Abstract
Entity—attribute—value (EAV) tables form the major component of several mainstream electronic patient record systems (EPRSs). Such systems have been optimized for real-time retrieval of individual patient data. Data warehousing, on the other hand, involves cross-patient data retrieval based on values of patient attributes, with a focus on ad hoc query. Attribute-centric query is inherently more difficult when data are stored in EAV form than when they are stored conventionally. The authors illustrate their approach to the attribute-centric query problem with ACT/DB, a database for managing clinical trials data. This approach is based on metadata supporting a query front end that essentially hides the EAV/non-EAV nature of individual attributes from the user. The authors' work does not close the query problem, and they identify several complex subproblems that are still to be solved.
A clinical patient record needs to store several thousand possible facts for patients across all specialties. For any given patient, however, the number of actual findings may not exceed a few dozen. A conventional (“orthodox”) database table design (one fact per column) is unsuitable for such data, because of database vendor limitations on the number of columns per table and the need to continually add new tables or columns whenever new facts need incorporation. Most mainstream electronic patient record systems (EPRSs) deal with this problem through the entity-attribute-value (EAV) representation. In this methodology (also referred to as row modeling), the fact descriptors (attributes) are treated as data, so that the addition of new facts does not make database restructuring necessary. An EAV table of patient information records an entity (e.g., the patient, clinical event, or date), the attribute, and the associated value of that attribute.
The following example, which does not describe physical implementation, illustrates the EAV concept. A “conventional” table of laboratory values would have patient ID and date followed by numerous columns for individual tests, such as hemoglobin, potassium, and alanine transaminase. Each column would contain values for the appropriate test. A particular row would record all tests done for a given patient at a particular date and time and would appear as follows:
(<patient XYZ>, 1/5/98 12:00 AM, 12.5 gm/dl, 4.9 Meq/L, 80 IU...)
Tests not done on that patient would have the corresponding columns empty (null). In an EAV design, the patient ID and date columns appear as before, but instead of numerous columns with the names of tests hard-coded, there would be only two more columns, “Lab TestName” (the attribute) and “Value.” Thus, to record lab tests for a patient, there would be quadruples of the following form:
(<patient XYZ>, 1/5/98 12:00 AM, “Hemoglobin,” 12.5 gm/dl)
(<patient XYZ>, 1/5/98 12:00 AM, “Potassium,” 4.9 Meq/L)
and so on. One row is created for each test performed.
An EAV design represents a column-to-row transformation, because each row of such a table stores one fact about a patient at a particular instant in time. Examples of EPRSs with a major EAV component include the 3M clinical data repository1 which is based on the pioneering HELP system,2,3 and the Columbia-Presbyterian data repository.4,5
Electronic patient record systems focus on real-time retrieval (as well as real-time updates or additions) of individual patient data for clinical decision support. In contrast, a clinical data warehouse focuses exclusively on data retrieval, in batch mode. The basic clinical warehouse operation involves identifying a set of patients matching a particular profile in terms of demographic criteria, clinical history and findings, laboratory parameters, or particular medical and surgical interventions. Once these patients are identified, data on particular attributes can be extracted for analysis. Alternatively, patients from this set can be recruited for a prospective study. In summary, EPRS queries can be described as primarily entity (patient)-centric and clinical warehouse queries as attribute-centric.
In a warehouse setting, ad hoc query capability is essential: Query performance is important, but less so than ease of query formulation. As we shall see, the problem of attribute-centric query is a complex one and far from being solved. This paper describes our approach to tackling the attribute-centric query problem in the context of ACT/DB,6 a client-server data-base for managing clinical trials data. Deployed at the Yale Cancer Center and the Yale Clinical Trials Office, ACT/DB currently uses an Oracle 7 server and Microsoft Access clients.
In this paper, we confine ourselves to relational database technology and to the relational component of existing EPRSs. (For efficient retrieval of individual patient events, systems such as the 3M clinical data repository, which use relational database engines, redundantly store patient event data using nonrelational representations such as ASN.1 structures. However, such representations cannot be used advantageously by an attribute-centric query process.)
Background
Issues Involved in the Attribute-centric Query of Clinical Databases
We first discuss the factors that make ad hoc query of complex clinical databases far more difficult than that of conventional databases.
The Heterogeneous Data Problem
In an operational clinical database, whether EPRS or warehouse, data are stored in both conventional and EAV forms (e.g., patient demographics are usually stored conventionally). Furthermore, all patient data are rarely stored in a single general-purpose EAV table. Special-purpose EAV tables (row-modeled tables) may store restricted kinds of homogeneous data—e.g., laboratory values, pharmacy orders and medications, billable surgical and medical interventions, and general clinical observations. In a special-purpose EAV table, the name of a laboratory test or medication is treated as an attribute.
As we will show, queries on EAV data are quite different from those on non-EAV data. For data display or export to analytic packages, it is essential to perform row-to-column transformation of EAV data as needed, so that all attributes are presented conventionally. For this purpose, one must keep track of which attribute is stored in which table and whether it is in EAV or conventional form. The larger the system, the more difficult the tracking task becomes. For example, the Columbia-Presbyterian Medical Center Medical Entities Dictionary currently tracks 43,000 attributes7; the 3M clinical data repository schema has approximately 100 EAV plus conventional patient data tables.
Complex Boolean Queries on EAV versus Conventional Data
Complex Boolean queries of a conventional table (for example, “identify all patients where sex is female AND age >=35 AND city = `New York”') can be created easily by database neophytes through graphic front ends such as GQL and Microsoft's MS Query. The EAV equivalent is significantly more complicated, because each attribute-value pair for a patient is stored as a separate row in a table. When performing a similar complex Boolean query on a single EAV table, the conceptual AND, OR, and NOT operations must be translated into the row-based operations of set union, set intersection, and set difference, respectively.
The set operations, which are more complex as well as relatively inefficient, have received little support from database vendors because of their relative infrequency in traditional business applications. While specified in ANSI SQL-92, many well-regarded engines (e.g., Microsoft/Sybase SQL Server) do not directly support intersection and difference. One must achieve the equivalent result through self-joins (if the same EAV table is used more than once) and nested subqueries, respectively. This requires relatively advanced structured query language (SQL) skills. Self-joins, for example, employ the same table under multiple aliases, which are temporary names used to avoid ambiguity.8
Support for SQL-92, even if universally available, would help only SQL programmers. It would not translate into improved end-user query productivity, discussed later under “Challenges.” This is because set operations do not readily translate into a visual/graphic metaphor. In commercial “visual” query tools such as MS Query and GQL (which are intended for end-user deployment), for example, unions and nested subqueries lack graphic counterparts and have to be achieved by switching to “programmer mode” and writing explicit SQL.
Limits on the Number of Tables or Aliases per Query
Consider a query that asks for, say, 20 attributes on a set of patients (specified by Social Security number or medical record unit number). If all the data were in a conventional table, one would simply select those 20 columns. In the EAV situation, one cannot simply write a single SQL SELECT statement involving a join of 20 aliases. Most database engines limit the number of tables or aliases per statement (e.g., Sybase's current limit is 169).
