Abstract
Autism spectrum disorder (ASD) affects ~ 2% of children in the United States. The etiology of ASD likely involves environmental factors triggering physiological abnormalities in genetically sensitive individuals. One of these major physiological abnormalities is mitochondrial dysfunction, which may affect a significant subset of children with ASD. Here we systematically review the literature on human studies of mitochondrial dysfunction related to ASD. Clinical aspects of mitochondrial dysfunction in ASD include unusual neurodevelopmental regression, especially if triggered by an inflammatory event, gastrointestinal symptoms, seizures, motor delays, fatigue and lethargy. Traditional biomarkers of mitochondrial disease are widely reported to be abnormal in ASD, but appear non-specific. Newer biomarkers include buccal cell enzymology, biomarkers of fatty acid metabolism, non-mitochondrial enzyme function, apoptosis markers and mitochondrial antibodies. Many genetic abnormalities are associated with mitochondrial dysfunction in ASD, including chromosomal abnormalities, mitochondrial DNA mutations and large-scale deletions, and mutations in both mitochondrial and non-mitochondrial nuclear genes. Mitochondrial dysfunction has been described in immune and buccal cells, fibroblasts, muscle and gastrointestinal tissue and the brains of individuals with ASD. Several environmental factors, including toxicants, microbiome metabolites and an oxidized microenvironment are shown to modulate mitochondrial function in ASD tissues. Investigations of treatments for mitochondrial dysfunction in ASD are promising but preliminary. The etiology of mitochondrial dysfunction and how to define it in ASD is currently unclear. However, preliminary evidence suggests that the mitochondria may be a fruitful target for treatment and prevention of ASD. Further research is needed to better understand the role of mitochondrial dysfunction in the pathophysiology of ASD.
Key points
| Clinical aspects of mitochondrial dysfunction in autism spectrum disorder (ASD) include unusual neurodevelopmental regression, especially if triggered by an inflammatory event, gastrointestinal symptoms, seizures, motor delays, fatigue and lethargy. |
| Many genetic abnormalities have been associated with mitochondrial dysfunction in ASD, including chromosomal abnormalities, mitochondrial DNA mutation and large-scale deletions, and mutations in both mitochondrial and non-mitochondrial nuclear genes. |
| Several environmental factors, including toxicants, microbiome metabolites and an oxidized microenvironment have been shown to modulate mitochondrial function in ASD tissues. |
Introduction
Autism spectrum disorder (ASD) affects ~ 2% of children in the United States. The cause(s) of ASD are still unknown, but evidence for a simple genetic defect is lacking [1]. The etiology of ASD likely involves environmental factors that affect cell signaling, metabolic, immune and epigenetic processes in genetically sensitive individuals [1, 2]. Of note, ASD is associated with physiological disturbances including abnormal redox and mitochondrial metabolism. In fact, between 5% and 80% of children with ASD demonstrate evidence of mitochondrial dysfunction, with many demonstrating novel types of mitochondrial dysfunction rather than classic mitochondrial disease (MD) [3–5]. This is in comparison to the general population where MD is believed to affect less than 0.1% of the population [6]. A systematic review of the literature on mitochondrial dysfunction in individuals with ASD was published in 2012 [3] based on the recognition of early reports of mitochondrial dysfunction in ASD [7–14], but an update to this systematic review has not been published in over 5 years, during which time both genetic and metabolic laboratory testing have advanced significantly and new techniques for measuring mitochondrial function have become accessible.
Methods
Search Strategy
A prospective protocol for this systematic review was developed a priori, and the search terms and selection criteria were chosen to capture all pertinent publications. A computer-aided search of PUBMED, Google Scholar, CINAHL, EmBase, Scopus and ERIC databases from inception through February 2018 was conducted to identify pertinent publications using the search terms ‘autism’, ‘autistic’, ‘Asperger’, ‘ASD’, ‘pervasive’, and ‘pervasive developmental disorder’ in all combinations with the terms ‘mitochondria’ OR ‘mitochondrial’ OR ‘lactic’ OR ‘lactate’ OR ‘pyruvate’ OR ‘pyruvic’ OR ‘ammonia’ OR ‘creatine kinase’ OR ‘oxidative phosphorylation’ OR ‘phosphorylation’ OR ‘carnitine’ OR ‘acyl-carnitine’ OR ‘fatty acid oxidation’ OR ‘alanine’ OR ‘respiratory chain’ OR ‘electron transport chain’ OR ‘ATP’ OR ‘adenosine.’ The references cited in identified publications were also searched to locate additional studies.
Study Selection
One reviewer (DAR) screened the titles and abstracts of all potentially relevant publications. Studies were initially included if they (1) involved individuals with ASD and (2) reported at least one finding that could indicate mitochondrial dysfunction. Abstracts or posters from conference proceedings were included if published in a peer-reviewed journal. After screening all records, 293 publications met inclusion criteria; two reviewers (DAR, REF) then independently reviewed these articles (Fig. 1). Articles were excluded if they:
Did not involve humans or human cells (e.g., animal models).
Did not present new or unique data (such as review articles or letters to the editor).
Presented duplicate data.
Reported biochemical markers related to a non-mitochondrial disorder or cellular mechanism.
Reported markers related to a known side effect of a medication (for example, elevated ammonia from valproic acid or rhabdomyolysis from olanzapine).
Fig. 1.
Flowchart of systematic review and study selection. ASD autism spectrum disorder
Overall, 220 studies were selected for this review. Other studies that support the discussion of MD in ASD are also referenced.
Summary of the Literature
Clinical Aspects
Only a limited number of studies have examined the clinical characteristics of children with ASD and MD. Weissman et al. [15] was one of the first compilations of clinical symptoms of children with ASD diagnosed with MD (ASD/MD). In their case series of 25 patients, they noted a high rate of non-neurological medical problems, including gastrointestinal (GI) dysfunction and prenatal or perinatal complications, constitutional symptoms such as excessive fatigability and exercise intolerance, early gross motor delay and unusual patterns of regression, including multiple regression and regression after 3 years of age.
Rossignol and Frye [3] reviewed the clinical characteristics of all the reported cases of children with ASD/MD and compared these to the clinical characteristics of the general ASD population (ASD/NoMD) as well as children with MD but not ASD (MD/NoASD). As compared to ASD/NoMD children, ASD/MD children had a higher rate of neurodevelopmental regression, seizures, gross motor delay and GI abnormalities. As compared to MD/NoASD, ASD/MD children demonstrated higher rates of fatigue and lethargy, ataxia, GI abnormalities and normal brain imaging and were less likely to have abnormal light microscopy. ASD/MD children were more likely to have elevated lactate than both MD/NoASD and ASD/NoMD groups.
Frye and Rossignol [16] pointed out that many of the clinical symptoms outlined in the Morava criteria [17], which set forth clinical diagnostic criteria for MD, overlap with characteristics of children with ASD, including developmental delays, seizures, neurodevelopmental regression, GI and endocrine abnormalities, familial recurrence and neuropathies.
In the evaluation of the etiology of a person with ASD, the presence of regression especially with recurrent episodes and multiple organ dysfunctions should prompt an extended evaluation for MD [18, 19]. Indeed, Shoffner et al. [20] demonstrated that most patients diagnosed by his center with ASD/MD experienced neurodevelopmental regression resulting in the development of ASD following an inflammatory event associated with a fever within the preceding 2 weeks.
Several case series have noted the association between mitochondrial dysfunction in ASD and epilepsy [15, 20]. Indeed, one of the first descriptions of the overlap of mitochondrial dysfunction and ASD was the HEADD syndrome, characterized by hypotonia, epilepsy, autism, and developmental delay [21]. These authors noted electron transport chain (ETC) complex deficiencies in subunits encoded by mitochondrial DNA (mtDNA) and that several patients demonstrated large-scale mtDNA deletions. Interestingly, these authors noted a high rate of ETC Complex III (C3) deficiency, which has been echoed in other case reports of children with ASD/MD and epilepsy [22, 23]. Another interesting case report linked mitochondrial dysfunction with ASD in a patient with Dravet syndrome, suggesting that even clearly genetically based epilepsy with ASD could have a mitochondrial component [24].
Lastly, one report suggests that children with ASD and classic MD are more likely to also have intellectual disabilities, suggesting early identification and treatment may be particularly useful for these children [25].
Biomarkers of Mitochondrial Disorders
Traditional Biomarkers of Mitochondrial Dysfunction
There are several biomarkers which are commonly used to identify individuals with MD, including lactate, pyruvate, alanine and creatine kinase. However, none of these biomarkers are very specific. In the 2012 meta-analysis of biomarkers in MD by Rossignol and Frye, several traditional biomarkers were elevated in many children with ASD. Using normal reference ranges, studies have demonstrated that ASD was associated with elevations in lactate, pyruvate, lactate-to-pyruvate ratio, alanine, creatine kinase, ammonia and aspartate aminotransferase (AST) and depression in carnitine [3]. Separate studies verified mean elevations in lactate and pyruvate as well as depressions in carnitine and ubiquinone in individuals with ASD as a group compared to contemporaneous controls [3]. Lactate, pyruvate, carnitine, creatine kinase, AST and alanine aminotransferase (ALT) all demonstrated significantly more variation in the ASD group as compared to contemporaneous controls.
Interestingly, elevation in lactate was the first indicator that children with ASD may have a mitochondrial disorder. First reported in 1985 before MD was well described in medicine, Dr Mary Coleman suggested that children with ASD might have a disorder of carbohydrate metabolism [26]. Since the meta-analysis in 2012, lactate continues to be reported as a potential biomarker for abnormal mitochondrial metabolism in children with ASD [27–30]. Despite these consistent elevations in lactate, it is important to consider that many factors can falsely elevate lactate, such as excessive muscle movement, which can occur if the child is struggling or because the tourniquet is left on too long. Thus, the search for alternative and more reliable biomarkers of mitochondrial dysfunction are being pursued for diagnosing MD in general as well as identifying children with ASD that might have mitochondrial dysfunction.
Other traditional biomarkers that have continued to be reported as abnormal in ASD include pyruvate [28], lactate-to-pyruvate ratio [31], creatine kinase [28, 30, 32], AST [27, 30], ALT [27] and ubiquinone [28, 33]. Other studies have reported abnormal urine citric acid cycle metabolites which are also biomarkers of mitochondrial dysfunction [34, 35].
Aside from elevations in alanine, some clinicians use the ratio of alanine to lysine as pioneered by Richard Kelley [36], and others have found abnormalities in other amino acids possibly related to mitochondrial metabolism [37]. In a study of 25 high-functioning males with ASD, plasma levels of arginine were elevated while 5-oxoproline was reduced [38]. A study of 60 families found elevations in plasma glutamate and aspartate [39]. Another study reported reduced plasma cysteine, tyrosine, serine, α-aminoadipic acid, carnosine and β-alanine and increased levels of glutamic acid, hydroxyproline, phosphoserine and β-amino-isobutyric acid [40]. Another study has reported elevation in γ-aminobutyric acid and glutamic acid in individuals with ASD [41]. Although plasma taurine was reported to be elevated in one study [38], another study suggested that this elevation was also found in matched related controls [39] and an another study found no differences in taurine as compared to sibling and parent controls [42]. The variability across studies in amino acid metabolites could be due to several factors. Indeed, the metabolism and transport of certain amino acids has been suggested to be abnormal in children with ASD. For example, increased transport of alanine and decreased transport of tyrosine across the cell membrane was found in fibroblast culture from 11 children with ASD [43], a genetic disorder of branched chain amino acid metabolism has been described in children with ASD [44], and abnormalities in transport of amino acids across the blood–brain barrier has also been implicated in ASD [45]. However, dietary intake and whether or not the sample was post-prandial or fasting have probably the most influence on plasma levels of amino acids.
Buccal Swab
Biochemical measurements of mitochondrial function can be difficult to obtain. Direct measurement of mitochondrial function by enzymology typically requires invasive biopsy, requiring anesthesia, which has risks, especially in individuals with MD. Biopsy also limits the ability to repeat the test over time to follow disease status. Goldenthal et al. [46] developed and validated the non-invasive buccal swab technique, showing that buccal tissue enzyme measurements correspond to measurements from muscle biopsy in individuals with MD. The buccal swab technique has been used to measure mitochondrial function in individuals with MD [46–48], genetic syndromes [48–50] and ASD [50–54].
Five studies have used the buccal swab technique in individuals with ASD. Four of these studies have examined citrate synthase (CS) as well as ETC Complex I (C1) and ETC Complex IV (C4) activity normalized to CS [51, 53, 54], while one study also measured ETC Complex II (C2) and C2 + C3 activity [49]. Goldenthal et al. [51] noted several unique characteristics of mitochondrial function in children with ASD who were not taking mitochondrial supplements for at least 2 weeks. First, C1 demonstrated a significantly higher variation in ASD participants as compared to a control population, with 12% and 7% of children with ASD demonstrating C1 activity lower and higher, respectively, than the control range. There were also a significant number of individuals with ASD who demonstrated abnormal C4 activity, with 28% and 3% demonstrating C4 activity lower and higher, respectively, than the control range. The authors note that individuals with ASD and seizures more often had a deficit in C4 activity. Lastly, Goldenthal et al. [51] reported that 65% of the individuals with ASD demonstrated a C1-to-C4 ratio outside of the control reference range and that individuals with more severe ASD were more likely to have a higher C1-to-C4 ratio.
In another large study, children with ASD who were not taking any supplements or medications that could interfere with mitochondrial function at the time of testing were examined using the buccal swab technique [53]. Variation in CS, C1 and C4 activity was greater in the ASD group as compared to the control range. Significant elevations in CS, C1 and C4 outside of the reference range were found in 22%, 11% and 8% of children with ASD, respectively, which were significantly more than what would be expected by chance. Significant depression in CS, C1 and C4 activity below the reference range was found in 3%, 28% and 3% of children with ASD, respectively, which was significant only for C1. The C1-to-C4 ratio was above and below the control reference ranges in 7% and 17% of children with ASD, respectively. Overall 62% of children with ASD were found to have some abnormality in mitochondrial enzyme activity. Most interestingly, this study examined the relationship between mitochondrial enzyme activity and measures of cognitive development and behavior. Specifically, childhood development, as measured by the Vineland Adaptive Behavior Scale (VABS), was related to both C1 and C4 activity. C4 activity demonstrates an inverted U-shaped relationship to VABS subscales such that both high and low C4 activity was related to poorer scores on the VABS. In contrast, C1 activity was linearly related to the VABS such that lower C1 activity was related to lower VABS scores and higher C1 activity was related to higher VABS scores. CS activity was found to be related to Social Responsiveness Scale scores.
The largest study measuring mitochondrial enzymatic activity using the buccal swab technique in children with ASD concentrated on whether there was a significant effect of various treatments for mitochondrial disorders on enzymatic function and introduced new biomarkers of mitochondrial function [54]. Overall, the study demonstrated that supplementation with folate, cobalamin, fatty acids and antioxidants influenced mitochondrial enzymatic activity as measurable with the buccal swab technique. Secondly, the study looked at the change in the linear relationship between the three enzymes measured with the buccal swab technique. Folate supplementation resulted in a more positive relationship between CS and C1 activity and C1 and C4 activity. Cobalamin supplementation resulted in a more positive correspondence between CS and C1 activity. The authors suggested that the more positive correlations between enzyme activities indicated increased ETC coupling.
A small pilot study of 11 children with ASD has shown a correspondence between change in ETC activity, as measured by the buccal swab technique, as a consequence of treatment with a mitochondrial cocktail, and improvement in behavior [55].
The last study using the buccal swab examined a genetic syndrome known as Phelan-McDermid syndrome (del22q13) that has a high comorbidity of ASD [49]. One interesting aspect of this study was the subset of children with C1 abnormalities. Those with higher than normal C1 activity demonstrated a developmental course more consistent with ASD, while those with below normal C1 activity demonstrated a developmental course more consistent with children with MD, suggesting that the subset of children with ASD were unique in demonstrating overactive C1 activity. Additional studies utilizing the buccal swab analysis in individuals with ASD would be helpful in expanding the work in this area.
Biomarkers of Abnormal Fatty Acid Metabolism
Several authors have reported biomarkers representing abnormalities in fatty acid metabolism in individuals with ASD [4, 56]. Clark-Taylor and Clark-Taylor [56] were the first to report fatty acid metabolism abnormalities associated with ASD. They reported a case of a child with elevations in unsaturated fatty acid metabolites C14:1 and C14:2 along with abnormalities in citric acid cycle, ammonia and cholesterol metabolism. The authors suggested that these abnormalities were the result of a defect in long-chain acyl-CoA dehydrogenase despite the fact that there is no convincing evidence for a deficiency in this enzyme in humans. ASD patients from Saudi Arabia were found to have elevations in saturated fatty acids and depressions in polyunsaturated fatty acids as compared to age-matched controls [57]. In another study, children with ASD from Egypt were found to have lower plasma levels of polyunsaturated fatty acids, except linoleic acid, as compared to healthy controls [58]. In a small study from Canada, all children with ASD were found to have elevations in polyunsaturated long-chain fatty acids and/or saturated very long-chain fatty acids as compared to age-matched controls [59]. The frontal cortex of patients with 15q11.2-q13 duplication and ASD was found to have lipofuscin deposits as compared to age-matched controls, potentially as a product of increased lipid peroxidation [60].
