Abstract
Melanopsin is a blue light-sensitive opsin photopigment involved in a range of non-image forming behaviours, including circadian photoentrainment and the pupil light response. Many naturally occurring genetic variants exist within the human melanopsin gene (OPN4), yet it remains unclear how these variants affect melanopsin protein function and downstream physiological responses to light. Here, we have used bioinformatic analysis and in vitro expression systems to determine the functional phenotypes of missense human OPN4 variants. From 1242 human OPN4 variants collated in the NCBI Short Genetic Variation database (dbSNP), we identified 96 that lead to non-synonymous amino acid substitutions. These 96 missense mutations were screened using sequence alignment and comparative approaches to select 16 potentially deleterious variants for functional characterisation using calcium imaging of melanopsin-driven light responses in HEK293T cells. We identify several previously uncharacterised OPN4 mutations with altered functional properties, including attenuated or abolished light responses, as well as variants demonstrating abnormal response kinetics. These data provide valuable insight into the structure–function relationships of human melanopsin, including several key functional residues of the melanopsin protein. The identification of melanopsin variants with significantly altered function may serve to detect individuals with disrupted melanopsin-based light perception, and potentially highlight those at increased risk of sleep disturbance, circadian dysfunction, and visual abnormalities.
Electronic supplementary material
The online version of this article (10.1007/s00018-018-2813-0) contains supplementary material, which is available to authorized users.
Keywords: Melanopsin, Single-nucleotide polymorphisms, Opsins, Photopigment
Introduction
Melanopsin (OPN4) is a blue light-sensitive opsin-type G-protein coupled receptor (GPCR) that is expressed within a small subset of retinal ganglion cells of the human retina [1–3], termed intrinsically photosensitive retinal ganglion cells (ipRGCs) [4–6]. These inner retina photoreceptors characteristically mediate a range of non-image forming (NIF) responses to light, including circadian photoentrainment [7–9], regulation of sleep [10–13], and the pupillary light response [14, 15]. However, recent evidence has revealed additional roles for melanopsin-based light perception in visual signalling pathways [16–19], and during development of visual [20] and non-visual systems [21]. Furthermore, the growing appreciation of how sleep and circadian disruption may contribute to the onset of symptoms of neurological and psychiatric conditions has highlighted the importance of the melanopsin system to human health and disease [22].
Like other genes, naturally occurring mutations and sequence changes are known to exist within the human melanopsin gene. The most common forms of genetic variation are single-nucleotide polymorphisms (SNPs), where an individual nucleotide base differs at a specific location within the genome in more than 1% of the population [23]. Substitutions that occur less frequently are classified as rare variants [24]. The majority of genetic variants occur in non-coding regions of genes or lead to synonymous amino acid substitutions [25]. However, genetic variation can also cause frameshifts, nonsense and missense mutations, leading to disruption or total loss of protein function and potentially causing disease. For example, mutations in rhodopsin are a leading cause of blindness, with over 150 rhodopsin mutations associated with human retinal disease [26, 27].
Both common SNPs and rare gene variants have been reported in the human melanopsin gene (OPN4) from whole-genome and exome sequencing projects, such as the 1000 Genomes project [28]. The OPN4 sequence variants identified to date are collated in the NCBI Short Genetic variation database (dbSNP) [29] and number over 1200 in total. Of these, 96 OPN4 variants reported in the dbSNP database lead to non-synonymous missense mutations, yet the functional consequences of these mutations on melanopsin activity, and the extent to which they may influence melanopsin-dependant behaviours in humans, remain largely undetermined. The exceptions are a series of studies examining the possible association of two OPN4 SNPs, P10L and T394I, with abnormal melanopsin-driven behaviours and higher frequency of seasonal affective disorder [30–35]. However, at present, the mechanisms by which OPN4 polymorphisms may influence melanopsin protein function are currently unclear.
Identification of OPN4 mutations that give rise to altered melanopsin function is an important step towards understanding the role of melanopsin in human physiology and behaviour. Yet, the structure–function relationships of melanopsin are still poorly defined. The majority of targeted mutagenesis studies of melanopsin have explored post-translational modification sites [36–40] and the retinal chromophore-binding pocket [41–43]. It is, therefore, difficult to predict the functional consequences of OPN4 variants based solely on existing knowledge of melanopsin structure–function.
All GPCRs, including melanopsin, share a common seven-transmembrane helical structure and many highly conserved functional domains [44, 45]. As such, studies of sequence homology with other GPCRs offer a valuable approach for predicting the impact of OPN4 variants. As one of the most-well characterised GPCRs, rhodopsin is especially useful as a model for structure–function relationships in GPCRs and, in particular, other opsin photopigments. Opsin photopigments, which consist of a vitamin A-derived retinal chromophore bound to an opsin protein moiety, have a conserved activation mechanism. Isomerisation of the chromophore after absorbing a photon causes a conformation change in the opsin protein from inactive to an active state, triggering activation of a G-protein signalling pathway and downstream phototransduction cascades [46]. The critical residues responsible for these functions in rhodopsin have been extensively documented based on crystal structures (first in [47], summarised in [48]), site-directed mutagenesis studies [49–53], and the identification of over 150 rhodopsin mutations associated with retinal disease [26, 27].
This comparative approach has its limits and is only valid for OPN4 variants that have equivalent residues within rhodopsin. While sequence homology is relatively high within transmembrane regions and other key conserved GPCR domains, overall rhodopsin and melanopsin share only 28% amino acid identity [45]. The N- and C-terminal domains of melanopsin are highly variable between different species, and melanopsin contains several extended insertions within the intracellular loops that are absent in rhodopsin. Furthermore, there are several functional differences between melanopsin and rhodopsin, including G-protein specificity ([54–57], reviewed in [58]) and chromophore regeneration mechanisms [55, 56, 59–61], suggesting that key functional residues may differ between these two opsins even within highly conserved domains. It is, therefore, necessary to directly investigate the functional properties of melanopsin genetic variants to determine their role in melanopsin activity.
Here, bioinformatic analysis of sequence alignments and comparative approaches were used to identify 16 missense OPN4 variants likely to result in loss-of-function phenotypes from the 96 known OPN4 missense mutations. Further functional characterisation of these 16 variants was completed using in vitro heterologous expression combined with calcium imaging of melanopsin-driven light responses. Using this approach, we identified a number of previously uncharacterised OPN4 variants with atypical functional properties. These data provide important insights into the key functional domains of the melanopsin protein and highlight individuals with potential increased risk of visual, sleep, and circadian dysfunction.
Materials and methods
Identification of OPN4 variants
Polymorphisms in the OPN4 gene (Gene ID 94233, transcript NM_033282.2) were identified from the NCBI Short Genetic Variation database (dbSNP) Build 140 [29]. The dbSNP is a large public database that collates simple genetic variations, including common SNPs and rare genetic variants. Each genetic variant in dbSNP is assigned a reference SNP ID (rs#), and includes information on the variant position, alleles, and validation status. Missense variants were identified and prioritised for further in vitro screening using criteria shown in Table 1. Melanopsin protein sequences from the following species were used for multiple alignment—Homo sapiens (NP_150598.1, NP_001025186.1), Pan troglodytes (XP_001135445.1, XP_001135533.1), Macaca mullata (XP_001088248.2), Canis familiaris (XP_853735.2), Bos taurus (NP_001179328.1), Felis catus (AAR36861), Mus musculus (NP_038915.1, NP_001122071.1), Rattus Norvegicus (NP_620215.1), Phosopus sungorus (AAU11506), Spalax ehrenbergi (CAO02487), Gallus gallus (ABX10832.1, ABX10833.1, ABX10834.1, NP_989956.1, ABX10831.1), Sminthopsis crassicaudata (ABD38715), Danio rerio (NP_001245153.1, ADN39430, NP_840074.1, NP_001243006.1, ADN39434.1), Podarcis siculus (AAY34941.2), Xenopus laevis (NP_001079143.1), and Brachiostoma belcheri (BAE00065). Multiple sequences for a given species represent different splice variants or distinct melanopsin genes, which are duplicated in non-mammals [62]. Protein sequences were aligned using multiple sequence alignment software MAFFT [63].
Table 1.
