Abstract
Between January and July 2017, lumpy skin disease (LSD) outbreaks were reported in cattle in Namibia. DNA was extracted from skin biopsies taken from 32 cattle, and the RNA polymerase 30 kDa subunit (RPO30) gene of the LSD virus (LSDV) was successfully amplified by PCR. Phylogenetic analysis revealed that the newly sequenced LSDV isolates from Namibia were identical to LSDV isolates identified previously in Burkina Faso, Egypt, Greece, Niger, Serbia and South Africa. Given that only unvaccinated herds were affected by LSD, it is recommended that the current vaccination programmes in Namibia be re-evaluated to allow nationwide coverage.
Lumpy skin disease (LSD) is a disease of cattle caused by lumpy skin disease virus (LSDV), a DNA virus belonging to the genus Capripoxvirus within the family Poxviridae [3]. LSD was first reported in Zambia in 1929 from which it spread south to southern African countries and north to Sudan. The first diagnosis of LSD outside Africa was in Israel in 1989, followed by reports from Bahrain, Kuwait, Oman, Yemen, Lebanon, Jordan and Turkey [4, 11]. In 2015, LSD was detected in Europe (i.e., Greece and the Balkans) [1, 8]. LSD has a substantial economic impact in affected regions, causing decreases in milk yield, abortion and infertility in cows, and a decreased growth rate in beef cattle [9]. Morbidity rates can vary between 1 and 20%, although outbreaks with rates as high as 50% have been reported [4]. The control of LSD can be achieved through vaccination, restriction of animal movement, and culling of infected and exposed animals [2, 8]. Although LSD has been endemic in Namibia for many years, there is no genetic information available on local LSDV isolates. The present study describes the first genetic characterization of LSDV in the country.
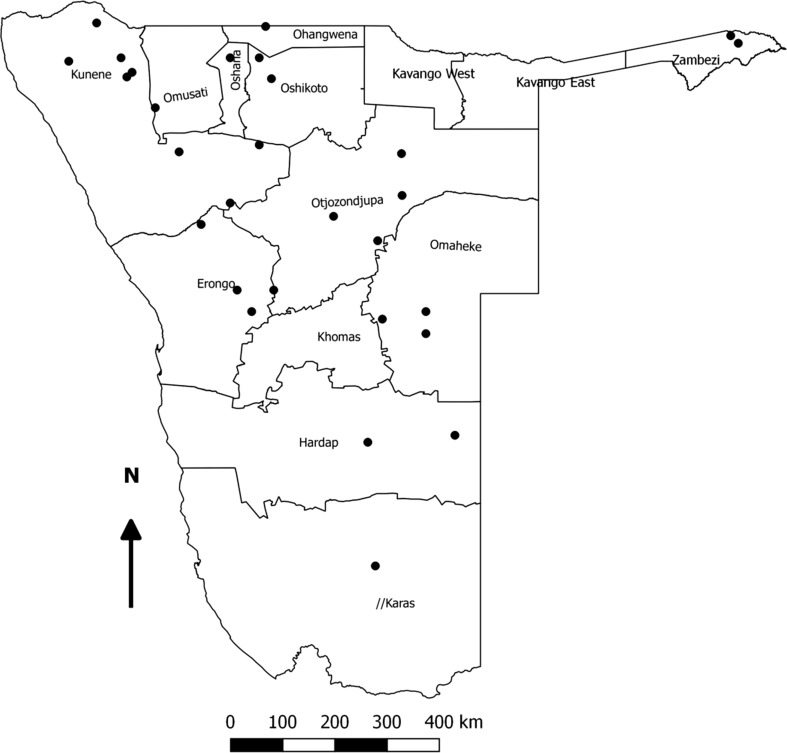
During the period under investigation (January to July 2017), there were 32 LSDV outbreaks affecting 10 out of the 14 regions of Namibia (Fig. 1): Omaeke (4 outbreaks), Otjozondjupa (4 outbreaks), Hardap (2 outbreaks), Oshikoto (2 outbreaks), Kunene (9 outbreaks), Zambesi (2 outbreaks), Oshana (1 outbreak), Ohangwena (1 outbreak), Karas (1 outbreak), and Erongo (6 outbreaks). One representative sample from each outbreak was included in this study. Skin nodule biopsies were collected aseptically from sick cattle, and the samples were sent, refrigerated, to the Central Veterinary Laboratory (CVL) of Windhoek. Upon arrival at the CVL, the samples were stored at – 20 °C until processing. DNA was extracted from the tissue homogenates using a Maxwell®16 Tissue DNA Purification Kit (Promega, Madison, WI, USA) with an elution volume of 300 μl following the manufacturer’s instructions. A commercial real-time PCR assay, Genesig® Advance Kit LSDV116 RNA polymerase subunit (Primerdesigntm Ltd, Chandler’s Ford, UK), was used to confirm the presence