Frye [4] reported the results of metabolic work-ups of 133 consecutive patients evaluated in a medically based ASD clinic using a standardized metabolic screening algorithm which included screening for fatty acid oxidation defects [61, 62]. Abnormalities found in fatty acid oxidation were unique: short-chain and long-chain acyl-carnitines were elevated, but medium-chain acyl-carnitines were normal. Further review of 213 patients with ASD who underwent a metabolic evaluation in a medically based ASD clinic [5] found that 17% showed consistent elevations in short and long acyl-carnitines (CESLAC), with C4OH, C14 and C16:1 being statistically significantly elevated above the upper limit of normal. Overall there was no clear, consistent genetic abnormality, and the individuals with these abnormalities had a particularly high rate of neurodevelopmental regression. Muscle biopsies showed a partial defect in C1 and C1 + C3. Glutathione (GSH) metabolism abnormalities were also found. Interestingly, this same pattern of acyl-carnitine and GSH abnormalities was independently reported in the rodent propionic acid (PPA) model of ASD [63–65]. Theoretically, PPA may be overproduced by the overrepresented species of Clostridia found in the GI tract of some children with ASD [66–68] and could result in mitochondrial dysfunction. This finding could also explain the association between ASD and children with propionic acidemia, an inborn error of metabolism [69]. However, other studies have suggested that children with ASD and fatty acid abnormalities demonstrate a decrease in PPA in their blood [57].
Abnormalities in fatty acid metabolism may help account for the relative deficiency in carnitine associated with ASD [58, 70], and the fact that mutations in the trimethyllysine hydroxylase epsilon (TMLHE) gene, the first enzyme in carnitine biosynthesis, is a risk factor for ASD [71]. In addition, children with ASD in general [72–74] and those with the TMLHE mutations specifically [75] benefit from supplementation with L-carnitine. Clearly, further research will be needed to better understand the significance of these fatty acid abnormalities in ASD and whether they are truly linked to disruptions in the enteric microbiome.
Other Novel Biomarkers
Researchers from King Saud University in Saudi Arabia have been particularly active in the identification and discovery of novel biomarkers of mitochondrial dysfunction and have used receiver operating characteristic (ROC) analysis to identify combinations of biomarkers which may be diagnostic of children with ASD and mitochondrial dysfunction [28, 76, 77]. Novel biomarkers found to be potentially useful include lactate dehydrogenase [28], lactate oxidase, pyruvate kinase and hexokinase [78], Na+/K+ ATPase [76, 79], caspase 3 [77] and caspase 7 [28]. Although promising, many of these studies have limited samples sizes and use different combinations of biomarkers from study to study, making these studies interesting but preliminary. Selecting the most promising biomarkers and investigating them on a large sample of ASD subjects and appropriate typically developing and developmentally delayed controls will be necessary to validate this approach.
Anti-mitochondrial antibodies are a recently described biomarker. In one study of 54 children with ASD from Saudi Arabia, antibodies to the mitochondrial M2 subtype antigen were found in 52% of children with ASD (percentage positive in controls not reported), and titers of this antibody were higher in children with ASD, particularly those with more severe ASD, as compared to age- and gender-matched control participants [80]. In an independent study, antibodies against type 2 mitochondrial antigen were found to be increased in children with ASD from Greece as compared to typically developing controls [81]. In these samples, mtDNA was found to be increased in the serum from the children with ASD as compared to controls. The authors suggested that increased levels of the peptide neurotensin, which was found to be increased in children with ASD in other studies, induces the release of mtDNA, which then acts as an immune trigger.
Other potential biomarkers include growth differentiation factor 15 (GDF15) and fibroblast growth factor 21 (FGF21), which are newer mitochondrial biomarkers that possess a high specificity for MD [82]. They have yet to be studied in ASD patients.
Genetic Aspects of Mitochondrial Disorders in Autism Spectrum Disorder (ASD)
Although it has been estimated that the genetic etiology of ASD may account for up to 40% of cases [83, 84] and whole exome sequencing (WES) and chromosomal microarray analysis (CMA) studies have reported yields up to 30% [85] and 26% [86], respectively, separate clinical studies have failed to confirm this high rate of genetic disorders in children with ASD. For example, a study from Canada found that 9.3% and 8.4% of children with ASD received a molecular diagnosis using WES and CMA, respectively, resulting in only 15.8% of children with ASD receiving a molecular diagnosis [84].
The moderate rate of a molecular diagnosis is at odds with the high heritability rate associated with ASD; for example, there is a 70–90% concordance rate for monozygotic twins and up to 10% for dizygotic twins and a 25-fold increased prevalence of ASD in siblings of ASD children [87]. Thus, it is likely that the etiology of ASD is multifactorial and influenced by a complex interplay between the inherited genome and environmental effects, some of which may be related to the maternal environment [88, 89]. Epigenetic interactions which modulate the expression of nuclear [90] and mitochondrial [91] genes can be influenced by environmental factors and may also play a crucial role in ASD. These interactions must surely modulate mitochondrial function, influencing neurodevelopment. Indeed, alterations in multiple bioenergetic and metabolic genes required for mitochondrial function may lead to abnormalities in cerebral activity, resulting in cognitive and behavioral abnormalities characteristic of ASD [92].
Mitochondrial genes can be affected by copy number variations (CNVs) or regions of homozygosity (ROH). Chromosomal regions affected by CNVs can contain genes associated with mitochondrial function and neurodevelopmental disorders. For instance, a 7q31.1 deletion/duplication disrupts the IMMP2L gene encoding an inner mitochondrial membrane protease-like protein required for processing of cytochromes inside mitochondria and is implicated in ASD [93]. The IMMP2L–DOCK4 gene region on chromosome 7 plays a role in ASD susceptibility [94], and IMMP2L deletions have been demonstrated to have an association with ASD [95]. However, deleterious point mutations in IMMP2L were not identified in a significant number of ASD patients in other studies [96]. Mitochondrial dysfunction was also demonstrated in ASD patients with other CNVs, including 15q11-q13 duplication [22, 60], 5q14.3 deletion [48], 22q13 duplication or Phelan–McDermid syndrome [49, 97] as well as chromosomal disorders such as Down syndrome [98] and X- and Y-chromosome loci rearrangements [99]. An interesting example is Phelan-McDermid syndrome, which is typically caused by a microdeletion in the 22q13 region. Although much research has concentrated on SHANK3, which is important for synaptic function, six mitochondrial genes are also present in this region and may account for symptoms associated with mitochondrial dysfunction [49].
ROH are regions that are identical in homologous regions of paired chromosomes. Small ROH reflect our common human inheritance; larger and more numerous ROH can occur because of chromosomal segments shared between consanguineous family members. ROH can include genes associated with mitochondrial function as well as cerebral synapses and neurotransmitters that are associated with ASD [100].
Primary mitochondrial disease (PMD) involves a genetic defect that results in an impairment of mitochondrial oxidative phosphorylation [101]. PMD is estimated to affect 5% of children with ASD [3], based upon three large studies [102–104]. Mutations in mtDNA are the most common genetic mutations associated with ASD/MD [3]. Numerous mtDNA point mutations [105, 106] and deletions [107] show association with ASD; large-scale mtDNA deletions have been associated with epilepsy [21]. Examples of mtDNA genes reported include MT-ATP6 [85, 108, 109], MT-ND5 [108], MT-CYB [110], MT-TK [111] and MT-TL1 [112], and MT-CO1 and MT-CO2 [113]. Mutations in mtDNA such as m.3260A > G that causes mitochondrial encephalomyopathy, lactic acidosis and stroke-like episodes (MELAS) have been associated with ASD [104, 112], as has primary Leber hereditary optic neuropathy mtDNA mutations [104]. ASD can be an early presentation of another MELAS mutation (m.3243A > G) [114], sometimes with a prominent manifestation of mtDNA depletion syndrome [115]. Kent et al. [116] found m.3243A > G to be a rare cause of isolated ASD. mtDNA lineages or haplogroups significantly contribute to overall ASD risk in some studies [117, 118], while other studies have not confirmed a major role for mtDNA variation in ASD susceptibility [119, 120]. Increased mtDNA copy number in leukocytes from children with ASD has been reported in six studies [121–126], while increased mtDNA damage and deletions in leukocytes from children with ASD have been reported in four studies [121, 122, 126, 127]. Interestingly, Wong et al. also found an increase in microdeletions of p53, which is a regulator of mtDNA integrity [126], leading others to suggest that changes in mtDNA may be an epiphenomenon of genetic abnormalities in nuclear DNA (nDNA) mutations [128].
nDNA gene mutations can also cause PMD. Mutations in NDUFA5 [129, 130], NDUFS4 [108], POLG [104] and SCO2 [104] have also been found to be associated with ASD. SNPs in the SLC25A12 gene have been reported by multiple authors to be strongly associated with ASD [131–143]. Mutations in SLC6A8, resulting in X-linked creatine transporter disorder including features of ASD, have been reported [144]. Heterozygous mutations in adjoining components in a multimeric complex or metabolic pathway may combine to lead to impairment through synergistic heterozygosity. This is well-described, but not yet for the mitochondrial–ASD relationship. Since these types of relationships are complex, bioinformatics platforms used to interpret WES data may not be able to detect these complex genetic interactions. In addition, novel variants of unknown significance can be difficult to interpret, and in some situations, a mutation may cause gain of function, and act in a dominant manner. A recently developed method called transmission and de novo association (TADA) analysis considers both transmitted and de novo mutations together in the same statistical model by weighting more detrimental mutations such as de novo loss-of-function mutation more heavily than an inherited loss-of-function mutation [145].
Genetic abnormalities have not been reported in most ASD/MD cases [3]. In addition, the number of children with ASD who have abnormal nutritional and mitochondrial biomarkers greatly outweighs the number who can be diagnosed with a PMD. This raises the possibility that many children with ASD may have secondary mitochondrial dysfunction (SMD). There are examples of SMD related to hereditary defects in non-mitochondrial diseases. For example, a pair of siblings with ASD-associated gene mutations in WDR45 and DEPDC5 were found to have evidence of mitochondrial dysfunction [50]. In another interesting case series of children with Dravet syndrome and SCN1A mutations, two cases were found to manifest mitochondrial dysfunction, and one of the cases had ASD [24]. In another case, lymphoblastoid cell lines (LCLs) derived from a child with ASD and a mutation in RPL10 were found to have changes in redox-sensitive components of energy metabolism [146]. Interestingly, candidate genes implicated in ASD have been found to be enriched in modules related to mitochondrial function [147]. These cases demonstrate the importance of advanced genetic testing combined with metabolic/mitochondrial evaluation in the workup of children with ASD [71].
Mutations in non-mitochondrial genes or environmental factors may be acting via epigenetic mechanisms in ASD [101, 148]. Down-regulation of genes of mitochondrial oxidative phosphorylation and varying gene expression related to myelination, inflammation and purinergic signaling have been previously identified in ASD patients [149]. Others have found upregulation of ribosomal, spliceosomal, and mitochondrial pathways and the down-regulation of neuroreceptor-ligand, immune response and calcium signaling pathways in gene expression profiles of ASD patients [150]. One meta-analysis of over 1000 microarray samples across 12 independent studies demonstrated that genes highly ranked with consistent changes in expression in the brain suggested modulation of mitochondrial function [151]. However, one study examining expression of the C1 75-kDa subunit in blood did not find consistent changes in children with ASD [152].
Other studies have examined the expression of genes in brain tissue in ASD. Depressed expression in ETC genes in the occipital and cerebellar areas and ETC and non-ETC genes in the cingulate, thalamus and frontal areas [153] has been reported. In addition, changes in genes that control mitochondrial dynamics have been noted in the temporal lobe in ASD [154]. Still others have demonstrated brain region-specific expression alterations in mitochondrial nDNA genes such as CMYA3, MTX2, SLC25A27, DNAJC19, DNM1L, LRPPRC, SLC25A12, SLC25A14, SLC25A24 and TOMM20 in ASD patients [131, 132].
Cellular Models of Mitochondrial Dysfunction in ASD
Evaluating Cellular Bioenergetics Using the Seahorse Bioscience XF96 Analyzer
One of the major advances in measuring mitochondrial function is the recently introduced Seahorse 96 XF Analyzer (Agilent Technologies, Santa Clara, CA, USA) that measures oxygen consumption rate (OCR) in real time in a 96-well plate in a broad range of intact living cell types [155, 156]. The assays performed on the Seahorse aid in the elucidation of mitochondrial enzyme defects, including oxidative phosphorylation [157], glycolytic [158] and fatty acid oxidation [159] pathway abnormalities. In our experiments, we use the 96 wells to run samples in quadruplicate, include matched groups on the same plate such as tissue from children with ASD, typically developing siblings and unrelated controls, as well as test several specific manipulations of the tissue samples.
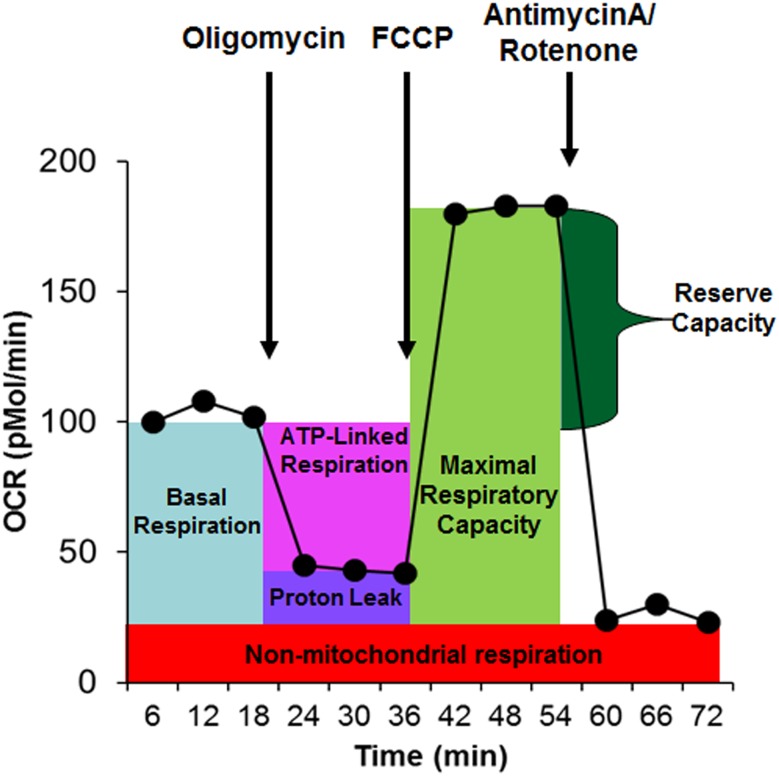
Each well contains four reagent ports which allow for the sequential injection of compounds. A schematic showing the measurement sequence of the mitochondrial assay is depicted in Fig. 2. ATP-linked respiration (ALR) represents ATP production and is determined by injecting the complex V inhibitor oligomycin. The remaining OCR represents proton-leak respiration (PLR) and non-mitochondrial respiration. Maximal respiratory capacity (MRC) is determined by injecting carbonyl cyanide-p-trifluoromethoxyphenyl-hydrazon (FCCP). A decrease in MRC is consistent with a deficit in mitochondrial biogenesis, mtDNA damage and/or ETC inhibition. Reserve capacity (RC) determines the threshold at which bioenergetic dysfunction occurs. Specifically, when RC is zero or negative the cell cannot satisfy a bioenergetic demand [160]. In addition, the extracellular acidification rate (ECAR), a reflection of lactate production, is also measured. From ECAR, glycolytic rate and glycolytic reserve are calculated. In addition, to obtain a measure of the relative utilization of oxidative versus glycolytic pathways, two measures are calculated. The oxidative to glycolytic ratio is calculated as basal OCR divided by basal ECAR, and the maximal oxidative capacity to glycolytic ratio is calculated as the MRC divided by the maximal glycolytic rate.
Fig. 2.