Criteria for identifying OPN4 variants for in vitro screening
| Variant property | Reasoning | Method | Criteria for inclusion |
|---|---|---|---|
| Validation status | dbSNP accepts submissions from a variety of sources of varying quality. Variants with multiple submissions are less likely to be false positives | Restricted analysis to “validated” variants obtained by non-computational methods and those with frequency data attached | Validated by multiple submissions, preferably including the 1000 Genomes project [28] |
| Conservation between OPN4 of different species | Highly conserved amino acids of OPN4 are more likely to be functionally important | Aligned 20 OPN4 amino acid sequences from 16 different species with multiple alignment using fast Fourier transform (MAFFT) software | Substitutions that occur at highly conserved OPN4 residues |
| Amino acid substitution properties | Substitutions that substantially change the biochemical properties of amino acids are more likely to affect protein function [64] | Compared the charge, hydrophobicity, bond formation, polarity and side chain size of amino acid substitutions using NCBI amino acid explorer [65] | Significant change in hydrophobicity, charge or polarity of amino acid |
| Location in protein | Mutations located in key functional domains, such as transmembrane helices or intracellular loops, are more likely to disrupt function [66] | Human OPN4 and bovine rhodopsin protein sequences were aligned and OPN4 domains defined based on rhodopsin structure [47] | Substitutions that occur in transmembrane helices or intracellular loops. |
| Functional role of equivalent residue in rhodopsin | Residues conserved between OPN4 and rhodopsin may perform similar functions, and show similar effects following mutation | Comparison to known rhodopsin mutations made following alignment of human OPN4 and bovine rhodopsin protein sequences | Evidence of functional effects in rhodopsin |
| Minor allele frequency (MAF) | The more frequent the minor allele, the greater potential relevance to the human population as a whole | Frequency data is reported in dbSNP, with MAF expressed as a value from 0 to 1 | Variants with higher MAF scores (> 0.01) |
A BLAST protein alignment [67] of human melanopsin (NP_150598.1) and bovine rhodopsin (NP_001014890.1) was used to define melanopsin protein domains and identify the equivalent position of the OPN4 variants within the rhodopsin protein (Figure S1). A literature search was then performed in NCBI PubMed (http://www.ncbi.nlm.nih.gov/pubmed/) and Google Scholar (http://www.scholar.google.co.uk/) to determine whether the equivalent residues are functionally significant in rhodopsin, based on analysis of crystal structure [47, 68, 69] and lists of rhodopsin mutations known to be associated with retinitis pigmentosa [26, 70].
Generation of OPN4 expression vectors
Single-nucleotide point mutations were introduced to pcDNA3.1 plasmid (Invitrogen) containing human melanopsin (NM_033282.2) with a 1D4 tag [57] using a Quikchange II XL site-directed mutagenesis kit (Stratagene). Mutagenesis primers are shown in Table S2. Successful introduction of mutations was confirmed by Sanger sequencing (Source Biosciences). Plasmid production and purification was performed using standard techniques.
Cell culture and transient transfection
HEK293T cells (ATCC) were cultured in DMEM (Sigma) with 10% foetal bovine serum (Life Technologies), 2 mM l-glutamine (Sigma), and 1% (v/v) penicillin/streptomycin (Sigma). Cells were maintained in a humidified incubator at 37 °C with 5% CO2, fed fresh media every 2–3 days and passaged before reaching confluence. 24 h after seeding into multi-well plates (see below), cells were placed in antibiotic-free DMEM with 10% foetal bovine serum and 2 mM l-glutamine, and transiently transfected using Genejuice transfection reagent (Merck Millipore) according to the manufacturer’s instructions with a 1:3 ratio of DNA (µg) to Genejuice (µl).
Immunocytochemistry
HEK293T cells were seeded into 6-well plates containing 13 mm glass coverslips at ~ 1 × 105 cells per well and transfected as described above. 48 h after transfection, cells were fixed with 4% methanol-free paraformaldehyde (Thermo Scientific) in phosphate buffered saline (PBS) for 10 min at room temperature and immunostained as described previously [71]. A rabbit polyclonal anti-human melanopsin antibody (H-300, Santa Cruz Biotechnology, 1:400) was incubated for 1 h at room temperature. Secondary antibody was donkey anti-rabbit IgG conjugated to Alexa 568 (Life Technologies, 1:200). After final wash step, nuclear counterstaining was performed with 0.5 μg/ml DAPI for 10 min at room temperature. Coverslips were then mounted onto glass microscope slides using Prolong Gold anti-fade mounting media (Life Technologies). Fluorescence images were collected using an inverted LSM 710 laser scanning confocal microscope (Zeiss) and Zen 2009 image acquisition software (Zeiss). Individual channels were collected sequentially. Laser lines for excitation were 405 and 561 nm, with emissions collected between 440–480 and 580–625 nm for blue and red fluorescence respectively. Images were collected using a 40× objective with images collected every 1.0 µm in the Z axis. Global enhancement of brightness and contrast was performed using ZenLite 2011 software (Zeiss) and applied equally to all images.
Fluorescent calcium imaging
HEK293T cells were seeded into 24-well plates at 4 × 104 cells per well and transfected with pcDNA3.1 OPN4-1D4 plasmid 24 h later. 24 h after transfection, cells were incubated with 5 µM Fluo-4 AM calcium indicator dye (Life Technologies), 0.015% Pluronic F-127 (Life Technologies), 2.5 µM probenecid (Life Technologies), and 20 µM 9-cis retinal (Sigma) for 20 min at 37 °C. Following dye loading, cells were incubated for a further 10 min at 37 °C in DMEM with 2.7 µM probenecid and then transferred to Hank’s buffered saline solution without additional calcium (Gibco) prior to calcium imaging. All steps were conducted under dim red light (610 ± 10 nm, 3.02 × 1011 photons/cm2/s in the working area). Calcium imaging was performed using a FLUOstar Omega plate reader (BMG Labtech) at room temperature. Total fluorescence values from each well were collected every 2 s for a total of 120 s, with individual wells imaged sequentially. Each data point was generated by averaging fluorescence values collected from 200 repeated 5 ms flashes of light generated by the plate reader’s internal xenon flash bulb with excitation and emission filters of 485 nm (12 nm bandwidth) and 520 nm (30 nm bandwidth), respectively. These multiple flashes of excitation light resulted in a near continuous illumination of cells (485 ± 6 nm, 1.21 × 1014 photons/cm2/s) and were sufficient to activate the melanopsin photopigment without further light stimulation.
Each 24-well plate contained four technical replicates of five different OPN4 variants and wild-type OPN4 positive control. Values for the four technical replicates of each plasmid obtained from individual plates were averaged to produce a single biological replicate for each sample. A total of six biological replicates (each the result of a separate transfection) were performed for each OPN4 variant or control. For analysis, biological replicates were first normalised to baseline (first recorded fluorescence value) for response amplitude comparisons (Baseline = 0, ΔF/F0), then normalised to maximum for comparing response kinetics (Baseline = 0, maximum = 1, ΔF/Fmax). As no statistical difference was observed in response properties of OPN4 WT replicates from different plates (Figure S2), the OPN4 variants were compared to pooled OPN4 WT data for subsequent comparisons.
Statistical analysis
Statistical tests were conducted using SPSS 22.0 (IBM). Unless stated otherwise, all comparisons between groups were tested using one-way between-subjects analysis of variance (ANOVA) with genotype as the independent variable. For one-way ANOVA, a post hoc Dunnett’s test was used to explore significant main effects of genotype, using OPN4 WT as the control group. A significance threshold of p ≤ 0.05 was used. In all figures, asterisk (*) indicates p < 0.05, **p < 0.01, and ***p < 0.001. Error bars show standard error of mean. Where error bars are smaller than data symbols, error bars are not shown.
Results
Identification and selection of 16 OPN4 variants for in vitro screening
At the time this study was conducted 1242 genetic variants of the OPN4 gene were identified in the dbSNP database (build 140), of which 96 lead to non-synonymous amino acid substitutions. A detailed description of these 96 OPN4 missense mutants is provided in Supplementary Table 1. Using sequence alignment and comparative approaches a subset of these 96 OPN4 variants were prioritised for functional screening in vitro using six main selection criteria, including validation status, conservation of residues in melanopsin across 16 different species, biochemical properties of the amino acid substitution, location of the mutation within key functional domains, functional role of the equivalent residue in rhodopsin (if present), and the overall frequency of the variant within the population. A detailed description of the criteria and methods used for selecting variants is shown in Table 1. OPN4 variants did not have to meet all criteria to be included for further analysis, with preference given to variants with altered amino acid biochemical properties and those occurring at sites highly conserved amongst melanopsin sequences of different species. Based on the selection criteria, 16 OPN4 variants were classified as being likely to result in change of function phenotypes and were selected for further in vitro functional testing. The location of the 16 OPN4 variants within the human melanopsin protein is shown in Fig. 1. Details of how each variant matched the criteria for selection is outlined in Table 2.
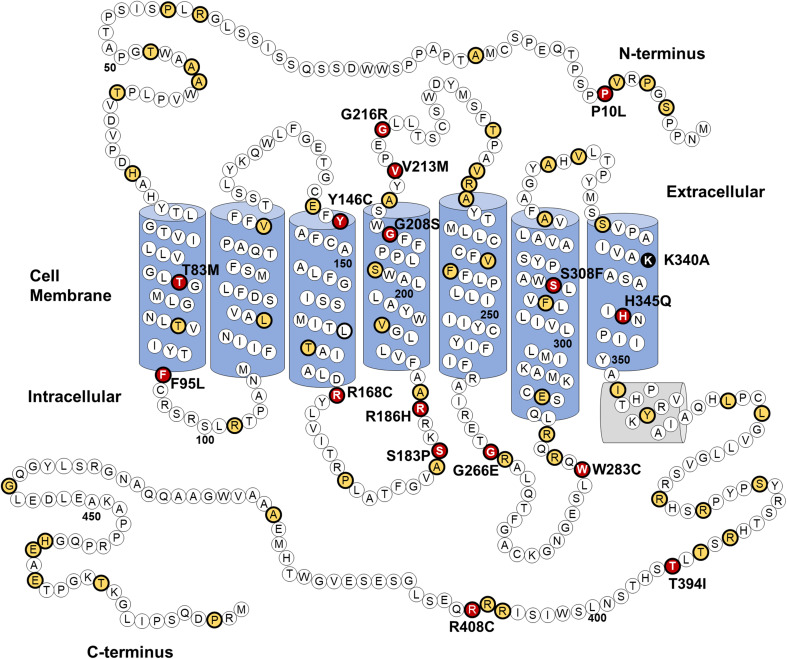
Fig. 1.
