Abstract
Purpose
Retinitis pigmentosa (RP) is a genetically heterogeneous trait with autosomal-recessive (ar) inheritance underlying 50% of genetic disease cases. Sixty-one arRP genes have been identified, and recently, DHX38 has been reported as a potential candidate gene for arRP with only a single family reported with a variant of unknown significance. We identified a missense variant in DHX38 that co-segregates with the arRP phenotype in two Pakistani families confirming the involvement of DHX38 in the etiology of early-onset RP.
Methods
Exome sequencing was performed using two DNA samples from affected members of Pakistani families (MA88 and MA157) with early onset arRP. Sanger sequencing of DNA samples from all family members confirmed the segregation of candidate variant within both families.
Results
A novel missense DHX38 variant c.971G>A; p.(Arg324Gln) was identified which segregates with the arRP phenotype and yielded a logarithm of the odds (LOD) score of 5.0 and 4.3 for families MA88 and MA157, respectively. This variant is predicted to be conserved and deleterious by several bioinformatics tools.
Conclusions
We identified a second deleterious DHX38 variant that segregates with arRP in two families, providing additional evidence that DHX38 is involved in RP etiology. DHX38 encodes for pre-mRNA splicing factor PRP16, which is important in catalyzing pre-mRNA splicing.
Keywords: DHX38, early onset autosomal recessive retinitis pigmentosa, cataract, exome sequencing, linkage analysis, mendelian disease, retinal dystrophy
Retinal dystrophy (RD) is one of many eye diseases that affect retinal structure and function. The most common type of RD is retinitis pigmentosa (RP; MIM268000). The prevalence of RP is estimated at 1 in 4000 individuals worldwide.1 RP symptoms include early-onset night blindness followed by the development of tunnel vision and slow progressive decrease in central vision. For some RP patients, progression leads to complete blindness. Additionally, RP patients can develop cataracts as the disease progresses.
RP is characterized by both genetic and clinical heterogeneity, with more than 90 genes identified of which 61 underlie autosomal-recessive RP (arRP; in the public domain, https://sph.uth.edu/retnet/). DHX38 was recently suggested as a new candidate gene involved in arRP. In the previous study, a missense variant c.995G>A; p.(Gly332Asp) in DHX38 was identified in a small consanguineous Pakistani family (A) that segregates with arRP logarithm of the odds (LOD Score = 2.65) suggesting that DHX38 could be a novel gene involved in arRP.2 This variant of uncertain significance (VUS) is not reported in Genome Aggregation Database (gnomAD) and is predicted to be damaging.
DHX38 was considered to be a gene of uncertain significance for arRP, because besides p.Gly332Asp, no other potentially pathogenic variants have been reported.
DHX38, which encodes DEAH (Asp-Glu-Ala-His) box polypeptide 38 (NM_014003.3), is located on chromosome 16q22.2 and contains 26 protein-coding exons. The gene encodes an ATP-dependent RNA helicase, pre-mRNA processing factor 16 (PRP16). Human ATPase PRP16 interacts with the spliceosome after the first splicing catalytic reaction and removes the lariat and joins the exons.3
We identified a second missense DHX38 variant, c.971G>A; p.(Arg324Gln), through genome-wide genotyping, homozygosity mapping, linkage analysis, and exome and Sanger sequencing using DNA samples from two arRP consanguineous Pakistani families (MA88 and MA157). This conserved and deleterious missense variant segregates with the arRP phenotype and produced LOD scores of 5.0 and 4.3 for families MA88 and MA157, respectively, when parametric linkage analysis was performed.
Materials and Methods
Patients
The institutional review boards of the Quaid-i-Azam University and the Baylor College of Medicine and Affiliated Hospitals approved the study. The study protocol was conducted according to the Tenets of the Declaration of Helsinki. Written informed consent was obtained from all participating members of the MA88 and MA157 families affected with arRP that were ascertained from two different villages of Bagh district in Azad Kashmir of Pakistan.
