Abstract
Background
Trastuzumab emtansine (T-DM1) is approved for the treatment of patients with human epidermal growth factor receptor 2 (HER2)-positive advanced breast cancer (ABC), and has high efficacy. However, some patients exhibit primary resistance to T-DM1, and thus methods that can predict resistance in clinical practice are needed. Genomic analysis of circulating tumor DNA (ctDNA) in plasma is a non-invasive and reproducible method. This study aimed to predict primary resistance to T-DM1 by combining genomic analysis of ctDNA and other clinicopathological features of patients with HER2-positive ABC.
Methods
The study population comprised 34 patients with HER2-positive ABC who had been treated with T-DM1. Correlations between clinicopathological characteristics of patients and primary resistance to T-DM1 were examined, and HER2 gene copy number and PIK3CA gene mutations were analyzed using plasma ctDNA samples obtained from 16 patients before T-DM1 administration.
Results
Among the 34 patients, nine (26.5%) had progressive disease at the first efficacy analysis; these patients were considered to have primary resistance to T-DM1. No significant difference was found in the rate of primary resistance to T-DM1 between groups. Among 16 patients whose ctDNA was analyzed, four showed primary resistance to T-DM1. These four patients showed negative HER2 gene amplification in ctDNA and were ER-positive and/or PR-positive by immunohistochemistry.
Conclusions
HER2 gene amplification in ctDNA and ER and PR status may predict primary resistance to T-DM1. A liquid biopsy before the initiation of T-DM1 treatment could be a non-invasive way to predict whether a patient would exhibit primary resistance to T-DM1.
Keywords: HER2, Trastuzumab emtansine (T-DM1), Primary resistance, Circulating tumor DNA (ctDNA)
Introduction
T-DM1 is an antibody–drug conjugate that combines the human epidermal growth factor receptor type 2 (HER2)-targeted antitumor properties of trastuzumab with the maytansinoid, DM1, a potent microtubule-disrupting agent, joined by a stable linker [1]. T-DM1 significantly improved progression-free survival (PFS) and overall survival (OS) compared to the combination of capecitabine and lapatinib in the phase III EMILIA study [2], and has become a standard treatment for patients with HER2-positive ABC previously treated with trastuzumab and taxanes in accordance with ASCO guidelines [3]. In the phase III MARIANNE trial, patients with previously untreated HER2-positive locally advanced or metastatic breast cancer were randomized to trastuzumab plus a taxane (docetaxel or paclitaxel), T-DM1 plus placebo, or T-DM1 plus pertuzumab. Both T-DM1-containing regimens showed non-inferiority, but not superiority, in terms of PFS over trastuzumab plus a taxane. The objective response rate was 67.9% with trastuzumab plus a taxane, 59.7% with T-DM1 plus placebo, and 64.2% with T-DM1 plus pertuzumab [4]. These results implied that a significant proportion of patients experience disease progression without any signs of tumor regression or stabilization of disease despite early treatment with T-DM1, and the primary resistance rate to T-DM1 is probably as high as 30%. In contrast, patients whose tumors responded to T-DM1 tended to have durable responses with favorable toxicity profiles and QOL compared to the comparator [4]. Thus, if patients with tumors that are primarily resistant to T-DM1 are excluded, the drug could have a greater therapeutic index.
Several mechanisms of resistance to HER2-directed therapies have been described in preclinical studies, including the accumulation of 95HER2, truncated ERBB2 receptors which lack an extracellular trastuzumab-binding domain [5], loss of PTEN which activates the phosphoinositide 3-kinase (PI3K) pathway [6], mutations in phosphoinositide-3-kinase, catalytic, alpha polypeptide (PIK3CA) [7, 8], heregulin-dependent-HER2/HER3 signaling [9, 10], and signaling through insulin-like growth factor 1 receptor [11]. However, low expression of HER2 mRNA or HER2 protein is the only clinically definite biomarker related to low efficacy of T-DM1 [12, 13]. Methods of predicting drug resistance that are feasible in clinical settings are highly desired.
Cell-free DNA (cfDNA) refers to fragmented DNA from cells that circulate in blood [14–16]. cfDNA in cancer patients include tumor-derived DNA (circulating tumor DNA: ctDNA), which can harbor tumor-specific genetic changes. Genomic analysis of ctDNA in plasma is non-invasive and reproducible, and can be used to elucidate mechanisms underlying drug resistance.
This study aimed to predict primary resistance to T-DM1 by combining genomic analysis of ctDNA and other clinicopathological features of patients with HER2-positive ABC.