The Seahorse assay measures mitochondrial function by monitoring the change in the oxygen consumption rate (OCR) as various reagents are injected into the sample of living tissue. Three measures of OCRs are obtained over an 18-min period to determine mitochondrial activity. Reagents are added to determine several parameters of mitochondrial activity. Basal respiration is initially determined as the difference between baseline OCR and non-mitochondrial OCR. Oligomycin, which is a complex V inhibitor, is added to determine the portion of basal respiration that is ATP-linked respiration and proton-leak respiration. Carbonyl cyanide-p-trifluoromethoxyphenyl-hydrazon (FCCP), a protonophore, is added to collapse the inner membrane gradient, driving the mitochondria to respire at its maximal rate. This determines maximal respiratory capacity. Antimycin A and rotenone, which are inhibitors of complex III and I, are added to stop mitochondrial respiration to determine the non-mitochondrial respiration. Reserve capacity is calculated as the difference between basal respiration and maximal respiratory capacity
Primary Immune Cells
To date there are only a few published reports of aberrant mitochondrial function in peripheral blood cells from individuals with ASD [50, 121, 122, 161]. Giulivi et al. found decreased activity of C1, C2 and C4 in 80%, 60% and 30%, respectively, of lymphocytes derived from ten ASD subjects, as compared to ten control children [121]. The same group subsequently reported a significantly lower OCR as well as decreased C1 and C2 activity in granulocytes from the same cohort of children with ASD as compared to controls [122]. Both lymphocytes and granulocytes from the children with ASD exhibited significantly higher mitochondrial production of H2O2 as compared to controls [121, 122].
Our group has used primary cells to look at mitochondrial function in individual patients. In a recent case report, Drs Rose and Frye examined mitochondrial function in peripheral blood mononuclear cells (PBMCs) using the Seahorse analyzer in two siblings with ASD, each with a novel mutation in a different gene [50]. Our group compared these findings to mitochondrial function measured in typically developing children. PBMCs from the sibling who carried a mutation in the WDR45 gene exhibited increased mitochondrial respiration linked to ATP production, while PBMCs from the sibling who carried a DEPDC5 gene mutation exhibited increased PLR and a reduction in respiration linked to ATP production. Importantly, increased mitochondrial respiration in PBMCs from the affected sibling with the WDR45 mutation reflected significant elevations in several ETC complexes measured in contemporaneous muscle and skin biopsies, while decreased mitochondrial respiration in PBMCs from the sibling with the DEPDC5 mutation reflected decreased C4 activity as measured by the buccal swab technique.
In addition to these functional studies, several groups have reported surrogate markers of mitochondrial dysfunction in peripheral blood cells from children with ASD. Pecorelli et al. demonstrated ultrastructural changes, including densely packed and irregularly arranged cristae, in PBMC mitochondria from children with ASD [161]. Several groups have reported increased mtDNA abnormalities in peripheral blood cells in ASD [121–127].
Lymphoblastoid Cell Lines
LCLs are the major cellular model of mitochondrial dysfunction in ASD. LCLs are B cells transformed by the Epstein-Barr virus. They are created in a cell culture laboratory from fresh blood samples or obtained from numerous biorepositories. LCLs exhibit expression of genes in a wide range of metabolic pathways that is specific to the individual from whom the cells were derived [162].
In 2009, James et al. first reported abnormal mitochondrial function in LCLs derived from children with ASD [163]. Specifically, exposure to a nitric oxide donor induced a greater mitochondrial membrane potential reduction in ASD LCLs as compared to control LCLs, demonstrating a hypersensitivity to nitrosative stress in the ASD LCL mitochondria. ASD LCLs also demonstrated increased free radical production and more oxidized cellular and mitochondrial GSH pools, consistent with redox abnormalities reported in children with ASD [164–167] and elevations in reactive nitrogen species in mitochondria from individuals with ASD [168].
A series of studies by Drs Rose and Frye using the ASD LCL model has demonstrated abnormal mitochondrial bioenergetics in a subgroup of LCLs from children with ASD as compared to LCLs from unaffected siblings as well as unaffected unrelated children and adults [169–171]. The abnormal ASD LCL subgroup exhibits increased OCR associated with all Seahorse parameters, including ALR, PLR, MRC and RC [160, 172]. The LCLs derived from children with autistic disorder (AD) are classified into two groups: those with normal mitochondrial function (AD-N) and those with atypical mitochondrial function (AD-A) [170, 171, 173]. The AD-A LCLs have respiratory rates approximately twice that of control and AD-N LCLs [170, 171, 173]. These metabolic groupings are consistent and reproducible [169–171, 174–176]. We recently demonstrated that this alteration in respiration is associated with more severe repetitive behaviors [171]. We believe that this increase in respiratory rate may be a protective adaptation designed to resist environmental stressors, perhaps because of previous exposure to environmental toxicants [170, 176].
Several other groups have also described mitochondrial dysfunction in the ASD LCL model. A metabolic profiling study of 87 ASD LCLs revealed abnormal tryptophan metabolism in ASD as compared to control LCLs [177]. Tryptophan metabolism is one of several pathways leading to the production of nicotinamide adenine dinucleotide (NAD), a critical energy carrier for the ETC. Bu et al. [178] demonstrated severe mitochondrial dysfunction in LCLs derived from ten Chinese Han male children with ASD. Specifically, compared to LCLs from unrelated, unaffected control children, ASD LCLs exhibited decreased mitochondrial membrane potential and decreased activities of C1 and C3, as well as increased intracellular and mitochondrial reactive oxygen species (ROS) generation and increased mitochondrial-mediated apoptosis. In addition, the ASD LCLs were found to have significantly reduced expression of peroxisome proliferator-activated receptor γ coactivator 1-α (PGC-1α), a major regulator of mitochondrial function, and sirtuin-1 (SIRT1), a metabolic sensor; PGC-1α overexpression improved all mitochondrial abnormalities. In another cohort of LCLs established from Chinese Han individuals with ASD, Zhang et al. [179] examined ETC activities and reported decreased activity of C1 in two out of five ASD LCLs harboring mutations in the AMPD1 gene, which codes for adenosine monophosphate (AMP) deaminase.
Mitochondrial Dysfunction in the Brain in ASD
Since the brain has a high metabolic demand and is especially dependent on mitochondrial function, abnormalities in mitochondria function would be expected to cause brain dysfunction. Studies provide support for mitochondrial dysfunction in the brain of individuals with ASD. Magnetic resonance spectroscopy (MRS) using both 31P and 1H techniques have examined energy metabolites in the brain of individuals with ASD. 31P-MRS has reported abnormal energy metabolites in frontal cortex [180, 181]. Some 1H-MRS studies have found a reduction in N-acetylaspartate (NAA) in the global white and gray matter and the parietal, anterior cingulate and cerebellum [182] associated with ASD, while others have not been able to detect differences in NAA [183]. Another study in adults with Asperger syndrome has shown an increase in NAA and choline in the prefrontal cortex, with concentrations of these metabolites correlating with obsessive behavior and social function, respectively [184]. An interesting meta-analysis using meta-regression suggests that these discrepancies may be due to an age-related decline in brain NAA specific to those with ASD [185].
Studies using 1H-MRS have found an increase in lactate in the cingulate gyrus, subcortical gray matter nuclei, corpus callosum, superior temporal gyrus, and pre- and post-central gyri, with this abnormality being more common in adults with ASD [186], while others have not found any increase in lactate in individuals with ASD [187]. MRS studies on lactate in the brain are not without controversy. Some have suggested that the fact that lactate was found more often found in adults with ASD may reflect the notion that lactate was associated with comorbidities such as anxiety rather than related to the true etiology of ASD [188]. The original authors did not agree and suggested that perhaps worsening of mitochondrial function with age or ascertainment bias in the recruitment of adults with ASD might be a more likely explanations [189]. Others have pointed out that early MRS techniques may not be powerful enough to determine lactate abnormalities, especially because such early techniques are not uncommonly negative even in individuals with known MD, thus potentially explaining the negative findings; the lack of contemporaneous controls with MD make the interpretation of negative findings even more problematic [190]. The authors of the original study pointed out that their use of propofol anesthesia could increase the sensitivity of their study by provoking mitochondrial dysfunction in the patients with an underlying MD [191].
ETC function has also been directly measured in post-mortem brain tissue from individuals with ASD. ETC function has been reported to be depressed in frontal [192], temporal [154, 192] and cerebellar [192] areas from ASD-derived brain tissue. Other studies noted decreases in the activity of non-ETC mitochondrial enzymes (aconitase, pyruvate dehydrogenase) in frontal [193], temporal [173] and cerebellar [173] tissue derived from children with ASD.
Mitochondrial Dysfunction in the Gastrointestinal Tract in ASD
GI symptoms are prominent in ASD and in patients with MD, separately, and multiple GI disorders are known to be caused by MD [194]. Thus, Rose et al. [195] examined mitochondrial function in rectal and cecum biopsies from ten children with ASD and compared them to ten children with Crohn’s disease and ten neurotypical children with non-specific GI complaints using a single-blind, case–control design. Except for C2, the protein quantity of all ETC complexes was found to be higher in the cecum as compared to the rectum in ASD samples when compared to other groups. For both rectal and cecum biopsies, ASD samples demonstrated higher C1 activity, but not C4 or CS activity, compared to other groups. The authors suggested that this represented a unique pathophysiology of the GI symptoms in children with ASD and proposed that because most abnormalities were localized to the cecum, there may be a role for imbalances in the microbiome in children with ASD that drives mitochondrial dysfunction in the GI tract. Another explanation is that mitochondrial dysfunction could influence the microbiome.
Evidence of Environmental Influences of Mitochondrial Function in ASD
Despite decades of research focused on the genetic basis of ASD, the minority of ASD cases can be attributed to single-gene or chromosome defects [196]. The majority of ASD cases likely result from a complex interplay of polygenetic and environmental factors [88, 89]. Mitochondrial dysfunction is one of the most compelling mechanisms of gene–environment interactions, as mitochondria are influenced directly by exogenous environmental stressors, and secondarily through intrinsic factors such as ROS, inflammatory mediators and local metabolic modulators such as the enteric microbiome. The effect of intrinsic and extrinsic environmental factors on mitochondrial function in ASD has been examined by exposing LCLs to various agents.
Intrinsic Microenvironmental Stressors
The Mitochondrial Oxidative Stress Test (MOST)
Two major metabolic abnormalities associated with ASD are mitochondrial dysfunction and oxidative stress. The interconnection between these two metabolic abnormalities is well known: oxidative stress causes mitochondrial dysfunction and dysfunctional mitochondria produce ROS. ROS is also a byproduct of normal mitochondrial function. Additionally, both intrinsic and extrinsic stressors can cause detrimental effects by increasing ROS and/or reducing mitochondrial function.
The interaction between mitochondrial function and oxidative stress is especially pertinent to the pathophysiology of ASD. In children with ASD, the reduced form of GSH, the major intracellular antioxidant responsible for maintaining redox homeostasis and reducing ROS in the cytosol and mitochondria, is usually deficient [163, 166, 167, 173]. Our laboratory has shown that, in ASD, oxidized glutathione disulfide (GSSG) is elevated and proteins and DNA show oxidative damage in PBMCs [164] and post-mortem brain samples [173]. Other groups have verified GSH abnormalities in post-mortem brain samples [197, 198] and have shown oxidative damage to mtDNA [127] and lipids [199] in individuals with ASD. Epigenetic changes related to redox abnormalities [200, 201] have been found in individuals with ASD, including in studies in our laboratory [201]. In addition, several studies have suggested that oxidative stress biomarkers may be diagnostic for individuals with ASD [28, 31, 76, 202], and one study has demonstrated that ASD/MD children have a different profile of redox metabolism abnormalities as compared to ASD/NoMD children [203].
Thus, to better understand the interplay between these two major metabolic abnormalities, we developed a method to systematically manipulate ROS during mitochondrial function testing. We call this the Mitochondrial Oxidative Stress Test (MOST). Excessive ROS can lead to a depletion of mitochondrial RC and can activate mechanisms designed to protect the mitochondria from oxidative damage. Hill et al. [172] showed that acute increases in ROS deplete RC and that cell viability is reduced once RC is exhausted. A decrease in RC is linked to aging [204], heart disease [205], and neurodegenerative disorders [206, 207].
Thus, because ROS is systemically increased in vitro we concentrated on the changes in RC. To increase oxidative stress in vitro we utilized 1,4-naphthoquinone (DMNQ), an agent that generates intracellular superoxide and hydrogen peroxide similar to that generated by nicotinamide adenine dinucleotide phosphate oxidase in vivo and does not directly deplete thiols [160]. To determine the change in mitochondrial response with increasing ROS, cells are exposed to one of several DMNQ concentrations (5, 10, 12.5 and 15 μM) for 1 h before the Seahorse assay. The standard mitochondrial stress test is completed and the change in RC with increasing concentrations of DMNQ is measured. This change measures the rate of loss of RC with increased physiological stress. Cells that have a greater loss of RC at lower DMNQ concentrations are considered to have mitochondria that are more vulnerable to physiological stress.
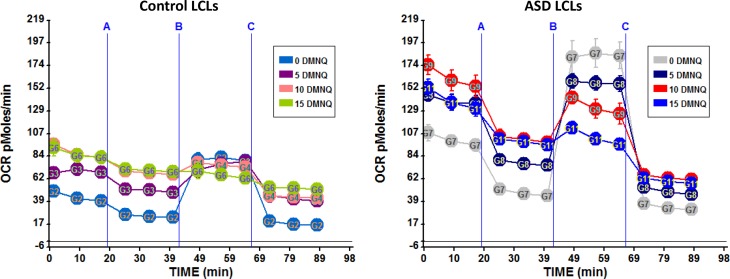
The effect of the MOST on the mitochondrial assay can be seen in Fig. 3. One of the significant findings from the MOST was found for the subset of LCLs found to have an apparent increased mitochondrial capacity to produce ATP, called AD-A LCLs in Sect. 3.4.3. The AD-A LCL subgroup exhibits an atypical mitochondrial response to increased DMNQ, with a precipitous decline in RC as ROS increases [169, 170]. In Fig. 3, we see that the AD-A LCLs have a high basal respiratory rate as well as a higher MRC as compared to the control LCLs. However, the LCLs exposed to higher concentrations of DMNQ demonstrated a smaller difference between the basal respiratory rate and the MRC such that at the highest DMNQ concentration, the MRC is below the basal respiratory rate, indicating a negative RC, which is consistent with mitochondrial exhaustion and collapse of cellular physiology.
Fig. 3.
The Mitochondrial Oxidative Stress Test (MOST) Seahorse assay results for lymphoblastoid cell lines (LCLs) derived from children with autism spectrum disorder (ASD) or age-matched controls. LCLs are exposed to one of three concentrations of an agent that increases oxidative stress known as 1,4-naphthoquinone (DMNQ) or not exposed to DMNQ (i.e., DMNQ 0). Reagents for the Seahorse assay are added at time points A, B and C as previously outlined in Fig. 2. Notice the higher overall respiratory rate of the ASD LCLs as compared to the control LCLs and the greater change in these values as DMNQ concentration is increased. OCR oxygen consumption rate
Oxidative Stress and Mitochondrial Dysfunction Correspond in the ASD Brain
Some studies have implicated an association between mitochondrial dysfunction and oxidative stress in the brain tissue of children with ASD. Mitochondrial function was found to be decreased in the temporal lobe (BA 21) of individuals with ASD, associated with an increase in oxidative DNA damage and a decrease in superoxide dismutase 2 activity [154]. In another study, increased oxidative stress and decreased ETC activity was found in the cerebellum, frontal cortex and temporal cortex of children with ASD [192]. One study found increased oxidative damage to mitochondrial proteins along with increased C4 activity in the superior temporal gyrus (BA 41/42 or 22) of individuals with ASD [208]. Lastly, another study found that aconitase activity was negatively correlated with the GSH redox ratio in the cerebellum and the temporal lobe (BA 22) of individuals with ASD [173].
Extrinsic Environmental Factors
Ethylmercury is an environmental toxin known to deplete GSH and induce oxidative stress and mitochondrial dysfunction. Using our LCL model, we demonstrated that acute exposure to ethylmercury induces a greater reduction in ALR, MRC and RC in the AD-A subgroup of LCLs as compared to control LCLs [174]. Pretreatment of the ethylmercury-sensitive subgroup with N-acetyl-cysteine to increase GSH normalized baseline respiratory parameters and blunted the exaggerated ethylmercury-induced RC depletion [174]. Similarly, Sharpe et al. [209] reported that a subset of ASD LCLs and their unaffected siblings exhibited mitochondrial hypersensitivity to ethylmercury using a surrogate assay of mitochondrial function known as 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT) reduction.
Trichloroethylene (TCE) is an environmental toxicant and common environmental pollutant that has been linked to neurobehavioral and immune abnormalities as well as to Parkinson’s disease and ASD [210–213]. Using our LCL model, we exposed ASD and control LCLs to trichloroacetaldehyde hydrate (TCAH), an in vivo TCE metabolite, either alone or followed by an acute exposure to DMNQ, an ROS generator [176]. TCAH exposure by itself in control and ASD LCLs resulted in a similar decline in mitochondrial respiration. However, for the AD-A subset of ASD LCLs as well as control LCLs, acute ROS exposure following TCAH exposure attenuated the decline in mitochondrial respiration, indicating that mitochondrial protective pathways may have been activated by acute ROS, and that these protective pathways had been primed for activation in an AD-A subset of LCLs by TCAH.