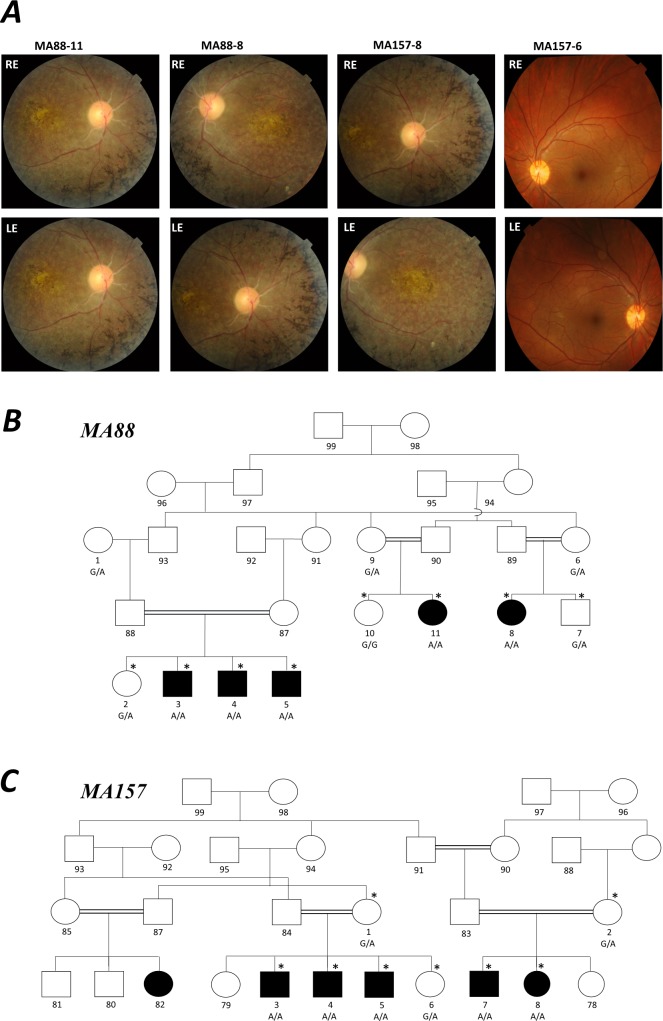
Family history and clinical information were obtained from both MA88 and MA157 family members (Fig. A and Table). Fundoscopy was performed for every affected family member who participated in the study from pedigrees MA88 and MA157 and unaffected family member (MA157-6; Figs. A–C). Vision tests and examinations were also performed on unaffected members from both families, MA88-2, MA88-7, MA88-10, MA157-2, and MA157-6. Peripheral blood samples were collected from MA88 (N = 11) and MA157 (N = 8) family members. DNA was isolated from the collected blood samples using a standard phenol-chloroform extraction protocol.4
Figure.
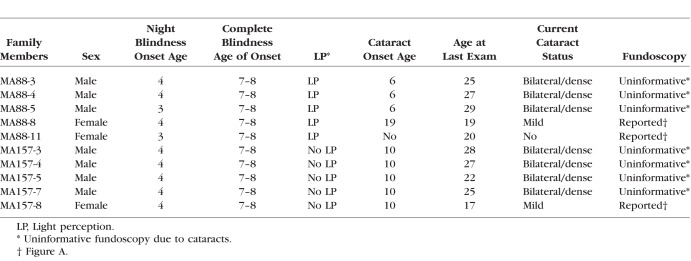
Genetic and clinical findings for arRP Pakistani families, MA88 and MA157. All individuals from family MA88 with a DNA sample underwent whole-genome genotyping except MA88-5 who was ascertained after genotyping was completed. MA88-3 and MA157-7 were selected to undergo exome sequencing. (A) Fundoscopy images obtained from MA88-11 (20 years old), MA88-8 (19 years old), and MA157-8 (17 years old) display clustered areas of intraretinal pigment on the periphery, macular-atrophy, and attenuation of arteries, while fundoscopy for unaffected MA157-6 (21 years old) was normal. Fundoscopy images for the males who all had bilateral cataracts are not displayed, because it was not possible to view the retina, optic disc, or macula. Pedigree drawing (B) of family MA88 and (C) family MA157. Squares represent males and circles females with filled symbols representing individuals with arRP and clear figures representing unaffected family members. Double lines denote consanguineous marriages. The asterisk symbol (*) above individuals from both families MA88 and MA157 indicates members who were examined and vision tests were performed. Below each family member for which a DNA sample was available is shown their DHX38 c.971G>A genotype.