Such limits are enforced because memory requirements and execution times go up nonlinearly with the number of aliases. (For example, in our informal tests on a high-end PC with 128 MB of RAM, a five-table/alias join yielding a one-million-row result was performed with Microsoft Access in less than a minute. A 20-table join (with much smaller tables) that was expected to yield only 50 result rows never completed even when run overnight in Access, and Oracle for Windows NT on a 64 MB machine did not perform any better.) We later describe how the program code discussed in the present paper works around such limits.
The Missing Data Problem
Consider the 20-attribute scenario in the above example, where some attributes have not currently been recorded for particular patients. With a conventional table, the query would not need to be modified in any way, and all patient records would appear in the output with the missing attributes as blanks (nulls) in the appropriate columns. In the EAV context, however, missing data introduce a complication: With standard (inner) joins of the EAV tables, any patient records with at least one missing attribute will not appear in the result table.
With EAV tables, forcing display of patient records with partial data requires full outer joins.10 A full outer join specifies that both tables on either side of a join should have all their rows preserved. However, while SQL-92 specifies the syntax for full outer joins, this is not directly supported by the major database vendors currently. To achieve an equivalent result requires multiple steps, each involving intricate programming.
Failure of Static (Precompilation) Strategies
Given these issues, it is clear that, without special tools, the design of SQL queries for heterogeneous clinical databases requires hand-crafted code written by persons who are experts in both SQL and the database schema. It is possible to achieve some performance gains by precompiling this code into database-specific “stored procedures” that can then be executed as needed. Unfortunately, this solution can be fragile. A database schema is never static: Attributes are often migrated across tables to improve efficiency or simplify the schema. For example, the number of data tables in the 3M clinical data repository may decrease in future versions, with more facts being migrated to a general-purpose “observations” table (S. M. Huff, personal communication). Consequently, carefully formulated procedures written for an older version of the schema will have to be rewritten to work with the newer version.
In any case, SELECT statements involving unions, self-joins, and nested subqueries (which are the basis of set operations) are difficult for the database engine to optimize.11 Therefore, any performance gains of precompiled SQL over dynamically interpreted SQL are expected to be modest.
Challenges
Given programmers with enough expertise in SQL and schema knowledge, and enough time to write, debug, and maintain queries and stored procedures, answers can always be extracted from a heterogeneous clinical database. However, we now encounter two major hurdles in the accessibility of the data:
The people who understand the data best (in terms of clinical science or epidemiology) are not the ones most qualified to query it.
Even competent SQL programmers must spend much time exploring the data dictionary to determine the location and nature of individual attributes and dealing with the intricacies of data extraction in the EAV context. The error-proneness and tedium of the code that they must compose manually contribute to a backlog with respect to the outputs that end users want from the system.
As is well known, the traditional strength of relational database technology over nonrelational approaches has been “empowerment” of relatively nonsophisticated users with respect to ad hoc query (e.g., with graphic tools). However, in the case of heterogeneous clinical databases, such empowerment is illusory because of the complexity of the coding required. Some form of electronic assistance is clearly necessary to improve the ease (and hence throughput) of query composition. In ACT/DB, the component that assists attribute-centric query is called Query Kernel (QK). Before we go on to QK's operation, we first consider how the basic EAV architecture can be adapted to improve the efficiency of attribute-centric query.
Efficiency Considerations for Attribute-centric EAV Schemas: Datatypes
Some EPRSs use a single EAV table to store all values as strings, whether the values are intrinsically text, numeric, or date. (The PTXT strings of the original HELP system were of this nature.) This is perfectly adequate for EPRS applications involving simple look-up of a patient's data. For attribute-centric query based on values of attributes (e.g., abnormally high values for a particular laboratory test), however, such a design is unsuitable. This is because ASCII strings representing numbers have a different sort order from true numbers (i.e., an index on the value field is essentially useless). In addition, if statistical aggregates are to be computed, further inefficiency results because the strings need to be converted to numbers on the fly.* If, instead, values are stored in their intrinsic data type instead of as strings, it is possible to achieve fast searches by creating compound indexes on a combination of the attribute ID and the value. Other clinical researchers have also improved query efficiency by converting data into its intrinsic type,12 although their work does not address EAV representation.
Designed for attribute-centric query, ACT/DB uses six general-purpose EAV tables, depending on whether the data type of the value is integer, real, date, short string (less than 256 characters), long text, or binary data such as images. (The last two data types cannot, of course, be indexed.) The ACT/DB database also introduces enumerated, ordinal, and Boolean data types as subtypes of the integer data type (i.e., they are stored in the same table). An enumerated data type is a range of integers, each being associated with a descriptive phrase—e.g., “Type of Transfusion” may be 1-Whole Blood, 2-Plasma, 3-RBC, 4-WBC, 5-Platelet, or 6-Other. An ordinal data type is similar, except that the numbers can be compared for relative magnitude—e.g., “Tumor Progression” may be 0-Cure, 1-Decrease, 2-No Change, or 3-Increase. Boolean data types allow only two values, “True” and “False.”
The concept of data types is critical to ACT/DB because their use assists in determining the semantic correctness of a query formulation. Thus, aggregate functions such as average and standard deviation are permissible only for numeric attributes, and comparison operators other than equality and inequality cannot be used for enumerated attributes.
Relational databases permit strong typing of columns in conventional tables, and advances in newer systems are aimed at encouraging and enhancing such typing, e.g., through the use of domains or object classes.13 Such power should not be discarded in the EAV context by converting data to least-common-denominator string form if the only expected benefit is simplification of the task of programming storage management.
Previous Work Addressing Attribute-centric Query
Most ad hoc query of clinical databases has focused on patient-centric queries. The Web-accessible Columbia-Presbyterian Medical Center Query Builder,14 for example, works off the medical center's data repository and generates both Health Level 7 and Arden Syntax15 for several categories of data, e.g., patient demographics, laboratory values, medications, and diagnoses. Detailed descriptions of true clinical data warehouses (which store multiple kinds of clinical data, possibly combined with nonclinical data, within the same database) have not been published so far. The Intermountain Health Care Data Warehouse Project deploys a number of separate “data marts,” each containing a particular category of data.16 (The difference between a mart and a warehouse is one of scale and scope. A data mart, which contains a subset of the enterprise's data, is specialized for answering queries specific to that subset. A warehouse contains all the organization's data.)
