Abstract
The selective impact of pathogen epidemics on host defenses can be strong but remains transient. By contrast, life-history shifts can durably and continuously modify the balance between costs and benefits of immunity, which arbitrates the evolution of host defenses. Their impact on the evolutionary dynamics of host immunity, however, has seldom been documented. Optimal investment into immunity is expected to decrease with shortening lifespan, because a shorter life decreases the probability to encounter pathogens or enemies. Here, we document that in natural populations of Arabidopsis thaliana, the expression levels of immunity genes correlate positively with flowering time, which in annual species is a proxy for lifespan. Using a novel genetic strategy based on bulk-segregants, we partitioned flowering time-dependent from -independent immunity genes and could demonstrate that this positive covariation can be genetically separated. It is therefore not explained by the pleiotropic action of some major regulatory genes controlling both immunity and lifespan. Moreover, we find that immunity genes containing variants reported to impact fitness in natural field conditions are among the genes whose expression covaries most strongly with flowering time. Taken together, these analyses reveal that natural selection has likely assorted alleles promoting lower expression of immunity genes with alleles that decrease the duration of vegetative lifespan in A. thaliana and vice versa. This is the first study documenting a pattern of variation consistent with the impact that selection on flowering time is predicted to have on diversity in host immunity.
Keywords: immunity; flowering time; lifespan; Arabidopsis thaliana, transcriptomics; trade-off; pleiotropy; bulk-segregant sequencing
Introduction
The ability of organisms to defend against pathogens is a major determinant of survival in natural populations (Parker and Gilbert 2004; Chisholm et al. 2006; Lee and Mazmanian 2010). Pathogens have long been suspected to impose a fast evolution of the host immune system and the “Red Queen” Hypothesis is nowadays a keystone of evolutionary biology (Van Valen 1973; Liow et al. 2011). Evidence that pathogens drive the molecular evolution of host defense systems has been accumulating in an array of plant and animal systems (Bergelson et al. 2001; de Meaux and Mitchell-Olds 2003; Moeller and Tiffin 2005; Laine et al. 2011; Maekawa et al. 2011; Ravensdale et al. 2011; Dybdahl et al. 2014; Karasov et al. 2014; Siddle and Quintana-Murci 2014; Parker et al. 2015; Metzger et al. 2016).
Yet, the possible impact of changes in ecology on the evolution of defense systems should also be considered as they may durably change the exposure of hosts to pathogens. Invasive species, for example, owe much of their success to the release from pathogen and pest pressures (Mitchell and Power 2003; Mitchell et al. 2010). Similarly, shifts in life history can alter the balance between costs and benefits of host defense systems (Herms and Mattson 1992). Shifting from perennial to annual life cycles, or evolving from a winter-annual to summer-annual cycling occurs frequently across plant phylogenies (Garnier 1992; Michaels et al. 2003; Franks et al. 2007; Tank and Olmstead 2008; Matthew Ogburn and Edwards 2015; Kiefer et al. 2017). The reduction in lifespan that follows such life history changes concomitantly reduces the probability to encounter ennemies (Jokela et al. 2000). As a matter of fact, woody plant species with longer lifespan often display stronger herbivore defenses (Endara and Coley 2011). As a consequence, immunity and lifespan are expected to coevolve.
Arabidopsis thaliana populations offer an optimal model for catching the coevolution of life history and immunity in the act. A. thaliana has become over the last decade a powerful model system to address ecological questions at the genetic level (Mitchell-Olds and Schmitt 2006; Bergelson and Roux 2010; Roux and Bergelson 2016). Experiments in common gardens have been performed to describe the architecture of natural variation in fitness and to infer geographic distributions of locally adaptive mutations (Fournier-Level et al. 2011, 2016; Hancock et al. 2011). Analyses of mutants and recombinant inbred lines (RIL) have allowed reconstructing the contribution of phenotypes to fitness (Wilczek et al. 2009; Chiang et al. 2013; Fournier-Level et al. 2013). Secondary chemical compounds were shown to have evolved to deter predominant herbivores in natural populations (Brachi et al. 2013; Kerwin et al. 2015). Clinal variation along the latitudinal range of the species reveals how phenotypes expressed along the life cycle are jointly shaped by natural selection (Lasky 2012; Debieu et al. 2013; Vidigal et al. 2016).
A. thaliana is arguably one of the species for which we have the largest amount of genetic and phenotypic information on both immune reactions against pathogens and variation in the duration of the vegetative lifespan. As such, it is an optimal model system for assessing the impact of life history changes, which modify plant vegetative lifespan, on the evolution of the immunity system. Indeed, in annual (monocarpic) species, which grow and reproduce only once, flowering time marks the end of the vegetative growth phase. Seed production in monocarpic species is terminated by senescence and death, so that flowering time provides a good proxy for lifespan. In A. thaliana, it has been scored in a number of conditions (Brachi et al. 2010; Sasaki et al. 2015; Roux and Bergelson 2016) and flowering time changes are often locally adaptive (Le Corre 2005; Toomajian et al. 2006; Montesinos-Navarro et al. 2011; Debieu et al. 2013; Li et al. 2014; Hu et al. 2017). Natural variation in flowering time can thus be used to investigate the impact of lifespan changes on host defenses.