Patients and methods
Patients and data collection
Electronic medical records at Kindai University Hospital were used to identify breast cancer patients who received T-DM1 from May 1, 2015 to April 30, 2017 at the Department of Surgery or Department of Medical Oncology. There were 34 eligible patients and no patient was excluded. Therefore, 34 patients were included in the study. The following data were collected: patient demographics, including date of birth and sex; diagnoses; and tumor characteristics, including stage (TNM classification, 7th edition), pathological diagnosis, estrogen receptor (ER) status, progesterone receptor (PR) status, and HER2 status. Hormonal receptor-positive samples were defined as those in which ≥ 10% of cells were positive by immunohistological staining. HER2 positivity was assessed by immunohistochemistry (IHC). In cases with IHC2+, the fluorescent in situ hybridization (FISH) test was performed. Treatment history was also collected, including information on neoadjuvant or adjuvant treatment regimens, chemotherapy and hormonal therapy before T-DM1 for metastatic disease and tumor response, and number and type of chemotherapy for metastatic breast cancer. Tumor response to treatment was determined by the treating physician using Response Evaluation Criteria in Solid Tumors (RECIST). In cases of recurrent metastatic disease, the first chemotherapy administered was defined as first-line chemotherapy, whereas in cases of primary stage IV disease, overall first chemotherapy was defined as first-line chemotherapy. Maintenance anti-HER2 therapy was not counted as a treatment. For example, paclitaxel plus trastuzumab followed by trastuzumab was counted as one regimen.
The date of T-DM1 treatments and tumor response to T-DM1 using computed tomography (CT) and RECIST v1.1 [17] were recorded. A patient was considered to have primary resistance to T-DM1 when progressive disease (PD) was recorded in the first evaluation of T-DM1 response.
This study was approved by the ethics committee of Kindai University Faculty of Medicine. Written informed consent is mandatory for collecting ctDNA at our institution. However, patients were given the opportunity to opt-out of ctDNA collection and provide only information via a website.
Circulating tumor DNA analysis
HER2 gene copy number and PIK3CA gene mutations were analyzed using ctDNA from 16 patients who provided written informed consent before T-DM1 administration. Seven milliliters of venous blood were collected before the patients were administered T-DM1. EDTA-treated blood samples were centrifuged and plasma supernatant was stored at − 80 °C until analysis. DNA was purified from plasma with the cobas® ctDNA Sample Preparation Kit (Roche Diagnostics, Pleasanton, CA). The copy number of HER2 and PIK3CA mutations was determined using the QX100 Droplet Digital PCR System in accordance with the manufacturer’s instructions (Bio-Rad, Hercules, CA, USA). The relative HER2 copy number ratio was calculated relative to the control Elongation factor Tu GTP-binding domain 2 (EFTUD2), as previously described [18].
Primer sequences used in the HER2 copy number analysis were as follows: HER2 forward, 5′-ACAACCAAGTGAGGCAGGTC-3′; HER2 reverse, 5′-GTATTGTTCAGCGGGTCTCC-3′; HER2 probe, 5′-/56-FAM/AGGCACCCA/ZEN/GCTCTTTGAGGACAAC/3IABkFQ/-3′; EFTUD2 forward, 5′-GGTCTTGCCAGACACCAAAG-3′; EFTUD2 reverse, 5′-TGAGAGGACACACGCAAAAC-3′; EFTUD2 probe, 5′-/5HEX/TCCAGGTAG/ZEN/GACATCCTTTGGCTTT/3IABkFQ/-3′ [18].
PCR was performed using the following cycling conditions: 95 °C for 10 min, 40 cycles of 94 °C for 30 s and 60 °C for 60 s, followed by enzyme deactivation at 98 °C for 10 min. For the PIK3CA mutation assay, primers and probes for detecting PIK3CA mutations E542K, E545K, and H1047R were purchased from Bio-Rad. PCR was performed using the following cycling conditions: 95 °C for 10 min, 40 cycles of 94 °C for 30 s and 55 °C for 60 s, followed by enzyme deactivation at 98 °C for 10 min. After thermal cycling, plates were transferred to a Droplet reader. Digital PCR data were analyzed using the QuantaSoft analytical software package (Bio-Rad). The copy number of each gene was estimated from the Poisson distribution. Positive HER2 amplification with digital PCR was defined as a HER2 ratio (HER2/EFTUD2 copy number ratio) of 1.25, as described in a previous study [18].
Minimum detection limit of the assay and determination of cut-off values
The digital PCR method showed minimum detection limits of 0.03% for PIK3CA E542K, 0.03% for E545K, and 0.03% for H1047R using plasmid DNA (data not shown). For plasma samples, cut-off values were determined using samples from healthy volunteers. The cut-off value for plasma ctDNA samples was set at 3 copies for all PIK3CA mutations, because the background noise in plasma ctDNA from 10 healthy volunteers was exactly zero. For HER2 copy number, the cut-off value for plasma ctDNA samples was set at 1.25, as described in a previous study [18].
Statistical analysis
Descriptive statistics were used to examine demographics of patients and tumor characteristics. Fisher’s exact test was used to compare the rate of primary resistance to T-DM1 in two independent groups. Time to progression (TTP), which was defined as the number of days from T-DM1 initiation to objective disease progression, was estimated by the Kaplan–Meier method. An unstratified log-rank test was used to compare TTP between groups.
All tests were two tailed, with p < 0.05 considered statistically significant. All survey data were coded and analyzed with the use of standard EZR (Saitama Medical Center, Jichi Medical University), a graphical user interface for R (The R Foundation for Statistical Computing, version 2.13.0). More precisely, it is a modified version of R commander designed to add statistical functions frequently used in biostatistics [19].