OPN4 genetic variants selected for in vitro screening. Location of 96 naturally occurring non-synonymous amino acid substitutions (thick black outline) in the human OPN4 protein, of which 16 (red) were screened using immunocytochemistry and calcium imaging of melanopsin-driven light responses. Location of variants not screened in vitro (yellow) and the K340A control mutation (black) is also shown. Secondary structure based on homology with bovine rhodopsin [47]
Table 2.
Properties of OPN4 variants selected for in vitro screening
| Mutation | Conservation between species | Amino acid properties | Protein position | MAF | Equivalent rhodopsin residue | Functional role of equivalent residue in rhodopsin |
|---|---|---|---|---|---|---|
|
P10La rs2675703 |
Not conserved | Both non-polar and hydrophobic | NT | 0.08 (T) | Not conserved | |
|
T83M rs202029105 |
Conserved in Opn4M, l/V/L in Opn4X | Polar to non-polar | TM1 | 0.0002 (T) | L50 | Part of chromophore channel [72] |
|
F95L rs573102269 |
Conserved in all species | Loss of large side chain. Both hydrophobic and non-polar | TM1 | 0.0002 (C) | T62 | H-bond between TM1 and TM2 [73] |
|
Y146C rs200099863 |
Conserved in all species | Both polar and similar hydrophobicity | TM3 | 0.0002 (G) | E113 | Schiff Base Counterion [ 49 ] |
|
R168Cb rs143641898 |
Conserved in all species | Positive charge to polar. Increased hydrophobicity | TM3 / IL2 | 0.0005 (T) | R135 c | Conserved DRY motif [ 47 ] |
|
S183P rs151123640 |
Conserved in most species (T in Amphioxus) | Polar to non-polar | IL2 | 0.0002 (C) | E150c | Required for normal membrane trafficking [74] |
|
R186Hb rs141089672 |
Conserved in most species (K in Chicken and Zebrafish) | Both positive charge, increase in hydrophobicity | IL2 | 0.0005 (A) | Not conserved | |
|
G208S rs549998450 |
Conserved in all species | Non-polar to polar. Introduce large side chain | TM4 EL2 | 0.0004 (A) | G174 c | Unknown |
|
V213M rs202171086 |
Conserved in Opn4M, l/V in Opn4X | Both hydrophobic and non polar | EL2 | 0.0002 (A) | I179 | Unknown |
|
G216R rs201432667 |
Conserved in all species | Non-polar to polar, loss of hydrophobicity | EL2 | 0.0002 (A) | G182 c | Part of H-bond within EL2 [ 75 ] |
|
G266E rs576858032 |
Conserved in mammals, S/G/N in non-mammals | Non-polar to negative charge. Introduce large side chain and decrease hydrophobicity | IL3 | 0.0002 (A) | A234 | Arrestin Binding Site [76] |
|
W283C rs145634412 |
Not conserved | Non-polar to polar, loss of large side chain | IL3 | 0.001 (T) | Not Conserved | |
|
S308F rs559392371 |
Conserved in all species | Positive charge to polar. Similar hydrophobicity | TM6 | 0.001 (T) | C264 | Part of xWxPY motif [77, 78] |
|
H345Q rs184720512 |
Conserved in Opn4M, Y in Opn4X | Polar to non-polar, increase hydrophobicity | TM7 | 0.0002 (G) | Y301 | H-bond between TM2 and 7 [73] |
|
T394Ia rs1079610 |
Conserved as S or T in all species | Polar to non-polar, increase hydrophobicity | CT | 0.25 (C) | Not Conserved | |
|
R408C rs199878852 |
Conserved in mammals, not conserved in non-mammals | Positive charge to polar | CT | 0.0002 (T) | Not Conserved |
Highlighted in bold are properties of each variant that met criteria for further investigation. Minor allele is shown in brackets beneath MAF score. All variants were validated by multiple submissions and 1000 Genomes project [28], unless stated otherwise
aVariants with published data
bVariants validated by multiple submissions only
cEquivalent residue in rhodopsin is mutated in individuals with retinitis pigmentosa [70]
Overall, only five of the 96 non-synonymous mutations identified, P10L, T394I, G444D, R406W, and L365V, occur in more than 1% of the population and can be considered polymorphic. SNPs G444D, R406W, and L365V are located in poorly conserved domains of the melanopsin protein and produce only minor changes in biochemical properties of amino acids (Fig. 1). These substitutions were, therefore, deemed unlikely to significantly affect melanopsin function and were excluded from further analysis. Indeed, the majority of missense variants identified were rejected due to their location at sites poorly conserved amongst melanopsins, suggesting a non-critical role in melanopsin function. Given the suggested impact of OPN4 SNPs P10L and T394I on human behaviour [30–35] and their comparatively high minor allele frequency (MAF), both P10L and T394I were selected for further in vitro functional testing.
Notably, we identified several OPN4 variants that occur at highly conserved sites known to be critical for normal rhodopsin and GPCR function, including the conserved E/DRY motif required for G-protein activation [79], the vertebrate opsin counterion in transmembrane helix 3 which maintains stability of the chromophore when bound to opsin [49], and the conserved xWxPY opsin motif in transmembrane helix 6 [77, 78], which is essential for the opsin confirmation change necessary to trigger the phototransduction cascade. Mutation of these highly conserved sites severely disrupts protein function [49, 80, 81] and may, therefore, result in similar loss-of-function phenotypes in melanopsin. The minor allele frequency of variants located within these highly conserved sites indicates that these potentially deleterious mutations are rare (MAF < 0.01, Table 2).
We also identified a number of OPN4 variants located in the less conserved regions of the N-terminus, C-terminus, and intracellular loops of melanopsin, for which there are no homologous residues in rhodopsin—which may represent variants at sites with melanopsin-specific functional properties. These included P10L, R186H, W283C, T394I, and R408C. In addition to the 16 OPN4 variants selected for in vitro screening, the OPN4 K340A mutant was generated to act as a negative control during calcium imaging assays. This construct contains a targeted site-specific mutation (i.e., one not found naturally in the human genome) at the retinal chromophore-binding site rendering melanopsin non-functional [41, 42].
Cellular localisation and membrane trafficking of mutant OPN4 protein
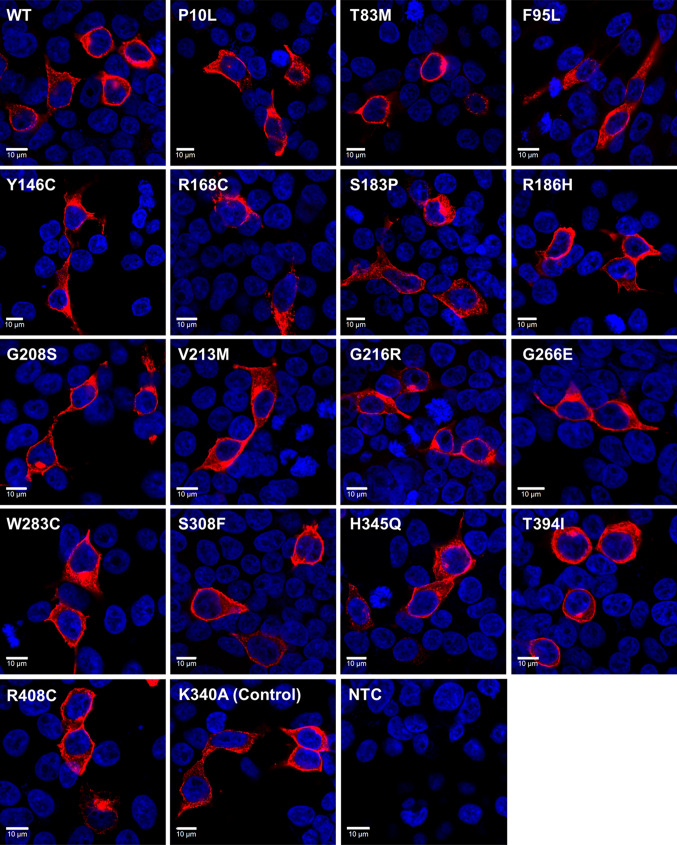
In the first instance, the subcellular localisation and membrane trafficking of selected OPN4 variants was examined using immunocytochemistry following transient transfection in HEK293T cells (Fig. 2). Overall, levels of OPN4 protein staining were similar between the different variants. All variants appeared to be trafficked normally and were expressed at the plasma membrane, with no obvious differences observed in overall levels of expression or cellular localisation between OPN4 WT and any OPN4 variants examined. Notably, we did not observe any evidence of increased protein aggregation in the endoplasmic reticulum for any OPN4 variant tested, as may be expected for mutations affecting tertiary structure and protein folding [26, 82].
Fig. 2.