The immune system has also been intensively studied in this species, revealing multiple layers of defenses, ranging from basal immunity, which is sufficient to control most microbes, to severe reactions that actively defeat virulent pathogens (Jones and Dangl 2006). Strain-specific immunity components are likely to be linked in their evolution to the virulence specificity of co-occurring pathogens (de Meaux and Mitchell-Olds 2003; Moeller and Tiffin 2005; Roux and Bergelson 2016). Recent fluctuations in the composition of the pathogen population may therefore affect the specific components of immunity targeted by these epidemics and thereby mask or blur the long-term impact of lifespan modifications. To minimize this effect and to highlight the impact of lifespan variation, we took a genomics approach and examined how flowering time covaries with expression levels of genes with an experimentally supported function in immunity. These approximately 700 genes jointly reflect a broad spectrum of traits, which, when their expression increases have a positive effect on immunity (Eulgem 2005; Vetter et al. 2012; Boccara et al. 2014). We test below whether their expression level, a proxy for their effectiveness, covariates with flowering time, a proxy for lifespan in the field and further examine the roles played by demographic history and pleiotropy in shaping patterns of covariation.
Results
Positive Covariation between Expression Levels of Immunity Genes and the Timing of Flowering in Swedish A. thaliana Populations
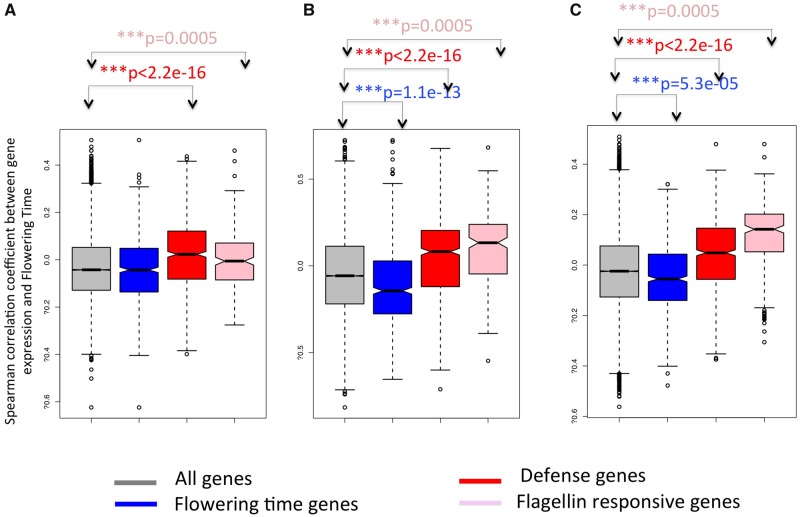
We first focused on a set of 138 genotypes originating from Sweden because high quality data were available for both genome-wide expression profiles and flowering time estimates (Dubin et al. 2015; Sasaki et al. 2015). These two studies were part of a single experiment in which flowering time and gene expression were characterized at both 16°C and 10°C under long day conditions in growth chambers. We focused on the data collected at 16°C and computed Spearman correlation coefficients between the expression level of each gene and flowering time. Of 22,686 genes, for which expression levels could be quantified, 1,374 (6%) were significantly correlated with flowering time under a 5% false discovery rate (FDR). We first verified that genes annotated for their function in flowering time were among the genes whose expression correlates with the phenotype. Overall, genes with an experimentally validated function in flowering time in the genome were not enriched among those genes (6.9% of 630 genes at FDR 0.05, hypergeometric test, P = 0.19), yet the two well-known regulators of flowering time, FLOWERING LOCUS C and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS-1 (FLC and SOC1, Spearman correlation ρ=0.50 and −0.62, FDR-corrected P = 2.87e-6 and P = 7e-12, respectively) were the two most strongly correlated genes. In addition, using the R-package TopGO, we examined patterns of functional enrichment among genes that tended to be more expressed in early flowering genotypes. Many functional gene ontology (GO) categories related to cell differentiation and growth were enriched (supplementary table 1, Supplementary Material online) and the GO category “regulation of flower development” was among the most overrepresented (P = 8.00e-14, supplementary table 1, Supplementary Material online). This observation confirmed the biological relevance of the data set examined.