Table.
Phenotype Details and the Age of Onset for the Affected Family Members From MA88 and MA157
Homozygosity Mapping and Linkage Analysis
Genome-wide genotyping using the Infinium HumanCoreExome Chip (Illumina, San Diego, CA, USA), which contains approximately 550-K single-nucleotide polymorphism (SNP) markers with a mean intermarker distance of 5.5 kb, was performed using 10 DNA samples (4 affected and 6 unaffected) from MA88 family. One additional affected family member (MA88-5) was collected after genome-wide genotyping was completed. The SNP genotype data were analyzed using homozygosity mapping and linkage analysis. Homozygosity mapping was performed using affected and unaffected family members using HomozygosityMapper.5 Two-point and multipoint linkage analyses were performed using Superlink6 assuming a completely penetrant autosomal recessive mode of inheritance with a disease allele frequency of 5.0 × 10−6. Marker allele frequencies were estimated from founders and reconstructed founder genotypes from pedigree MA88 and other Pakistani pedigrees, which were genotyped at the same time. To obtain genetic map distances, for multipoint linkage analysis, interpolation was performed using the Rutgers-Combined Linkage maps.7 For family MA157 whole-genome genotyping was not performed, but an affected family member was selected for exome sequencing. For both families two-point linkage analysis was performed for the DHX38 variant, c.971G>A; p.(Arg324Gln) by analyzing data from the 11 and eight members from family MA88 and MA157, respectively, using a minor allele frequency of 5.0 × 10−6 for the alternative allele of c.971G>A.
Exome Sequencing, Filtering, and Analysis
DNA samples from one affected male from each family (MA88-3 and MA157-7) were selected for exome sequencing. Sequence capture was performed using the Roche NimbleGen SeqCap EZ Human Exome Library v.2.0 (∼37-Mb target, Basel, Switzerland). Sequencing to a median read depth of 77× for MA88-3 and 90× for MA157 was performed on a HiSeq4000 (Illumina). Fastq files were aligned to the hg19 human reference sequence using Burrows-Wheeler Aligner (BWA)8 to generate demultiplexed BAM file. Variant detection and calling were performed using the Genome Analysis Toolkit (GATK).9 The variant call file (VCF) was annotated using ANNOVAR10 and Variant Effect Predictor (VEP).11 Variant filtering was performed using Gemini,12 prioritizing variants with a minor allele frequency less than 0.001 in every ancestry group within Genome Aggregation Database (gnomAD)13 and are also predicted to be deleterious and conserved by multiple bioinformatics tools (Polyphen2, Harvard Medical School, Boston, MA, USA; Provian, J. Craig Venter Institute, La Jolla, CA, USA; SIFT, Fred Hutchinson Cancer Research Center, Seattle, WA, USA; Mutation taster, Universitätsmedizin, Berlin, Germany; GERP++ RS, Stanford University, Stanford, CA, USA; and phyloP, Universitätsmedizin, Berlin, Germany). Variants that lie within regions of linkage and homozygosity mapped in MA88 family were prioritized. The exome data from MA88-3 and MA157-7 were also analyzed by examining all variants regardless of frequency within and surrounding the 16q22 region, which contains DHX38, to determine if they shared a haplotype in common.14
Segregation Validation by Sanger Sequencing
To verify co-segregation of DHX38 variant c.971G>A; p.(Arg324Gln) in families MA88 and MA157, forward and reverse PCR primers were designed. Standard conditions were used to perform PCR reactions. The amplified PCR products were treated with ExoSAP-IT PCR Product Cleanup Reagent (ThermoFisher Scientific, Sugar Land, TX, USA). Sequencing was run on an ABI 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) using the BigDye-Terminator v3.1 cycle sequencing kit (Applied Biosystems). Sanger sequencing was performed using available DNA samples from families MA88 (N = 11) and MA157 (N = 8).