Data warehousing technology is relatively new. Because of its initial deployment in business applications, textbooks describing warehouse design, such as Kimball,17 completely ignore the problem of general-purpose EAV data tables, which store heterogeneous facts in a single table. By heterogeneous, we mean that the same table stores disparate facts such as history findings, examination findings, and laboratory results. Here, the nature and number of facts that will be stored for a given entity (the patient) cannot be predicted in advance—they depend on a particular patient's ailment. The problem of numerous and varying attributes per entity appears to be unique to databases that reflect rapidly evolving or highly heterogeneous domains.†
Design Objectives
Query Kernel aims to address the problems of attribute-centric query identified earlier. It is designed to improve productivity by facilitating the creation and execution of syntactically and semantically correct queries through a graphical user interface (GUI). While it is aimed primarily at analysts or biostatisticians who are familiar with data analysis but not necessarily experts in SQL, experienced SQL programmers can use it productively as well. The basic premise of QK is that, from the user's viewpoint, it should not matter whether an attribute's data are stored in general-purpose EAV tables, special-purpose EAV tables, or conventional tables. It should be possible to freely select any combination of attributes, and responsibility for generating the correct code (performing set operations, creating full outer joins and smaller intermediate sets, etc.) should be borne by QK and not by the user.
System Description
The ACT/DB Query Kernel: Principles and Metadata
In order to meet its design objectives, QK uses a data dictionary (metadata) that is the foundation of both the user interface and code generation. Before describing the structure of the metadata, we discuss an important component of it—namely, the description of where a particular attribute is located.
Location of Data: Views
Data within ACT/DB are stored in several related tables. For example, the general-purpose EAV tables do not actually store the patient ID, study ID, and date/times of a patient event. This information is stored in an event header table, which has a machine-generated unique identifier (event ID) as primary key. The general-purpose EAV tables have an event ID field that points to this event record.
For the purpose of an attribute-centric query, the schema of a database can be simplified through the creation of a series of “views.” Views are a standard relational database mechanism for combining parts of different tables and presenting them as though the combination were a single table. Query Kernel uses a set of views to locate patient data for individual attributes. The requirements of these views are as follows:
Every view should combine entity information (patient ID, time stamps, etc.) with attribute and value information. For EAV views, the attribute and value will be in two separate columns, while for conventional views, they happen to be the same column.
Every attribute in the system must be associated with one, and only one, view.
There can be any number of instances of a given attribute per patient, either because there are multiple data points per patient or as a consequence of the join operation used to create the view. For example, one view in ACT/DB combines basic patient demographics (name, date of birth, sex, etc.) with study enrollment data (primary diagnosis, date of informed consent, etc.). If a given patient is enrolled in multiple studies, there will be multiple instances of (identical) demographic data for that patient.
The advantage of defining such views is that they are much fewer in number than the tables on which they are based. For example, QK uses only nine views—six general-purpose EAV views corresponding to each of the data-type-specific tables, two views for special-purpose EAV tables (patient selection criteria and onprotocol therapies), and the demographics-enrollment view mentioned above. This number is not expected to grow in the foreseeable future. (ACT/DB uses the two relatively small special-purpose EAV tables only because they serve to test code that will possibly be ported to other systems, where such tables are used extensively, e.g., for laboratory values and medications. It is not reasonable to expect the administrators of such systems to undertake the nontrivial task of moving all data from these tables into general-purpose EAV tables just for the benefit of a query tool. Instead, the query tool must adapt itself to the attributes and data, as and where it finds them.)
Structure of the Metadata
The metadata record the following information for each attribute.
The name of the attribute, a description, and its datatype. Name and description are used for keyword searching of the metadata to select individual attributes.
The event type. This is the nature of the time-oriented event (none, instant, period) that it represents. Event type determines the number of time stamps associated with that attribute: zero, one, or two, respectively. Basic demographic data are not time-stamped. Most attributes are of the instant variety, having a single time stamp recording the time of the event. A few attributes are of the period variety, with two time stamps representing the start and end of the event.
The name of the view where it occurs, and whether this table or view represents an EAV or conventional form of storage.
The names of the attribute and value fields in this view, and the attribute ID (typically a long integer identifier) for this particular attribute, if EAV. (For example, all general-purpose EAV views in ACT/DB have the attribute column name “Question_ID” and the value column name “Value.”) If the attribute is non-EAV, the attribute ID is null and the attribute and value fields are identical.
If the data type is Enumerated or Ordinal, the ID of the controlled-vocabulary group (“Choice Set”) that defines the set of integers for that attribute, with their associated descriptive phrases. This information is used to populate the contents of a pull-down list when the user chooses an enumerated or ordinal attribute. This way, the user can work, as far as possible, with the symbolic (i.e., meaningful) representations of the integers rather than with their internal representations.
If it happens to be a laboratory test, a lab test ID pointing to the laboratory tests table. The metadata also record whether the normal range for this attribute is age-dependent and whether it is sex-dependent. (This is used to retrieve queries specified in terms of “normal” ranges for age and sex, as described later.)
Information specifying the higher-order grouping of the attribute within forms used in the study. (Since the same attribute can be used in multiple studies, this information is retrieved only if we are performing study-specific data extracts.)
Population of the Metadata
The number of attributes in a system can be very large. Clearly, if the metadata required by QK for every attribute had to be entered by hand, considerable manual effort would be involved. Query Kernel therefore captures its metadata from other parts of the ACT/DB system. Every production EAV system must maintain a controlled vocabulary of attributes, and ACT/DB is no exception. Special-purpose EAV data tables are also managed with their own controlled vocabulary tables—e.g., for laboratory tests or medications, where the names of medications or tests are treated as attributes. Metadata capture works as follows:
For conventional (non-EAV) views that do not use controlled vocabulary items, QK uses the host database engine's built-in system dictionary to capture all the column definitions in each view.
For EAV views, QK records against each view the name of the associated controlled vocabulary table and the names of the attribute ID field (a long integer), the attribute name field, and the attribute description field (if available) in this table.
Currently, the metadata are not updated automatically (e.g., whenever new attributes are added to the system's controlled-vocabulary tables or migrated from one place to another). Instead, it is re-created on demand through a button-click. The volume of the metadata is currently small enough (around 1,000 attributes) that re-creation takes less than 15 seconds. If, however, the number of attributes grows by an order of magnitude or more, we will have to devise a more efficient means of synchronizing the metadata with the contents of their source vocabularies. One such means may be through the creation of an “intelligent software agent”18 to monitor changes to the state of the controlled vocabulary tables and make the required adjustments to the metadata tables.
Output Operations
The QK kernel code is used for two related but distinct operations—true ad hoc query, which may or may not be study-specific, and creation of study-specific data extracts on the client for export to statistics packages or generation of formatted hard-copy reports. As will be discussed shortly, the output of the first operation can feed into the second. The primary difference between the two operations is that a single data-extract specification creates a single output file (a database table on the client) generally containing a large number of attributes (typically 50 or more). The values in the output are the original raw data only. A single ad hoc query operation, on the other hand, can create zero, one, or multiple output tables in a single operation, and the output can contain statistical aggregates, raw values, or both.
We first discuss the principles and interface of the ad hoc query operation.