Results
Patients’ characteristics and response to T-DM1
A total of 34 patients with HER2-positive metastatic breast cancer who had been treated with T-DM1 were included in this study. Plasma samples were obtained from 16 patients who provided written informed consent for ctDNA analysis. Demographic and disease characteristics of the study population are summarized in Table 1. Only 26.5% of patients had primary stage IV breast cancer. HER2 status of the primary site was IHC 3+ in 30 patients (88.2%) and IHC 2+ and FISH positive in four patients (11.8%). Of these 34 patients, 52.9% had ER-positive and/or PR-positive breast cancer. Fifty percent of patients were administered T-DM1 as first- or second-line chemotherapy for metastatic disease. All but one patient had received T-DM1 in the neoadjuvant, adjuvant, or metastatic setting. Table 1 also summarizes prior therapies (before T-DM1) for metastatic disease.
Table 1.
Demographic and disease characteristics of the study population (N = 34)
| Characteristics | No. (%) |
|---|---|
| Age, years | |
| Median (range) | 62 (38–84) |
| Stage IV or recurrent | |
| Stage IV | 9 (26.5) |
| Recurrent | 25 (73.5) |
| Pathology | |
| Invasive ductal carcinoma | 32 (94.1) |
| Medullary carcinoma | 1 (2.9) |
| Poorly differentiated adenocarcinoma | 1 (2.9) |
| HER2 status of primary site | |
| IHC 3+ | 30 (88.2) |
| IHC 2+ and FISH+ | 4 (11.8) |
| Hormone receptor status | |
| ER and/or PR positive | 18 (52.9) |
| ER and PR negative | 16 (47.1) |
| Neoadjuvant or adjuvant chemotherapy | |
| Anthracycline followed by taxane + trastuzumab | 8 (23.5) |
| Taxane + trastuzumab | 4 (11.8) |
| Trastuzumab without chemotherapy | 2 (5.9) |
| Anthracycline followed by trastuzumab | 1 (2.9) |
| Anthracycline followed by taxane | 1 (2.9) |
| Taxane | 1 (2.9) |
| UFT + trastuzumab | 1 (2.9) |
| CMF | 1 (2.9) |
| Doxifluridine | 1 (2.9) |
| Neoadjuvant or adjuvant endocrine therapy | |
| Yes | 10 (29.4) |
| Prior lines of chemotherapy for metastatic disease before T-DM1 | |
| 0 | 2 (5.9) |
| 1 | 15 (44.1) |
| 2 | 4 (11.8) |
| 3 | 6 (17.6) |
| 4 | 3 (8.8) |
| ≥ 5 | 4 (11.8) |
| Type of prior therapy for metastatic disease before T-DM1 | |
| Trastuzumab | 29 (85.3) |
| Taxane | 20 (58.8) |
| Pertuzumab | 15 (44.1) |
| Capecitabine | 15 (44.1) |
| Hormonal therapy | 12 (35.3) |
| Lapatinib | 11 (32.4) |
| Vinorelbine | 7 (20.6) |
| S-1 | 4 (11.7) |
| Eribulin | 3 (8.8) |
| Cyclophosphamide | 2 (5.8) |
| Others | 4 (11.7) |
IHC immunohistochemistry; FISH fluorescent in situ hybridization; CMF cyclophosphamide, methotrexate, and fluorouracil
All 34 patients were available for efficacy analysis. Best response to T-DM1 is shown in Table 2. The response rate was 26.5%. Nine patients (26.5%) had progressive disease at the first efficacy analysis, and were considered to have primary resistance to T-DM1.
Table 2.
Best response to T-DM1
| Response | No. of patients (%) |
|---|---|
| CR + PR | 9 (26.5) |
| CR | 2 (5.9) |
| PR | 7 (20.6) |
| SD | 13 (38.2) |
| PD | 9 (26.5) |
| Unknown | 3 (8.8) |
| Total | 34 |
CR complete response, PR partial response, SD stable disease, PD progressive disease
HER2 copy number gain and PIK3CA mutations in plasma samples
Results of HER2 copy number and PIK3CA mutation analyses of plasma samples are summarized in Table 3. For both the HER2 copy number assay and PIK3CA mutation assay, all plasma ctDNA samples could be evaluated, since the copy number of the reference gene (EFTUD2) was more than 100. HER2 copy number gains were observed in 5 of 16 patients (31%). The PIK3CA E545K mutation was detected in 2 of 16 patients (13%), while the PIK3CA E542K mutation and H1047R mutation were not detected.
Table 3.