Bisphenol A (BPA) is a common chemical used to make plastics. Human exposure to BPA is considered widespread. Kaur et al. [214] exposed LCLs from ASD and unaffected sibling pairs to BPA. BPA induced ROS and decreased mitochondrial membrane potential in both ASD and unaffected sibling LCLs. When unaffected twin and non-twin siblings were analyzed separately, BPA-induced ROS production was greater in ASD and unaffected twin siblings as compared to non-twin siblings. BPA exposure upregulated mtDNA copy number in LCLs from unaffected twin siblings as compared to non-twin siblings. mtDNA in ASD LCLs was not examined.
The Enteric Microbiome Environment in ASD
ASD is also associated with environmental factors such as enteric microbiome-derived metabolites that may alter mitochondrial function by mechanisms other than oxidative stress [215]. Alterations in the gut microbiome have been associated with ASD [194, 216], and a major mechanism by which enteric bacteria influence host physiology is through the production of short-chain fatty acids. Clostridia spp., a major short-chain fatty acid producer, repeatedly has been found to be overrepresented in the ASD GI tract [217, 218]. Our ASD LCL model has been instrumental in gaining a better understanding of the impact of short-chain fatty acids on mitochondrial function in ASD.
PPA is a ubiquitous short-chain fatty acid which is a major fermentation product of the enteric microbiome. PPA is particularly interesting because it is a natural component of normal intermediary metabolism, being the end product of odd-chain fatty acid oxidation. PPA induces ASD-like behaviors in rodents [63, 219] and causes abnormalities in metabolic, immune and neurophysiological systems [63, 219]; PPA also regulates genes in human cell lines [220]. As mentioned above, similar redox and fatty acid metabolic abnormalities are found in both children with ASD and the PPA induced rodent model of ASD [5]. Thus, PPA is an interesting molecule as it is directly involved in metabolism as an intermediate and can modulate gene expression.
Exposing control and ASD LCLs to various concentrations of PPA for 24–48 h revealed dose and exposure time-dependent effects on mitochondrial function. ASD LCLs demonstrated an enhancement of mitochondrial function with PPA exposure as indicated by an increase in RC as compared to control LCLs [175]. However, ROS exposure negated the positive PPA effect on RC in ASD LCLs. Indeed, in the context of increased ROS, instead of enhancing RC in ASD LCLs, PPA resulted in a greater decrease in RC in ASD LCLs as compared to control LCLs exposed to PPA and ROS. Thus, it appeared that the effect of PPA on mitochondrial function was context dependent as its ability to facilitate or demean mitochondrial function was dependent on the redox microenvironment.
Butyrate is another short-chain fatty acid primarily produced by the enteric microbiome which can be a fuel source and modulate gene expression. Exposure of LCLs to butyrate for 24–48 h had differential effects on mitochondrial function in control and ASD LCLs [221]. Butyrate increased respiration in the AD-A LCLs, while decreasing mitochondrial respiration in control LCLs. When ROS was increased in vitro, butyrate prevented a diminutive effect of ROS on mitochondrial respiration in all LCLs. At the highest concentration investigated (1 mM), butyrate increased the expression of genes involved in mitochondrial fission (DRP1, FIS1), mitophagy (PINK1, LC3, PTEN), oxidative stress (UCP2, NRF2, SOD2), energy metabolism, such as mammalian target of rapamycin (mTOR), AMPK and SIRT3, as well as genes known to be involved in response to physiological stress, such as PGC1α. These findings suggest that butyrate has a protective effect on the mitochondria.
Treatments of Mitochondrial Dysfunction in ASD
There is only one study published to date that examined the behavioral effects of a customized mitochondrial supplement in ASD children with mitochondrial dysfunction. Legido et al. [55] in an open-label study treated 11 children with ASD and abnormal C1 and/or C4 activity with a mitochondrial cocktail containing carnitine, coenzyme Q10 and α-lipoic acid. Three months of treatment reduced the C1-to-C4 ratio as well as improved several behavior scales, including lethargy and inappropriate speech subscales of the Aberrant Behavior Checklist. Three months after withdrawal of the treatment, the lethargy and inappropriate speech subscales significantly worsened.
As previously mentioned, in a relatively large study, we have examined whether various treatments for mitochondrial disorders influenced enzymatic activity in children with ASD using the buccal swab technique [54]. Overall, the study demonstrated that folate, cobalamin, fatty acids and antioxidant supplementation increased mitochondrial enzymatic activity and folate and cobalamin influenced that relationship between enzyme activity, suggesting increased ETC coupling.
As we have previously reviewed, although most studies have not selected individuals with ASD and mitochondrial dysfunction for treatment with supplements that target the mitochondria, several studies have demonstrated that mitochondrial supplements may be helpful in children with ASD [73]. For example, carnitine deficiency appears to be common in children with ASD [58, 70]. Two double-blind, placebo-controlled studies demonstrated improvement in ASD symptoms with carnitine supplementation [74], with some improvements directly related to the change in blood carnitine levels [72]. In another study, reduced NAD and ribose appeared to improve metabolic biomarkers in children with ASD and symptoms of mitochondrial dysfunction [222]. Two studies have reported behavioral improvements in children with ASD using ubiquinol [223] and coenzyme Q10 [224].
Discussion
Defining Mitochondrial Dysfunction in ASD
Major criteria that define classic MD (i.e., PMD) include unequivocal genetic mutations, severe depressions (i.e., < 30%) of ETC function or syndromic presentation [16]. However, repeated studies have suggested that genetic defects are found only in a minority of children with ASD, including a minority of children with ASD/MD [3], and many case reports and series have described only moderate, rather than severe, deficiencies in ETC activity [5, 225]. In classic MD, the mitochondria are thought to be severely dysfunctional, with significant depression in mitochondrial respiration. Criteria such as the modified Walker’s criteria reflect this notion and are commonly used to diagnose MD. Many genetic and non-genetic disorders that can cause SMD may respond to treatments that target the mitochondria [101].
Perhaps more striking is the fact that ETC activity in muscle [225, 226], skin [5], buccal epithelium [51, 52, 227] and brain [208] has been documented to be significantly increased, rather than decreased, in some individuals with ASD. This is consistent with our in vitro data showing elevated mitochondrial respiration in LCLs derived from children with ASD [169–171, 174–176, 228]. We recently demonstrated that this alteration in respiration is associated with more severe repetitive behaviors [171]. We believe that this increase in respiratory rate may be an adaptation designed to resist chronic exposure to toxicants associated with ASD, perhaps because of previous exposure to environmental toxicants [170, 176]. We have also demonstrated that this increased respiratory rate results in an increased susceptibility of the mitochondrial to acute in vitro increases in ROS [169, 170], and we have reported that this subset of ASD LCLs respond differently to environmental exposures associated with ASD [174–176, 221]. Further research will be needed to better understand this unique alteration in mitochondrial physiology.
Thus, the current data points to possible non-genetic defects or changes causing abnormal mitochondrial function as well as unique types of mitochondrial dysfunction in individuals with ASD [5, 226]. As mitochondrial dysfunction in ASD appears to be qualitatively and quantitatively different from what occurs in classic MD, it is important to consider how to define mitochondrial dysfunction in ASD. We have suggested that other criteria, such as the Morava criteria, which is clinically based, may be more appropriate for diagnosing MD in children with ASD [16]. However, it is clear we need a better definition on what degree of mitochondrial dysfunction is abnormal in ASD and how it is linked to symptomology. We have demonstrated that changes in ETC activity are associated with changes in behavior and development on a continuum [53] and that treating mitochondrial abnormalities in children with ASD has positive benefits for ASD-related behaviors [55]. In addition, we have demonstrated that certain common supplements positively modulate mitochondrial activity in children with ASD [54]. Nevertheless, it is not clear how to define these variations in mitochondrial function as clearly abnormal or how to decide which children will most likely respond to treatment for mitochondrial dysfunction. Clearly, further research is needed in this potentially fruitful area.
Possible Links to Clinical Aspects of ASD
There are several clinical aspects of ASD that may be associated with abnormalities in mitochondrial function, including abnormalities in immune function, cerebral folate deficiency, autonomic abnormalities and prenatal brain abnormalities.
Autoimmune diseases are diagnosed significantly more often among children with ASD than among controls [229] and among their family members [230]. In addition, some children with ASD also have been documented to have immunoglobulin deficiency [231]. Several studies have shown that immune cells in children with ASD demonstrate mitochondrial dysfunction [3, 121, 169, 228, 232], and recent studies have documented that proper mitochondrial function is essential for immune regulation [233] and mitochondrial dysfunction is associated with immunodeficiencies [234]. Thus, it is very possible that mitochondrial dysfunction could lead to the immune abnormalities seen in ASD. Alternatively, autoimmunity causes inflammatory responses that result in catabolism and high rates of ATP consumption from the release of proinflammatory cytokines [235]. Thus, it is very possible that an initial autoimmune process can drive mitochondrial dysfunction. Indeed, mitochondrial dysfunction may ensue during an inflammatory process. This could result in developmental regression from typically development to an ASD phenotype, as reported by Shoffner et al. [20]. Inflammation can affect epigenetics that involve both nDNA and mtDNA. This represents yet another mechanism by which the immune system may adversely affect the mitochondria. Finally, the effect of anti-mitochondrial antibodies reported to be associated with ASD [80, 81] on mitochondrial function is yet to be explored in detail. This may provide an interesting connection between the immune system and the mitochondria.
About 15 years ago, Ramaekers and Blau described a case series of children with normal neurodevelopment during early infancy followed by neurodevelopmental regression. These children had low concentrations of folate in their cerebrospinal fluid, but normal concentrations in their blood [236]. This newly described neurodevelopmental disorder was named cerebral folate deficiency (CFD) syndrome to signify deficiency of folate specific to the brain. Early reports found that some children with CFD also had ASD [237], with further studies suggesting that CFD was linked to children with low-functioning ASD who also had neurological abnormalities [238]. Recent studies have suggested that CFD is found in about a quarter of children with ASD [239]. CFD is caused by dysfunction of the folate receptor α, the major transport mechanism for folate transport into the brain, which can be caused by folate receptor α autoantibodies and/or mitochondrial dysfunction. Cases of ASD with mitochondrial dysfunction have been reported in CFD [240]. Thus, the connection between mitochondrial dysfunction, CFD and ASD may be very important and has the potential to be one of the major pathophysiological mechanisms that drives cognitive and behavioral abnormalities. Most significant is the fact that CFD is treatable and potentially reversible with leucovorin calcium. Given the recent studies that have demonstrated that leucovorin calcium is beneficial to behavior and language in children with ASD [241, 242], as well as the fact that folates appear to positively modulate mitochondrial function [54, 243], this should be considered a very promising area of therapeutics in ASD.
Other abnormalities that may be connected to mitochondrial dysfunction are prenatal abnormalities and autonomic dysfunction. Patients with ASD have demonstrated abnormalities of brain growth and development including hypoplasia of the corpus callosum, cerebellar hypoplasia, failures of synaptic development including dendritic sprouting and axonal branching, among others. These abnormalities may reflect impaired mitochondrial function, energetics of the microtubule system, and cell motility in the development of the nervous system. Weissman et al. reported prenatal abnormalities in their case series of children with ASD/MD [15]. Dysautonomia, such as excessive dilatation of the pupils and tachycardia, are commonly seen in children with ASD [244]. Autonomic dysfunction has been associated with mitochondrial problems [245] and has been reported in a child with ASD and mitochondrial myopathy [246]. Some have pointed to a potential connection between nitric oxide, mitochondrial dysfunction and dysautonomia in ASD [247]. Thus, further studies are needed to investigate the potential connection between these clinical entities and mitochondrial function in ASD.
Studying Environmental Agents Affecting Mitochondrial Function
As has been demonstrated by several groups, including ours, LCLs derived from individuals with ASD are the primary cellular model to study mitochondrial dysfunction in ASD [3, 121, 169, 228, 232]. Furthermore, LCLs from individuals with ASD paired with LCLs from unaffected twin and non-twin siblings make an excellent model to examine the mitochondrial effects of environmental factors associated with ASD and whether or not these factors can induce mitochondrial adaptations that result in vulnerabilities leading to dysfunction [174–176, 221, 248]. Given the importance of the mitochondria in environmental exposures and the fact that environmental exposures most likely have a considerable influence in the etiology of ASD, further research into the connection between environmental factors and mitochondrial dysfunction in ASD is clearly needed.
Novel Markers for Measuring Mitochondrial Function in ASD
One exciting recent development is the introduction of new biomarkers that may be quite helpful in identifying mitochondrial dysfunction. The buccal swab technique non-invasively measures mitochondrial enzymatic activity and calculates the relative activity of enzymes, by using a ratio or examining the linear relationship between enzyme activities. Other techniques such as using respirometry or enzymology on primary immune cells are other promising techniques for identifying mitochondrial dysfunction in individuals with ASD. Furthermore, novel blood-based markers have been investigated and can discriminate children with ASD from healthy controls [28, 76–79]. Abnormalities in mitochondrial function also appear to include alterations in fatty acid metabolism, including a unique pattern of acyl-carnitine elevation linked to a partial C1 deficiency that may have a connection to modulatory influences from the microbiome [5]. Clearly, these biomarkers remain a promising area, but results are still preliminary. Further development of these and other techniques are needed to help better identify and define individuals with ASD and mitochondrial abnormalities.
Conclusions
Mitochondrial dysfunction appears to be closely associated with at least a subset of patients with ASD. The etiology of mitochondrial dysfunction as well as how to define it in individuals with ASD is not clear at this time. Preliminary studies suggest that the mitochondria may be a fruitful target for treatment and prevention of ASD, but further research to better understand the role of the mitochondria in the pathophysiology of ASD is needed.
Acknowledgements
We thank the families and individuals with ASD that participated in the Autism Genetic Research Exchange and the studies at the National Institutes of Mental Health for providing cell lines to us for research. We would like to thank the supporters of our research studies, including the Arkansas Biosciences Institute (Little Rock, AR, USA), the Jonty Foundation (St. Paul, MN), the Autism Research Institute (San Diego, CA), the Gupta Family Foundation (Atherton, CA), the Jane Bostford Johnson Foundation (New York, NY), the Jager Family Foundation (Chicago, IL) and the Phoenix Children’s Hospital Foundation (Phoenix, AZ).
Funding
There was no funding for this article.
Conflict of Interest
The authors (SR, DN, DR, MG, SK and RF) have no conflicts of interest to declare.