Ad Hoc Query
Query Kernel's ad hoc query can best be understood by analogy. Consider a simple flat file containing a patient ID and a large number of attributes. To extract data on a subset of patients from such a file, the basic operation (as phrased in SQL) is:
SELECT Patient_ID <attributes to be displayed>
FROM Data_table
WHERE <patient selection criteria based on attributes>
The ad hoc query process in effect creates the illusion of a single patient data table containing a very large number of attributes, with their associated time stamps. The user specifies selection criteria and (optionally) output attributes. Query Kernel uses the selection criteria to identify a set (subgroup) of patients. If no output attributes are specified (i.e., only patient ID is desired), no output tables are created, and the set of IDs is saved under a user-specified name for later use. Such a set is typically used in the data extraction operation, where extraction can be limited to a particular set of patients. (The default is all patients in the study.)
If one or more output attributes are specified, then one or more output tables are created. All attributes without time stamps and all statistical aggregates (which yield a single value per patient) are placed in a single table, one row per patient, along with the patient IDs. Each attribute value that is time-stamped is placed in its own separate table, along with patient IDs and associated time-stamp values. Query Kernel segregates time-stamped raw attribute data into individual tables because multiple values occur per patient, and the number of values per patient (or per attribute) is generally not constant.
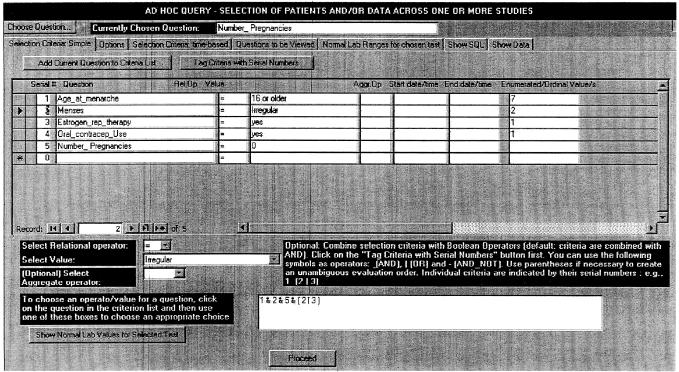
The ad hoc query interface is shown in ▶. This is based on a “Tab Control” with multiple pages. We now describe each of the components.
Figure 1.
Specifying selection criteria in the Ad Hoc Query form. This and the next figure deal with patients in a prospective breast cancer study. The selection criteria are patients who had menarche at age 16 years or later, no pregnancies, irregular menses, and a history of either estrogen replacement therapy or oral contraceptive use. This is specified by the compound selection criterion “1 & 2 & 5 & (3 | 4)” in the lower middle of the figure, where the numbers refer to individual selection criteria. The rightmost column in the figure, “Enumerated/Ordinal Values,” is used to display (and manipulate, if necessary) the internal integers corresponding to symbolic phrases for such data types. Thus, the number 2 corresponds to the symbolic phrase “irregular” for the attribute “menses.”
Choosing a Question‡
Clicking on the “Choose Question” button (top left) opens a dialog box (not shown) where the user specifies one or more keywords with wild-card patterns if desired, optionally combined in complex Boolean fashion. (The graphical interface actually generates the wild cards and Boolean expression based on the user's selections.) The user next brings up a list of matching attributes with a button click and selects one of them. This attribute, the “chosen question,” can be used repeatedly in the selection criteria as well as in the output attributes. (For example, one may display both means and standard deviations for values of a particular laboratory test.) When an attribute is added to the criterion list, the attribute's metadata are stored with the criterion, to facilitate the task of later code generation.
The user does not need to know whether the attribute is stored in conventional or EAV form. Thus, the conventional attributes “Last Name” and “Race” are available, as are the EAV attributes “Blood Hemoglobin” and “Serum Potassium.” When an attribute is selected, its associated metadata are captured and stored but are not shown to the user.
Selection Criteria
Each selection criterion (also shown in ▶) is identified by the following fields:
A serial number, used when criteria are to be combined in complex Boolean fashion, as discussed later.
The attribute/question name.
A relational operator, chosen from the pull-down box below the criterion list. If no operator is chosen, it defaults to equality. The available choices are determined by the data type of the current attribute. For example, only “=” and “< >” are available for Boolean and enumerated attributes. For string data types, the operator LIKE, which allows wild cards, is also available.
-
A value, usually entered by the user, except for Boolean, enumerated, and ordinal data types, where the user can choose from the Value pull-down box. This box's contents change dynamically to hold a show a list of the descriptive phrases associated with the “Choice Set” of the attribute for the currently selected criterion, along with their corresponding internally used integers. When the user chooses a phrase from the box, the corresponding integer value is saved for later use (shown in the Enumerated/Ordinal Values column of ▶). In addition, for attributes that are laboratory tests, the choice “normal” becomes available. (We discuss the use of “normals” later.)
A series of values can also be specified as comma-separated items, and a range can be specified as two values separated by a tilde. These cause QK to generate SQL code containing IN and BETWEEN clauses, respectively.
To use IN or BETWEEN for enumerated or ordinal data types, the field in the Enumerated/Ordinal Values column can be edited directly, e.g., by typing in a list of integers with separating commas. This is the only instance where the user must work with the internal rather than the symbolic representation of the data type elements. For these data types, the integers are used instead of the symbolic phrases because symbolic phrases may contain commas (e.g., “multiorgan disease, both sides of diaphragm” for a Hodgkin's disease grading) that could confuse the program's parser.
-
An aggregate function (optional). This lets the user specify criteria based on aggregates. For example, to identify patients having a consistently high serum alanine transaminase value, one might specify the aggregate MIN, the relational operator “>,” and the value “NORMAL.”§
The aggregates supported are MIN, MAX, COUNT, AVG, STD (standard deviation), EARLIEST, and LATEST. The last two, which are not part of SQL, have been borrowed from the latest draft specification of the Arden Syntax for medical logic modules.19 They refer to the chronologically first and last values of an attribute (when multiple values are present because of repeated sampling) and are meaningful only for time-stamped attributes when starting and ending date/times have been specified.
As in the case of relational operators, the list of available aggregate functions is constrained by the data type of the current criterion's attribute (e.g., AVG and STD are available only for numeric attributes). If no aggregate operator is supplied, a patient will be selected if even a single value for that patient matches the criterion.
Starting and ending date/times (optional). These may be specified to limit the search to a chronologic range.
Combining Criteria
To combine criteria, their serial numbers are used, along with the Boolean operators & (AND), | (OR), and - (AND-NOT, for set differences) and parentheses to disambiguate complex expressions as necessary (e.g., (1 & (2 | 3)) - 4). If no compound criterion is specified, all criteria are ANDed with each other.
Output Attributes
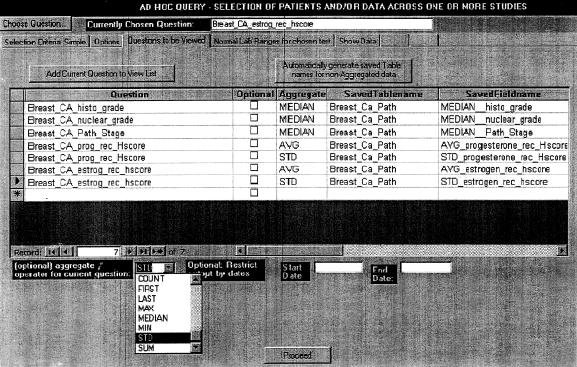
The page where output attributes are specified is shown in ▶. Each output attribute is defined by the following properties:
Figure 2.