HER2 copy number gain and PIK3CA mutations in plasma samples
| No. | Relative quantitation of HER2 gene | PIK3CA E542K-MUT | PIK3CA E542K-WT | PIK3CA E545K-MUT | PIK3CA E545K-WT | PIK3CA H1047R-MUT | PIK3CA H1047R-WT |
|---|---|---|---|---|---|---|---|
| (copies) | |||||||
| 1 | 0.91 | 0 | 686 | 0 | 726 | 0 | 674 |
| 2 | 1.1 | 1.6 | 344 | 0 | 390 | 1.6 | 398 |
| 3 | 0.95 | 0 | 396 | 0 | 480 | 0 | 402 |
| 4 | 0.93 | 0 | 202 | 0 | 222 | 0 | 186 |
| 5 | 1.02 | 0 | 552 | 0 | 592 | 0 | 632 |
| 6 | 1.11 | 0 | 1264 | 18a | 1174 | 0 | 1210 |
| 7 | 1.11 | 0 | 814 | 0 | 864 | 2.4 | 818 |
| 8 | 1.06 | 0 | 612 | 0 | 642 | 0 | 508 |
| 9 | 2.14a | 0 | 1066 | 0 | 1000 | 0 | 990 |
| 10 | 4.23a | 0 | 572 | 0 | 628 | 0 | 556 |
| 11 | 0.93 | 1.8 | 4000 | 4.6a | 3860 | 0 | 4140 |
| 12 | 1.45a | 0 | 480 | 0 | 522 | 0 | 482 |
| 13 | 7.56a | 0 | 1680 | 1.6 | 1620 | 0 | 1480 |
| 14 | 1.23 | 0 | 144 | 0 | 220 | 0 | 248 |
| 15 | 1.82a | 0 | 502 | 0 | 552 | 0 | 570 |
| 16 | 1.14 | 0 | 512 | 0 | 540 | 0 | 544 |
HER2 human epidermal growth factor receptor 2; PIK3CA phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha; WT wild type; MUT mutant
aGreater than cut-off value
Predictors of primary resistance to T-DM1
The rate of primary resistance to T-DM1 was compared between the following groups: recurrent vs. primary stage IV, ER-positive and/or PR-positive vs. ER-negative and PR-negative, T-DM1 as second-line therapy or earlier vs. T-DM1 as later than second-line therapy, prior treatment with anthracycline vs. no prior treatment with anthracycline, prior treatment with pertuzumab vs. no prior treatment with pertuzumab, prior treatment with lapatinib vs. no prior treatment with lapatinib, HER2 gene amplification positive vs. negative, and PIK3CA mutation positive vs. negative (Table 4). Among these, those with ER-positive and/or PR-positive disease were more likely to have primary resistance to T-DM1 than those with ER-negative and PR-negative disease, but the difference between the two subsets was not significant (35.3% vs. 17.6%, p = 0.438). Diseases treated with T-DM1 as second-line therapy exhibited primary resistance to T-DM1 less often than those treated with T-DM1 as third- or later-line therapy, but the difference was not significant (17.6% vs. 35.3%, p = 0.438). Diseases with HER2 gene amplification were less likely to have primary resistance to T-DM1 compared to those without HER2 gene amplification, but this difference also was not significant (0% vs. 36.4%, p = 0.245). Both patients with PIK3CA mutations showed primary resistance to T-DM1, while only two of 14 patients without PIK3CA mutations showed primary resistance to T-DM1 (100% vs. 14.3%, p = 0.05).
Table 4.
Clinicopathological features and primary resistance to T-DM1
| Characteristics | No. of patients (%) | p value |
|---|---|---|
| All patients | 34 | |
| Primary resistance to T-DM1 | 9 (26.5) | |
| De novo stage IV or recurrent | 1 | |
| De novo stage IV | 9 | |
| Primary resistance to T-DM1 | 2 (22.2) | |
| Recurrent | 25 | |
| Primary resistance to T-DM1 | 7 (28.0) | |
| Hormone receptor status | 0.438 | |
| ER and/or PR positive | 17 | |
| Primary resistance to T-DM1 | 6 (35.3) | |
| ER and PR negative | 17 | |
| Primary resistance to T-DM1 | 3 (17.6) | |
| T-DM1 as second-line therapy or earlier | 17 | 0.438 |
| Primary resistance to T-DM1 | 3 (17.6) | |
| T-DM1 as later than second-line therapy | 17 | |
| Primary resistance to T-DM1 | 6 (35.3) | |
| Prior anthracycline | 1 | |
| Yes | 11 | |
| Primary resistance to T-DM1 | 3 (27.3) | |
| No | 23 | |
| Primary resistance to T-DM1 | 6 (26.1) | |
| Prior pertuzumab | 0.462 | |
| Yes | 15 | |
| Primary resistance to T-DM1 | 5 (33.3) | |
| No | 19 | |
| Primary resistance to T-DM1 | 4 (21.1) | |
| Prior lapatinib | 1 | |
| Yes | 11 | |
| Primary resistance to T-DM1 | 3 (27.3) | |
| No | 23 | |
| Primary resistance to T-DM1 | 6 (26.1) | |
| HER2 gene amplification in ctDNA | 0.245 | |
| Positive | 5 | |
| Primary resistance to T-DM1 | 0 (0) | |
| Negative | 11 | |
| Primary resistance to T-DM1 | 4 (36.4) | |