Melanopsin variants show normal membrane localisation. Heterologous expression of pcDNA3.1 OPN4 WT and OPN4 variants in HEK293T cells labelled with anti-OPN4 antibody (red) and DAPI nuclear stain (blue). Scale bar 10 µm
Functional characterisation of OPN4 variants using fluorescent calcium imaging
We next used in vitro fluorescent calcium imaging methods to characterise the functional properties of each of the 16 selected OPN4 variants. In line with the previous reports, light stimulation (485 ± 6 nm, 1.21 × 1014 photons/cm2/s) led to a rapid elevation of intracellular calcium levels within HEK293T cells transfected with OPN4 WT (ΔF/F0, Fig. 3) [37, 38, 57], consistent with the activation of a Gαq/11 signalling cascade and release of calcium from intracellular stores [41, 42, 83].
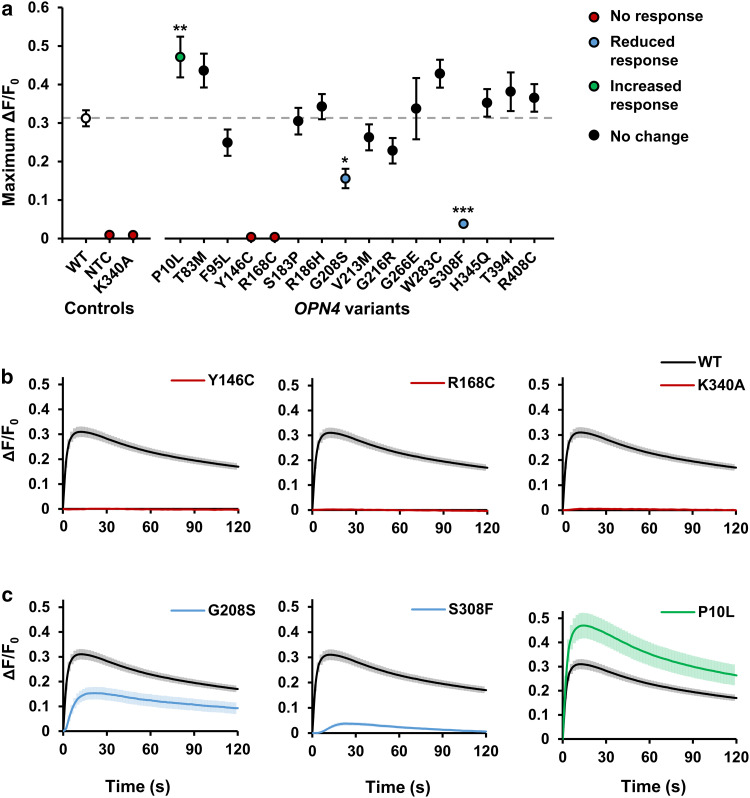
Fig. 3.
Melanopsin variants show abnormal intracellular calcium responses to light. Intracellular calcium levels of HEK293T cells transiently transfected with pcDNA3.1 OPN4 WT and OPN4 variants were monitored using the fluorescent calcium indicator Fluo4-AM. Melanopsin-driven light responses were triggered by the first light exposure used for fluorescent imaging (485 ± 6 nm). a Mean response amplitude (maximum ΔF/F0) for OPN4 WT and each OPN4 variant tested. Dashed grey line shows OPN4 WT response. b, c Traces showing the kinetics of intracellular calcium responses recorded from b non-functional OPN4 variants (red) and c OPN4 variants with significantly attenuated (blue) or elevated (green) intracellular calcium responses compared to OPN4 WT (black). N = 6 biological replicates for all groups except OPN4 WT (N = 24). Asterisk indicates significant Dunnett’s post hoc test (p > 0.05) compared to OPN4 WT. NTC is no transfection control. Error bars show standard error of mean. Where error bars are smaller than symbol, error bars are not shown
Several OPN4 variants failed to show any detectable changes in intracellular calcium levels in response to light (Fig. 3a, b). This included the negative control mutation K340A, which is incapable of binding retinal chromophore [41, 42], but also OPN4 variants Y146C and R168C. The response amplitudes of these variants were indistinguishable from untransfected cells, F(2,15) = 2.31, p = 0.134 (One-way ANOVA), suggesting that these mutations render OPN4 protein non-functional.
The remaining OPN4 variants appeared to produce functional melanopsin protein capable of coupling to a Gαq/11 signalling pathway, as shown by light-induced elevations of intracellular calcium levels (Fig. 3a). Whilst the majority of OPN4 variants showed response amplitudes similar to that observed for OPN4 WT controls, there was a significant main effect of genotype on response amplitude for functional variants, F(14, 93) = 7.44, p < 0.001 (One-way ANOVA). A Dunnett’s post hoc test revealed this was driven by three OPN4 variants with significantly attenuated or elevated responses compared to OPN4 WT (Fig. 3c). Both OPN4 S308F (p < 0.001) and G208S (p = 0.011) showed a significant reduction in response amplitude compared to OPN4 WT, with this effect observed consistently across all biological replicates. Only one variant, SNP P10L, demonstrated an overall elevation of response amplitude (p = 0.009). However, the increased response of P10L relative to OPN4 WT was not consistently observed across all biological replicates.
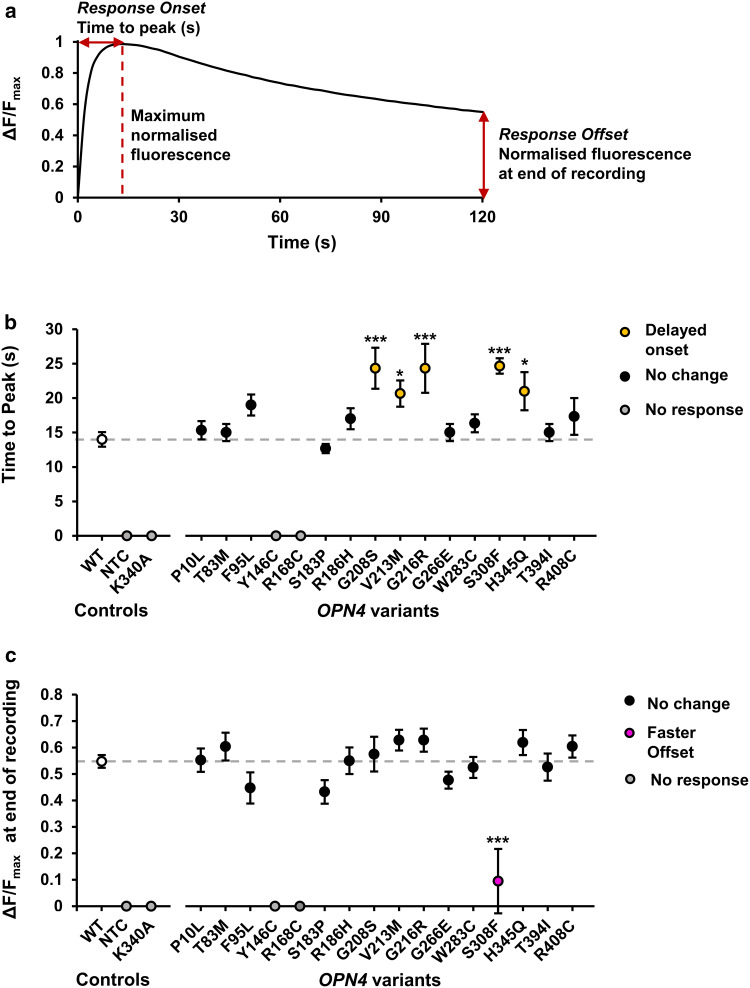
Comparison of response kinetics identified several OPN4 variants with modified response properties (Fig. 4). There was a significant main effect of genotype on rate of response onset, measured as time to peak fluorescence, F(14,93) = 4.73, p < 0.001 (One-way ANOVA), which a Dunnett’s test revealed was due to slower response onset in five OPN4 variants, G208S (p < 0.001), G216R (p < 0.001), S308F (p < 0.001), V213 M (p = 0.038), and H345Q (p = 0.025) (Fig. 4b). As a measure of response decay, normalised fluorescence values were compared at the end of the recordings (120 s after first light exposure). Only a single OPN4 variant, S308F, demonstrated a faster response offset compared to OPN4 WT controls (p < 0.001) (Fig. 4c) as determined using a Dunnett’s post hoc test to explore the significant main effect of genotype on response decay, F(14, 93) = 5.73, p < 0.0001 (one-way ANOVA). A summary of the functional phenotypes observed from all OPN4 variants tested in vitro is shown in Table 3.
Fig. 4.
OPN4 variants have abnormal response kinetics. a Parameters used to define response kinetics. Data were first normalised to baseline (Baseline = 0, ΔF/F0), then normalised to maximum for comparing response kinetics (Baseline = 0, maximum = 1, ΔF/Fmax). Response onset was measured as time to peak fluorescence (s). Response offset was measured as relative fluorescence recorded at end of recording (120 s after the first light exposure). b Mean response onset for each OPN4 variant tested. c Mean response offset for each OPN4 variant tested. Dashed grey line shows OPN4 WT response. N = 6 biological replicates for all OPN4 variants and N = 24 biological replicates for OPN4 WT control. Asterisk and triple asterisk represent significant post hoc Dunnett’s tests (p < 0.05 and p < 0.001 respectively) compared to OPN4 WT. NTC is no tranfection control. Error bars show standard error of the mean. Where error bars are smaller than symbol, error bars are not shown
Table 3.