Next, we tested whether immunity genes were enriched among genes whose expression correlated with the timing of flowering. Among genes with significant correlation with the phenotype, we observed a significant excess of immunity genes (8.6% of 691 genes at 5% FDR, hypergeometric test, P = 0.002). The distribution of correlation coefficients was also significantly skewed toward higher correlation coefficients for immunity genes (fig. 1A, Kolmogorov–Smirnov test, P < 2.2e-16). GO enrichment analysis showed that genes involved in GO “oxidation–reduction process” and “response to wounding” were among the most strongly enriched (P < 1e-30, P = 1.1e-19, respectively, supplementary table 1, Supplementary Material online). This first analysis revealed a pronounced pattern of positive covariation between flowering time and the expression of immunity genes.
Fig. 1.
—Distribution of Spearman correlation coefficients between expression levels of each expressed Arabidopsis thaliana gene and flowering time. Gray: All expressed genes; Blue: Genes annotated as flowering time genes (FT genes); Red: Genes annotated as immunity genes; Pink: Flagellin-responsive (FlaRe) genes (Navarro et al. 2004). (A) For 138 Swedish genotypes; (B) Analysis restricted to 51 Swedish genotypes showing correlated flowering time at 10°C and 16°C; (C) Species-wide sample of 52 genotypes. Distribution for each group of genes was compared with the genome-wide distribution (black double-head arrow) with a Kolmogorov–Smirnov test. P values are given in the color corresponding to the gene class. Spearman correlation coefficients were computed between expression levels of each of 23,511 expressed A. thaliana genes, reported in Durbin et al. (2015) for ninth leaf seedlings, and flowering time measured in the same condition for 51 genotypes originating from natural populations in Sweden (Sasaki et al. 2015). ***P < 0.001.
In laboratory conditions, genotypes with a strong requirement for vernalization tend to show a strong delay in flowering that often does not translate into late flowering in the field (Brachi et al. 2010; Li et al. 2014). Indeed, in the field plants often experience sufficient levels of cold to fulfill their vernalization requirement. In fact, only the 51 genotypes that advanced their flowering time at 16°C compared with 10°C (e.g., those that did not need low temperatures to induce flowering), showed a correlation in their flowering across temperatures (Sasaki et al. 2015). Flowering time variation across the latter subsample of genotypes may therefore allow a more accurate classification of genotypes with increasing vegetative lifespan. Among the 507 out of 22,686 (2.2%) genes that displayed a significant positive correlation with flowering time at 10% FDR across this restricted sample of genotypes, 16/630 genes were annotated for their function in flowering. As in the above, several known flowering time regulators were among the genes associated with flowering time, such as FLOWERING LOCUS C (FLC), GIGANTEA, FLOWERING PROMOTING FACTOR 1-LIKE PROTEIN 2 (FLP2) or even the genes PHYTOCHROME INTERACTING FACTOR 4 (PIF4) and PHYTOCHROME INTERACTING FACTOR 5 (PIF5), which had been associated with accelerated flowering (Andrés and Coupland 2012; Thines et al. 2014). Although the whole set of flowering time genes was not significantly enriched among genes correlating positively with the timing of flowering (2.5%, hypergeometric test, P = 0.24), they tended to be more highly expressed in early flowering genotypes (excess of negative correlations, Kolmogorov–Smirnov test, P = 1.16e-13, fig. 1B). The GO category “regulation of flower development” was even more overrepresented in this data set (P < 1e-30, supplementary table 1, Supplementary Material online). Higher expression of genes associated with the positive regulation of flowering was observed among early flowering genotypes. This further confirms that expression variation was correctly quantified.
We also observed that variation in immunity gene expression tended to correlate positively with variation in flowering time, after excluding vernalization-sensitive genotypes. In total expression of 28 of the 691 genes belonging to the immunity gene category correlated significantly with flowering at 10% FDR. They were significantly enriched (4%, 1.8-fold enrichment, hypergeometric test, P = 0.0009). Compared with the ensemble of expressed genes in the genome, they generally tended to be more highly expressed in late flowering genotypes (marked excess of positive correlations, Kolmogorov–Smirnov test, P < 2.2e-16, fig. 1B). GO enrichment analysis showed that genes involved in the GO categories “response to chitin” and “regulation of plant-type hypersensitive response” were the two most strongly enriched (both P < 1e-30, supplementary table 1, Supplementary Material online). We thus conclude that the correlation between expression of immunity genes and the timing of flowering is independent of allelic variation in vernalization requirements.
Positive Covariation of Immunity Gene Expression with Flowering Time Is Independent of Population Structure and Is Also Detected in a Second Sample of Broader Geographic Origin
Relatedness among individuals in the sample may drive the correlation between expression of immunity genes and the timing of flowering. In fact, flowering time in the Swedish lines is strongly associated with the demographic history of these populations and thus with their population structure (Dubin et al. 2015; Sasaki et al. 2015). We therefore also computed for each gene, the correlation between gene expression and flowering time with a mixed-model that included a kinship matrix for the 51 genotypes that lacked vernalization requirement (see Materials and Methods; Yu et al. 2006; Stich et al. 2008). This analysis revealed that, for immunity genes, the distribution of correlation coefficient estimates remained strongly skewed toward positive values, after population structure was accounted for (Kolmogorov–Smirnov test, P = 2.2e-16, supplementary fig. 1, Supplementary Material online). However, the whole set of immunity genes was no longer enriched among genes with a significant covariation with flowering time (5.2% vs. 5%, hypergeometric test, P = 0.6).