| PIK3CA mutation in ctDNA | 0.05 | |
| Mutation positive | 2 | |
| Primary resistance to T-DM1 | 2 (100) | |
| Mutation negative | 14 | |
| Primary resistance to T-DM1 | 2 (14.3) |
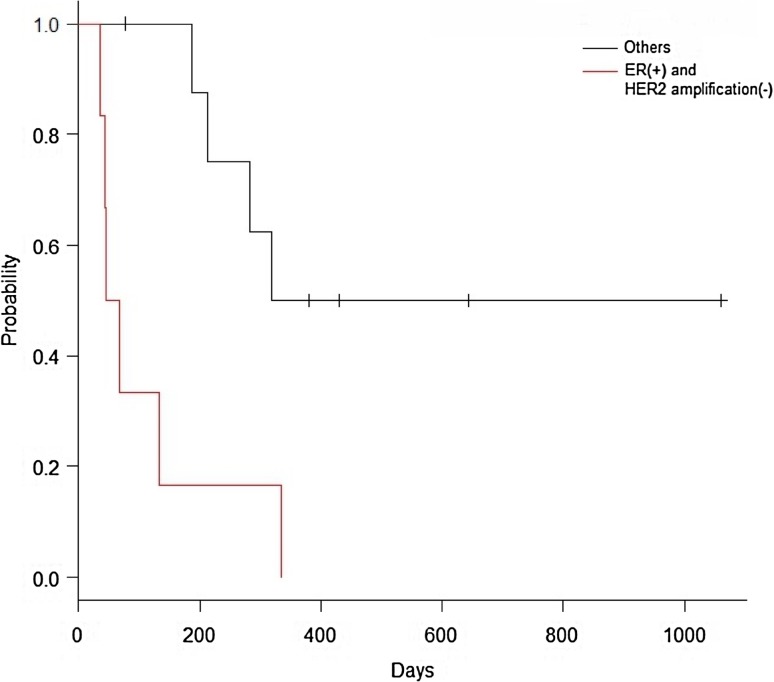
Among 16 patients whose ctDNA was analyzed, four showed primary resistance to T-DM1. These four patients showed negative HER2 gene amplification in ctDNA and were ER positive and/or PR positive (Table 5). Among the 16 patients, median TTP was 56.6 days (95%CI: 36-NA) for those with negative HER2 gene amplification in ctDNA and ER-positive and/or PR-positive tumors, and 318.0 days (95%CI: 187-NA) in patients with neither of these findings (p = 0.0037) (Fig. 1).
Table 5.
Rates of primary resistance to T-DM1 per HER2 gene amplification in ctDNA and hormone receptor status
| HER2 gene amplification in ctDNA | |||
|---|---|---|---|
| Positive (N/total) | Negative (N/total) | ||
| Hormone receptor status | ER and PR negative (N/total) | 0/4 | 0/5 |
| ER and/or PR positive (N/total) | 0/1 | 4/6 | |
N number of patients with primary resistance to T-DM1
Fig. 1.
Kaplan–Meier curve for time to progression (TTP). Sixteen patients whose ctDNA was analyzed were subjected to this analysis. Red line indicates patients with ER-positive and/or PR-positive breast cancer and negative HER2 amplification (n = 6), with a median TTP of 56.6 days (95% CI 36-NA). Black line indicates others (n = 10), with a median TTP of 318.0 days (95% CI 187-NA) (p = 0.0037). Whiskers indicate censored observations
Discussion
In this study, we investigated predictive markers for primary resistance to T-DM1. Negative HER2 gene amplification in ctDNA taken just before the initiation of T-DM1 treatment, ER positivity, and/or PR positivity are candidate negative predictive biomarkers. Our results suggest the potential importance of crosstalk between ER and HER2 pathways in the resistance to T-DM1 treatment.
In exploratory analysis, although not significant, breast cancer with ER and/or PR positivity, T-DM1 administered as third- or later-line therapy, and HER2 gene amplification negativity verified by ctDNA analysis tended to exhibit primary resistance to T-DM1. The lack of significance was likely due to a lack of statistical power. Further studies should be conducted to determine whether these factors are predictive factors using a larger population.
Among patients whose ctDNA was analyzed, all patients with breast cancer who had primary resistance to T-DM1 were ER positive and/or PR positive by IHC, and HER2 amplification-negative by analysis of ctDNA obtained just before initiation of T-DM1 treatment. TTP was significantly shorter in these patients compared to their counterparts. These data support previous studies that suggested the importance of crosstalk between ER and HER2 pathways in the resistance to anti-HER2 treatment. It is well known that patients with higher HER2 amplification have lower ER levels [20]. Moreover, crosstalk between ER and HER2 pathways is implicated in the resistance to hormonal therapy [21–23]. Preclinical and clinical data also suggest that crosstalk between ER and HER2 pathways is involved in the resistance to anti-HER2 treatment. For instance, one study reported that ER expression was significantly increased in xenograft tumors with acquired resistance to anti-HER2 therapy compared with untreated tumors in preclinical models [24]. In the phase III HERA trial, adjuvant trastuzumab followed by chemotherapy showed less benefit for HER2-positive breast cancer patients with ER-positive tumors and who had a low FISH ratio (≥ 2 to < 5) [25]. NeoALTTO, a phase III trial of neoadjuvant lapatinib with trastuzumab, showed a higher pathological complete response (pCR) rate in patients with ER-negative tumors than those with ER-positive tumors [26]. In CALGB40601, a phase III trial of neoadjuvant paclitaxel and trastuzumab with or without lapatinib, patients with ER-positive tumors showed lower rates of pCR compared to patients with ER-negative/HER2-positive tumors [27].