Summary of key features and phenotype of 16 screened OPN4 variants
| Variant | Key features | Observed phenotype (relative to OPN4 WT) | ||
|---|---|---|---|---|
| Normal | ||||
| T83M | Change in polarity | No difference | ||
| F95L | Conserved in all species | No difference | ||
| S183P | Conserved in most species change in polarity | No difference | ||
| R186H | Change in hydrophobicity | No difference | ||
| G266E | Change in charge and hydrophobicity | No difference | ||
| W283C | Change in polarity | No difference | ||
| T394I | SNP associated with abnormal NIF behaviour in humans. Conserved in all species as S/T | No difference | ||
| R408C | Change in charge | No difference | ||
| Non-functional | ||||
| Y146C | Conserved in all species | No response | ||
| R168C | Conserved in all GPCRs. Change in charge and hydrophobicity | No response | ||
| Abnormal function | Amplitude | Onset | Offset | |
| P10L | SNP associated with abnormal NIF behaviour in humans. | Elevated | No difference | No difference |
| G208S | Conserved in all species change in polarity | Attenuated | Delayed | No difference |
| V213M | Conserved in all species as l/V | No difference | Delayed | No difference |
| G216R | Conserved in all species change in polarity and hydrophobicity | No difference | Delayed | No difference |
| S308F | Conserved in all species change in charge | Attenuated | Delayed | Faster |
| H345Q | Change in polarity and hydrophobicity | No difference | Delayed | No difference |
Discussion
Here, we show that bioinformatic approaches using sequence alignment combined with in vitro calcium imaging provide a high-throughput method for identifying non-synonymous human OPN4 mutations which result in altered melanopsin signalling. By focusing on variants located at highly conserved sites or those leading to a significant change in amino acid biochemical properties, we prioritised 16 of 96 recorded missense OPN4 variants for in vitro screening. Immunocytochemistry demonstrated that all 16 melanopsin variants were successfully trafficked to the plasma membrane, while assessment of melanopsin-driven calcium responses identified several variants with abnormal responses to light.
Identification of OPN4 variants with abnormal functional phenotypes
Of the seven variants that showed abnormal calcium responses, four are particularly strong candidates for key functional residues in the melanopsin protein. Introduction of mutations Y146C and R168C in OPN4 abolished responses to light, and two further variants, S308F and G208S reliably demonstrated attenuated response amplitude and delayed onset compared to OPN4 WT controls. Furthermore, these deleterious variants occur at sites highly conserved between different classes of opsin (and GPCRs in general), providing likely mechanisms by which these mutations cause loss-of-function. For example, R168C is part of the E/DRY motif found in nearly all GPCRs. The “ionic lock” between these residues and a conserved negative-changed amino acid in transmembrane helix 6 (TM6) is necessary for maintaining the inactive conformation of GPCRs [47, 77, 79, 80]. S308F is part of the xWxPY motif located within TM6, acting as a “rotamer toggle switch” [77, 84], which facilitates rotation of TM6 into the active confirmation following photoisomerisation of retinal chromophore [77, 85]. Given that variation at these sites attenuates or abolishes melanopsin activity, it is highly likely that these motifs perform similar functions in melanopsin.
A third deleterious variant, Y146C, occurs at a conserved opsin site in TM3. In vertebrate opsins, this site represents the glutamic acid counterion, a negatively charged amino acid that counteracts the positive charge of the Schiff base that binds the chromophore to the opsin apoprotein allowing absorption of photons in the visible spectrum [49, 86]. In invertebrate opsins, this site is occupied by a conserved tyrosine, and the glutamic acid counterion is displaced to extracellular loop 2 [87]. Interestingly, melanopsin possesses an invertebrate-like conformation of residues at these two positions. It is, therefore, surprising that variation at the vertebrate counterion location causes a total loss of melanopsin function. This site cannot act as the melanopsin counterion as tyrosine is not negatively charged; however, it is possible that this site still critically interacts with the Schiff base and disrupts retinal binding or isomerisation when mutated.
Human OPN4 variants inform melanopsin structure–function relationships
Overall, we find that the most damaging OPN4 variants occur at highly conserved residues in the opsin chromophore-binding pocket. In contrast, substitutions in more variable protein domains have no observable effect even if they substantially alter hydrophobicity or charge of amino acid residues, such as R186H and G266E located in intracellular loops 2 and 3 respectively. Notably, none of the critical residues identified in the in vitro screen appear specific to melanopsin. In fact, of the five OPN4 variants tested for which there is no equivalent residue in rhodopsin, none appear to be important for normal melanopsin function.
Interestingly, neither OPN4 T394I nor P10L, two SNPs previously associated with abnormal and attenuated NIF behaviour in humans [30–35], showed melanopsin loss-of-function in vitro. Instead, we observed no difference between T394I and WT. Overall, we found significantly larger calcium responses for P10L compared to OPN4 WT, although this was not consistent between all biological replicates. Based on its position in the N-terminus and the loss of a proline, there has been some speculation that P10L has a similar effect on melanopsin as the P23H mutation in rhodopsin, which leads to severe protein misfolding [82, 88]. However, our cellular localisation data suggest this is not the case, with no evidence of protein misfolding observed for P10L. As such, there is currently no clear mechanism to explain how P10L might be associated with an increased risk of seasonal affective disorder as previously reported [33–35].
It remains possible that the T394I or P10L SNPs, as well as other OPN4 variants, may cause subtle phenotypes not captured by the in vitro calcium imaging screen used in this study. More detailed analysis using a range of light intensities and or stimulation protocols may reveal further phenotypes, such as reduced activation thresholds, altered responses to dim light, or changes in maximal responses elicited by saturating levels of light. However, it is also clear that in vitro cell line expression systems do not fully replicate the cellular environment of ipRGCs [58] and may, therefore, limit the study of OPN4 variants with roles in ipRGC-specific aspects of melanopsin protein function, such as regulatory motifs or post-translational modification sites. Examining the function of OPN4 variants following targeted expression into ipRGCs of the mouse retina, potentially via AAV delivery and Cre-lox based approaches [39, 40], would more closely replicate the native signalling environment of melanopsin proteins and permit in-depth functional analysis of melanopsin-specific functions, such as temporal integration, chromophore bistability, or contribution to multiple active states [60]. Nevertheless, functional characterisation of melanopsin variants in vivo is both time-consuming and expensive, and is, therefore, not suitable for screening large numbers of variants. The in vitro approach used here provides a relatively high-throughput and effective method for preliminary screening to identify OPN4 SNP variants with altered functional properties.
Implications of OPN4 variants for the human population
The identification of naturally occurring OPN4 missense mutations resulting in deleterious functional properties suggests that individuals with non-functional or severely impaired melanopsin activity may exist within the human population. Given the frequency of these deleterious variants, it would seem that individuals homozygous for these mutations (or carrying copies of two different deleterious variants) will be extremely rare. These observations alone suggest that a strong selective pressure exists to maintain normal melanopsin function. Therefore, an important consideration is whether humans either heterozygous or homozygous for these mutations will exhibit significant disruption of melanopsin-dependant behaviours. Screening such individuals for circadian disruption or abnormal non-image forming responses to light could help to determine the role of melanopsin in human physiology, and whether altered melanopsin light perception is associated with human disease.
However, based on our current understanding of the melanopsin system and studies of mice with genetically ablated melanopsin [7–14], we may expect that even highly deleterious OPN4 variants will only result in minimal behavioural phenotypes. In the healthy retina, melanopsin-expressing ipRGCs receive synaptic input from rods and cones and act as a conduit for passing outer retina light signals to NIF centres [89]. The overlapping roles of rod, cone, and melanopsin-driven light responses in ipRGC function likely minimises the behavioural effect of melanopsin missense mutations. For example, previous reports have demonstrated that the NIF responses to light of melanopsin knockout mice are typically attenuated, but not abolished—with modest deficits typically observed only under bright light conditions. However, it has also become increasingly clear that melanopsin provides a greater contribution to ipRGC function and NIF responses to light under more physiological conditions, including control of steady-state pupil size under constant environmental illumination [90].
Recent evidence has revealed additional roles for melanopsin within the classical visual system [16–20], with melanopsin-driven light responses contributing to brightness discrimination [91, 92], contrast sensitivity [17, 18], and adaptation of visual responses [93, 94], via both retrograde signalling within the retina and modulation of dopamine signalling pathways [19, 95–99], as well as through direct projections of ipRGCs to visual centres [16, 17]. Furthermore, people with abnormal melanopsin function could exhibit other symptoms related to the role of melanopsin during postnatal development, including the patterning of retina vasculature [100], and the refinement and segregation of retinogeniculate projections to visual areas of the brain [20, 21], potentially influencing visual acuity in adulthood [21]. It is, therefore, possible that humans possessing deleterious OPN4 variants may experience a range of behavioural symptoms related to abnormal melanopsin function, including circadian disruption, attenuated pupil constriction, reductions in contrast sensitivity, and attenuated adaptation of visual signalling pathways to changes in environmental light conditions.