Specifying output attributes for patients identified by the criteria of ▶. These are the medians of the histologic tumor grade, nuclear grade, and pathology grade (all of which are ordinal data types), and the average and standard deviation of both the estrogen receptor and progesterone receptor histologic scores. Tumors are studied at multiple times during the course of follow-up and may be collected from multiple sites (e.g., primary, recurrent primary, or metastatic). Since all the output attributes are aggregates (one value per patient), they will be placed in a single output table (given the name “Breast_Ca_Path”). The field names in this output table have been specified by typing in and are slightly more mnemonic than the default field names, which are created by concatenating the aggregate name and the attribute name.
The name of the attribute. As stated at the start of this discussion of “Ad Hoc Query,” the patient ID and time stamp fields will always be generated appropriately in the output, and so do not have to be specified.
-
An aggregate function, if desired. As stated earlier, an attribute may be used multiple times with different aggregate functions.
In addition to the functions mentioned under “Selection Criteria” above, the Median can also be specified. The median is preferable to the average for attributes with highly skewed frequency distributions (e.g., white blood cell count in cancer patients), and its omission from SQL (no relational database engine supports it) significantly weakens SQL's power for statistical analysis.
Query Kernel computes the medians for an attribute (grouped by patient ID) through SQL code described in the legend of ▶. (This code uses the “template” feature described later.) This code is tolerably fast but understandably less efficient than if “median” were a built-in SQL function. (The reason we do not allow the median to be specified in the selection criteria is that here every single instance of the attribute (i.e., for all patients in the system or study) needs to be processed. This may slow performance to unacceptable levels with large data sets.)
The name of the output table for each attribute and the name of the field in the table. Default names are suggested for each table and field, based on a combination of the attribute name and the aggregate, but these can be overridden by the user.
-
Whether the attribute is mandatory or optional. In an output set, some (mandatory) attributes may be critical to a particular analysis, and there is no point in listing data for patients who lack even a single value of that attribute. Other (optional) attributes would be useful if available, but it doesn't critically matter if they are not.
For example, suppose that drug XYZ is suspected of causing cholestatic jaundice. As a selection criterion, we specify patients who have received at least 20 doses of XYZ. The desired output attributes are the following laboratory tests: alkaline phosphatase, direct bilirubin, 5-nucleotidase, and leucine aminopeptidase. We may decide that patients lacking information on either of the first two parameters are not worth including in an analysis. The latter two tests, while useful for obstructive jaundice, are not always available because they are seldom performed for screening, so patients lacking data on either of these tests may still show up. Therefore, we designate the first two attributes as mandatory and the last two as optional.
Figure 3.
Template for computing the median values of an attribute (for each patient). The SQL code inserts sorted Patient_ID and attribute values into a table, TempMed1, that automatically creates sequentially numbered records (with the sequence number the RecID field). The maximum and minimum RecIDs for each patient are then determined, and the “middle” RecIDs computed. For patients with an odd number of values, there is a single “middle” value, while for patients with an even number of values, there are two “middle” values. The values corresponding to these RecIDs are then extracted into another table, TempMed2, and the median for each patient is determined by taking the average of the two values (for even numbers) or using the single value (for odd numbers).
Selection Criteria and Output Options
The output attribute values may be restricted by a range of dates. The user may limit the selection of patients to those enrolled in a specified study or set of studies by selecting studies from a list box. As described earlier, the user may save the set of patients identified by the selection criteria for later use. Enumerated or ordinal attributes may be displayed either in their symbolic form or in numeric form; the latter is more suitable for graphing (especially with ordinal attributes).
Display of Ad Hoc Query Data
The tables specified by the user are created on the client. These may be viewed directly by users experienced in the use of Microsoft Access. However, the list of tables already in the schema is quite large. As a convenience, QK provides a “show data” page where a list of all tables generated for the present query is displayed. After the user selects a table, its contents (with column headers) are displayed in spreadsheet form within a scrolling window. Output is ordered by patient, start of event, and end of event (where applicable).
Saving a Query and Running a Saved Query
A query specification can be saved for later reuse under a user-specified name. The specification is saved in three tables. A top-level table stores the query name and user-supplied description, along with settings such as whether a patient set is to be created, the Boolean expression that combines selection criteria, and the list of studies, if any, that can restrict the scope of the query. Two tables, which are related many-to-one to this table, store the individual selection criteria and the output attributes, respectively. The user can later load a saved query by name, optionally edit its specifications, and run it.
The ability to save queries and run them later is useful when the same set of queries need to be run repeatedly through a clinical trial. We also foresee saved queries as the basis for eventually implementing (non-real-time) event monitors within ACT/DB.
Code Generation
Query Kernel generates SQL based on the selection criteria, output attributes, and options specified by the user to retrieve data appropriately. Many of its operations use tables on the server and client with a predetermined structure: These tables hold intermediate results and are emptied prior to being reused.
While QK checks extensively for errors prior to code generation, semantic checking is nonetheless limited: Nonsensical selection criteria, such as “pregnant males,” will still pass through, although they will obviously not return any results.
Code generation in QK uses SQLGEN, a subroutine library to facilitate client-server development.20 The SQLGEN routines take care of such details as generating quotes around string values and omitting them from numeric values. They are extensively used, for example, in composing the WHERE clauses for individual selection criteria.
Identifying Patients on the Basis of Selection Criteria
For each individual selection criterion, QK generates and executes SQL code that creates a temporary table on the client, containing only matching patient IDs. Individual tables are then combined using set intersection, union, or difference, based on the contents of the Boolean expression, using a simple parser and stack evaluator.21 The stack evaluator is based on work done previously by the first author of this paper.22
Creation of Output Tables
Query Kernel segregates the output attributes into two sets—those that are guaranteed to occur once per patient (either because the attribute is not time-stamped or because an aggregate function is used) and those that are not. The former attributes are destined for a single table. The latter will go into their own individual tables, because (as stated earlier) the number of values per patient (and per attribute) will generally not be the same.
Query Kernel identifies all patient records that have at least one value for a mandatory parameter and saves them to a temporary table. All output tables are then filtered by removing records of patients who are not in this set. Also, the non-time-stamped output table is built up with inner joins for mandatory attributes, followed by left outer joins for optional attributes.
Templates
In order to perform many of its SQL generation tasks, QK uses templates. A template is SQL text that contains placeholders, which are replaced at run time by parameters passed to the template. For example, for the template “Select ∼1 from ∼2 where ∼3” and the parameters “Last_Name, First_Name,” “Patients,” and “Investigator = `Klein',” the template expands to “Select Last_Name, First_Name from Patients where Investigator = `Klein'.” A template generally contains multiple SQL statements, separated by semicolons (and is executed one statement at a time). It can also contain C-style comments, which are stripped off before execution of the SQL code. (Because numeric parameters are not mnemonic, comments are essential for the developer to document the template.)