We note that accounting for population structure also did not change the pattern of covariation between gene expression of flowering genes and timing of flowering itself. They showed a coefficient distribution that was strongly skewed toward negative values (Kolmogorov–Smirnov test, P = 2.2e-16) and were significantly overrepresented among genes with expression significantly associated with flowering time (8% vs. 5% at 5% FDR, hypergeometric test, P = 0.0005).
We further investigated whether the skew toward positive covariation between immunity gene expression and flowering time is limited to the regional subset of genotypes growing in Sweden or whether it is a feature of diversity that segregates across the whole range of the species. For this, we turned to a species-wide data set of gene expression variation collected in young seedlings (Schmitz et al. 2013). For 52 of these genotypes, the duration of vegetative growth had been determined under natural conditions in the field (Brachi et al. 2010). Although a skew toward negative correlation for flowering time genes was observed (Kolmogorov–Smirnov test, P = 5.3e-5, fig. 1C), the seedling of these earlier flowering genotypes did not yet express genes important for the formation of flower (supplementary table 1, Supplementary Material online).
Nevertheless, we again observed a strong skew toward positive correlation between immunity gene expression and flowering time, indicating that genotypes that will flower later expressed them at a higher level (Kolmogorov–Smirnov test, P < 2.2e-16, fig. 1C). Immunity genes were not particularly enriched among genes with significantly correlated expression and flowering time at 5% FDR (5% for both, hypergeometric test, P = 0.24). Yet, GO categories such as “response to chitin,” “respiratory burst involved in immunity response,” “response to wounding,” and “immunity response to fungus” were the four most strongly enriched functions among genes with highest Spearman correlation coefficients (all P < 1e-30, supplementary table 1, Supplementary Material online).
Contrasting genotypes of diverse flowering time (e.g., lifespan) revealed that, in natural populations, immunity genes tend to covary positively with this trait. The latter two analyses showed that this effect remained when population structure was accounted for and was also detectable in another gene expression data set and with a different set of genotypes.
A Bulk-Segregant Analysis Demonstrates That Covariation Is Not Due to Pleiotropic Effect of Flowering Time Control
The tendency of immunity genes to show expression levels correlating positively with flowering may be due to the pleiotropic action of regulatory genes that coregulate flowering time and immunity. In plants, the impact of development and growth regulators on defense systems is being increasingly recognized (Alcázar et al. 2011). There is evidence that flowering time and defense control each other (Korves and Bergelson 2003; Martinez et al. 2004; Whalen 2005; Develey-Riviere and Galiana 2007; Pagan et al. 2008; Pajerowska-Mukhtar et al. 2009; Fan et al. 2014; Jiménez-Góngora et al. 2015; Kerwin et al. 2015; Lozano-Durán and Zipfel 2015; Lyons et al. 2015; Davila Olivas et al. 2017). If so, the pattern we observed would not reflect the joint optimization of immunity and life history strategy but only the pleiotropic action of their regulators. We therefore asked to which extent flowering time and the expression of immune-related genes could be genetically separated and thus evolve independently.
We therefore designed an experiment to describe the level of pleiotropy of flowering time regulators on the expression of immunity genes. If such regulators control the pattern of covariation reported in figure 1, it should not be possible to separate variation in immunity gene expression from variation in flowering time in a segregating recombinant inbred population. We used the two genotypes Col-0 and Bur-0, which differ in flowering time (Simon et al. 2008) and were also reported to exhibit markedly distinct sensitivities to flagellin, with the later flowering genotype Bur-0 displaying stronger basal immunity (Vetter et al. 2012). We analyzed the transcriptomes of these two lines at 14 and 28 days after germination (see Materials and Methods) and found that the skews shown in figure 1 remain when the data set was reduced to the genes that differed in expression between these two lines (supplementary fig. 2, Supplementary Material online). This confirmed that these two genotypes could help identify immunity genes that share genetic regulators with flowering time.