Results
Clinical Findings
Members from both arRP families developed night blindness between 3 (MA88-5 and MA88-11) and 4 (MA88-3, MA88-4, MA88-8, MA157-3, MA157-4, MA157-5, MA157-7, and MA157-8) years of age and developed complete blindness between the ages of 7 and 8 (Table). Currently, all affected family members are older than 20 years of age. Affected members of MA88 are blind but retain light perception and affected MA157 members are blind with no light perception. The three affected males from family MA88 were diagnosed with bilateral cataracts at 6 years of age. Affected female family member MA88-8, displayed cataracts at 19 years of age; however, her affected female cousin MA88-11, did not have any sign of cataracts at 20 years of age. All affected MA157 family members (4 males and 1 female) started developing cataracts at approximately 10 years of age. For the three affected females (MA88-8, MA88-11, and MA157-8) fundoscopy displayed macular atrophy, attenuation of arteries, and clustered areas of intraretinal pigment on the periphery. Fundoscopy was also performed on the affected males from both families (MA88-3, MA88-4, MA88-5, and MA157-7) but due to bilateral cataracts it was not possible to view the retina, optic disc, or macula. Fundoscopy performed on the unaffected MA157-6 at 21 years of age showed a healthy macula with bright arteries. Unaffected family members were also examined, and vision tests were performed at 19 (MA88-10), 20 (MA88-2), 21 (MA88-7 and MA157-6), 51 (MA157-2), and 54 (MA157-1) years of age and their vision was within the normal range. Individuals MA88-6 and MA88-9 who are 43 and 41 years of age, respectively, self-reported to have no visional problems (Fig. A).
Homozygosity Mapping and Linkage Analysis
Analysis of the genotype array data for the four MA88 family members revealed a homozygous region of 10.76 Mb flanked by rs12931803 and rs17679567 (chr16: 66122316-7689012) on chromosome 16q21-q23.1. Multipoint linkage analysis with the markers in this region produced a LOD score of 4.8. Two-point linkage analysis with the DHX38 variant, c.971G>A; p.(Arg324Gln) produced a LOD score of 5.0 and 4.3 at theta = 0 for MA88 and MA157 families, respectively.
Filtering and Segregation Analysis
The exome data generated for individual 3 (MA88) contained 25,961 variants with alternative alleles in 12,502 genes and the data from individual 7 (MA157) had 32,462 variants with alternative alleles in 12,619 genes. The analysis of the exome data was performed by selecting homozygous protein coding variants with minor allele frequency (MAF) less than 0.001 in gnomAD for every population and that were predicted to be pathogenic by bioinformatics tools. Meeting these criteria, the exome data from individuals MA88-3 and MA157-7 had nine and seven variants, respectively, each in a separate gene. Only the c.971G>A; p.(Arg324Gln) DHX38 variant mapped to the 16q21-q23.1, region of linkage and homozygosity, which was identified by analyzing family MA88 genotyping data. Additionally, of the candidate variants identified for families MA88 and MA157 only the DHX38 NM_014003.3: g.72133641G>A, c.971G>A: p.(Arg324Gln) variant, which is in exon 8 was found in both exomes. Furthermore, DHX38 had previously been reported to be a gene of uncertain significance for arRP. To ensure that the variant within this gene was not a sequencing or an alignment error, the respective genomic region was examined in the exome data from MA88-3 and MA157-7 individuals by using the Integrative Genome Viewer (IGV).15 The variant had a high reads quality in both aligned exome data. Sanger sequencing was used to further validate the variant and established that it co-segregated with the arRP phenotype for both families MA88 and MA157. Affected arRP individuals are homozygous for the alternative allele and unaffected family members either heterozygous or wild-type genotype (Figs. B, C).
Analysis of single nucleotide variations (SNVs), using the exome data from MA88-3 and MA157-7, surrounding the causal variant DHX38 chr16:72133641:G>A identified a 15.7-Mb (based on 96 SNVs; chr16: 65038674-80718879) shared haplotype. The size of the haplotype is a strong indication that families MA88 and MA157 likely have a quite recent common ancestor.