Conclusion
In conclusion, we have identified several previously uncharacterised naturally occurring missense variants in the human OPN4 gene that result in melanopsin proteins with a significant loss-of-function phenotype. These data indicate that individuals with abnormal or abolished melanopsin activity may exist within the human population, and although rare, may be at increased risk of sleep and circadian disruption, as well as potential visual deficits. The growing availability of next-generation sequencing data is likely to lead to the identification of further rare genetic variants of the human OPN4 gene. The methods described here for identification and in vitro functional characterisation of deleterious OPN4 variants will be highly applicable for future investigations of OPN4 mutations and defining structure–function relationships of the melanopsin protein.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Data S1. Properties of 96 missense mutations in the OPN4 gene identified from dbSNP (Build 140). (XLSX 15 kb)
Figure S1. Amino acid alignment of human melanopsin (OPN4) and bovine rhodopsin (RHO). Protein domain boundaries (shown as vertical lines) are defined based on crystal structure of bovine rhodopsin [47]. TM = transmembrane helix, IL = intracellular loop, EL = extracellular loop. Highly variable N and C termini are excluded from the alignment. OPN4 variants selected for in vitro screening with equivalent rhodopsin residues are highlighted in yellow. Numbers show position of the first and last residues of each row (TIF 693 kb)
Figure S2. Comparison of OPN4 WT positive controls between plates confirms melanopsin-driven calcium responses are replicable. Intracellular calcium levels of HEK293T cells transiently transfected with pcDNA3.1 OPN4 WT as monitored using Fluo4-AM. (a) Kinetics of intracellular calcium normalised to baseline (first data point, ΔF/F0). (b) Mean response amplitude, measured as maximum ΔF/F0, is not significantly different between biological replicates, F(3,20) = 0.54, p = 0.659 (one-way ANOVA) (c-d) Kinetics of intracellular calcium normalised to baseline, then maximum (Baseline = 0, Maximum = 1, ΔF/Fmax) are not significantly different between biological replicates, assessed using (c) Mean time to peak ΔF/Fmax, F(3,20) = 0.79, p = 0.513 (one-way ANOVA) and (d) Mean ΔF/Fmax at end of recording, F(3,20) = 1.00, p =0.412 (one-way ANOVA). N = 6 biological replicates for each OPN4 WT plate. n.s. = not significant. Error bars = standard error of the mean (TIF 662 kb)
Table S1. Site-directed mutagenesis primers used for construction of OPN4 variant expression constructs. Base pair change from template sequence is shown in bold and underlined. (TIF 866 kb)
Funding
Funding was provided by Biotechnology and Biological Sciences Research Council (Grant no: BB/M009998/1) and Engineering and Physical Sciences Research Council (Grant no: 1350916).
Contributor Information
Steven Hughes, Phone: +44 (0)1865 618 669, Email: steven.hughes@ndcn.ox.ac.uk.
Mark W. Hankins, Phone: +44 (0)1865 618 661, Email: mark.hankins@eye.ox.ac.uk
References
- 1.Hannibal J, Christiansen AT, Heegaard S, et al. Melanopsin expressing human retinal ganglion cells: subtypes, distribution, and intraretinal connectivity. J Comp Neurol. 2017;525:1934–1961. doi: 10.1002/cne.24181. [DOI] [PubMed] [Google Scholar]
- 2.Nasir-Ahmad S, Lee SCS, Martin PR, Grunert U. Melanopsin-expressing ganglion cells in human retina: morphology, distribution, and synaptic connections. J Comp Neurol. 2017 doi: 10.1002/cne.24176. [DOI] [PubMed] [Google Scholar]
- 3.Liao H-W, Ren X, Peterson BB, et al. Melanopsin-expressing ganglion cells on macaque and human retinas form two morphologically distinct populations. J Comp Neurol. 2016;524:2845–2872. doi: 10.1002/cne.23995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Provencio I, Rodriguez IR, Jiang G, et al. A novel human opsin in the inner retina. J Neurosci. 2000;20:600–605. doi: 10.1523/JNEUROSCI.20-02-00600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hattar S, Liao HW, Takao M, et al. Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science. 2002;295:1065–1070. doi: 10.1126/science.1069609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berson DM, Dunn FA, Takao M. Phototransduction by retinal ganglion cells that set the circadian clock. Science. 2002;295:1070–1073. doi: 10.1126/science.1067262. [DOI] [PubMed] [Google Scholar]
- 7.Hattar S, Lucas RJ, Mrosovsky N, et al. Melanopsin and rod-cone photoreceptive systems account for all major accessory visual functions in mice. Nature. 2003;424:76–81. doi: 10.1038/nature01761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panda S, Sato TK, Castrucci AM, et al. Melanopsin (Opn4) requirement for normal light-induced circadian phase shifting. Science. 2002;298:2213–2216. doi: 10.1126/science.1076848. [DOI] [PubMed] [Google Scholar]
- 9.Ruby NF, Brennan TJ, Xie X, et al. Role of melanopsin in circadian responses to light. Science. 2002;298:2211–2213. doi: 10.1126/science.1076701. [DOI] [PubMed] [Google Scholar]
- 10.Tsai JW, Hannibal J, Hagiwara G, et al. Melanopsin as a sleep modulator: circadian gating of the direct effects of light on sleep and altered sleep homeostasis in Opn4(−/−) mice. PLoS Biol. 2009;7:e1000125. doi: 10.1371/journal.pbio.1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altimus CM, Guler AD, Villa KL, et al. Rods-cones and melanopsin detect light and dark to modulate sleep independent of image formation. Proc Natl Acad Sci USA. 2008;105:19998–20003. doi: 10.1073/pnas.0808312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lupi D, Oster H, Thompson S, Foster RG. The acute light-induction of sleep is mediated by OPN4-based photoreception. Nat Neurosci. 2008;11:1068–1073. doi: 10.1038/nn.2179. [DOI] [PubMed] [Google Scholar]
- 13.Pilorz V, Tam SKE, Hughes S, et al. Melanopsin regulates both sleep-promoting and arousal-promoting responses to light. PLoS Biol. 2016;14:e1002482. doi: 10.1371/journal.pbio.1002482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lucas RJ, Hattar S, Takao M, et al. Diminished pupillary light reflex at high irradiances in melanopsin-knockout mice. Science. 2003;299:245–247. doi: 10.1126/science.1077293. [DOI] [PubMed] [Google Scholar]
- 15.Gamlin PDR, McDougal DH, Pokorny J, et al. Human and macaque pupil responses driven by melanopsin-containing retinal ganglion cells. Vision Res. 2007;47:946–954. doi: 10.1016/j.visres.2006.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ecker JL, Dumitrescu ON, Wong KY, et al. Melanopsin-expressing retinal ganglion-cell photoreceptors: cellular diversity and role in pattern vision. Neuron. 2010;67:49–60. doi: 10.1016/j.neuron.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Estevez ME, Fogerson PM, Ilardi MC, et al. Form and function of the M4 cell, an intrinsically photosensitive retinal ganglion cell type contributing to geniculocortical vision. J Neurosci. 2012;32:13608–13620. doi: 10.1523/JNEUROSCI.1422-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt TM, Alam NM, Chen S, et al. A role for melanopsin in alpha retinal ganglion cells and contrast detection. Neuron. 2014;82:781–788. doi: 10.1016/j.neuron.2014.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang D-Q, Wong KY, Sollars PJ, et al. Intraretinal signaling by ganglion cell photoreceptors to dopaminergic amacrine neurons. Proc Natl Acad Sci USA. 2008;105:14181–14186. doi: 10.1073/pnas.0803893105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Renna JM, Weng S, Berson DM. Light acts through melanopsin to alter retinal waves and segregation of retinogeniculate afferents. Nat Neurosci. 2011;14:827–829. doi: 10.1038/nn.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chew KS, Renna JM, McNeill DS, et al. A subset of ipRGCs regulates both maturation of the circadian clock and segregation of retinogeniculate projections in mice. Elife. 2017 doi: 10.7554/eLife.22861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ksendzovsky A, Pomeraniec IJ, Zaghloul KA, et al. Clinical implications of the melanopsin-based non-image-forming visual system. Neurology. 2017;88:1282–1290. doi: 10.1212/WNL.0000000000003761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abecasis GR, Altshuler D, Auton A, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bush WS, Moore JH. Chapter 11: genome-wide association studies. PLoS Comput Biol. 2012;8:e1002822. doi: 10.1371/journal.pcbi.1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kitts A, Phan L, Ward M et al (2013) The Database of Short Genetic Variation (dbSNP). In: Bethesda MD (ed) The NCBI handbook, 2nd edn. National Center for Biotechnology Information (US). https://www.ncbi.nlm.nih.gov/books/NBK174586/. (internet). Accessed 1 Aug 2016