We designed a cost-effective approach to identify the genes whose expression variation cannot be separated from flowering time. We used 244 recombinant inbred lines (RILs) derived from a cross between the parents Bur-0 and Col-0, followed by >8 generations of selfing (Simon et al. 2008). We bulked RILs by their flowering time and characterized their transcriptomes at 14 and 28 days after germinations using RNA sequencing (see Materials and Methods). In RILs, the genomes of the parental genotypes are randomly shuffled by recombination. Because of this genetic property, RILs are commonly used to identify Quantitative Trait Loci (QTL), which are genomic regions underlying the genetic control of phenotypic variation. In our approach, this means that differences in gene expression between early- and late-flowering RILs reflect differences that are genetically associated with flowering time. The experimental strategy is described in supplementary figures 3 and 4, Supplementary Material online. This strategy does not allow characterizing the exact genetic architecture of gene expression variation, but it allows the identification of genes whose expression variation is controlled either by flowering-time regulators or by genes located in the genomic vicinity of these regulators. Thereafter, we named these genes Flowering-Time (FT)-dependent genes.
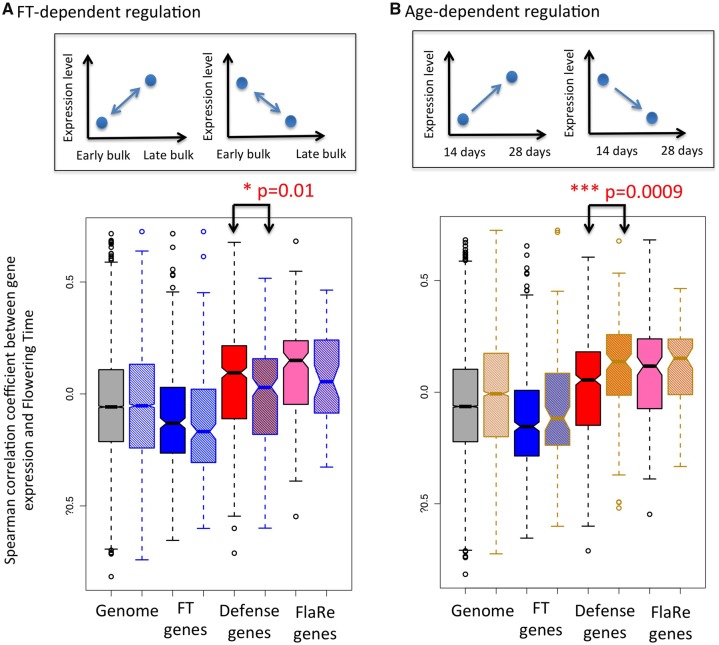
Of a total of 20,553 genes expressed in both the parental genotypes and RIL pools, 6,097 (29%) were differentially expressed between early and late flowering RIL pools, that is, FT-dependent. As expected, there was a strong excess of genes annotated as having a function in flowering time among FT-dependent genes (223/630–36%, hypergeometric test, P = 3.7e-5). This demonstrated that this strategy effectively highlighted genes whose expression is under the genetic control of flowering time regulators. By contrast, immunity genes were not overrepresented among FT-dependent genes. More so, they were clearly underrepresented among FT-dependent genes at the second time point of sampling (1.15-fold less abundant than expected by chance at day 28, hypergeometric test, P = 0.01). Only 19% of all immunity genes were FT-dependent. These genes, however, did not explain the skew toward positive covariation with flowering time reported in figure 1. Immunity genes, whose expression was not differently expressed between RIL pools (i.e., genes whose expression is not dependent on the regulators of flowering time), in fact, tended to be more skewed toward positive correlation coefficients than FT-dependent immunity genes (Kolmogorov–Smirnov test, P = 0.01, fig. 2A). We observed that FT-dependent flowering time genes did not shift significantly from the distribution of correlation in the rest of the genome (Kolmogorov–Smirnov test, P = 0.15, fig. 2A). Therefore, the excess of positive expression covariation with flowering time observed among immunity genes is most strongly driven by genes whose expression level was easily separated from variation in flowering by recombination.
Fig. 2.
—Distribution of Spearman correlation coefficients between gene expression level and flowering time. (A) Partition of genes controlled by flowering time (hatched boxes with blue border) versus independent from flowering time (uniform boxes with black border); (B) Partition of genes controlled by development (hatched boxes with orange border) versus independent from development (uniform boxes with black border). Inserts in the top of the figure illustrates how these gene classes were defined. Immunity genes that are not controlled by flowering time but controlled by development tend to have higher correlation coefficients of natural variation for expression with natural variation for flowering time. Gray: All expressed genes; Blue: Genes annotated as flowering time genes (FT genes); Red: Genes annotated as immunity genes; Pink: Flagellin-responsive (FlaRe) genes (Navarro et al. 2004). P values for Kolmogorov–Smirnov test comparing the distribution of genes within each category that are independent of or regulated by (A) flowering time or (B) age are shown when significant. Note that only 12 FlaRe genes are controlled by flowering time in our experiment. Spearman correlation coefficients were computed between expression levels of each of 23,511 expressed Arabidopsis thaliana genes, reported in Dubin et al. (2015) for ninth leaf seedlings, and flowering time measured in the same condition for 51 genotypes originating from natural populations in Sweden (Sasaki et al. 2015). *P < 0.05, ***P < 0.001.