Templates are a macro-substitution mechanism (similar to C's sprintf function) that get around some of the limitations of stored procedures, which are fairly rigid in what parameters can be passed to them. (For example, stored procedures will not allow table or field names to be passed as parameters, because these must be known at compile time, not run time.) Because templates are stored as data, they make the code generator easier to maintain and debug. Template processing is handled by the SQLGEN library. The template for median computation is illustrated in ▶.
Handling Laboratory Data: Defining “Normal”
When retrieving patient data for laboratory tests, one is often interested in identifying only patients whose values are in particular ranges (i.e., normal, below normal, or above normal) for one or more tests. The most important determinants of “normal range” for a test are the performing laboratory and the age and sex of the patient. (Query Kernel does not handle the physiologic conditions of pregnancy and lactation.)
When an attribute happens to be a laboratory test, ACT/DB tracks the performing laboratory ID in the event record. (This defaults to zero to indicate a “generic” laboratory.) The database stores a table of normal values for laboratory tests by age, sex, and laboratory. To identify all patients with a “high” value for a particular parameter, QK does the following:
The view containing the data (and the laboratory ID) is restricted by the attribute of interest.
The patient demographics table (which contains date of birth and sex) is optionally restricted by age and sex (e.g., we may be searching for female patients over 45 years of age). Age is determined by the difference in years between the date of birth and the date the laboratory test was performed (the “start of event” field in the view containing the data).
The laboratory test ranges table is restricted by the attribute/laboratory test of interest and optionally by age and sex criteria.
The three tables above are then joined, and the output is restricted to those rows where the value is greater than the upper range of normal.
The template that identifies patients based on an aggregate function applied to normal (e.g., AVG (attribute) > NORMAL) is illustrated in ▶.
Figure 4.
Template for identifying patients based on the concept of “normal” laboratory values. The template code joins three tables or views—the view containing the laboratory values, the patient (demographics) table, and the Lab _test_ranges table that contains normal values for each test, by laboratory ID and the age and sex of the subject. The join would return multiple rows per patient, so the output is restricted by use of the age and sex for each patient. The num-ber of rows from the patient demographics table may be further limited if the user has specified that only patients of a particular age range or sex are to be considered.
Currently, the handling of normal values by QK is limited in that it does not allow expressions such as “values that are two times the normal value.” For such purposes, values must be explicitly supplied as numbers. (The button labeled “Show Normal Values for Selected Test” on the Selection Criteria page shown in ▶ is designed to assist the user. When clicked it will go to the appropriate page and display the normal values for the current attribute arranged by age, sex, and laboratory.)
Query Kernel does not handle multiple units for the same laboratory test. This is not a significant drawback, because accredited laboratories are standardized with respect to test units.
Data Extracts
In data extraction, the user extracts all of the (typically) several hundred attributes measured in a study into as few output tables as possible. An extract is intended for export to analytic packages or reporting tools, which require that data be structured in conventional rather than EAV form. Whereas in ad hoc query the user picks one attribute at a time, in data extraction the user selects entire groups of questions through a different interface. (In ACT/DB, attributes are grouped by the study designer into functional and logical categories such as hematology and physical examination as part of the study protocol.)
Typical study designs require redundant sampling of certain attributes at multiple phases of the study, with some attributes being sampled more often than others. In addition, some patients may have more data than others because, for example, they require a greater number of chemotherapy cycles for an adequate response. This results in an unequal number of data points across attributes per patient. If one attempted to put the entire study data into a single output table structured in conventional form (one column per attribute), then in most cases that file would not be strictly rectangular. There would be numerous “missing values,” which were merely artifacts of the sampling process.
The simplest way to avoid such artifacts is to generate one output table per logical attribute group. This, however, can yield a large number of output files and complicates analysis that involves comparing attributes that lie in different output files. If we know that certain groups always occur together in a particular study (e.g., routine clinical chemistry, hematology, and physical examination are always gathered every cycle) then such co-occurring groups can be extracted together. (In ACT/DB, group co-occurrence information is part of the study protocol definition.) This way, the number of output files is minimized.
Data Structures and Code Generation
Query Kernel includes a batch facility that lets the user generate multiple extracts at a time. Each extract is defined by an extract name, a description, a flag indicating whether the data will be exported to a statistical package, the list of attributes to be exported, and an output table name. When the same set of extracts is run repeatedly for different subgroups of patients, the output table names can be changed for each run.
Attributes are segregated into four sets: attributes stored conventionally with and without time stamps, and attributes stored in EAV form with and without time stamps. (Conventional time-stamped attributes do not currently exist in ACT/DB, but the algorithm considers this possibility anyway because it may be ported to other systems.) For a given extract, one or more of these sets may be empty. The attributes of each set are merged to create a temporary table, and the temporary tables are then joined to create the final extract.
Conventional attributes are segregated by the view where they occur, so that all the desired attributes in a single view can be extracted through a single SQL SELECT statement. For EAV attributes, on the other hand, extraction initially occurs one attribute at a time.
Unlike the case of ad hoc query, full outer joins are used throughout the data extraction operation. As mentioned earlier, a single extract can have numerous attributes or fields (up to 255). To bypass the problem, mentioned earlier, of practical limits on the number of aliases per join, the extraction algorithm works as follows:
All combinations of patient ID (i.e., all patients in the trial or the subgroup) and phase ID for the set of selected attributes are generated using a “Cartesian join” mechanism8 and stored in a temporary server table.
For each attribute that must be extracted, this temporary table is “left outer joined” with a selection on the patient data view that holds the attribute and the result captured on the client. This way, even if information on a particular attribute has not been recorded at all, all combinations of patients and phases will still appear, with the attribute column holding null values. The result is a set of modestly sized temporary tables on the client with program-generated names, one table per attribute.
These tables are combined into one table using standard inner joins and a stepwise join that combines a maximum of five tables at a time and creates a new table. The resultant tables are then combined with each other, and so on, until finally only a single table is left. At each step, the number of new tables created is CEILING (N/5) where N was the number of tables created in the previous step. Thus, for an extract that will eventually contain 132 attributes, the number of new tables in each step are 27, 6, 2, and 1. At the end of each step, temporary tables created during the previous step are erased.
Status Report
Query Kernel was created some time after ACT/DB's initial production deployment. We have considerable practical experience with the data extract module, employing it on a regular basis. The ad hoc query module is in pilot deployment: We have tested it on the protocols within the system. We expect that the functionality and user interface of this module will evolve over the next year as it is used by an increasing number of our scientific collaborators at Yale and elsewhere. Some features may also change on the basis of user feedback.
Discussion
While the work embodied in QK does not represent any major conceptual breakthroughs, our implementation using sufficiently rich metadata to direct the ad hoc query of heterogeneous clinical databases appears to be original. Metadata as stored in the system data dictionary are used by all SQL-based database engines to process queries on conventional database architectures, and QK represents an extension of such functionality. An advantage of generating SQL dynamically through the metadata is that the user is protected against changes in the schema.