of capripoxvirus DNA in all of the 32 samples tested, with Ct values between 24 and 36 being recorded. Next, two pairs of primers CpRPO30-OL1F (5’-CAGCTGTTTGTTTACATTTGATTTTT-3’) and CpRPO30-OL1R (5’-TCGTATAGAAACAAGCCTTTAATAGA-3’) (pair 1), CpRPO30-OL2F (5’-TTTGAACACATTTTATTCCAAAAAG-3’) and CpRPO30-OL2R (5’-AACCTACATGCATAAACAGAAGC-3’) (pair 2) were used to amplify two overlapping fragments of the RPO30 gene to generate the full-length sequence of the gene [5]. (Molecular epidemiological analysis using the RPO30 gene is used to genotype capripoxviruses. The variability within each genotype is generally very low, so any nucleotide sequence variation is highly indicative of a true difference between isolates, i.e., vaccine strains versus field isolates). The PCR was conducted in a reaction volume of 25 µl containing 500 nM forward primer, 500 nM reverse primer, 0.2 mM dNTPs, 1x PCR buffer (QIAGEN), 2.5 U of Taq polymerase (QIAGEN) and 5 µl of template DNA. An initial denaturation at 95 °C for 4 min was followed by 40 cycles at 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 45 s, and then a final extension at 72 °C for 7 min. The expected positive PCR products of 554 bp (for pair 1 primers) and 520 bp (for pair 2 primers) were visualized on a 1.5% agarose gel for all of the 32 samples. The amplicons were purified using a Wizard SV Gel and PCR Clean-Up System (Promega) and sequenced commercially by LGC Genomics (Berlin, Germany). The RPO30 sequences from the 32 samples were edited and assembled using the Staden software package version 2.0.0b8. All of the generated sequences were submitted to GenBank with accession numbers MG757462 to MG757493. Multiple sequence alignments were performed using the ClustalW algorithm implemented in the BioEdit software package version 7.2.6 to compare the RPO30 gene sequences of the isolates involved in the outbreaks. Additional RPO30 gene sequences were retrieved from GenBank and included in the data set. For construction of phylogenetic trees, the neighbor-joining method in MEGA7 was used with the maximum composite likelihood nucleotide substitution model, the pairwise deletion option, and 1000 bootstrap replicates [6] (Table 1).
Fig. 1.
Geographic distribution of lumpy skin disease outbreaks reported in cattle in Namibia from January to July 2017
Table 1.
RPO30 sequences submitted to GenBank
| Sample ID | Accession number (GenBank) | Collection date | Place collected | Host |
|---|---|---|---|---|
| LSDV_NAM_30_1 | MG757462 | January 2017 | Omaeke | Cattle |
| LSDV_NAM_30_2 | MG757463 | January 2017 | Omaeke | Cattle |
| LSDV_NAM_211 | MG757464 | January 2017 | Otjozondjupa | Cattle |
| LSDV_NAM_212 | MG757465 | January 2017 | Otjozondjupa | Cattle |
| LSDV_NAM_415 | MG757466 | February 2017 | Otjozondjupa | Cattle |
| LSDV_NAM_416 | MG757467 | February 2017 | Otjozondjupa | Cattle |