Age-Regulated Immunity Genes Often Show Positive Covariation with Flowering Time
Immunity genes are often observed to change their activity with age and development (Barton and Boege 2017). Because we had sampled material at day 14 and day 28 after germination, we could also separate genes whose expression changed with age (here after named age-regulated genes) from genes with similar expression levels in 14- and 28-day-old plants (see Materials and Methods). Age-regulated genes were markedly more frequent among annotated immunity genes than among annotated flowering time genes (243/630–38% vs. 334/691—48%, for flowering-time and immunity genes, respectively, χ2 test, P = 7.2e-11). In A. thaliana, a so-called age-related resistance is activated in older A. thaliana plants, providing them with a immunity barrier against a broad spectrum of pathogens (Rusterucci et al. 2005). In agreement with our findings, the timing of age-related resistance had been reported not to stand under the direct control of flowering time (Wilson et al. 2013).
The subset of genes, whose expression variation in natural populations correlated with flowering time, were also enriched among age-regulated genes (hypergeometric test, P = 7.2e-11). Altogether, 4% (348/8,565) and 6% (498/7,935) of age-independent and age-regulated genes, respectively, were correlated with flowering time at 5% FDR. Immunity genes contributed significantly to this excess, because the expression levels of immunity genes that were age-regulated tended to show a strong skew toward positive correlation with flowering time in natural populations (fig. 2B, Kolmogorov–Smirnov test, P = 0.0009). Our analysis thus indicates that the tendency of immunity genes to covary positively with flowering time in natural population is 1) not explained by the genetic control of flowering time and 2) increased among genes whose expression is regulated by plant age.
Genes Activated by Elicitors of Basal Immunity Also Show an Excess of Positive Correlations with Flowering Time
In the above analyses, immunity levels were represented by a set of 731 genes annotated for functions related to immunity. To test whether this trend toward positive covariation between immunity gene expression and flowering time was limited to the set of genes defined by Gene Ontology categories, we analyzed an independent set of immunity-related genes: the 245 genes whose expression is activated in Arabidopsis seedlings upon perception of flagellin by the PAMP receptor kinase FLAGELLIN SENSING 2 (FLS2), hereafter named FlaRe genes (Navarro et al. 2004). FlaRe genes coordinate cellular and developmental responses to exposure of molecular signatures of bacteria. Only 10 FlaRe genes overlapped with the immunity-annotated genes used above. We observed that FlaRe genes were enriched among genes showing positive covariation with flowering time (fig. 1A–C, Kolmogorov–Smirnov test, P < 2.2e-16). This observation remained when accounting for population structure (supplementary fig. 1, Supplementary Material online, Kolmogorov–Smirnov test, P < 2.2e-16) and was also seen for flowering time measured in the field in a species-wide sample of genotypes (fig. 1C, Kolmogorov–Smirnov test, P < 2.2e-16). When partitioning genes according to whether or not they were FT-dependent or age-regulated, we observed that FT-dependence did not significantly change the distribution of correlation coefficients between FlaRe gene expression and flowering time across natural genotypes (fig. 2A and B, Kolmogorov–Smirnov test, P = 0.15 and P = 0.32, for FT-controlled and age-regulated genes, respectively). Nevertheless, FlaRe genes were significantly underrepresented among FT-dependent genes, especially at the second sampling time point (1.8-fold less frequent among flowering time controlled genes, hypergeometric test, P = 2.24e-05). By contrast, they were overrepresented among age-regulated genes (2.1-fold more frequent among age-regulated genes, hypergeometric test, P = 2.6e-08). Thus, the positive covariation reported in figure 1A–C is unlikely to result from the pleiotropic action of flowering time regulators on FlaRe genes. This suggests that, like for annotated immunity genes, alleles attenuating the expression of FlaRe genes were assorted with early flowering alleles in natural populations and vice versa.
Fitness-Associated Immunity Genes Show Higher Correlation Coefficients with Flowering Time
We further asked whether genes with fitness-relevant variation have expression levels that are more strongly assorted with variation in the timing of flowering. A reciprocal transplant experiment performed in 4 locations throughout Europe identified 866 nucleotide variants in the genome of A. thaliana that significantly associated with fitness differences manifested in natural conditions (Fournier-Level et al. 2011). Of these variants, 15 mapped to immunity genes and 17 to flowering genes. Association with fitness coincided with a skew toward higher correlation coefficients for immunity genes only (fig. 3, Kolmogorov–Smirnov test, D = 0.46, P = 0.014, and P > 0.05 for immunity and flowering time genes, respectively). One of the immunity genes (AT3G16720), which is activated upon exposure to the fungal PAMP chitin, was FT-dependent but it did not explain this pattern (Kolmogorov–Smirnov test, P = 0.028 without AT3G16720). Five of the immunity genes with FT-independent immune functions were age-regulated (AT1G18150, AT1G80840, AT4G01700, AT5G19510, AT5G57220) but this did not explain the pattern either (Kolmogorov–Smirnov test, P = 0.009 without these genes). Of the 245 FlaRe genes, 3 contained fitness-associated SNPs. These three genes were among the genes with highest correlation coefficients (AT1G19670: ρ=0.397, AT3G16720: ρ=0.282, AT4G38860: ρ=0.487). We thus observe that immunity genes that can be most relevant for fitness in natural populations of A. thaliana are also genes whose expression levels were most strongly assorted with alleles determining flowering time.