The ideas behind QK have been partially inspired by the concept of the “universal relation” of Ullman.23 This work was aimed at creating the illusion of a single table within a database and sparing the user the task of having to specify intertable joins during ad hoc query. The “universal relation” concept has since been partially discredited. For example, certain queries (e.g., those using self-joins) cannot be specified unambiguously without the use of explicit joins and aliases.24 Also, Ullman failed to anticipate the power of today's query GUIs (such as Microsoft's Visual-Query-By-Example technology) and did not consider EAV data, which adds many complications to the query process.
Unlike the “universal relation,” QK is not intended to provide a front end to any arbitrarily complex relational schema. It is constrained to operating on a single entity within the schema (in this case, the patient). It takes advantage of the fact that a clinical event has a very simple conceptual structure, in that a single event is associated with any number of findings described by attribute-value pairs.
The role of QK should be regarded as complementary to the use of analytic tools such as multidimensional database engines.25 The latter will work well on homogeneous tables such as laboratory tests or prescription/refill data but are not suited for heterogeneous tables such as clinical observations.
Limitations and Future Work
We must emphasize that the work described in this paper does not represent a complete and final solution to the heterogeneous data query problem. Outstanding issues that need to be addressed are discussed below.
Optimization of the Query Process
The current approach of QK to identifying a set of patients on the basis of compound selection criteria—extracting individual sets of patients on the basis of each individual criterion before combining them using a stack evaluator—is somewhat simplistic. Certain criteria (e.g., sex = female) result in relatively large intermediate sets. Because the intermediate result sets are downloaded on the client machine before they are processed further, this can stress the client-server communications link in a low-to-moderate-bandwidth setting.
It is possible to optimize this operation by generating SQL that judiciously combines multiple criteria in a single SELECT statement that operates on server tables so that the intermediate sets are guaranteed to be of modest size. For this strategy to work successfully, ACT/DB needs to maintain statistics that can be consulted by QK. In particular, counts of values for the attributes that are in EAV form need to be stored. The EAV design is essentially the database equivalent of a sparse-matrix representation, and some attributes are sparser than others. For example, a significant proportion of patients would have values for Hemoglobin, but a much smaller number would have values for anti-DNA antibodies. Similar statistics (such as the number of rows in tables) are used by existing database engines to perform “cost-based optimization” when processing queries on conventional tables. An interesting research direction will be to control the Oracle server's cost-based optimization by making QK generate “hints” along with SQL. (Hints are pseudocomments read by the SQL parser and passed to the optimizer.)
An alternative approach to stored statistics is to dynamically determine the potential row count of individual selection criteria before deciding whether they could be usefully combined. As the volume of data grows, the overhead of prior determination of row counts will be offset by increased efficiency when retrieving the actual data.
If the data within ACT/DB increase by a couple of orders of magnitude, such optimization will be mandated.
Enrichment with Semantic Information
Semantics can be used to enrich querying in two distinct ways. First, it is possible to prevent obviously nonsensical queries (e.g., identify pregnant males) through a set of rules. We are in the process of incorporating rules into ACT/DB for the purposes of cross-field validation, and queries can benefit from such rules as well. Of course, the question then arises as to how extensive rule coverage should be. After all, QK is intended for use by those who have a reasonable knowledge of clinical medicine.
Second, attributes often form a hierarchy, especially in the domain of disease and medications. For example, the attribute “Cephalosporin” represents a parent concept that can have the children “Cefazolin,” “Ceftriaxone,” and so forth. One might want to query the database for cephalosporin usage (or beta-lactam, or antibiotic usage) when the medication data records the actual drugs rather than the drug family.
Support of such queries requires the use of a medical entities dictionary. The National Library of Medicine's Unified Medical Language System26 has the foundations of a semantic network (where concepts are classified into broad categories). In addition, the UMLS records explicit links (such as parent-child relationships) between concepts as described by human data curators, as well as pair-wise frequency of co-occurrences of concepts as recorded by a computerized scan of biomedical literature. (Co-occurrences have been used for automated knowledge discovery through the clustering of algorithms to create “concept spaces.”27) We expect to eventually incorporate such a dictionary into ACT/DB, most likely by building on the existing infrastructure of the UMLS.
Support of Temporal Primitives for Query
Temporal databases are an active area of research. The only well-known clinical system employing a rich set of temporal functions (and handling period data robustly) is Das and Musen's CHRONUS system.28,29 (However, TimeLine SQL, the language created for CHRONUS, is significantly different from the draft ANSI version of SQL-Temporal, which is intended for incorporation into SQL-3.30,31) Support of period data is not yet part of the latest draft version of Arden Syntax but is planned for a future release.19
It is clear that QK would benefit enormously from the ability to specify temporal relationships between individual selection criteria (e.g., that a particular symptom occurred within a certain time period after administration of a particular medication). Such ability would result in output that is more concise and would enhance the specificity of queries. Adding such support is not a trivial undertaking: Our long-term goal is to combine the functionality of SQL-Temporal with Arden Syntax (the latter has a few useful functions not defined in the former).
Limitations of the Graphic Metaphor
The operations performed by QK are not possible through SQL generation alone. In many cases, the execution of procedural subroutines using the client programming language (Visual Basic for Applications) is interspersed with that of generated SQL code. We have not tried to create a brand-new programming language that would seek to conceal the difference between EAV and non-EAV data; the GUI takes care of that. However, a GUI, while easier to use than a language, is ultimately less expressive than the latter (“what you see is all you get”). Query Kernel employs only a subset of SQL; for example, expressions involving mathematical functions other than the statistical aggregates are not available.
We have deliberately limited ourselves in this matter and avoided reinventing the wheel. The output created by QK is not intended to be the final output that all users could ever want. In many cases, the output of QK is only the starting point for some other operation, such as a cosmeticized report, statistical analysis, graphing, or further querying—in all cases, using standard, off-the-shelf tools. However, it is possible that certain relatively modest programming enhancements will result in a significant increase in functionality. (For example, the aggregate functions have been implemented because they are, in most cases, part of SQL itself, and users/programmers who manipulate conventional tables expect to be able to use them.) Greater testing of QK with more users will determine the nature of such enhancements.
While QK does not close the attribute-centric query problem, many of its existing features should prove instructive to researchers who are grappling with similar problems.
Acknowledgments
Dr. Vincent de Vita, Director, Yale Cancer Center, first identified the need for a clinical trials database. Dr. Perry Miller, Director, Yale Center for Medical Informatics, defined the need for a robust means of handling laboratory values generated from multiple laboratories. Dr. Lee Schacter of the Yale Cancer Center, Dr. John Fisk of Vanderbilt University, and Dr. Rajani Nadkarni of Medical Oncology Associates, New Haven, provided valuable inputs on the specifications of the Query Kernel. The Principal Investigator for the Breast Cancer Protocol used to illustrate Figures ▶ and ▶ is Dr. Michael Reiss of the Yale Cancer Center.