HER2 inhibition may enhance ER expression on tumor cells and result in T-DM1 resistance. If this hypothesis is true, then combination therapy with T-DM1 and hormonal agents may be effective in overcoming T-DM1 resistance in HER2-positive and ER-positive breast cancer. In contrast to our findings, however, in the subgroup analysis of TH3RESA, a phase III trial of T-DM1 in patients with previously treated HER2-positive metastatic breast cancer, T-DM1 was effective for both ER-positive and ER-negative breast cancer [28]. Since hormone receptors and HER2 status can change during the course of treatment [29], hormone receptor status should also be evaluated just before treatment initiation.
In the present study, the frequency of PIK3CA mutations in ctDNA was too low for comparison between the PIK3CA mutation and wild-type groups. There has been controversy regarding the utility of PIK3CA mutations as a biomarker for HER2-positive breast cancer. PIK3CA mutations have been reported to be a prognostic factor for worse outcome in patients treated with pertuzumab, trastuzumab, and docetaxel, or capecitabine and lapatinib; however, this was not the case in T-DM1-treated patients [12, 30].
HER2 amplification was negative in 11 of 16 patients, and the concordance rate between HER2 positivity (IHC3+ or IHC2+ and FISH positive) in archival tissue and HER2 amplification in ctDNA was only 31%. The discordance in mutation profiles between ctDNA and tissue samples is generally small but evident in the setting of various cancers [31–33]. This is possibly attributed to the following three factors: time between tumor tissue sampling and blood, tumor heterogeneity, and biological factors that affect ctDNA shedding. Anti-HER2 therapy performed before liquid biopsy might contribute to changes in HER2 status. Changes in HER2 status in breast cancer after trastuzumab-based systemic therapy have been observed in several studies. One study showed that, while 17 of 23 patients with HER2 3+ (IHC) breast cancer retained HER2 3 + status, 2 and 4 patients showed HER2 2+ (IHC) and 1 + or 0 (IHC), respectively, at the primary site after neoadjuvant trastuzumab-based therapy [34]. Another study reported that 10 of 34 (43%) breast cancer patients with positive HER2 amplification by FISH at baseline became FISH negative after trastuzumab-based systemic therapy [35]. Furthermore, in a prospective study, 37% of patients with HER2-positive metastatic breast cancer (3 + by IHC or 2 + by IHC and chromogenic in situ hybridization (CISH) positive at the primary or metastatic site) lost their HER2 overexpression and were scored 0 or + 1 by IHC or negative on the CISH test after trastuzumab-based therapy; HER2 status was unchanged in the remaining cases [36]. Liquid biopsy may thus allow for non-invasive, real-time monitoring of changes in tumor biology.
This study has some limitations. First, the study was a retrospective exploratory analysis of a small number of patients at one institution. A sample size that is too small can reduce statistical power, and for this reason, we cannot draw definitive conclusion. Second, the distribution of demographic and disease characteristics differs from that of the current general population of breast cancer patients in Japan. For example, about half of our patients were heavily pre-treated before being approved for T-DM1. Moreover, five patients (14.7%) did not receive a trastuzumab-containing treatment prior to T-DM1 in the metastatic setting (three patients were administered lapatinib as anti-HER2 treatment and two received T-DM1 as first-line therapy).
In conclusion, our findings suggest that HER2 gene amplification in ctDNA and ER and PR status may predict primary resistance to T-DM1. A liquid biopsy just before the initiation of T-DM1 treatment could be a non-invasive way to predict whether a patient would exhibit primary resistance to T-DM1. Future prospective studies with a larger study population to evaluate these potential biomarkers are warranted.
Acknowledgements
We are grateful to the patients and their families who participated in our survey.
Conflict of interest
JT is an advisor of EISAI Co. Ltd and ASAHI-KASEI Co. Ltd. JT has received honoraria from Taiho Pharmaceutical Co., Ltd., EISAI Co. Ltd, Kyowa Hakko Kirin Co. Ltd, Novartis Pharma K.K., Chugai Pharmaceutical Co. Ltd, and AstraZeneca K.K., and remunerations from Novartis Pharma K.K. and Daiichi Sankyo Co., Ltd. Other authors declare that they have no conflicts of interest.