Fig. 3.
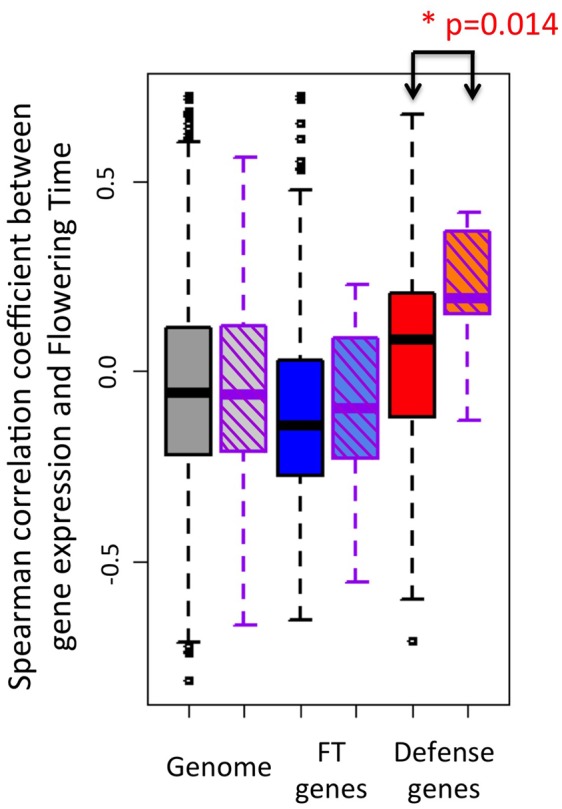
—Distribution of Spearman correlation coefficients between gene expression level and flowering time. All expressed genes—uniform boxes with black border—versus genes with fitness-associated SNPs in Fournier-Level et al. (2011)—hatched boxes with purple border. Gray: All expressed genes; Blue: Genes annotated as flowering time genes (FT genes); Red: Genes annotated as immunity genes. Immunity genes that carry SNPs associating with fitness tend to have higher correlation coefficients of natural variation for expression with natural variation for flowering time. P values for Kolmogorov–Smirnov test comparing the distribution for genes within each category are shown when significant. Spearman correlation coefficients were computed between expression levels of each of 23,511 expressed Arabidopsis thaliana genes, reported in Durbin et al. (2015) for ninth leaf seedlings, and flowering time measured in the same condition for 51 genotypes originating from natural populations in Sweden (Sasaki et al. 2015). *P < 0.05.
Discussion
Evidence for Concerted Evolution of Immunity and Flowering Time in A. thaliana
Our analyses reveal that, in A. thaliana, individuals with a shorter vegetative lifespan tend to express immunity genes at a lower level. The bulk analysis of early- and late-flowering RILs shows that this pattern of covariation results from the combination of independent alleles controlling immunity gene expression and flowering time in natural populations, because these alleles could be separated in the segregating recombinant offspring of an early- and a late-flowering genotype. Because covariation is also 1) robust to the demographic history of the populations and 2) particularly pronounced for immunity-gene variants that associate with fitness, our analyses suggest that this allelic combination is assembled by natural selection. This pattern is confirmed by the examination of genes annotated with a function in immunity and genes observed to respond to elicitation by the common bacterial elicitor flagellin. Our data further suggest that much of the positive covariation between immunity gene expression and flowering depends on plant age. This factor is of recognized importance in plant immunity (Alcázar et al. 2011; Carella et al. 2015; Lozano-Durán and Zipfel 2015) and also very well documented in ecological studies (Barton and Boege 2017). Based on our findings, it is tempting to speculate that variation in age-dependent regulation of immunity may mediate the covariation we report.
Covariation between Immunity and Flowering Time Is Not Explained by Variation in Vernalization Requirements
Flowering time variation depends on seasonal fluctuations, on the timing of germination and on the genetics of its control (Lempe et al. 2005; Balasubramanian et al. 2006; Korves et al. 2007; Burghardt et al. 2015; Hu et al. 2017). Genotypes with a strong vernalization requirement, which are thought to have an obligate winter annual strategy, contribute strongly to the variation reported in the literature because they display much delayed flowering in the laboratory (Lempe et al. 2005; Li et al. 2014; Sasaki et al. 2015). The pattern we report, however, is not due to the assortment of immunity gene expression variants with alleles imposing a strong vernalization requirement. Indeed, since we observed that the pattern of covariation between immunity gene expression and flowering time is magnified in plants whose flowering is not accelerated by cold exposure (fig. 1A and B), this pattern is not driven by the genotypes requiring vernalization. In addition, the same pattern of covariation is observed in a global sample of ecotypes, whose flowering time was scored in an outdoor common garden experiment, where plants were naturally vernalized (fig. 1C). Therefore, we believe that the flowering time measures we used here do capture some of the natural lifespan variation. Future studies will have to confirm that flowering time variation scales with average differences in the lifespan expressed at the location of origin of each genotype.