- 26.Mendes HF, van der Spuy J, Chapple JP, Cheetham ME. Mechanisms of cell death in rhodopsin retinitis pigmentosa: implications for therapy. Trends Mol Med. 2005;11:177–185. doi: 10.1016/j.molmed.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 27.Athanasiou D, Aguila M, Bellingham J, et al. The molecular and cellular basis of rhodopsin retinitis pigmentosa reveals potential strategies for therapy. Prog Retin Eye Res. 2017 doi: 10.1016/j.preteyeres.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Auton A, Brooks LD, Durbin RM, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee S, Hida A, Tsujimura S, et al. Association between melanopsin gene polymorphism (I394T) and pupillary light reflex is dependent on light wavelength. J Physiol Anthropol. 2013;32:16. doi: 10.1186/1880-6805-32-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee S-I, Hida A, Kitamura S, et al. Association between the melanopsin gene polymorphism OPN4*Ile394Thr and sleep/wake timing in Japanese university students. J Physiol Anthropol. 2014;33:9. doi: 10.1186/1880-6805-33-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Higuchi S, Hida A, Tsujimura S, et al. Melanopsin gene polymorphism I394T is associated with pupillary light responses in a dose-dependent manner. PLoS ONE. 2013;8:e60310. doi: 10.1371/journal.pone.0060310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roecklein KA, Rohan KJ, Duncan WC, et al. A missense variant (P10L) of the melanopsin (OPN4) gene in seasonal affective disorder. J Affect Disord. 2009;114:279–285. doi: 10.1016/j.jad.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roecklein K, Wong P, Ernecoff N, et al. The post illumination pupil response is reduced in seasonal affective disorder. Psychiatry Res. 2013;210:150–158. doi: 10.1016/j.psychres.2013.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roecklein K, Wong P, Franzen PL, et al. Melanopsin gene variations interact with season to predict sleep onset and chronotype. Chronobiol Int. 2012;29:1036–1047. doi: 10.3109/07420528.2012.706766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fahrenkrug J, Falktoft B, Georg B, Rask L. N-linked deglycosylated melanopsin retains its responsiveness to light. Biochemistry. 2009;48:5142–5148. doi: 10.1021/bi900249n. [DOI] [PubMed] [Google Scholar]
- 37.Blasic JRJ, Lane Brown R, Robinson PR. Light-dependent phosphorylation of the carboxy tail of mouse melanopsin. Cell Mol Life Sci. 2012;69:1551–1562. doi: 10.1007/s00018-011-0891-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blasic JRJ, Matos-Cruz V, Ujla D, et al. Identification of critical phosphorylation sites on the carboxy tail of melanopsin. Biochemistry. 2014;53:2644–2649. doi: 10.1021/bi401724r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mure LS, Hatori M, Zhu Q, et al. Melanopsin-encoded response properties of intrinsically photosensitive retinal ganglion cells. Neuron. 2016;90:1016–1027. doi: 10.1016/j.neuron.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Somasundaram P, Wyrick GR, Fernandez DC, et al. C-terminal phosphorylation regulates the kinetics of a subset of melanopsin-mediated behaviors in mice. Proc Natl Acad Sci USA. 2017;114:2741–2746. doi: 10.1073/pnas.1611893114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melyan Z, Tarttelin EE, Bellingham J, et al. Addition of human melanopsin renders mammalian cells photoresponsive. Nature. 2005;433:741–745. doi: 10.1038/nature03344. [DOI] [PubMed] [Google Scholar]
- 42.Kumbalasiri T, Rollag MD, Isoldi MC, et al. Melanopsin triggers the release of internal calcium stores in response to light. Photochem Photobiol. 2007;83:273–279. doi: 10.1562/2006-07-11-RA-964. [DOI] [PubMed] [Google Scholar]
- 43.Tsukamoto H, Kubo Y, Farrens DL, et al. Retinal attachment instability is diversified among mammalian melanopsins. J Biol Chem. 2015;290:27176–27187. doi: 10.1074/jbc.M115.666305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenbaum DM, Rasmussen SGF, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Provencio I, Jiang G, De Grip WJ, et al. Melanopsin: an opsin in melanophores, brain, and eye. Proc Natl Acad Sci USA. 1998;95:340–345. doi: 10.1073/pnas.95.1.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith SO. Structure and activation of the visual pigment rhodopsin. Annu Rev Biophys. 2010;39:309–328. doi: 10.1146/annurev-biophys-101209-104901. [DOI] [PubMed] [Google Scholar]
- 47.Palczewski K, Kumasaka T, Hori T, et al. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 48.Deupi X. Relevance of rhodopsin studies for GPCR activation. Biochim Biophys Acta. 2014;1837:674–682. doi: 10.1016/j.bbabio.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 49.Sakmar TP, Franke RR, Khorana HG. Glutamic acid-113 serves as the retinylidene Schiff base counterion in bovine rhodopsin. Proc Natl Acad Sci USA. 1989;86:8309–8313. doi: 10.1073/pnas.86.21.8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin SW, Kochendoerfer GG, Carroll KS, et al. Mechanisms of spectral tuning in blue cone visual pigments. Visible and raman spectroscopy of blue-shifted rhodopsin mutants. J Biol Chem. 1998;273:24583–24591. doi: 10.1074/jbc.273.38.24583. [DOI] [PubMed] [Google Scholar]
- 51.Franke RR, Konig B, Sakmar TP, et al. Rhodopsin mutants that bind but fail to activate transducin. Science. 1990;250:123–125. doi: 10.1126/science.2218504. [DOI] [PubMed] [Google Scholar]
- 52.Karnik SS, Ridge KD, Bhattacharya S, Khorana HG. Palmitoylation of bovine opsin and its cysteine mutants in COS cells. Proc Natl Acad Sci USA. 1993;90:40–44. doi: 10.1073/pnas.90.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hwa J, Reeves PJ, Klein-Seetharaman J, et al. Structure and function in rhodopsin: further elucidation of the role of the intradiscal cysteines, Cys-110, -185, and -187, in rhodopsin folding and function. Proc Natl Acad Sci USA. 1999;96:1932–1935. doi: 10.1073/pnas.96.5.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qiu X, Kumbalasiri T, Carlson SM, et al. Induction of photosensitivity by heterologous expression of melanopsin. Nature. 2005;433:745–749. doi: 10.1038/nature03345. [DOI] [PubMed] [Google Scholar]
- 55.Panda S, Nayak SK, Campo B, et al. Illumination of the melanopsin signaling pathway. Science. 2005;307:600–604. doi: 10.1126/science.1105121. [DOI] [PubMed] [Google Scholar]
- 56.Graham DM, Wong KY, Shapiro P, et al. Melanopsin ganglion cells use a membrane-associated rhabdomeric phototransduction cascade. J Neurophysiol. 2008;99:2522–2532. doi: 10.1152/jn.01066.2007. [DOI] [PubMed] [Google Scholar]
- 57.Bailes HJ, Lucas RJ. Human melanopsin forms a pigment maximally sensitive to blue light (λmax ≈ 479 nm) supporting activation of G(q/11) and G(i/o) signalling cascades. Proc Biol Sci. 2013;280:20122987. doi: 10.1098/rspb.2012.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hughes S, Hankins MW, Foster RG, Peirson SN. Melanopsin phototransduction: slowly emerging from the dark. Prog Brain Res. 2012;199:19–40. doi: 10.1016/B978-0-444-59427-3.00002-2. [DOI] [PubMed] [Google Scholar]
- 59.Mure LS, Cornut P-L, Rieux C, et al. Melanopsin bistability: a fly’s eye technology in the human retina. PLoS ONE. 2009;4:e5991. doi: 10.1371/journal.pone.0005991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Emanuel AJ, Do MTH. Melanopsin tristability for sustained and broadband phototransduction. Neuron. 2015;85:1043–1055. doi: 10.1016/j.neuron.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sexton TJ, Golczak M, Palczewski K, Van Gelder RN. Melanopsin is highly resistant to light and chemical bleaching in vivo. J Biol Chem. 2012;287:20888–20897. doi: 10.1074/jbc.M111.325969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bellingham J, Chaurasia SS, Melyan Z, et al. Evolution of melanopsin photoreceptors: discovery and characterization of a new melanopsin in nonmammalian vertebrates. PLoS Biol. 2006;4:e254. doi: 10.1371/journal.pbio.0040254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cai Z, Tsung EF, Marinescu VD, et al. Bayesian approach to discovering pathogenic SNPs in conserved protein domains. Hum Mutat. 2004;24:178–184. doi: 10.1002/humu.20063. [DOI] [PubMed] [Google Scholar]
- 65.Bulka B, desJardins M, Freeland SJ. An interactive visualization tool to explore the biophysical properties of amino acids and their contribution to substitution matrices. BMC Bioinformatics. 2006;7:329. doi: 10.1186/1471-2105-7-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Small KM, Tanguay DA, Nandabalan K, et al. Gene and protein domain-specific patterns of genetic variability within the G-protein coupled receptor superfamily. Am J Pharmacogenomics. 2003;3:65–71. doi: 10.2165/00129785-200303010-00008. [DOI] [PubMed] [Google Scholar]
- 67.Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 68.Okada T, Ernst OP, Palczewski K, Hofmann KP. Activation of rhodopsin: new insights from structural and biochemical studies. Trends Biochem Sci. 2001;26:318–324. doi: 10.1016/S0968-0004(01)01799-6. [DOI] [PubMed] [Google Scholar]