Bioinformatic Evaluation of DHX38 c.971G>A; p.(Arg324Gln)
The DHX38 c.971G>A; p.(Arg324Gln) variant found in family MA88 was reported in gnomAD at a very low frequency 4.1 × 10−6, with only one alternative allele that has been reported (Latino population) in a total of 246,008 haplotypes. This variant has a CADD C-score16 of 34 and was predicted as probably damaging and deleterious by several additional bioinformatic tools (Polyphen2 HVAR score = 0.999, PROVEAN score = −3.78, SIFT score = 0.01, Mutation Taster = disease causing). The amino acid residue is also highly conserved in 95 vertebrate species (GERP++ RS score = 4.69 and phyloP score = 9.24).
Discussion
In this report, we identified a novel missense variant c.971G>A; p.(Arg324Gln) within DHX38 that segregates within two Pakistani families MA88 and MA157 with early-onset arRP. The novel arRP missense variant was identified using genome-wide genotyping, linkage analysis, homozygosity mapping, and exome and Sanger sequencing using DNA samples from family MA88 and by exome and Sanger sequencing of DNA samples from MA157. In silico prediction tools indicate that this missense variant is likely pathogenic. This variant segregates perfectly with the arRP phenotype in families MA88 and MA157 and was not reported in 30,766 south Asian haplotypes in gnomAD. This is the second arRP variant reported in DHX38, which is classified as likely pathogenic17 suggesting that DHX38 is a novel gene underlying severe early-onset arRP.
DHX38 encodes PRP16, which belongs to the subfamily of DEAH box proteins. DEAH box proteins are involved in a diversity of cellular processes essential in the modification of RNA secondary structure,18 pre-mRNA splicing, and spliceosome assembly.19 The PRP16 is an RNA-dependent ATPase that interacts with spliceosome during the second splicing step. The cryo-EM structure of the spliceosome after the first catalytic step reveals a restructuring of several RNA and protein domains mediated by PRP16. The movement of the branched intron away from the catalytic center is initiated by PRP16.20 This conformational rearrangement, induced by the ATPase Prp16, is suggested to be crucial in the fidelity of splicing.21
The human PRP16 protein domains have high homology to yeast protein, especially in the c-terminal where identity is 51%. Human PRP16 protein identity with yeast diverges in the N-terminus22 but the exact role of this domain is not well understood. The variants reported in family A (Gly332Asp)2 and families MA88 and MA157 p.(Arg324Glu) are both localized in the N-terminal region of PRP16 protein and thus indicate their importance for the normal protein function. This led to the assumption that missense variants at Arg324 and Gly332 might lead to the misfolding of PRP16, which can probably influence the conformational rearrangement of the spliceosome. Although Prp16 is supposed to act at a distance by pulling substrate RNA from the catalytic core of the spliceosome,23 a conformational change may prevent it from promoting accurate splicing.
Families MA88 and MA157 were ascertained from two different villages of the Bagh district in Azad Kashmir of Pakistan. Although they have no knowledge of being related, the exome data suggested that they share a common ancestor. Family MA157 has two branches that are related by marriage, it was also reported by the family that these two branches share a common ancestor, but the exact relationship is unknown.
Interestingly, the three DHX38 families display early-onset night blindness (i.e., by 5 years of age). Members of Pakistani family A and family MA157 who are homozygous for p.(Gly332Asp) and p.(Arg324Glu), respectively, were completely blind (no light perception) by 20 years of age. Family A fundus photographs displayed a developed macular coloboma. On the other hand, MA88 family members who are homozygous for p.(Arg324Glu), were reported to be blind by the age of 7 and still have light perception in their early twenties. Unlike families MA88 and MA157, none of the members of family A had cataracts.