References
- 1.Lewis Phillips GD, Li G, Dugger DL, et al. Targeting HER2-positive breast cancer with trastuzumab-DM1, an antibody-cytotoxic drug conjugate. Cancer Res. 2008;68:9280–9290. doi: 10.1158/0008-5472.CAN-08-1776. [DOI] [PubMed] [Google Scholar]
- 2.Verma S, Miles D, Gianni L, et al. Trastuzumab emtansine for HER2-positive advanced breast cancer. N Engl J Med. 2012;367:1783–1791. doi: 10.1056/NEJMoa1209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giordano SH, Temin S, Kirshner JJ, et al. Systemic therapy for patients with advanced human epidermal growth factor receptor 2-positive breast cancer: American Society of Clinical Oncology clinical practice guideline. J Clin Oncol. 2014;32:2078–2099. doi: 10.1200/JCO.2013.54.0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paul Anthony Ellis CHB, Wolfgang Eiermann, et al. Phase III, randomized study of trastuzumab emtansine (T-DM1) ± pertuzumab (P) vs trastuzumab + taxane (HT) for first-line treatment of HER2-positive MBC: primary results from the MARIANNE study. J Clin Oncol 2015; 33 (suppl; abstr 507):203.
- 5.Scaltriti M, Rojo F, Ocana A, et al. Expression of p95HER2, a truncated form of the HER2 receptor, and response to anti-HER2 therapies in breast cancer. J Natl Cancer Inst. 2007;99:628–638. doi: 10.1093/jnci/djk134. [DOI] [PubMed] [Google Scholar]
- 6.Nagata Y, Lan KH, Zhou X, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell. 2004;6:117–127. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 7.Berns K, Horlings HM, Hennessy BT, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 8.Kataoka Y, Mukohara T, Shimada H, et al. Association between gain-of-function mutations in PIK3CA and resistance to HER2-targeted agents in HER2-amplified breast cancer cell lines. Ann Oncol. 2010;21:255–262. doi: 10.1093/annonc/mdp304. [DOI] [PubMed] [Google Scholar]
- 9.Lee-Hoeflich ST, Crocker L, Yao E, et al. A central role for HER3 in HER2-amplified breast cancer: implications for targeted therapy. Cancer Res. 2008;68:5878–5887. doi: 10.1158/0008-5472.CAN-08-0380. [DOI] [PubMed] [Google Scholar]
- 10.Nonagase Y, Yonesaka K, Kawakami H, et al. Heregulin-expressing HER2-positive breast and gastric cancer exhibited heterogeneous susceptibility to the anti-HER2 agents lapatinib, trastuzumab and T-DM1. Oncotarget. 2016;7:84860–84871. doi: 10.18632/oncotarget.12743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nahta R, Yuan LX, Zhang B, et al. Insulin-like growth factor-I receptor/human epidermal growth factor receptor 2 heterodimerization contributes to trastuzumab resistance of breast cancer cells. Cancer Res. 2005;65:11118–11128. doi: 10.1158/0008-5472.CAN-04-3841. [DOI] [PubMed] [Google Scholar]
- 12.Baselga J, Lewis Phillips GD, Verma S, et al. Relationship between tumor biomarkers and efficacy in EMILIA, a Phase III study of trastuzumab emtansine in HER2-positive metastatic breast cancer. Clin Cancer Res. 2016;22:3755–3763. doi: 10.1158/1078-0432.CCR-15-2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burris HA, Rugo HS, Vukelja SJ, et al. Phase II study of the antibody drug conjugate trastuzumab-DM1 for the treatment of human epidermal growth factor receptor 2 (HER2)-positive breast cancer after prior HER2-directed therapy. J Clin Oncol. 2011;29:398–405. doi: 10.1200/JCO.2010.29.5865. [DOI] [PubMed] [Google Scholar]
- 14.Shaw JA, Stebbing J. Circulating free DNA in the management of breast cancer. Ann Transl Med. 2014;2:3. doi: 10.3978/j.issn.2305-5839.2013.06.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang DH, Ensor JE, Liu ZB, et al. Cell-free DNA as a molecular tool for monitoring disease progression and response to therapy in breast cancer patients. Breast Cancer Res Treat. 2016;155:139–149. doi: 10.1007/s10549-015-3635-5. [DOI] [PubMed] [Google Scholar]
- 16.Nakauchi C, Kagara N, Shimazu K, et al. Detection of TP53/PIK3CA mutations in cell-free plasma DNA from metastatic breast cancer patients using next generation sequencing. Clin Breast Cancer. 2016;16:418–423. doi: 10.1016/j.clbc.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 17.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 18.Gevensleben H, Garcia-Murillas I, Graeser MK, et al. Noninvasive detection of HER2 amplification with plasma DNA digital PCR. Clin Cancer Res. 2013;19:3276–3284. doi: 10.1158/1078-0432.CCR-12-3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanda Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Trans. 2013;48:452–458. doi: 10.1038/bmt.2012.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konecny G, Pauletti G, Pegram M, et al. Quantitative association between HER-2/neu and steroid hormone receptors in hormone receptor-positive primary breast cancer. J Natl Cancer Inst. 2003;95:142–153. doi: 10.1093/jnci/95.2.142. [DOI] [PubMed] [Google Scholar]
