweig SA, Watson DK, et al. Loss of expression and function of SOCS3 is an early event in HNSCC: altered subcellular localization as a possible mechanism involved in proliferation, migration and invasion. PLoS One. 2012;7:45197. doi: 10.1371/journal.pone.0045197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Calarco A, Pinto F, Pierconti F, Sacco E, Marrucci E, et al. Role of SOCS3 evaluated by immunohistochemical analysis in a cohort of patients affected by prostate cancer: preliminary results. Urologia. 2012;79:4–8. doi: 10.5301/RU.2012.9392. [DOI] [PubMed] [Google Scholar]
- 69.Hebenstreit D, Wirnsberger G, Horejs-Hoeck J, Duschl A. Signaling mechanisms, interaction partners, and target genes of STAT6. Cytokine Growth Factor Rev. 2006;17:173–188. doi: 10.1016/j.cytogfr.2006.01.004. [DOI] [PubMed] [Google Scholar]