References
- 1.Frye RE, Rossignol DA. Identification and treatment of pathophysiological comorbidities of autism spectrum disorder to achieve optimal outcomes. Clin Med Insights Pediatr. 2016;10:43–56. doi: 10.4137/CMPed.S38337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rossignol DA, Frye RE. A review of research trends in physiological abnormalities in autism spectrum disorders: immune dysregulation, inflammation, oxidative stress, mitochondrial dysfunction and environmental toxicant exposures. Mol Psychiatry. 2012;17(4):389–401. doi: 10.1038/mp.2011.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossignol DA, Frye RE. Mitochondrial dysfunction in autism spectrum disorders: a systematic review and meta-analysis. Mol Psychiatry. 2012;17(3):290–314. doi: 10.1038/mp.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frye RE. Biomarker of abnormal energy metabolism in children with autism spectrum disorder. North Am J Med Sci. 2012;5:141–147. [Google Scholar]
- 5.Frye RE, Melnyk S, Macfabe DF. Unique acyl-carnitine profiles are potential biomarkers for acquired mitochondrial disease in autism spectrum disorder. Transl Psychiatry. 2013;3:e220. doi: 10.1038/tp.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parikh S, Goldstein A, Karaa A, Koenig MK, Anselm I, Brunel-Guitton C, et al. Patient care standards for primary mitochondrial disease: a consensus statement from the Mitochondrial Medicine Society. Genet Med. 2017 doi: 10.1038/gim.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castro-Gago M, Blanco-Barca O, Gomez-Lado C, Pintos-Martinez E, Campos-Gonzalez Y, Eiris-Punal J. Association between autistic spectrum and mitochondrial pathology. Rev Neurol. 2008;47(1):52–53. [PubMed] [Google Scholar]
- 8.Holtzman D. Autistic spectrum disorders and mitochondrial encephalopathies. Acta Paediatr. 2008;97(7):859–860. doi: 10.1111/j.1651-2227.2008.00883.x. [DOI] [PubMed] [Google Scholar]
- 9.Oliveira G, Ataide A, Marques C, Miguel TS, Coutinho AM, Mota-Vieira L, et al. Epidemiology of autism spectrum disorder in Portugal: prevalence, clinical characterization, and medical conditions. Dev Med Child Neurol. 2007;49(10):726–733. doi: 10.1111/j.1469-8749.2007.00726.x. [DOI] [PubMed] [Google Scholar]
- 10.Tsao CY, Mendell JR. Autistic disorder in 2 children with mitochondrial disorders. J Child Neurol. 2007;22(9):1121–1123. doi: 10.1177/0883073807306266. [DOI] [PubMed] [Google Scholar]
- 11.Chugani DC, Sundram BS, Behen M, Lee ML, Moore GJ. Evidence of altered energy metabolism in autistic children. Prog Neuropsychopharmacol Biol Psychiatry. 1999;23(4):635–641. doi: 10.1016/s0278-5846(99)00022-6. [DOI] [PubMed] [Google Scholar]
- 12.Enns GM. Autistic features part of global neurologic syndrome in children who have mitochondrial disease. AAP News. 2008;29(7):20. [Google Scholar]
- 13.Guevara-Campos J, Gonzalez-Guevara L, Briones P, Lopez-Gallardo E, Bulan N, Ruiz-Pesini E, et al. Autism associated to a deficiency of complexes III and IV of the mitochondrial respiratory chain. Invest Clin. 2010;51(3):423–431. [PubMed] [Google Scholar]
- 14.Guevara-Campos J, Gonzalez-Guevara L, Puig-Alcaraz C, Cauli O. Autism spectrum disorders associated to a deficiency of the enzymes of the mitochondrial respiratory chain. Metab Brain Dis. 2013;28(4):605–612. doi: 10.1007/s11011-013-9419-x. [DOI] [PubMed] [Google Scholar]
- 15.Weissman JR, Kelley RI, Bauman ML, Cohen BH, Murray KF, Mitchell RL, et al. Mitochondrial disease in autism spectrum disorder patients: a cohort analysis. PLoS One. 2008;3(11):e3815. doi: 10.1371/journal.pone.0003815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frye RE, Rossignol DA. Mitochondrial dysfunction can connect the diverse medical symptoms associated with autism spectrum disorders. Pediatr Res. 2011;69(5 Pt 2):41R–47R. doi: 10.1203/PDR.0b013e318212f16b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morava E, van den Heuvel L, Hol F, de Vries MC, Hogeveen M, Rodenburg RJ, et al. Mitochondrial disease criteria: diagnostic applications in children. Neurology. 2006;67(10):1823–1826. doi: 10.1212/01.wnl.0000244435.27645.54. [DOI] [PubMed] [Google Scholar]
- 18.Haas RH. Autism and mitochondrial disease. Dev Disabil Res Rev. 2010;16(2):144–153. doi: 10.1002/ddrr.112. [DOI] [PubMed] [Google Scholar]
- 19.Schaefer KA, Toral MA, Velez G, Cox AJ, Baker SA, Borcherding NC, et al. Calpain-5 expression in the retina localizes to photoreceptor synapses. Invest Ophthalmol Vis Sci. 2016;57(6):2509–2521. doi: 10.1167/iovs.15-18680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shoffner J, Hyams L, Langley GN, Cossette S, Mylacraine L, Dale J, et al. Fever plus mitochondrial disease could be risk factors for autistic regression. J Child Neurol. 2010;25(4):429–434. doi: 10.1177/0883073809342128. [DOI] [PubMed] [Google Scholar]
- 21.Fillano JJ, Goldenthal MJ, Rhodes CH, Marin-Garcia J. Mitochondrial dysfunction in patients with hypotonia, epilepsy, autism, and developmental delay: HEADD syndrome. J Child Neurol. 2002;17(6):435–439. doi: 10.1177/088307380201700607. [DOI] [PubMed] [Google Scholar]
- 22.Filipek PA, Juranek J, Smith M, Mays LZ, Ramos ER, Bocian M, et al. Mitochondrial dysfunction in autistic patients with 15q inverted duplication. Ann Neurol. 2003;53(6):801–804. doi: 10.1002/ana.10596. [DOI] [PubMed] [Google Scholar]
- 23.Poling JS, Frye RE, Shoffner J, Zimmerman AW. Developmental regression and mitochondrial dysfunction in a child with autism. J Child Neurol. 2006;21(2):170–172. doi: 10.2310/7010.2006.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Craig AK, de Menezes MS, Saneto RP. Dravet syndrome: patients with co-morbid SCN1A gene mutations and mitochondrial electron transport chain defects. Seizure. 2012;21(1):17–20. doi: 10.1016/j.seizure.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 25.Guevara-Campos J, Gonzalez-Guevara L, Cauli O. Autism and intellectual disability associated with mitochondrial disease and hyperlactacidemia. Int J Mol Sci. 2015;16(2):3870–3884. doi: 10.3390/ijms16023870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coleman M, Blass JP. Autism and lactic acidosis. J Autism Dev Disord. 1985;15(1):1–8. doi: 10.1007/BF01837894. [DOI] [PubMed] [Google Scholar]
- 27.Karim M, Begum S, Shahzadi S. Serum lactate, AST, ALT in male autistic children in Bangladesh. J Bangladesh Soc Physiol. 2016;10(2):56–60. [Google Scholar]
- 28.Khemakhem AM, Frye RE, El-Ansary A, Al-Ayadhi L, Bacha AB. Novel biomarkers of metabolic dysfunction is autism spectrum disorder: potential for biological diagnostic markers. Metab Brain Dis. 2017;32(6):1983–1997. doi: 10.1007/s11011-017-0085-2. [DOI] [PubMed] [Google Scholar]
- 29.Laszlo A, Horvath E, Eck E, Fekete M. Serum serotonin, lactate and pyruvate levels in infantile autistic children. Clin Chim Acta. 1994;229(1–2):205–207. doi: 10.1016/0009-8981(94)90243-7. [DOI] [PubMed] [Google Scholar]
- 30.Shahjadi S, Khan AS, Ahmed MU. Mitochondrial dysfunction in early diagnosed autism spectrum disorder children. J Dhaka Med Coll. 2017;26(1):43–47. [Google Scholar]
- 31.Essa MM, Guillemin GJ, Waly MI, Al-Sharbati MM, Al-Farsi YM, Hakkim FL, et al. Increased markers of oxidative stress in autistic children of the Sultanate of Oman. Biol Trace Elem Res. 2012;147(1–3):25–27. doi: 10.1007/s12011-011-9280-x. [DOI] [PubMed] [Google Scholar]
- 32.Lv MN, Zhang H, Shu Y, Chen S, Hu YY, Zhou M. The neonatal levels of TSB, NSE and CK-BB in autism spectrum disorder from Southern China. Transl Neurosci. 2016;7(1):6–11. doi: 10.1515/tnsci-2016-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mousavinejad E, Ghaffari MA, Payami S, Lamuchi-Deli N, Ashtary-Larky D. Coenzyme-Q10 deficiency and stress oxidative in children with autism spectrum disorders. J Neurol Neurorehabil Res. 2017;2(2):25–29. [Google Scholar]
- 34.Shaw W. Elevated urinary glyphosate and clostridia metabolites with altered dopamine metabolism in triplets with autistic spectrum disorder or suspected seizure disorder: a case study. Integr Med (Encinitas). 2017;16(1):50–57. [PMC free article] [PubMed] [Google Scholar]
- 35.Shaw W, Kassen E, Chaves E. Increased urinary excretion of analogs of Krebs cycle metabolites and arabinose in two brothers with autistic features. Clin Chem. 1995;41(8 Pt 1):1094–1104. [PubMed] [Google Scholar]
- 36.Kelley RI. Evaluation and treatment of patients with autism and mitochondrial disease. 2009. http://www.epidemicanswers.org/wp-content/uploads/2010/05/Dr.-Richard-Kelly-Autism_Mitochondrial_Disease.pdf. Accessed 29 June 2010.
- 37.Mussap M, Noto A, Fanos V. Metabolomics of autism spectrum disorders: early insights regarding mammalian-microbial cometabolites. Expert Rev Mol Diagn. 2016;16(8):869–881. doi: 10.1080/14737159.2016.1202765. [DOI] [PubMed] [Google Scholar]
- 38.Kuwabara H, Yamasue H, Koike S, Inoue H, Kawakubo Y, Kuroda M, et al. Altered metabolites in the plasma of autism spectrum disorder: a capillary electrophoresis time-of-flight mass spectroscopy study. PLoS One. 2013;8(9):e73814. doi: 10.1371/journal.pone.0073814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreno H, Borjas L, Arrieta A, Sáez L, Prassad A, Estévez J, et al. Heterogeneidad clínica del síndrome autista: un estudio en sesenta familias [Clinical heterogeneity of the autistic syndrome: a study of 60 families] Invest Clin. 1992;33(1):13–31. [PubMed] [Google Scholar]
- 40.Zaki MM, Abdel-Al H, Al-Sawi M. Assessment of plasma amino acid profile in autism using cation-exchange chromatography with postcolumn derivatization by ninhydrin. Turk J Med Sci. 2017;47(1):260–267. doi: 10.3906/sag-1506-105. [DOI] [PubMed] [Google Scholar]
- 41.Abu Shmais GA, Al-Ayadhi LY, Al-Dbass AM, El-Ansary AK. Mechanism of nitrogen metabolism-related parameters and enzyme activities in the pathophysiology of autism. J Neurodev Disord. 2012;4(1):4. doi: 10.1186/1866-1955-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park E, Cohen I, Gonzalez M, Castellano MR, Flory M, Jenkins EC, et al. Is taurine a biomarker in autistic spectrum disorder? Adv Exp Med Biol. 2017;975:3–16. doi: 10.1007/978-94-024-1079-2_1. [DOI] [PubMed] [Google Scholar]
- 43.Fernell E, Karagiannakis A, Edman G, Bjerkenstedt L, Wiesel FA, Venizelos N. Aberrant amino acid transport in fibroblasts from children with autism. Neurosci Lett. 2007;418(1):82–86. doi: 10.1016/j.neulet.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 44.Novarino G, El-Fishawy P, Kayserili H, Meguid NA, Scott EM, Schroth J, et al. Mutations in BCKD-kinase lead to a potentially treatable form of autism with epilepsy. Science (New York, NY) 2012;338(6105):394–397. doi: 10.1126/science.1224631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tarlungeanu DC, Deliu E, Dotter CP, Kara M, Janiesch PC, Scalise M et al. Impaired amino acid transport at the blood brain barrier is a cause of autism spectrum disorder. Cell. 2016;167(6):1481–94 e18. 10.1016/j.cell.2016.11.013. [DOI] [PMC free article] [PubMed]
- 46.Goldenthal MJ, Kuruvilla T, Damle S, Salganicoff L, Sheth S, Shah N, et al. Non-invasive evaluation of buccal respiratory chain enzyme dysfunction in mitochondrial disease: comparison with studies in muscle biopsy. Mol Genet Metab. 2012;105(3):457–462. doi: 10.1016/j.ymgme.2011.11.193. [DOI] [PubMed] [Google Scholar]
- 47.Yorns WR, Jr, Valencia I, Jayaraman A, Sheth S, Legido A, Goldenthal MJ. Buccal swab analysis of mitochondrial enzyme deficiency and DNA defects in a child with suspected myoclonic epilepsy and ragged red fibers (MERRF) J Child Neurol. 2012;27(3):398–401. doi: 10.1177/0883073811420870. [DOI] [PubMed] [Google Scholar]
- 48.Ezugha H, Goldenthal M, Valencia I, Anderson CE, Legido A, Marks H. 5q14.3 deletion manifesting as mitochondrial disease and autism: case report. J Child Neurol. 2010;25(10):1232–1235. doi: 10.1177/0883073809361165. [DOI] [PubMed] [Google Scholar]
- 49.Frye RE, Cox D, Slattery J, Tippett M, Kahler S, Granpeesheh D, et al. Mitochondrial dysfunction may explain symptom variation in Phelan-McDermid syndrome. Sci Rep. 2016;6:19544. doi: 10.1038/srep19544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burger BJ, Rose S, Bennuri SC, Gill PS, Tippett ML, Delhey L, et al. Autistic siblings with novel mutations in two different genes: insight for genetic workups of autistic siblings and connection to mitochondrial dysfunction. Front Pediatr. 2017;5:219. doi: 10.3389/fped.2017.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goldenthal MJ, Damle S, Sheth S, Shah N, Melvin J, Jethva R, et al. Mitochondrial enzyme dysfunction in autism spectrum disorders; a novel biomarker revealed from buccal swab analysis. Biomark Med. 2015;9(10):957–965. doi: 10.2217/bmm.15.72. [DOI] [PubMed] [Google Scholar]
- 52.Legido A, Jethva R, Goldenthal MJ. Mitochondrial dysfunction in autism. Semin Pediatr Neurol. 2013;20(3):163–175. doi: 10.1016/j.spen.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 53.Delhey L, Kilinc EN, Yin L, Slattery J, Tippett M, Wynne R, et al. Bioenergetic variation is related to autism symptomatology. Metab Brain Dis. 2017;32(6):2021–2031. doi: 10.1007/s11011-017-0087-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Delhey LM, Nur Kilinc E, Yin L, Slattery JC, Tippett ML, Rose S, et al. The effect of mitochondrial supplements on mitochondrial activity in children with autism spectrum disorder. J Clin Med. 2017 doi: 10.3390/jcm6020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Legido A, Goldenthal M, Garvin B, Damle S, Corrigan K, Connell J, et al. Effect of a combination of carnitine, coenzyme Q10 and alpha-lipoic acid (MitoCocktail) on mitochondrial function and neurobehavioral performance in children with autism spectrum disorder (P3.313) Neurology. 2018;90(15 Supplement):P3.313. [Google Scholar]
- 56.Clark-Taylor T, Clark-Taylor BE. Is autism a disorder of fatty acid metabolism? Possible dysfunction of mitochondrial beta-oxidation by long chain acyl-CoA dehydrogenase. Med Hypotheses. 2004;62(6):970–975. doi: 10.1016/j.mehy.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 57.El-Ansary AK, Bacha AG, Al-Ayahdi LY. Plasma fatty acids as diagnostic markers in autistic patients from Saudi Arabia. Lipids Health Dis. 2011;10:62. doi: 10.1186/1476-511X-10-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mostafa GA, El-Gamal HA, El-Wakkad ASE, El-Shorbagy OE, Hamza MM. Polyunsaturated fatty acids, carnitine and lactate as biological markers of brain energy in autistic children. Int J Child Neuropsychiatry. 2005;2(2):179–188. [Google Scholar]
- 59.Pastural E, Ritchie S, Lu Y, Jin W, Kavianpour A, Khine Su-Myat K, et al. Novel plasma phospholipid biomarkers of autism: mitochondrial dysfunction as a putative causative mechanism. Prostaglandins Leukot Essent Fatty Acids. 2009;81(4):253–264. doi: 10.1016/j.plefa.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 60.Frackowiak J, Mazur-Kolecka B, Schanen NC, Brown WT, Wegiel J. The link between intraneuronal N-truncated amyloid-beta peptide and oxidatively modified lipids in idiopathic autism and dup(15q11.2-q13)/autism. Acta Neuropathol Commun. 2013;1:61. doi: 10.1186/2051-5960-1-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Munnich A, Rustin P. Clinical spectrum and diagnosis of mitochondrial disorders. Am J Med Genet. 2001;106(1):4–17. doi: 10.1002/ajmg.1391. [DOI] [PubMed] [Google Scholar]