Among the 90 known RP genes, nine genes encode for splicing factors or components of splicing machinery, with seven of these genes underlying autosomal-dominant RP and two genes, ARL2BP24 and DHX38 causing arRP. The observed phenotypic variability is not unusual for RP and has also been reported in other splicing factor genes causing retinal dysfunction. For example, for the spliceosomal protein PRPF8, which underlies autosomal-dominant RP recent phenotype–genotype correlation studies on a large number of RP patients from different pedigrees that segregate a variety of missense PRPF8 variants, displayed significant clinical severity and age of onset differences.25,26 Intrafamilial variable RP phenotypes have also been previously described.27 Intrafamilial variability in families MA88 and MA157 were also observed. For both families, all arRP males developed cataracts by the age of 10, while the females with arRP were either diagnosed with less severe cataracts later (i.e., between 17 and 20 years or did not develop cataracts; i.e., by 20 years of age).
In conclusion, we identified a novel missense variant in DHX38 that co-segregates with the arRP phenotype in two Pakistani families. The arRP families provide additional evidence for the involvement of DHX38 in arRP. Further genetic, functional, and structural studies are needed to reveal the function of the N-terminus region of PRP16 protein and its implication in excising the lariat intron and joining the exons during the second step of the splicing process.
Acknowledgments
The authors thank the family members who participated in this study.
Supported by grants from the Higher Education Commission, Islamabad under National Research Program for Universities (NRPU-420; MA; Islamabad, Pakistan). Genotyping and Sequencing were provided by the University of Washington Center for Mendelian Genomics (UW-CMG) and were funded by the National Human Genome Research Institute and the National Heart, Lung and Blood Institute grant HG006493 (DAN, MJB, and SML; Bethesda, MD, USA).
Disclosure: Z. Latif, None; I. Chakchouk, None; I. Schrauwen, None; K. Lee, None; R.L.P. Santos-Cortez, None; I. Abbe, None; A. Acharya, None; A. Jarral, None; I. Ali, None; E. Ullah, None; M.N. Khan, None; G. Ali, None; T.H. Tahir, None; M.J. Bamshad, None; D.A. Nickerson, None; W. Ahmad, None; M. Ansar, None; S.M. Leal, None
Appendix
Members of the University of Washington Center for Mendelian Genomics (UWCMG) Study Group
Michael J. Bamshad,1,2 Suzanne M. Leal,3 Deborah A. Nickerson,1 Peter Anderson,1 Marcus Annable,1 Elizabeth E. Blue,1 Kati J. Buckingham,1 Imen Chakchouk,3 Jennifer Chin,1 Jessica X Chong,1 Rodolfo Cornejo Jr.,1 Colleen P. Davis,1 Christopher Frazar,1 Martha Horike-Pyne,1 Gail P. Jarvik,1 Eric Johanson,1 Ashley N. Kang,1 Tom Kolar,1 Stephanie A. Krauter,1 Colby T. Marvin,1 Sean McGee,1 Daniel J. McGoldrick,1 Karynne Patterson,1 Sam W. Phillips,1 Jessica Pijoan,1 Matthew A. Richardson,1 Peggy D. Robertson,1 Isabelle Schrauwen,3 Krystal Slattery,1 Kathryn M. Shively,1 Joshua D. Smith,1 Monica Tackett,1 Alice E. Tattersall,1 Marc Wegener,1 Jeffrey M. Weiss,1 Marsha M. Wheeler,1 Qian Yi,1 and Di Zhang3
1University of Washington, Seattle, Washington, United States
2Seattle Children's Hospital, Seattle, Washington, United States
3Baylor College of Medicine, Houston, Texas, United States
References
- 1.Hamel C. Retinitis pigmentosa. Orphanet J Rare Dis. 2006;1:40. doi: 10.1186/1750-1172-1-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ajmal M, Khan MI, Neveling K, et al. A missense mutation in the splicing factor gene DHX38 is associated with early-onset retinitis pigmentosa with macular coloboma. J Med Genet. 2014;51:444–448. doi: 10.1136/jmedgenet-2014-102316. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, Guthrie C. PRP16, a DEAH-box RNA helicase, is recruited to the spliceosome primarily via its nonconserved N-terminal domain. RNA. 1998;4:1216–1229. [PMC free article] [PubMed] [Google Scholar]