This work was supported in part by grant G08-LM-05583 from the National Library of Medicine and by departmental grants from the Yale Cancer Center, the Yale Clinical Trials Office, and the Yale Department of Anesthesiology.
Footnotes
Arden Syntax medical logic modules, which operate on EPRS data, use a rich set of statistical operators. Because they operate on a single patient's data at a time, any inefficiency resulting from string-to-number conversion is not particularly noticeable. However, it is quite another proposition to scan an entire database looking for all patients whose minimum serum alanine transaminase values were above a particular value (e.g., patients presenting with chronic active hepatitis).
Many business data tables are row-modeled and can be regarded as special-purpose EAV tables. In a Sales table, for example, the names of product items are not hard-coded in columns any more than are the names of medications in a prescriptions table. Instead, we record a product ID or medication ID. However, the number of attributes that describe a given Sale—customer, product category, quantity purchased, discount, etc.—is fixed and unvarying for the most part. That is, a Sales table, while row-modeled, is homogeneous.
Note that “question” and “attribute” are synonymous, but non-informaticians are more comfortable with the former term.
For criteria without aggregates, QK generates a standard SELECT... FROM... WHERE statement. Criteria with aggregates cause appending of a GROUP BY... HAVING clause. Structured query language has been greatly criticized for its non-orthogonality when aggregate functions are involved, and QK hides such non-orthogonality from the user.
References
- 1.The 3M Clinical Data Repository. Murray, Utah: 3M Health Information Systems, 1998.
- 2.Huff SM, Haug DJ, Stevens LE, Dupont CC, Pryor TA. HELP the next generation: a new client-server architecture. Proc 18th Symp Comput Appl Med Care. 1994: 271-5. [PMC free article] [PubMed]
- 3.Huff SM, Berthelsen CL, Pryor TA, Dudley AS. Evaluation of a SQL model of the HELP patient database. Proc 15th Symp Comput Appl Med Care. 1991: 386-90. [PMC free article] [PubMed]
- 4.Friedman C, Hripcsak G, Johnson S, Cimino J, Clayton P. A generalized relational schema for an integrated clinical patient database. Proc 14th Symp Comput Appl Med Care. 1990: 335-9.
- 5.Johnson S, Cimino J, Friedman C, Hripcsak G, Clayton P. Using metadata to integrate medical knowledge in a clinical information system. Proc 14th Symp Comput Appl Med Care. 1990: 340-4.
- 6.Nadkarni PM, Brandt C, Frawley S, et al. ACT/DB: a client-server database for managing entity-attribute-value clinical trials data. J Am Med Inform Assoc. 1998;5(2): 139-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cimino JJ, Clayton PD, Hripcsak G, Johnson SB. Knowledge-based approaches to the maintenance of a large controlled medical terminology. J Am Med Inform Assoc. 1994;1: 35-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Date CJ, McGoveran D. A guide to the SQL standard. Reading, Mass.: Addison-Wesley, 1992.
- 9.Sybase Inc. System 11 Reference. Vol 1. Emeryville, Calif.: Sybase, 1996.
- 10.Melton J, Simon AR. Understanding the new SQL: a complete guide. San Mateo, Calif.: Morgan Kaufman, 1993.
- 11.Celko J. Everything you know is wrong. DBMS Magazine. 1996;9(9): 18-20. [Google Scholar]
- 12.Prather JC, Lobach DF, Hales JW, Hage ML, Fehrs SJ, Hammond WE. Converting a legacy system database into relational format to enhance query efficiency. Proc AMIA Annu Fall Symp. 1995: 372-6. [PMC free article] [PubMed]
- 13.Darwen H, Date C. Introducing the Third Manifesto. Database Programming and Design. 1995;8(1): 10-14. [Google Scholar]
- 14.Wilcox A, Hripcsak G. CPMC Query Builder. Available at: http://www.cpmc.columbia.edu/arden/qbr/. Accessed Sep 1998.
- 15.Hripcsak G. Writing Arden Syntax Medical Logic Modules. Comput Biol Med. 1994;24(5): 331-63. [DOI] [PubMed] [Google Scholar]
- 16.Wang P, Pryor TA, Narus S, Hardman R, Deavila M. The Web-enabled IHC enterprise data warehouse for clinical process improvement and outcomes measurement. Proc AMIA Annu Fall Symp. 1997: 1028.
- 17.Kimball R. The Data Warehousing Toolkit. New York: John Wiley, 1997.
- 18.McKie S. Software agents: application intelligence goes undercover. DBMS Magazine. 1995;8(4): 56-60. [Google Scholar]
- 19.Ludemann P, Wilcox A, Hripcsak G. Standard specification for defining and sharing modular health knowledge bases (Arden Syntax for Medical Logic Modules), version 4. Philadelphia, Pa.: American Society for Testing Materials, 1997. Also available at: http://www.cpmc.columbia.edu/arden/.
- 20.Nadkarni PM, Cheung KH. SQLGEN: an environment for rapid client-server database application development. Comput Biomed Res. 1995;28(12): 479-99. [PubMed] [Google Scholar]
- 21.Aho AV, Sethi R, Ullman JD. Section 2.3: Syntax-Directed Translation. Compilers: Principles, Techniques, Tools. Reading, Mass.: Addison-Wesley, 1988: 33-40.
- 22.Nadkarni PM. Concept Locator: A client-server application for retrieval of UMLS Metathesaurus concepts through complex Boolean query. Comput Biomed Res. 1997;30: 323-36. [DOI] [PubMed] [Google Scholar]
- 23.Ullman JD. Principles of database and knowledge-base systems. Rockville, Md.: Computer Science Press, 1989.
- 24.Date CJ. Selected Database Readings, 1985-1989. 7th ed. Reading, Mass.: Addison-Wesley, 1990.
- 25.Elkins SB. Open OLAP. DBMS Magazine. 1998;11(4): 34-8. [Google Scholar]
- 26.Lindberg DAB, Humphreys BL, McCray AT. The Unified Medical Language System. Methods Inform Med. 1993;32: 281-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen H, Ng TD, Martinez J, Schatz BR. Concept space approach to addressing the vocabulary problem in scientific information retrieval: an experiment on the worm community system. J Am Soc Inform Sci. 1997;48(1): 17-31. [Google Scholar]
- 28.Das AK, Musen MA. A temporal query system for protocol-directed decision support. Methods Inform Med. 1994;33(4): 358-70. [PubMed] [Google Scholar]
- 29.Das AK, Musen MA. A comparison of the temporal expressiveness of three database query methods. Proc 19th Annu Symp Comput Appl Med Care. 1995: 331-7. [PMC free article] [PubMed]
- 30.Snodgrass RT, Boehlen MH, Jensen CS, Steiner A. Adding Transaction Time to SQL Temporal: ANSI Experts' Contribution. Geneva, Switzerland: International Organization for Standardization, 1996.
- 31.Snodgrass RT, Boehlen MH, Jensen CS, Steiner A. Adding Valid Time to SQL Temporal: ANSI Experts' Contribution. Geneva, Switzerland: International Organization for Standardization, 1996.