- 62.Mitochondrial Medicine Society’s Committee on D, Haas RH, Parikh S, Falk MJ, Saneto RP, Wolf NI et al The in-depth evaluation of suspected mitochondrial disease. Mol Genet Metab. 2008;94(1):16–37. doi: 10.1016/j.ymgme.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.MacFabe DF, Cain DP, Rodriguez-Capote K, Franklin AE, Hoffman JE, Boon F, et al. Neurobiological effects of intraventricular propionic acid in rats: possible role of short chain fatty acids on the pathogenesis and characteristics of autism spectrum disorders. Behav Brain Res. 2007;176(1):149–169. doi: 10.1016/j.bbr.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 64.Thomas RH, Meeking MM, Mepham JR, Tichenoff L, Possmayer F, Liu S, et al. The enteric bacterial metabolite propionic acid alters brain and plasma phospholipid molecular species: further development of a rodent model of autism spectrum disorders. J Neuroinflammation. 2012;9:153. doi: 10.1186/1742-2094-9-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thomas RH, Foley KA, Mepham JR, Tichenoff LJ, Possmayer F, MacFabe DF. Altered brain phospholipid and acylcarnitine profiles in propionic acid infused rodents: further development of a potential model of autism spectrum disorders. J Neurochem. 2010;113(2):515–529. doi: 10.1111/j.1471-4159.2010.06614.x. [DOI] [PubMed] [Google Scholar]
- 66.De Angelis M, Piccolo M, Vannini L, Siragusa S, De Giacomo A, Serrazzanetti DI, et al. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS One. 2013;8(10):e76993. doi: 10.1371/journal.pone.0076993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Macfabe D. Autism: metabolism, mitochondria, and the microbiome. Glob Adv Health Med. 2013;2(6):52–66. doi: 10.7453/gahmj.2013.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang L, Christophersen CT, Sorich MJ, Gerber JP, Angley MT, Conlon MA. Elevated fecal short chain fatty acid and ammonia concentrations in children with autism spectrum disorder. Dig Dis Sci. 2012;57(8):2096–2102. doi: 10.1007/s10620-012-2167-7. [DOI] [PubMed] [Google Scholar]
- 69.Witters P, Debbold E, Crivelly K, Vande Kerckhove K, Corthouts K, Debbold B, et al. Autism in patients with propionic acidemia. Mol Genet Metab. 2016;119(4):317–321. doi: 10.1016/j.ymgme.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 70.Filipek PA, Juranek J, Nguyen MT, Cummings C, Gargus JJ. Relative carnitine deficiency in autism. J Autism Dev Disord. 2004;34(6):615–623. doi: 10.1007/s10803-004-5283-1. [DOI] [PubMed] [Google Scholar]
- 71.Celestino-Soper PB, Violante S, Crawford EL, Luo R, Lionel AC, Delaby E, et al. A common X-linked inborn error of carnitine biosynthesis may be a risk factor for nondysmorphic autism. Proc Natl Acad Sci USA. 2012;109(21):7974–7981. doi: 10.1073/pnas.1120210109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Geier DA, Kern JK, Davis G, King PG, Adams JB, Young JL, et al. A prospective double-blind, randomized clinical trial of levocarnitine to treat autism spectrum disorders. Med Sci Monit. 2011;17(6):PI15–PI23. doi: 10.12659/MSM.881792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Frye RE, Rossignol DA. Treatments for biomedical abnormalities associated with autism spectrum disorder. Front Pediatr. 2014;2:66. doi: 10.3389/fped.2014.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fahmy SF, El-hamamsy MH, Zaki OK, Badary OA. l-Carnitine supplementation improves the behavioral symptoms in autistic children. Res Autism Spectr Disord. 2013;7(1):159–166. [Google Scholar]
- 75.Ziats MN, Comeaux MS, Yang Y, Scaglia F, Elsea SH, Sun Q, et al. Improvement of regressive autism symptoms in a child with TMLHE deficiency following carnitine supplementation. Am J Med Genet Part A. 2015;167A(9):2162–2167. doi: 10.1002/ajmg.a.37144. [DOI] [PubMed] [Google Scholar]
- 76.El-Ansary A, Bjorklund G, Chirumbolo S, Alnakhli OM. Predictive value of selected biomarkers related to metabolism and oxidative stress in children with autism spectrum disorder. Metab Brain Dis. 2017;32(4):1209–1221. doi: 10.1007/s11011-017-0029-x. [DOI] [PubMed] [Google Scholar]
- 77.El-Ansary AK, Ben Bacha AG, Al-Ayadhi LY. Proinflammatory and proapoptotic markers in relation to mono and di-cations in plasma of autistic patients from Saudi Arabia. J Neuroinflammation. 2011;8:142. doi: 10.1186/1742-2094-8-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.El-Ansary A, Al-Daihan S, Al-Dabas A, Al-Ayadhi L. Activities of key glycolytic enzymes in the plasma of Saudi autistic patients. Open Access J Clin Trials. 2010;2010(2):49–57. [Google Scholar]
- 79.Al-Mosalem OA, El-Ansary A, Attas O, Al-Ayadhi L. Metabolic biomarkers related to energy metabolism in Saudi autistic children. Clin Biochem. 2009;42(10–11):949–957. doi: 10.1016/j.clinbiochem.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 80.Iqbal M, Bashir S, Al-Ayadhi L. Prevalence of antimitochondrial antibodies in autism spectrum subjects. Future Neurol. 2015;10(3):203–209. [Google Scholar]
- 81.Zhang B, Angelidou A, Alysandratos KD, Vasiadi M, Francis K, Asadi S, et al. Mitochondrial DNA and anti-mitochondrial antibodies in serum of autistic children. J Neuroinflammation. 2010;7:80. doi: 10.1186/1742-2094-7-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Finsterer J, Zarrouk-Mahjoub S. Biomarkers for detecting mitochondrial disorders. J Clin Med. 2018 doi: 10.3390/jcm7020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schaefer GB, Mendelsohn NJ, Professional P, Guidelines C. Clinical genetics evaluation in identifying the etiology of autism spectrum disorders: 2013 guideline revisions. Genet Med. 2013;15(5):399–407. doi: 10.1038/gim.2013.32. [DOI] [PubMed] [Google Scholar]
- 84.Tammimies K, Marshall CR, Walker S, Kaur G, Thiruvahindrapuram B, Lionel AC, et al. Molecular diagnostic yield of chromosomal microarray analysis and whole-exome sequencing in children with autism spectrum disorder. JAMA. 2015;314(9):895–903. doi: 10.1001/jama.2015.10078. [DOI] [PubMed] [Google Scholar]
- 85.Rossi M, El-Khechen D, Black MH, Farwell Hagman KD, Tang S, Powis Z. Outcomes of diagnostic exome sequencing in patients with diagnosed or suspected autism spectrum disorders. Pediatr Neurol. 2017;70(34–43):e2. doi: 10.1016/j.pediatrneurol.2017.01.033. [DOI] [PubMed] [Google Scholar]
- 86.Oikonomakis V, Kosma K, Mitrakos A, Sofocleous C, Pervanidou P, Syrmou A, et al. Recurrent copy number variations as risk factors for autism spectrum disorders: analysis of the clinical implications. Clin Genet. 2016;89(6):708–718. doi: 10.1111/cge.12740. [DOI] [PubMed] [Google Scholar]
- 87.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9(5):341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hallmayer J, Cleveland S, Torres A, Phillips J, Cohen B, Torigoe T, et al. Genetic heritability and shared environmental factors among twin pairs with autism. Arch Gen Psychiatry. 2011;68(11):1095–1102. doi: 10.1001/archgenpsychiatry.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sandin S, Lichtenstein P, Kuja-Halkola R, Larsson H, Hultman CM, Reichenberg A. The familial risk of autism. JAMA. 2014;311(17):1770–1777. doi: 10.1001/jama.2014.4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hall L, Kelley E. The contribution of epigenetics to understanding genetic factors in autism. Autism. 2014;18(8):872–881. doi: 10.1177/1362361313503501. [DOI] [PubMed] [Google Scholar]
- 91.Chen C, Chen Y, Guan MX. A peep into mitochondrial disorder: multifaceted from mitochondrial DNA mutations to nuclear gene modulation. Protein Cell. 2015;6(12):862–870. doi: 10.1007/s13238-015-0175-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Smith M, Flodman PL, Gargus JJ, Simon MT, Verrell K, Haas R, et al. Mitochondrial and ion channel gene alterations in autism. Biochim Biophys Acta. 2012;1817(10):1796–1802. doi: 10.1016/j.bbabio.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gimelli S, Capra V, Di Rocco M, Leoni M, Mirabelli-Badenier M, Schiaffino MC, et al. Interstitial 7q31.1 copy number variations disrupting IMMP2L gene are associated with a wide spectrum of neurodevelopmental disorders. Mol Cytogenet. 2014;7:54. doi: 10.1186/s13039-014-0054-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Maestrini E, Pagnamenta AT, Lamb JA, Bacchelli E, Sykes NH, Sousa I, et al. High-density SNP association study and copy number variation analysis of the AUTS1 and AUTS5 loci implicate the IMMP2L-DOCK4 gene region in autism susceptibility. Mol Psychiatry. 2010;15(9):954–968. doi: 10.1038/mp.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang Y, Liu Y, Zarrei M, Tong W, Dong R, Wang Y, et al. Association of IMMP2L deletions with autism spectrum disorder: a trio family study and meta-analysis. Am J Med Genet B Neuropsychiatr Genet. 2018;177(1):93–100. doi: 10.1002/ajmg.b.32608. [DOI] [PubMed] [Google Scholar]
- 96.Petek E, Schwarzbraun T, Noor A, Patel M, Nakabayashi K, Choufani S, et al. Molecular and genomic studies of IMMP2L and mutation screening in autism and Tourette syndrome. Mol Genet Genomics. 2007;277(1):71–81. doi: 10.1007/s00438-006-0173-1. [DOI] [PubMed] [Google Scholar]
- 97.Frye RE. Mitochondrial disease in 22q13 duplication syndrome. J Child Neurol. 2012;27(7):942–949. doi: 10.1177/0883073811429858. [DOI] [PubMed] [Google Scholar]
- 98.Lintas C, Sacco R, Persico AM. Genome-wide expression studies in autism spectrum disorder, Rett syndrome, and Down syndrome. Neurobiol Dis. 2012;45(1):57–68. doi: 10.1016/j.nbd.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 99.Smith M, Spence MA, Flodman P. Nuclear and mitochondrial genome defects in autisms. Ann N Y Acad Sci. 2009;1151:102–132. doi: 10.1111/j.1749-6632.2008.03571.x. [DOI] [PubMed] [Google Scholar]
- 100.Adi A, Tawil B, Aldosari M, Shinwari J, Nester M, Aldhalaan H, et al. Homozygosity analysis in subjects with autistic spectrum disorder. Mol Med Rep. 2015;12(2):2307–2312. doi: 10.3892/mmr.2015.3663. [DOI] [PubMed] [Google Scholar]
- 101.Niyazov DM, Kahler SG, Frye RE. Primary mitochondrial disease and secondary mitochondrial dysfunction: importance of distinction for diagnosis and treatment. Mol Syndromol. 2016;7(3):122–137. doi: 10.1159/000446586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Oliveira G, Diogo L, Grazina M, Garcia P, Ataide A, Marques C, et al. Mitochondrial dysfunction in autism spectrum disorders: a population-based study. Dev Med Child Neurol. 2005;47(3):185–189. doi: 10.1017/s0012162205000332. [DOI] [PubMed] [Google Scholar]
- 103.Correia C, Coutinho AM, Diogo L, Grazina M, Marques C, Miguel T, et al. Brief report: high frequency of biochemical markers for mitochondrial dysfunction in autism: no association with the mitochondrial aspartate/glutamate carrier SLC25A12 gene. J Autism Dev Disord. 2006;36(8):1137–1140. doi: 10.1007/s10803-006-0138-6. [DOI] [PubMed] [Google Scholar]
- 104.Scaglia F, Zhang S, Tan Z, Fouladi N, Schmitt E, Wong L-J, editors. Prevalence of autism spectrum disorders in subjects with definite diagnosis of mitochondrial cytopathies. In: The American Society of Human Genetics 59th Annual Meeting; 2009; Honolulu, Hawaii.
- 105.Wang Y, Picard M, Gu Z. Genetic evidence for elevated pathogenicity of mitochondrial DNA heteroplasmy in autism spectrum disorder. PLoS Genet. 2016;12(10):e1006391. doi: 10.1371/journal.pgen.1006391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mousavizadeh K, Askari M, Arian H, Gourjipour F, Nikpour AR, Tavafjadid M, et al. Association of human mtDNA mutations with autism in Iranian patients. J Res Med Sci. 2013;18(10):926. [PMC free article] [PubMed] [Google Scholar]
- 107.Varga NA, Pentelenyi K, Balicza P, Gezsi A, Remenyi V, Harsfalvi V, et al. Mitochondrial dysfunction and autism: comprehensive genetic analyses of children with autism and mtDNA deletion. Behav Brain Funct. 2018;14(1):4. doi: 10.1186/s12993-018-0135-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Patowary A, Nesbitt R, Archer M, Bernier R, Brkanac Z. Next generation sequencing mitochondrial DNA analysis in autism spectrum disorder. Autism Res. 2017;10(8):1338–1343. doi: 10.1002/aur.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Piryaei F, Houshmand M, Aryani O, Dadgar S, Soheili ZS. Investigation of the mitochondrial ATPase 6/8 and tRNA(Lys) genes mutations in autism. Cell J. 2012;14(2):98–101. [PMC free article] [PubMed] [Google Scholar]
- 110.Frye RE. Novel cytochrome B gene mutations causing mitochondrial disease in autism. J Ped Neuro. 2012;10(1):35–40. [Google Scholar]
- 111.Graf WD, Marin-Garcia J, Gao HG, Pizzo S, Naviaux RK, Markusic D, et al. Autism associated with the mitochondrial DNA G8363A transfer RNA(Lys) mutation. J Child Neurol. 2000;15(6):357–361. doi: 10.1177/088307380001500601. [DOI] [PubMed] [Google Scholar]
- 112.Connolly BS, Feigenbaum AS, Robinson BH, Dipchand AI, Simon DK, Tarnopolsky MA. MELAS syndrome, cardiomyopathy, rhabdomyolysis, and autism associated with the A3260G mitochondrial DNA mutation. Biochem Biophys Res Commun. 2010;402(2):443–447. doi: 10.1016/j.bbrc.2010.10.060. [DOI] [PubMed] [Google Scholar]
- 113.Avdjieva-Tzavella D, Mihailova S, Lukanov C, Naumova E, Simeonov E, Tincheva R, et al. Mitochondrial DNA mutations in two Bulgarian children with autistic spectrum disorders. Balkan J Med Genet. 2012;15(2):47–54. doi: 10.2478/bjmg-2013-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Serajee FJ, Zhang H, Huq A. Prevalence of common mitochondrial point mutations in autism. Neuropediatrics. 2006;37(Suppl 1):S127. [Google Scholar]
- 115.Pons R, Andreu AL, Checcarelli N, Vila MR, Engelstad K, Sue CM, et al. Mitochondrial DNA abnormalities and autistic spectrum disorders. J Pediatr. 2004;144(1):81–85. doi: 10.1016/j.jpeds.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 116.Kent L, Lambert C, Pyle A, Elliott H, Wheelwright S, Baron-Cohen S, et al. The mitochondrial DNA A3243A>G mutation must be an infrequent cause of Asperger syndrome. J Pediatr. 2006;149(2):280–281. doi: 10.1016/j.jpeds.2005.08.060. [DOI] [PubMed] [Google Scholar]
- 117.Kent L, Gallagher L, Elliott HR, Mowbray C, Chinnery PF. An investigation of mitochondrial haplogroups in autism. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(6):987–989. doi: 10.1002/ajmg.b.30687. [DOI] [PubMed] [Google Scholar]
- 118.Chalkia D, Singh LN, Leipzig J, Lvova M, Derbeneva O, Lakatos A, et al. Association between mitochondrial DNA haplogroup variation and autism spectrum disorders. JAMA Psychiatry. 2017;74(11):1161–1168. doi: 10.1001/jamapsychiatry.2017.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hadjixenofontos A, Schmidt MA, Whitehead PL, Konidari I, Hedges DJ, Wright HH, et al. Evaluating mitochondrial DNA variation in autism spectrum disorders. Ann Hum Genet. 2013;77(1):9–21. doi: 10.1111/j.1469-1809.2012.00736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Alvarez-Iglesias V, Mosquera-Miguel A, Cusco I, Carracedo A, Perez-Jurado LA, Salas A. Reassessing the role of mitochondrial DNA mutations in autism spectrum disorder. BMC Med Genet. 2011;12:50. doi: 10.1186/1471-2350-12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Giulivi C, Zhang YF, Omanska-Klusek A, Ross-Inta C, Wong S, Hertz-Picciotto I, et al. Mitochondrial dysfunction in autism. JAMA. 2010;304(21):2389–2396. doi: 10.1001/jama.2010.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Napoli E, Wong S, Hertz-Picciotto I, Giulivi C. Deficits in bioenergetics and impaired immune response in granulocytes from children with autism. Pediatrics. 2014;133(5):e1405–e1410. doi: 10.1542/peds.2013-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Budd MA, Calli K, Samson L, Bowes J, Hsieh AYY, Forbes JC, et al. Blood mitochondrial DNA content in HIV-exposed uninfected children with autism spectrum disorder. Viruses. 2018 doi: 10.3390/v10020077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chen S, Li Z, He Y, Zhang F, Li H, Liao Y, et al. Elevated mitochondrial DNA copy number in peripheral blood cells is associated with childhood autism. BMC Psychiatry. 2015;15:50. doi: 10.1186/s12888-015-0432-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yoo HJ, Park M, Kim SA. Difference in mitochondrial DNA copy number in peripheral blood cells between probands with autism spectrum disorders and their unaffected siblings. World J Biol Psychiatry. 2017;18(2):151–156. doi: 10.1080/15622975.2016.1234069. [DOI] [PubMed] [Google Scholar]