- 4.Yanamandra G, Lee ML. Isolation and characterization of human DNA in agarose block. Gene Anal Tech. 1989;6:71–74. doi: 10.1016/0735-0651(89)90018-6. [DOI] [PubMed] [Google Scholar]
- 5.Seelow D, Schuelke M, Hildebrandt F, Nürnberg P. HomozygosityMapper–an interactive approach to homozygosity mapping. Nucleic Acids Res. 2009;37:W593–599. doi: 10.1093/nar/gkp369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silberstein M, Tzemach A, Dovgolevsky N, Fishelson M, Schuster A, Geiger D. Online system for faster multipoint linkage analysis via parallel execution on thousands of personal computers. Am J Hum Genet. 2006;78:922–935. doi: 10.1086/504158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matise TC, Chen F, Chen W, et al. A second-generation combined linkage physical map of the human genome. Genome Res. 2007;17:1783–1786. doi: 10.1101/gr.7156307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McLaren W, Gil L, Hunt SE, et al. The Ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paila U, Chapman BA, Kirchner R, Quinlan AR. GEMINI: integrative exploration of genetic variation and genome annotations. PLoS Comput Biol. 2013;9:e1003153. doi: 10.1371/journal.pcbi.1003153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palotie A, Kiezun A, Moonshine AL, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith J, Coop G, Stephens M, Novembre J. Estimating time to the common ancestor for a beneficial allele. Mol Biol Evol. 2017;35:1003–1017. doi: 10.1093/molbev/msy006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med Off J Am Coll Med Genet. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bleichert F, Baserga SJ. The long unwinding road of RNA helicases. Mol Cell. 2007;27:339–352. doi: 10.1016/j.molcel.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 19.Hegele A, Kamburov A, Grossmann A, et al. Dynamic protein-protein interaction wiring of the human spliceosome. Mol Cell. 2012;45:567–580. doi: 10.1016/j.molcel.2011.12.034. [DOI] [PubMed] [Google Scholar]
- 20.Bertram K, Agafonov DE, Liu W-T, et al. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature. 2017;542:318–323. doi: 10.1038/nature21079. [DOI] [PubMed] [Google Scholar]
- 21.Konarska MM, Query CC. Insights into the mechanisms of splicing: more lessons from the ribosome. Genes Dev. 2005;19:2255–2260. doi: 10.1101/gad.1363105. [DOI] [PubMed] [Google Scholar]
- 22.Zhou Z, Reed R. Human homologs of yeast prp16 and prp17 reveal conservation of the mechanism for catalytic step II of pre-mRNA splicing. EMBO J. 1998;17:2095–2106. doi: 10.1093/emboj/17.7.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Semlow DR, Blanco MR, Walter NG, Staley JP. Spliceosomal DEAH-Box ATPases remodel pre-mRNA to activate alternative splice sites. Cell. 2016;164:985–998. doi: 10.1016/j.cell.2016.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davidson AE, Schwarz N, Zelinger L, et al. Mutations in ARL2BP, encoding ADP-ribosylation-factor-like 2 binding protein, cause autosomal-recessive retinitis pigmentosa. Am J Hum Genet. 2013;93:321–329. doi: 10.1016/j.ajhg.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Escher P, Passarin O, Munier FL, Tran VH, Vaclavik V. Variability in clinical phenotypes of PRPF8-linked autosomal dominant retinitis pigmentosa correlates with differential PRPF8/SNRNP200 interactions. Ophthalmic Genet. 2018;(39):80–86. doi: 10.1080/13816810.2017.1393825. [DOI] [PubMed] [Google Scholar]
- 26.Maubaret CG, Vaclavik V, Mukhopadhyay R, et al. Autosomal dominant retinitis pigmentosa with intrafamilial variability and incomplete penetrance in two families carrying mutations in PRPF8. Invest Ophthalmol Vis Sci. 2011;52:9304–9309. doi: 10.1167/iovs.11-8372. [DOI] [PubMed] [Google Scholar]
- 27.Parmeggiani F, Milan E, Costagliola C, et al. Macular coloboma in siblings affected by different phenotypes of retinitis pigmentosa. Eye. 2004;18:421–428. doi: 10.1038/sj.eye.6700689. [DOI] [PubMed] [Google Scholar]