- 21.Hurtado A, Holmes KA, Geistlinger TR, et al. Regulation of ERBB2 by oestrogen receptor-PAX2 determines response to tamoxifen. Nature. 2008;456:663–666. doi: 10.1038/nature07483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Massarweh S, Osborne CK, Creighton CJ, et al. Tamoxifen resistance in breast tumors is driven by growth factor receptor signaling with repression of classic estrogen receptor genomic function. Cancer Res. 2008;68:826–833. doi: 10.1158/0008-5472.CAN-07-2707. [DOI] [PubMed] [Google Scholar]
- 23.Montemurro F, Di Cosimo S, Arpino G. Human epidermal growth factor receptor 2 (HER2)-positive and hormone receptor-positive breast cancer: new insights into molecular interactions and clinical implications. Ann Oncol. 2013;24:2715–2724. doi: 10.1093/annonc/mdt287. [DOI] [PubMed] [Google Scholar]
- 24.Giuliano M, Hu H, Wang YC, et al. Upregulation of ER signaling as an adaptive mechanism of cell survival in HER2-positive breast tumors treated with anti-HER2 therapy. Clin Cancer Res. 2015;21:3995–4003. doi: 10.1158/1078-0432.CCR-14-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loi S, Dafni U, Karlis D, et al. Effects of estrogen receptor and human epidermal growth factor receptor-2 levels on the efficacy of trastuzumab: a secondary analysis of the HERA trial. JAMA Oncol. 2016;2:1040–1047. doi: 10.1001/jamaoncol.2016.0339. [DOI] [PubMed] [Google Scholar]
- 26.Baselga J, Bradbury I, Eidtmann H, et al. Lapatinib with trastuzumab for HER2-positive early breast cancer (NeoALTTO): a randomised, open-label, multicentre, phase 3 trial. Lancet. 2012;379:633–640. doi: 10.1016/S0140-6736(11)61847-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carey LA, Berry DA, Cirrincione CT, et al. Molecular heterogeneity and response to neoadjuvant human epidermal growth factor receptor 2 targeting in CALGB 40601, a randomized phase III trial of paclitaxel plus trastuzumab with or without lapatinib. J Clin Oncol. 2016;34:542–549. doi: 10.1200/JCO.2015.62.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krop IE, Kim SB, Martin AG, et al. Trastuzumab emtansine versus treatment of physician’s choice in patients with previously treated HER2-positive metastatic breast cancer (TH3RESA): final overall survival results from a randomised open-label phase 3 trial. Lancet Oncol. 2017;18:743–754. doi: 10.1016/S1470-2045(17)30313-3. [DOI] [PubMed] [Google Scholar]
- 29.Lindstrom LS, Karlsson E, Wilking UM, et al. Clinically used breast cancer markers such as estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2 are unstable throughout tumor progression. J Clin Oncol. 2012;30:2601–2608. doi: 10.1200/JCO.2011.37.2482. [DOI] [PubMed] [Google Scholar]
- 30.Baselga J, Cortés J, Im S-A, et al. Biomarker analyses in CLEOPATRA: a phase III, placebo-controlled study of pertuzumab in human epidermal growth factor receptor 2–positive, first-line metastatic breast cancer. J Clin Oncol. 2014;32:3753–3761. doi: 10.1200/JCO.2013.54.5384. [DOI] [PubMed] [Google Scholar]
- 31.Kim S-B, Dent R, Wongchenko MJ, et al. Concordance between plasma-based and tissue-based next-generation sequencing in LOTUS. Lancet Oncol 18: e638. [DOI] [PubMed]
- 32.Chae YK, Davis AA, Carneiro BA, et al. Concordance between genomic alterations assessed by next-generation sequencing in tumor tissue or circulating cell-free DNA. Oncotarget. 2016;7:65364–65373. doi: 10.18632/oncotarget.11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chae YK, Davis AA, Jain S, et al. Concordance of genomic alterations by next-generation sequencing in tumor tissue versus circulating tumor DNA in breast cancer. Mol Cancer Ther. 2017;16:1412–1420. doi: 10.1158/1535-7163.MCT-17-0061. [DOI] [PubMed] [Google Scholar]
- 34.Burstein HJ, Harris LN, Gelman R, et al. Preoperative therapy with trastuzumab and paclitaxel followed by sequential adjuvant doxorubicin/cyclophosphamide for HER2 overexpressing stage II or III breast cancer: a pilot study. J Clin Oncol. 2003;21:46–53. doi: 10.1200/JCO.2003.03.124. [DOI] [PubMed] [Google Scholar]
- 35.Hurley J, Doliny P, Reis I, et al. Docetaxel, cisplatin, and trastuzumab as primary systemic therapy for human epidermal growth factor receptor 2-positive locally advanced breast cancer. J Clin Oncol. 2006;24:1831–1838. doi: 10.1200/JCO.2005.02.8886. [DOI] [PubMed] [Google Scholar]
- 36.Pectasides D, Gaglia A, Arapantoni-Dadioti P, et al. HER-2/neu status of primary breast cancer and corresponding metastatic sites in patients with advanced breast cancer treated with trastuzumab-based therapy. Anticancer Res. 2006;26:647–653. [PubMed] [Google Scholar]