- 126.Wong S, Napoli E, Krakowiak P, Tassone F, Hertz-Picciotto I, Giulivi C. Role of p53, mitochondrial DNA deletions, and paternal age in autism: a case-control study. Pediatrics. 2016 doi: 10.1542/peds.2015-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Napoli E, Wong S, Giulivi C. Evidence of reactive oxygen species-mediated damage to mitochondrial DNA in children with typical autism. Mol Autism. 2013;4(1):2. doi: 10.1186/2040-2392-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Golzio C, Katsanis N. Mitochondrial copy number as a biomarker for autism? Pediatrics. 2016 doi: 10.1542/peds.2016-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Marui T, Funatogawa I, Koishi S, Yamamoto K, Matsumoto H, Hashimoto O, et al. The NADH-ubiquinone oxidoreductase 1 alpha subcomplex 5 (NDUFA5) gene variants are associated with autism. Acta Psychiatr Scand. 2011;123(2):118–124. doi: 10.1111/j.1600-0447.2010.01600.x. [DOI] [PubMed] [Google Scholar]
- 130.Villafuerte S. Suggestive evidence on the genetic link between mitochondria dysfunction and autism. Acta Psychiatr Scand. 2011;123(2):95. doi: 10.1111/j.1600-0447.2010.01654.x. [DOI] [PubMed] [Google Scholar]
- 131.Anitha A, Nakamura K, Thanseem I, Yamada K, Iwayama Y, Toyota T, et al. Brain region-specific altered expression and association of mitochondria-related genes in autism. Mol Autism. 2012;3(1):12. doi: 10.1186/2040-2392-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Blasi F, Bacchelli E, Carone S, Toma C, Monaco AP, Bailey AJ, et al. SLC25A12 and CMYA3 gene variants are not associated with autism in the IMGSAC multiplex family sample. Eur J Hum Genet. 2006;14(1):123–126. doi: 10.1038/sj.ejhg.5201444. [DOI] [PubMed] [Google Scholar]
- 133.Aoki Y, Cortese S. Mitochondrial aspartate/glutamate carrier SLC25A12 and autism spectrum disorder: a meta-analysis. Mol Neurobiol. 2016;53(3):1579–1588. doi: 10.1007/s12035-015-9116-3. [DOI] [PubMed] [Google Scholar]
- 134.Chien WH, Wu YY, Gau SS, Huang YS, Soong WT, Chiu YN, et al. Association study of the SLC25A12 gene and autism in Han Chinese in Taiwan. Prog Neuropsychopharmacol Biol Psychiatry. 2010;34(1):189–192. doi: 10.1016/j.pnpbp.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 135.Durdiakova J, Warrier V, Baron-Cohen S, Chakrabarti B. Single nucleotide polymorphism rs6716901 in SLC25A12 gene is associated with Asperger syndrome. Mol Autism. 2014;5(1):25. doi: 10.1186/2040-2392-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lepagnol-Bestel AM, Maussion G, Boda B, Cardona A, Iwayama Y, Delezoide AL, et al. SLC25A12 expression is associated with neurite outgrowth and is upregulated in the prefrontal cortex of autistic subjects. Mol Psychiatry. 2008;13(4):385–397. doi: 10.1038/sj.mp.4002120. [DOI] [PubMed] [Google Scholar]
- 137.Liu J, Yang A, Zhang Q, Yang G, Yang W, Lei H, et al. Association between genetic variants in SLC25A12 and risk of autism spectrum disorders: an integrated meta-analysis. Am J Med Genet B Neuropsychiatr Genet. 2015;168B(4):236–246. doi: 10.1002/ajmg.b.32304. [DOI] [PubMed] [Google Scholar]
- 138.Ramoz N, Reichert JG, Smith CJ, Silverman JM, Bespalova IN, Davis KL, et al. Linkage and association of the mitochondrial aspartate/glutamate carrier SLC25A12 gene with autism. Am J Psychiatry. 2004;161(4):662–669. doi: 10.1176/appi.ajp.161.4.662. [DOI] [PubMed] [Google Scholar]
- 139.Ramoz N, Cai G, Reichert JG, Silverman JM, Buxbaum JD. An analysis of candidate autism loci on chromosome 2q24-q33: evidence for association to the STK39 gene. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(7):1152–1158. doi: 10.1002/ajmg.b.30739. [DOI] [PubMed] [Google Scholar]
- 140.Rabionet R, McCauley JL, Jaworski JM, Ashley-Koch AE, Martin ER, Sutcliffe JS, et al. Lack of association between autism and SLC25A12. Am J Psychiatry. 2006;163(5):929–931. doi: 10.1176/ajp.2006.163.5.929. [DOI] [PubMed] [Google Scholar]
- 141.Segurado R, Conroy J, Meally E, Fitzgerald M, Gill M, Gallagher L. Confirmation of association between autism and the mitochondrial aspartate/glutamate carrier SLC25A12 gene on chromosome 2q31. Am J Psychiatry. 2005;162(11):2182–2184. doi: 10.1176/appi.ajp.162.11.2182. [DOI] [PubMed] [Google Scholar]
- 142.Silverman JM, Buxbaum JD, Ramoz N, Schmeidler J, Reichenberg A, Hollander E, et al. Autism-related routines and rituals associated with a mitochondrial aspartate/glutamate carrier SLC25A12 polymorphism. Am J Med Genet B Neuropsychiatr Genet. 2008;147(3):408–410. doi: 10.1002/ajmg.b.30614. [DOI] [PubMed] [Google Scholar]
- 143.Turunen JA, Rehnstrom K, Kilpinen H, Kuokkanen M, Kempas E, Ylisaukko-Oja T. Mitochondrial aspartate/glutamate carrier SLC25A12 gene is associated with autism. Autism Res. 2008;1(3):189–192. doi: 10.1002/aur.25. [DOI] [PubMed] [Google Scholar]
- 144.Aydin HI. Creatine transporter deficiency in two brothers with autism spectrum disorder. Indian Pediatr. 2018;55(1):67–68. [PubMed] [Google Scholar]
- 145.De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515(7525):209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chiocchetti AG, Haslinger D, Boesch M, Karl T, Wiemann S, Freitag CM, et al. Protein signatures of oxidative stress response in a patient specific cell line model for autism. Mol Autism. 2014;5(1):10. doi: 10.1186/2040-2392-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mahfouz A, Ziats MN, Rennert OM, Lelieveldt BP, Reinders MJ. Shared pathways among autism candidate genes determined by co-expression network analysis of the developing human brain transcriptome. J Mol Neurosci. 2015;57(4):580–594. doi: 10.1007/s12031-015-0641-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Esparham AE, Smith T, Belmont JM, Haden M, Wagner LE, Evans RG, et al. Nutritional and metabolic biomarkers in autism spectrum disorders: an exploratory study. Integr Med (Encinitas). 2015;14(2):40–53. [PMC free article] [PubMed] [Google Scholar]
- 149.Ginsberg MR, Rubin RA, Falcone T, Ting AH, Natowicz MR. Brain transcriptional and epigenetic associations with autism. PLoS One. 2012;7(9):e44736. doi: 10.1371/journal.pone.0044736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kong SW, Shimizu-Motohashi Y, Campbell MG, Lee IH, Collins CD, Brewster SJ, et al. Peripheral blood gene expression signature differentiates children with autism from unaffected siblings. Neurogenetics. 2013;14(2):143–152. doi: 10.1007/s10048-013-0363-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ch’ng C, Kwok W, Rogic S, Pavlidis P. Meta-analysis of gene expression in autism spectrum disorder. Autism Res. 2015;8(5):593–608. doi: 10.1002/aur.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Taurines R, Thome J, Duvigneau JC, Forbes-Robertson S, Yang L, Klampfl K, et al. Expression analyses of the mitochondrial complex I 75-kDa subunit in early onset schizophrenia and autism spectrum disorder: increased levels as a potential biomarker for early onset schizophrenia. Eur Child Adolesc Psychiatry. 2010;19(5):441–448. doi: 10.1007/s00787-009-0074-z. [DOI] [PubMed] [Google Scholar]
- 153.Anitha A, Nakamura K, Thanseem I, Matsuzaki H, Miyachi T, Tsujii M, et al. Downregulation of the expression of mitochondrial electron transport complex genes in autism brains. Brain Pathol. 2013;23(3):294–302. doi: 10.1111/bpa.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tang G, Gutierrez Rios P, Kuo SH, Akman HO, Rosoklija G, Tanji K, et al. Mitochondrial abnormalities in temporal lobe of autistic brain. Neurobiol Dis. 2013;54:349–361. doi: 10.1016/j.nbd.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Perez J, Hill BG, Benavides GA, Dranka BP, Darley-Usmar VM. Role of cellular bioenergetics in smooth muscle cell proliferation induced by platelet-derived growth factor. Biochem J. 2010;428(2):255–267. doi: 10.1042/BJ20100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hill BG, Higdon AN, Dranka BP, Darley-Usmar VM. Regulation of vascular smooth muscle cell bioenergetic function by protein glutathiolation. Biochim Biophys Acta. 2010;1797(2):285–295. doi: 10.1016/j.bbabio.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Dranka BP, Benavides GA, Diers AR, Giordano S, Zelickson BR, Reily C, et al. Assessing bioenergetic function in response to oxidative stress by metabolic profiling. Free Radical Biol Med. 2011;51(9):1621–1635. doi: 10.1016/j.freeradbiomed.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ferrick DA, Neilson A, Beeson C. Advances in measuring cellular bioenergetics using extracellular flux. Drug Discov Today. 2008;13(5–6):268–274. doi: 10.1016/j.drudis.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 159.Rogers GW, Nadanaciva S, Swiss R, Divakaruni AS, Will Y. Assessment of fatty acid beta oxidation in cells and isolated mitochondria. Current protocols in toxicology / editorial board, Mahin D Maines. 2014;60:25 3 1–19. 10.1002/0471140856.tx2503s60. [DOI] [PubMed]
- 160.Dranka BP, Hill BG, Darley-Usmar VM. Mitochondrial reserve capacity in endothelial cells: the impact of nitric oxide and reactive oxygen species. Free Radical Biol Med. 2010;48(7):905–914. doi: 10.1016/j.freeradbiomed.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pecorelli A, Cervellati F, Belmonte G, Montagner G, Waldon P, Hayek J, et al. Cytokines profile and peripheral blood mononuclear cells morphology in Rett and autistic patients. Cytokine. 2016;77:180–188. doi: 10.1016/j.cyto.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 162.Sie L, Loong S, Tan EK. Utility of lymphoblastoid cell lines. J Neurosci Res. 2009;87(9):1953–1959. doi: 10.1002/jnr.22000. [DOI] [PubMed] [Google Scholar]
- 163.James SJ, Rose S, Melnyk S, Jernigan S, Blossom S, Pavliv O, et al. Cellular and mitochondrial glutathione redox imbalance in lymphoblastoid cells derived from children with autism. FASEB J. 2009;23(8):2374–2383. doi: 10.1096/fj.08-128926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Rose S, Melnyk S, Trusty TA, Pavliv O, Seidel L, Li J, et al. Intracellular and extracellular redox status and free radical generation in primary immune cells from children with autism. Autism Res Treat. 2012;2012:986519. doi: 10.1155/2012/986519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.James SJ, Cutler P, Melnyk S, Jernigan S, Janak L, Gaylor DW, et al. Metabolic biomarkers of increased oxidative stress and impaired methylation capacity in children with autism. Am J Clin Nutr. 2004;80(6):1611–1617. doi: 10.1093/ajcn/80.6.1611. [DOI] [PubMed] [Google Scholar]
- 166.James SJ, Melnyk S, Jernigan S, Cleves MA, Halsted CH, Wong DH, et al. Metabolic endophenotype and related genotypes are associated with oxidative stress in children with autism. Am J Med Genet B Neuropsychiatr Genet. 2006;141B(8):947–956. doi: 10.1002/ajmg.b.30366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Melnyk S, Fuchs GJ, Schulz E, Lopez M, Kahler SG, Fussell JJ, et al. Metabolic imbalance associated with methylation dysregulation and oxidative damage in children with autism. J Autism Dev Disord. 2012;42(3):367–377. doi: 10.1007/s10803-011-1260-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chauhan A, Essa MM, Muthaiyah B, Brown WT, Chauhan V. Mitochondrial abnormalities in lymphoblasts from autism: P1-01-03. J Neurochem. 2009;109(Suppl. 1):273. [Google Scholar]
- 169.Rose S, Frye RE, Slattery J, Wynne R, Tippett M, Melnyk S, et al. Oxidative stress induces mitochondrial dysfunction in a subset of autistic lymphoblastoid cell lines. Transl Psychiatry. 2014;4:e377. doi: 10.1038/tp.2014.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Rose S, Frye RE, Slattery J, Wynne R, Tippett M, Pavliv O, et al. Oxidative stress induces mitochondrial dysfunction in a subset of autism lymphoblastoid cell lines in a well-matched case control cohort. PLoS One. 2014;9(1):e85436. doi: 10.1371/journal.pone.0085436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Rose S, Bennuri SC, Wynne R, Melnyk S, James SJ, Frye RE. Mitochondrial and redox abnormalities in autism lymphoblastoid cells: a sibling control study. FASEB J. 2017;31(3):904–909. doi: 10.1096/fj.201601004R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hill BG, Dranka BP, Zou L, Chatham JC, Darley-Usmar VM. Importance of the bioenergetic reserve capacity in response to cardiomyocyte stress induced by 4-hydroxynonenal. Biochem J. 2009;424(1):99–107. doi: 10.1042/BJ20090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Rose S, Melnyk S, Pavliv O, Bai S, Nick TG, Frye RE, et al. Evidence of oxidative damage and inflammation associated with low glutathione redox status in the autism brain. Transl Psychiatry. 2012;2:e134. doi: 10.1038/tp.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Rose S, Wynne R, Frye RE, Melnyk S, James SJ. Increased susceptibility to ethylmercury-induced mitochondrial dysfunction in a subset of autism lymphoblastoid cell lines. J Toxicol. 2015;2015:573701. doi: 10.1155/2015/573701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Frye RE, Rose S, Chacko J, Wynne R, Bennuri SC, Slattery JC, et al. Modulation of mitochondrial function by the microbiome metabolite propionic acid in autism and control cell lines. Transl Psychiatry. 2016;6(10):e927. doi: 10.1038/tp.2016.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Frye RE, Rose S, Wynne R, Bennuri SC, Blossom S, Gilbert KM, et al. Oxidative stress challenge uncovers trichloroacetaldehyde hydrate-induced mitoplasticity in autistic and control lymphoblastoid cell lines. Sci Rep. 2017;7(1):4478. doi: 10.1038/s41598-017-04821-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Boccuto L, Chen CF, Pittman AR, Skinner CD, McCartney HJ, Jones K, et al. Decreased tryptophan metabolism in patients with autism spectrum disorders. Mol Autism. 2013;4(1):16. doi: 10.1186/2040-2392-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bu X, Wu LuX, Yang L, Xu X, Wang J, et al. Role of SIRT1/PGC-1alpha in mitochondrial oxidative stress in autistic spectrum disorder. Neuropsychiatr Dis Treat. 2017;13:1633–1645. doi: 10.2147/NDT.S129081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zhang L, Ou J, Xu X, Peng Y, Guo H, Pan Y, et al. AMPD1 functional variants associated with autism in Han Chinese population. Eur Arch Psychiatry Clin Neurosci. 2015;265(6):511–517. doi: 10.1007/s00406-014-0524-6. [DOI] [PubMed] [Google Scholar]
- 180.Minshew NJ, Goldstein G, Dombrowski SM, Panchalingam K, Pettegrew JW. A preliminary 31P MRS study of autism: evidence for undersynthesis and increased degradation of brain membranes. Biol Psychiat. 1993;33(11–12):762–773. doi: 10.1016/0006-3223(93)90017-8. [DOI] [PubMed] [Google Scholar]
- 181.Golomb BA, Erickson LC, Scott-Van Zeeland AA, Koperski S, Haas RH, Wallace DC, et al. Assessing bioenergetic compromise in autism spectrum disorder with 31P magnetic resonance spectroscopy: preliminary report. J Child Neurol. 2014;29(2):187–193. doi: 10.1177/0883073813498466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ipser JC, Syal S, Bentley J, Adnams CM, Steyn B, Stein DJ. 1H-MRS in autism spectrum disorders: a systematic meta-analysis. Metab Brain Dis. 2012;27(3):275–287. doi: 10.1007/s11011-012-9293-y. [DOI] [PubMed] [Google Scholar]
- 183.Fayed N, Modrego PJ. Comparative study of cerebral white matter in autism and attention-deficit/hyperactivity disorder by means of magnetic resonance spectroscopy. Acad Radiol. 2005;12(5):566–569. doi: 10.1016/j.acra.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 184.Murphy DG, Critchley HD, Schmitz N, McAlonan G, Van Amelsvoort T, Robertson D, et al. Asperger syndrome: a proton magnetic resonance spectroscopy study of brain. Arch Gen Psychiatry. 2002;59(10):885–891. doi: 10.1001/archpsyc.59.10.885. [DOI] [PubMed] [Google Scholar]
- 185.Aoki Y, Kasai K, Yamasue H. Age-related change in brain metabolite abnormalities in autism: a meta-analysis of proton magnetic resonance spectroscopy studies. Transl Psychiatry. 2012;2:e69. doi: 10.1038/tp.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Goh S, Dong Z, Zhang Y, DiMauro S, Peterson BS. Mitochondrial dysfunction as a neurobiological subtype of autism spectrum disorder: evidence from brain imaging. JAMA Psychiatry. 2014;71(6):665–671. doi: 10.1001/jamapsychiatry.2014.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Corrigan NM, Shaw DW, Richards TL, Estes AM, Friedman SD, Petropoulos H, et al. Proton magnetic resonance spectroscopy and MRI reveal no evidence for brain mitochondrial dysfunction in children with autism spectrum disorder. J Autism Dev Disord. 2012;42(1):105–115. doi: 10.1007/s10803-011-1216-y. [DOI] [PubMed] [Google Scholar]