| LSDV_NAM_871 | MG757468 | March 2017 | Hardap | Cattle |
| LSDV_NAM_872 | MG757469 | March 2017 | Oshikoto | Cattle |
| LSDV_NAM_941 | MG757470 | March 2017 | Kunene | Cattle |
| LSDV_NAM_1001 | MG757471 | March 2017 | Oshana | Cattle |
| LSDV_NAM_1169 | MG757472 | March 2017 | Oshikoto | Cattle |
| LSDV_NAM_1172 | MG757473 | March 2017 | Ohangwena | Cattle |
| LSDV_NAM_1429 | MG757474 | April 2017 | Omaeke | Cattle |
| LSDV_NAM_1467 | MG757475 | April 2017 | Zambesi | Cattle |
| LSDV_NAM_1468 | MG757476 | April 2017 | Zambesi | Cattle |
| LSDV_NAM_1689 | MG757477 | April 2017 | Kunene | Cattle |
| LSDV_NAM_1690 | MG757478 | April 2017 | Kunene | Cattle |
| LSDV_NAM_1691 | MG757479 | April 2017 | Kunene | Cattle |
| LSDV_NAM_1701 | MG757480 | April 2017 | Omaeke | Cattle |
| LSDV_NAM_2190 | MG757481 | May 2017 | Erongo | Cattle |
| LSDV_NAM_2191_1 | MG757482 | May 2017 | Erongo | Cattle |
| LSDV_NAM_2191_2 | MG757483 | May 2017 | Erongo | Cattle |
| LSDV_NAM_2191_3 | MG757484 | May 2017 | Erongo | Cattle |
| LSDV_NAM_2226 | MG757485 | May 2017 | Erongo | Cattle |
| LSDV_NAM_2229 | MG757486 | May 2017 | Erongo | Cattle |
| LSDV_NAM_2864 | MG757487 | June 2017 | Kunene | Cattle |
| LSDV_NAM_3166 | MG757488 | June 2017 | Karas | Cattle |
| LSDV_NAM_3303 | MG757489 | June 2017 | Kunene | Cattle |
| LSDV_NAM_3306 | MG757490 | June 2017 | Kunene | Cattle |
| LSDV_NAM_3469 | MG757491 | June 2017 | Hardap | Cattle |
| LSDV_NAM_3540 | MG757492 | June 2017 | Kunene | Cattle |
| LSDV_NAM_3742 | MG757493 | June 2017 | Kunene | Cattle |
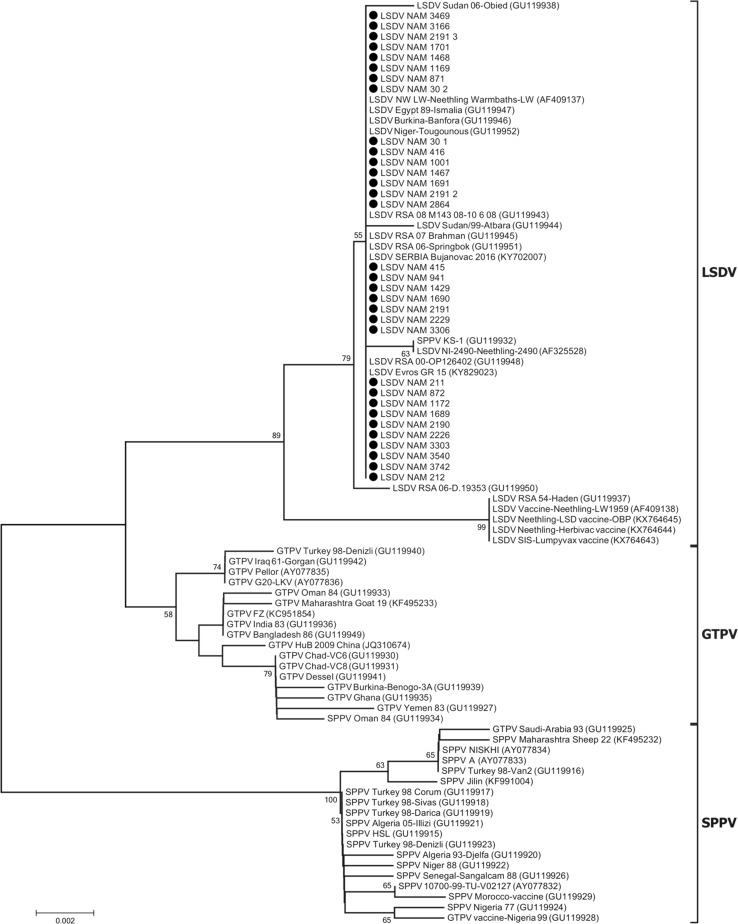
From the phylogenetic analysis of the RPO30 gene, it can be clearly seen that all of the biopsy samples collected in Namibia contained LSDV DNA and that the sequences were identical to each other (Fig. 2). In addition, they were identical to previously identified LSDVs from several countries both in Africa (Burkina Faso, Egypt, Niger and South Africa) and Europe (Greece and Serbia). Importantly, the RPO30 gene sequences of the Namibian field isolates were clearly different from those of the LSDV vaccines used in this country. In Namibia, three commercial vaccines are used, two of which contain cell-adapted strains of the so-called Neethling strain (Fig. 2, KX764644 and KX764645), and the third, an attenuated South African LSDV field isolate (Fig. 2, KX764643) [11]. It is possible that the LSDVs identified in Namibia and those in neighbouring South Africa share a common origin, but in order to confirm this, full-genome sequencing and further molecular epidemiological studies in the region are required. A recent characterization of the P32 gene of LSD samples in Zimbabwe identified two genetically distinct viruses circulating in the country [7]. A similar comparative study should be undertaken for other LSDV isolates collected in southern Africa in order to get a clearer picture of virus circulation at a regional level.