- 69.Scheerer P, Park JH, Hildebrand PW, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 70.Rakoczy EP, Kiel C, McKeone R, et al. Analysis of disease-linked rhodopsin mutations based on structure, function, and protein stability calculations. J Mol Biol. 2011;405:584–606. doi: 10.1016/j.jmb.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 71.Pires SS, Hughes S, Turton M, et al. Differential expression of two distinct functional isoforms of melanopsin (Opn4) in the mammalian retina. J Neurosci. 2009;29:12332–12342. doi: 10.1523/JNEUROSCI.2036-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schadel SA, Heck M, Maretzki D, et al. Ligand channeling within a G-protein-coupled receptor. The entry and exit of retinals in native opsin. J Biol Chem. 2003;278:24896–24903. doi: 10.1074/jbc.M302115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Okada T, Fujiyoshi Y, Silow M, et al. Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography. Proc Natl Acad Sci USA. 2002;99:5982–5987. doi: 10.1073/pnas.082666399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pulagam LP, Palczewski K. Electrostatic compensation restores trafficking of the autosomal recessive retinitis pigmentosa E150K opsin mutant to the plasma membrane. J Biol Chem. 2010;285:29446–29456. doi: 10.1074/jbc.M110.151407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun X, Agren H, Tu Y. Functional water molecules in rhodopsin activation. J Phys Chem B. 2014;118:10863–10873. doi: 10.1021/jp505180t. [DOI] [PubMed] [Google Scholar]
- 76.Raman D, Osawa S, Weiss ER. Binding of arrestin to cytoplasmic loop mutants of bovine rhodopsin. Biochemistry. 1999;38:5117–5123. doi: 10.1021/bi9824588. [DOI] [PubMed] [Google Scholar]
- 77.Deupi X, Kobilka B. Activation of G protein-coupled receptors. Adv Protein Chem. 2007;74:137–166. doi: 10.1016/S0065-3233(07)74004-4. [DOI] [PubMed] [Google Scholar]
- 78.Angel TE, Chance MR, Palczewski K. Conserved waters mediate structural and functional activation of family A (rhodopsin-like) G protein-coupled receptors. Proc Natl Acad Sci USA. 2009;106:8555–8560. doi: 10.1073/pnas.0903545106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rovati GE, Capra V, Neubig RR. The highly conserved DRY motif of class A G protein-coupled receptors: beyond the ground state. Mol Pharmacol. 2007;71:959–964. doi: 10.1124/mol.106.029470. [DOI] [PubMed] [Google Scholar]
- 80.Vogel R, Mahalingam M, Ludeke S, et al. Functional role of the “ionic lock”—an interhelical hydrogen-bond network in family A heptahelical receptors. J Mol Biol. 2008;380:648–655. doi: 10.1016/j.jmb.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 81.Goncalves JA, South K, Ahuja S, et al. Highly conserved tyrosine stabilizes the active state of rhodopsin. Proc Natl Acad Sci USA. 2010;107:19861–19866. doi: 10.1073/pnas.1009405107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Noorwez SM, Kuksa V, Imanishi Y, et al. Pharmacological chaperone-mediated in vivo folding and stabilization of the P23H-opsin mutant associated with autosomal dominant retinitis pigmentosa. J Biol Chem. 2003;278:14442–14450. doi: 10.1074/jbc.M300087200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hughes S, Jagannath A, Hickey D, et al. Using siRNA to define functional interactions between melanopsin and multiple G Protein partners. Cell Mol Life Sci. 2015;72:165–179. doi: 10.1007/s00018-014-1664-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shi L, Liapakis G, Xu R, et al. Beta2 adrenergic receptor activation. Modulation of the proline kink in transmembrane 6 by a rotamer toggle switch. J Biol Chem. 2002;277:40989–40996. doi: 10.1074/jbc.M206801200. [DOI] [PubMed] [Google Scholar]
- 85.Bhattacharya S, Hall SE, Vaidehi N. Agonist-induced conformational changes in bovine rhodopsin: insight into activation of G-protein-coupled receptors. J Mol Biol. 2008;382:539–555. doi: 10.1016/j.jmb.2008.06.084. [DOI] [PubMed] [Google Scholar]
- 86.Shichida Y, Matsuyama T. Evolution of opsins and phototransduction. Philos Trans R Soc Lond B Biol Sci. 2009;364:2881–2895. doi: 10.1098/rstb.2009.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Terakita A, Koyanagi M, Tsukamoto H, et al. Counterion displacement in the molecular evolution of the rhodopsin family. Nat Struct Mol Biol. 2004;11:284–289. doi: 10.1038/nsmb731. [DOI] [PubMed] [Google Scholar]
- 88.Olsson JE, Gordon JW, Pawlyk BS, et al. Transgenic mice with a rhodopsin mutation (Pro23His): a mouse model of autosomal dominant retinitis pigmentosa. Neuron. 1992;9:815–830. doi: 10.1016/0896-6273(92)90236-7. [DOI] [PubMed] [Google Scholar]
- 89.Güler AD, Ecker JL, Lall GS, et al. Melanopsin cells are the principal conduits for rod-cone input to non-image-forming vision. Nature. 2008;453:102–105. doi: 10.1038/nature06829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Keenan WT, Rupp AC, Ross RA, et al. A visual circuit uses complementary mechanisms to support transient and sustained pupil constriction. Elife. 2016 doi: 10.7554/eLife.15392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Brown TM, Tsujimura S-I, Allen AE, et al. Melanopsin-based brightness discrimination in mice and humans. Curr Biol. 2012;22:1134–1141. doi: 10.1016/j.cub.2012.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brown TM, Allen AE, al-Enezi J, et al. The melanopic sensitivity function accounts for melanopsin-driven responses in mice under diverse lighting conditions. PLoS One. 2013;8:e53583. doi: 10.1371/journal.pone.0053583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Milosavljevic N, Allen AE, Cehajic-Kapetanovic J, Lucas RJ. Chemogenetic Activation of ipRGCs Drives Changes in Dark-Adapted (Scotopic) Electroretinogram. Invest Ophthalmol Vis Sci. 2016;57:6305–6312. doi: 10.1167/iovs.16-20448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Allen AE, Storchi R, Martial FP, et al. Melanopsin-driven light adaptation in mouse vision. Curr Biol. 2014;24:2481–2490. doi: 10.1016/j.cub.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Newkirk GS, Hoon M, Wong RO, Detwiler PB. Inhibitory inputs tune the light response properties of dopaminergic amacrine cells in mouse retina. J Neurophysiol. 2013;110:536–552. doi: 10.1152/jn.00118.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Atkinson CL, Feng J, Zhang D-Q. Functional integrity and modification of retinal dopaminergic neurons in the rd1 mutant mouse: roles of melanopsin and GABA. J Neurophysiol. 2013;109:1589–1599. doi: 10.1152/jn.00786.2012. [DOI] [PubMed] [Google Scholar]
- 97.Prigge CL, Yeh P-T, Liou N-F, et al. M1 ipRGCs Influence Visual Function through Retrograde Signaling in the Retina. J Neurosci. 2016;36:7184–7197. doi: 10.1523/JNEUROSCI.3500-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Semo M, Coffey P, Gias C, Vugler A. Retrograde Melanopsin Signaling Increases With Age in Retinal Degenerate Mice Lacking Rods and the Majority of Cones. Invest Ophthalmol Vis Sci. 2016;57:115–125. doi: 10.1167/iovs.15-17609. [DOI] [PubMed] [Google Scholar]
- 99.Zhao X, Wong KY, Zhang D-Q. Mapping physiological inputs from multiple photoreceptor systems to dopaminergic amacrine cells in the mouse retina. Sci Rep. 2017;7:7920. doi: 10.1038/s41598-017-08172-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rao S, Chun C, Fan J, et al. A direct and melanopsin-dependent fetal light response regulates mouse eye development. Nature. 2013;494:243–246. doi: 10.1038/nature11823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Properties of 96 missense mutations in the OPN4 gene identified from dbSNP (Build 140). (XLSX 15 kb)
Figure S1. Amino acid alignment of human melanopsin (OPN4) and bovine rhodopsin (RHO). Protein domain boundaries (shown as vertical lines) are defined based on crystal structure of bovine rhodopsin [47]. TM = transmembrane helix, IL = intracellular loop, EL = extracellular loop. Highly variable N and C termini are excluded from the alignment. OPN4 variants selected for in vitro screening with equivalent rhodopsin residues are highlighted in yellow. Numbers show position of the first and last residues of each row (TIF 693 kb)
Figure S2. Comparison of OPN4 WT positive controls between plates confirms melanopsin-driven calcium responses are replicable. Intracellular calcium levels of HEK293T cells transiently transfected with pcDNA3.1 OPN4 WT as monitored using Fluo4-AM. (a) Kinetics of intracellular calcium normalised to baseline (first data point, ΔF/F0). (b) Mean response amplitude, measured as maximum ΔF/F0, is not significantly different between biological replicates, F(3,20) = 0.54, p = 0.659 (one-way ANOVA) (c-d) Kinetics of intracellular calcium normalised to baseline, then maximum (Baseline = 0, Maximum = 1, ΔF/Fmax) are not significantly different between biological replicates, assessed using (c) Mean time to peak ΔF/Fmax, F(3,20) = 0.79, p = 0.513 (one-way ANOVA) and (d) Mean ΔF/Fmax at end of recording, F(3,20) = 1.00, p =0.412 (one-way ANOVA). N = 6 biological replicates for each OPN4 WT plate. n.s. = not significant. Error bars = standard error of the mean (TIF 662 kb)
Table S1. Site-directed mutagenesis primers used for construction of OPN4 variant expression constructs. Base pair change from template sequence is shown in bold and underlined. (TIF 866 kb)