- 188.Dager SR, Corrigan NM, Shaw DW. Brain lactate as a potential biomarker for comorbid anxiety disorder in autism spectrum disorder. JAMA Psychiatry. 2015;72(2):190. doi: 10.1001/jamapsychiatry.2014.2419. [DOI] [PubMed] [Google Scholar]
- 189.Peterson BS, Goh S, Dong Z. Brain lactate as a potential biomarker for comorbid anxiety disorder in autism spectrum disorder-reply. JAMA Psychiatry. 2015;72(2):190–191. doi: 10.1001/jamapsychiatry.2014.2425. [DOI] [PubMed] [Google Scholar]
- 190.Rossignol DA, Frye RE. Substantial problems with measuring brain mitochondrial dysfunction in autism spectrum disorder using magnetic resonance spectroscopy. J Autism Dev Disord. 2012;42(4):640–642. doi: 10.1007/s10803-011-1276-z. [DOI] [PubMed] [Google Scholar]
- 191.Dager SR, Corrigan NM, Estes A, Shaw DWW. Further commentary on mitochondrial dysfunction in autism spectrum disorder: assessment and treatment considerations. J Autism Dev Disord. 2011;42(4):643–646. [Google Scholar]
- 192.Chauhan A, Gu F, Essa MM, Wegiel J, Kaur K, Brown WT, et al. Brain region-specific deficit in mitochondrial electron transport chain complexes in children with autism. J Neurochem. 2011;117(2):209–220. doi: 10.1111/j.1471-4159.2011.07189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Gu F, Chauhan V, Kaur K, Brown WT, LaFauci G, Wegiel J, et al. Alterations in mitochondrial DNA copy number and the activities of electron transport chain complexes and pyruvate dehydrogenase in the frontal cortex from subjects with autism. Transl Psychiatry. 2013;3:e299. doi: 10.1038/tp.2013.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Frye RE, Rose S, Slattery J, MacFabe DF. Gastrointestinal dysfunction in autism spectrum disorder: the role of the mitochondria and the enteric microbiome. Microb Ecol Health Dis. 2015;26:27458. doi: 10.3402/mehd.v26.27458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Rose S, Bennuri SC, Murray KF, Buie T, Winter H, Frye RE. Mitochondrial dysfunction in the gastrointestinal mucosa of children with autism: a blinded case-control study. PLoS One. 2017;12(10):e0186377. doi: 10.1371/journal.pone.0186377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Schaefer GB, Mendelsohn NJ. Genetics evaluation for the etiologic diagnosis of autism spectrum disorders. Genet Med. 2008;10(1):4–12. doi: 10.1097/GIM.0b013e31815efdd7. [DOI] [PubMed] [Google Scholar]
- 197.Chauhan A, Audhya T, Chauhan V. Brain region-specific glutathione redox imbalance in autism. Neurochem Res. 2012;37(8):1681–1689. doi: 10.1007/s11064-012-0775-4. [DOI] [PubMed] [Google Scholar]
- 198.Rossignol DA, Frye RE. Evidence linking oxidative stress, mitochondrial dysfunction, and inflammation in the brain of individuals with autism. Front Physiol. 2014;5:150. doi: 10.3389/fphys.2014.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Damodaran LP, Arumugam G. Urinary oxidative stress markers in children with autism. Redox Rep Commun Free Radical Res. 2011;16(5):216–222. doi: 10.1179/1351000211Y.0000000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Waly MI, Hornig M, Trivedi M, Hodgson N, Kini R, Ohta A, et al. Prenatal and postnatal epigenetic programming: implications for GI, immune, and neuronal function in autism. Autism Res Treat. 2012;2012:190930. doi: 10.1155/2012/190930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.James SJ, Shpyleva S, Melnyk S, Pavliv O, Pogribny IP. Complex epigenetic regulation of engrailed-2 (EN-2) homeobox gene in the autism cerebellum. Transl Psychiatry. 2013;3:e232. doi: 10.1038/tp.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Howsmon DP, Kruger U, Melnyk S, James SJ, Hahn J. Classification and adaptive behavior prediction of children with autism spectrum disorder based upon multivariate data analysis of markers of oxidative stress and DNA methylation. PLoS Comput Biol. 2017;13(3):e1005385. doi: 10.1371/journal.pcbi.1005385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Frye RE, Delatorre R, Taylor H, Slattery J, Melnyk S, Chowdhury N, et al. Redox metabolism abnormalities in autistic children associated with mitochondrial disease. Transl Psychiatry. 2013;3:e273. doi: 10.1038/tp.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Desler C, Hansen TL, Frederiksen JB, Marcker ML, Singh KK, Juel Rasmussen L. Is there a link between mitochondrial reserve respiratory capacity and aging? J Aging Res. 2012;2012:192503. doi: 10.1155/2012/192503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Sansbury BE, Jones SP, Riggs DW, Darley-Usmar VM, Hill BG. Bioenergetic function in cardiovascular cells: the importance of the reserve capacity and its biological regulation. Chem Biol Interact. 2011;191(1–3):288–295. doi: 10.1016/j.cbi.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Nicholls DG. Spare respiratory capacity, oxidative stress and excitotoxicity. Biochem Soc Trans. 2009;37(Pt 6):1385–1388. doi: 10.1042/BST0371385. [DOI] [PubMed] [Google Scholar]
- 207.Yadava N, Nicholls DG. Spare respiratory capacity rather than oxidative stress regulates glutamate excitotoxicity after partial respiratory inhibition of mitochondrial complex I with rotenone. J Neurosci. 2007;27(27):7310–7317. doi: 10.1523/JNEUROSCI.0212-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Palmieri L, Papaleo V, Porcelli V, Scarcia P, Gaita L, Sacco R, et al. Altered calcium homeostasis in autism-spectrum disorders: evidence from biochemical and genetic studies of the mitochondrial aspartate/glutamate carrier AGC1. Mol Psychiatry. 2010;15(1):38–52. doi: 10.1038/mp.2008.63. [DOI] [PubMed] [Google Scholar]
- 209.Sharpe MA, Gist TL, Baskin DS. B-lymphocytes from a population of children with autism spectrum disorder and their unaffected siblings exhibit hypersensitivity to thimerosal. J Toxicol. 2013;2013:801517. doi: 10.1155/2013/801517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Blossom SJ, Cooney CA, Melnyk SB, Rau JL, Swearingen CJ, Wessinger WD. Metabolic changes and DNA hypomethylation in cerebellum are associated with behavioral alterations in mice exposed to trichloroethylene postnatally. Toxicol Appl Pharmacol. 2013;269(3):263–269. doi: 10.1016/j.taap.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Blossom SJ, Gilbert KM. Exposure to a metabolite of the environmental toxicant, trichloroethylene, attenuates CD4+ T cell activation-induced cell death by metalloproteinase-dependent FasL shedding. Toxicol Sci. 2006;92(1):103–114. doi: 10.1093/toxsci/kfj212. [DOI] [PubMed] [Google Scholar]
- 212.Martinez TN, Greenamyre JT. Toxin models of mitochondrial dysfunction in Parkinson’s disease. Antioxid Redox Signal. 2012;16(9):920–934. doi: 10.1089/ars.2011.4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Rossignol DA, Genuis SJ, Frye RE. Environmental toxicants and autism spectrum disorders: a systematic review. Transl Psychiatry. 2014;4:e360. doi: 10.1038/tp.2014.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Kaur K, Chauhan V, Gu F, Chauhan A. Bisphenol A induces oxidative stress and mitochondrial dysfunction in lymphoblasts from children with autism and unaffected siblings. Free Rad Biol Med. 2014;76:25–33. doi: 10.1016/j.freeradbiomed.2014.07.030. [DOI] [PubMed] [Google Scholar]
- 215.Slattery J, MacFabe DF, Frye RE. The significance of the enteric microbiome on the development of childhood disease: a review of prebiotic and probiotic therapies in disorders of childhood. Clin Med Insights Pediatr. 2016;10:91–107. doi: 10.4137/CMPed.S38338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Rosenfeld CS. Microbiome disturbances and autism spectrum disorders. Drug Metab Dispos. 2015;43(10):1557–1571. doi: 10.1124/dmd.115.063826. [DOI] [PubMed] [Google Scholar]
- 217.Williams BL, Hornig M, Buie T, Bauman ML, Cho Paik M, Wick I, et al. Impaired carbohydrate digestion and transport and mucosal dysbiosis in the intestines of children with autism and gastrointestinal disturbances. PLoS One. 2011;6(9):e24585. doi: 10.1371/journal.pone.0024585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Finegold SM, Molitoris D, Song Y, Liu C, Vaisanen ML, Bolte E, et al. Gastrointestinal microflora studies in late-onset autism. Clin Infect Dis. 2002;35(Suppl 1):S6–S16. doi: 10.1086/341914. [DOI] [PubMed] [Google Scholar]
- 219.MacFabe DF, Cain NE, Boon F, Ossenkopp KP, Cain DP. Effects of the enteric bacterial metabolic product propionic acid on object-directed behavior, social behavior, cognition, and neuroinflammation in adolescent rats: relevance to autism spectrum disorder. Behav Brain Res. 2011;217(1):47–54. doi: 10.1016/j.bbr.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 220.Nankova BB, Agarwal R, MacFabe DF, La Gamma EF. Enteric bacterial metabolites propionic and butyric acid modulate gene expression, including CREB-dependent catecholaminergic neurotransmission, in PC12 cells–possible relevance to autism spectrum disorders. PLoS One. 2014;9(8):e103740. doi: 10.1371/journal.pone.0103740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Rose S, Bennuri SC, Davis JE, Wynne R, Slattery JC, Tippett M, et al. Butyrate enhances mitochondrial function during oxidative stress in cell lines from boys with autism. Transl Psychiatry. 2018;8(1):42. doi: 10.1038/s41398-017-0089-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Freedenfeld S, Hamada K, Audhya T, Adams J. Biochemical effects of ribose and NADH therapy in children with autism. Autism Insights. 2011;3:3–13. [Google Scholar]
- 223.Gvozdjakova A, Kucharska J, Ostatnikova D, Babinska K, Nakladal D, Crane FL. Ubiquinol improves symptoms in children with autism. Oxidative Med Cell Longev. 2014;2014:798957. doi: 10.1155/2014/798957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Mousavinejad E, Ghaffari MA, Riahi F, Hajmohammadi M, Tiznobeyk Z, Mousavinejad M. Coenzyme Q10 supplementation reduces oxidative stress and decreases antioxidant enzyme activity in children with autism spectrum disorders. Psychiatry Res. 2018;265:62–69. doi: 10.1016/j.psychres.2018.03.061. [DOI] [PubMed] [Google Scholar]
- 225.Frye RE. Novel cytochrome B gene mutations causing mitochondrial disease in autism. J Pediatr Neurol. 2012;10:35–40. [Google Scholar]
- 226.Frye RE, Naviaux RK. Autistic disorder with complex IV overactivity: a new mitochondrial syndrome. Journal of Pediatric Neurology. 2011;9:427–434. [Google Scholar]
- 227.Frye RE, Slattery J. The potential role of nitrous oxide in the etiology of autism spectrum disorder. Transl Psychiatry. 2016;6:e812. doi: 10.1038/tp.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Rose S, Frye RE, Slattery J, Wynne R, Tippett M, Melnyk S, et al. Oxidative stress induces mitochondrial dysfunction in a subset of autistic lymphoblastoid cell lines. Transl Psychiatry. 2015;5:e526. doi: 10.1038/tp.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Zerbo O, Leong A, Barcellos L, Bernal P, Fireman B, Croen LA. Immune mediated conditions in autism spectrum disorders. Brain Behav Immun. 2015;46:232–236. doi: 10.1016/j.bbi.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Comi AM, Zimmerman AW, Frye VH, Law PA, Peeden JN. Familial clustering of autoimmune disorders and evaluation of medical risk factors in autism. J Child Neurol. 1999;14(6):388–394. doi: 10.1177/088307389901400608. [DOI] [PubMed] [Google Scholar]
- 231.Gupta S, Aggarwal S, Heads C. Dysregulated immune system in children with autism: beneficial effects of intravenous immune globulin on autistic characteristics. J Autism Dev Disord. 1996;26(4):439–452. doi: 10.1007/BF02172828. [DOI] [PubMed] [Google Scholar]
- 232.Napoli E, Ross-Inta C, Wong S, Hung C, Fujisawa Y, Sakaguchi D, et al. Mitochondrial dysfunction in Pten haplo-insufficient mice with social deficits and repetitive behavior: interplay between Pten and p53. PLoS One. 2012;7(8):e42504. doi: 10.1371/journal.pone.0042504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Weinberg SE, Sena LA, Chandel NS. Mitochondria in the regulation of innate and adaptive immunity. Immunity. 2015;42(3):406–417. doi: 10.1016/j.immuni.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Parvaneh N, Quartier P, Rostami P, Casanova JL, de Lonlay P. Inborn errors of metabolism underlying primary immunodeficiencies. J Clin Immunol. 2014;34(7):753–771. doi: 10.1007/s10875-014-0076-6. [DOI] [PubMed] [Google Scholar]
- 235.Morris G, Berk M. The many roads to mitochondrial dysfunction in neuroimmune and neuropsychiatric disorders. BMC Med. 2015;13:68. doi: 10.1186/s12916-015-0310-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ramaekers VT, Blau N. Cerebral folate deficiency. Dev Med Child Neurol. 2004;46(12):843–851. doi: 10.1017/s0012162204001471. [DOI] [PubMed] [Google Scholar]
- 237.Ramaekers VT, Hansen SI, Holm J, Opladen T, Senderek J, Hausler M, et al. Reduced folate transport to the CNS in female Rett patients. Neurology. 2003;61(4):506–515. doi: 10.1212/01.wnl.0000078939.64774.1b. [DOI] [PubMed] [Google Scholar]
- 238.Ramaekers VT, Blau N, Sequeira JM, Nassogne MC, Quadros EV. Folate receptor autoimmunity and cerebral folate deficiency in low-functioning autism with neurological deficits. Neuropediatrics. 2007;38(6):276–281. doi: 10.1055/s-2008-1065354. [DOI] [PubMed] [Google Scholar]
- 239.Shoffner J, Trommer B, Thurm A, Farmer C, Langley WA, 3rd, Soskey L, et al. CSF concentrations of 5-methyltetrahydrofolate in a cohort of young children with autism. Neurology. 2016;86(24):2258–2263. doi: 10.1212/WNL.0000000000002766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Frye RE, Naviaux RK. Autistic disorder with complex IV overactivity: a new mitochondrial syndrome. J Ped Neuro. 2011;9(4):427–434. [Google Scholar]
- 241.Frye RE, Sequeira JM, Quadros EV, James SJ, Rossignol DA. Cerebral folate receptor autoantibodies in autism spectrum disorder. Mol Psychiatry. 2013;18(3):369–381. doi: 10.1038/mp.2011.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Frye RE, Slattery J, Delhey L, Furgerson B, Strickland T, Tippett M, et al. Folinic acid improves verbal communication in children with autism and language impairment: a randomized double-blind placebo-controlled trial. Mol Psychiatry. 2018;23(2):247–256. doi: 10.1038/mp.2016.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Morscher RJ, Ducker GS, Li SH, Mayer JA, Gitai Z, Sperl W, et al. Mitochondrial translation requires folate-dependent tRNA methylation. Nature. 2018;554(7690):128–132. doi: 10.1038/nature25460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Ming X, Patel R, Kang V, Chokroverty S, Julu PO. Respiratory and autonomic dysfunction in children with autism spectrum disorders. Brain Dev. 2016;38(2):225–232. doi: 10.1016/j.braindev.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 245.Zelnik N, Axelrod FB, Leshinsky E, Griebel ML, Kolodny EH. Mitochondrial encephalomyopathies presenting with features of autonomic and visceral dysfunction. Pediatr Neurol. 1996;14(3):251–254. doi: 10.1016/0887-8994(96)00046-x. [DOI] [PubMed] [Google Scholar]
- 246.Brown BD, Rais T. Autism in the son of a woman with mitochondrial myopathy and dysautonomia: a case report. Innov Clin Neurosci. 2015;12(9–10):29–32. [PMC free article] [PubMed] [Google Scholar]
- 247.Fluegge K. In regard to “Autism in the son of a woman with mitochondrial myopathy and dysautonomia: a case report”. Innov Clin Neurosci. 2016;13(5–6):13–14. [PMC free article] [PubMed] [Google Scholar]
- 248.Frye RE, Nankova B, Bhattacharyya S, Rose S, Bennuri SC, MacFabe DF. Modulation of immunological pathways in autistic and neurotypical lymphoblastoid cell lines by the enteric microbiome metabolite propionic acid. Front Immunol. 2017;8:1670. doi: 10.3389/fimmu.2017.01670. [DOI] [PMC free article] [PubMed] [Google Scholar]