Fig. 2.
Thirty-two complete sequences of the LSDV RPO30 gene aligned and analyzed by neighbour joining using MEGA7 software [6]. Numbers indicate the bootstrap values calculated from 1000 bootstrap replicates. Black dots correspond to the newly sequenced samples from Namibia (accession numbers MG757462 to MG757493). LSDV stands for lumpy skin disease virus, SPPV for sheep pox virus, and GTPV for goat pox virus
This study has confirmed that LSDV is still present and distributed across the whole country despite vaccination programmes. Importantly, it was observed that only unvaccinated herds were affected by the disease, which suggests that the present vaccination strategy in Namibia requires reevaluation to improve coverage and participation by farmers. It is known that the use of live attenuated vaccines is an effective way to control the spread of LSD, although good protection can only be achieved if sufficient herd immunity (over 80%) is maintained by carrying out annual vaccinations [10].
Funding
This work was supported through funding from the African Renaissance Funds (ARF) and from the Directorate of Veterinary Services, Ministry of Agriculture, Water and Forestry of Namibia.
Conflict of interest
The authors declare no conflicts of interest.
Ethical approval
This article does not contain animal studies.
References
- 1.Agianniotaki EI, Mathijs E, Vandenbussche F, Tasioudi KE, Haegeman A, Iliadou P, Chaintoutis SC, Dovas CI, Van Borm S, Chondrokouki ED, De Clercq K. Complete genome sequence of the lumpy skin disease virus isolated from the first reported case in Greece in 2015. Genome Announc. 2017;20:5. doi: 10.1128/genomeA.00550-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayelet G, Abate Y, Sisay T, Nigussie H, Gelaye E, Jemberie S, Asmare K. Lumpy skin disease: preliminary vaccine efficacy assessment and overview on outbreak impact in dairy cattle at Debre Zeit, central Ethiopia. Antiviral Res. 2013;98:261–265. doi: 10.1016/j.antiviral.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Buller RM, Arif BM, Black DN, Dumbell KR, Esposito JJ, Lefkowitz EJ, Tripathy DN. Family poxviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus taxonomy: classification and nomenclature of viruses. Eighth report of the international committee on taxonomy of viruses. San Diego: Elsevier Academic Press; 2005. pp. 117–133. [Google Scholar]
- 4.Davies FG. Lumpy skin disease of cattle: a growing problem in Africa and the Near East. World Anim Rev. 1991;68:37–42. [Google Scholar]
- 5.Gelaye E, Belay A, Ayelet G, Jenberie S, Yami M, Loitsch A, Tuppurainen E, Grabherr R, Diallo A, Lamien CE. Capripox disease in Ethiopia: genetic differences between field isolates and vaccine strain, and implications for vaccination failure. Antiviral Res. 2015;119:28–35. doi: 10.1016/j.antiviral.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mafirakureva P, Saidi B, Mbanga J. Incidence and molecular characterisation of lumpy skin disease virus in Zimbabwe using the P32 gene. Trop Anim Health Prod. 2017;49:47–54. doi: 10.1007/s11250-016-1156-9. [DOI] [PubMed] [Google Scholar]
- 8.Mercier A, Arsevska E, Bournez L, Bronner A, Calavas D, Cauchard J, Falala S, Caufour P, Tisseuil C, Lefrançois T, Lancelot R. Spread rate of lumpy skin disease in the Balkans, 2015–2016. Transbound Emerg Dis. 2017 doi: 10.1111/tbed.12624. [DOI] [PubMed] [Google Scholar]
- 9.Tuppurainen ES, Oura CA. Review: lumpy skin disease: an emerging threat to Europe, the Middle East and Asia. Transbound Emerg Dis. 2012;59:40–48. doi: 10.1111/j.1865-1682.2011.01242.x. [DOI] [PubMed] [Google Scholar]
- 10.Tuppurainen ES, Venter EH, Shisler JL, Gari G, Mekonnen GA, Juleff N, Lyons NA, De Clercq K, Upton C, Bowden TR, Babiuk S, Babiuk LA. Review: capripoxvirus diseases: current status and opportunities for control. Transbound Emerg Dis. 2017;64:729–745. doi: 10.1111/tbed.12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wainwright S, El Idrissi A, Mattioli R, Tibbo M, Njeumi F, Raizman E. Emergence of lumpy skin disease in the Eastern Mediterranean Basin countries. Empres Watch. 2013;29:1–6. [Google Scholar]