Positive Covariation between Lifespan and Immunity Suggests Cascading Effect of Flowering Time Adaptation on Immunity Evolution
Two alternative scenarios may lead to concerted evolution of flowering time and immunity. First, in conditions where disease pressure is high, both shorter lifespan and stronger immunity can be expected to be advantageous, in order to simultaneously minimize the probability of attack, and maximize the probability of survival in case of attack. Under such scenario, negative covariation between immunity and lifespan is expected. Alternatively, if lifespan is evolving under evolutionary forces independent of disease pressure, a reduced probability to encounter pathogens will favor mutations transferring energy allocated to immunity into energy allocated to growth. Indeed, defensive functions are known to be costly for the organism (Lochmiller and Deerenberg 2000; Purrington 2000). As a consequence the allocation into immunity is predicted to decrease where shorter lifespan evolves. Under this second scenario, a pattern of positive covariation is expected between immunity and lifespan.
The pattern of covariation we report here for immunity versus flowering time is indeed positive and thus lends support to the second scenario. Local adaptation of flowering time is well documented in A. thaliana (Le Corre 2005; Toomajian et al. 2006; Méndez-Vigo et al. 2011; Brachi et al. 2013; Debieu et al. 2013; Li et al. 2014; Burghardt et al. 2015; Vidigal et al. 2016). In addition, several studies support the idea that increased basal level in immunity components improves immunity (Vetter et al. 2012; Boccara et al. 2014). At the same time, variants involved in the surveillance systems directed against pathogenic virulence factors were shown to incur substantial fitness costs (Tian et al. 2003; but see also MacQueen et al. 2016) and variation in basal immunity was negatively correlated with plant growth (Vetter et al. 2012). Our results are thus compatible with an evolutionary scenario in which local adaptation of flowering time has cascading effect on immunity, possibly because a reduction of the plant’s lifespan increases the cost/benefit ratio of immunity. This may also explain why genes involved in local adaptation in China are enriched among both flowering time and immunity genes (Zou et al. 2017).
However, a positive pattern of covariation could also arise even if the two traits evolve independently. Indeed, it is possible that populations where early flowering is advantageous coincide with populations where disease pressure is lower and vice versa. We cannot formally exclude that this scenario does not apply, because too little is known about variation in disease pressure in A. thaliana natural populations. Several elements, however, indicate it is unlikely. First, the rapid cycling genotypes are more frequent at intermediate latitudes, where summers are mild and wet (Lempe et al. 2005; Debieu et al. 2013). Since these conditions are also favorable to diseases, it is unlikely that higher disease pressure is found in areas where delayed flowering is more adaptive. Second, it is unlikely that this pattern may be due to herbivore enemies. Indeed, more severe herbivory damage has been observed on early flowering A. thaliana individuals grown in the field (Weinig et al. 2003). This seems to be common in plant species and should select for higher defense among early flowering genotypes (Carmona et al. 2011). Third, such scenario would assume that variation in disease pressure does not alter the trade-off between survival and reproductive output. This trade-off, however, is central in many models explaining the evolution of the timing of flowering in monocarpic plant species (Mitchell-Olds 1996; Metcalf and Mitchell-Olds 2009; Ashworth et al. 2016).
Our results are therefore compatible with a scenario, in which adaptation of life history traits has a cascading effect on the evolution of immunity in A. thaliana. These findings do not contradict evidence that a tug of war characterizes the evolution of pathogen-specific components of immunity (Tellier and Brown 2007; Roux and Bergelson 2016). Indeed, by examining the basal expression level of a large set of genes involved in the immune reaction, the impact of durable selective forces on general immunity levels can be detected. This approach circumvents the potentially confounding signature left by a recent epidemics on strain-specific R-genes. Indeed, testing phenotypic variation in disease resistance across genotypes with different life-history alleles would probably reveal variation in gene-for-gene resistance, but the pervasive impact of selection fine-tuning energetic costs associated with immunity strategies would remained masked.
Interspecific differences in the investment in defence against herbivory has been often associated with differences in lifespan and growth rate (Endara and Coley 2011; Kooyers et al. 2017). Future studies will also have to examine whether a similar evolutionary trend has emerged in species that have reshaped their life history to decrease overall vegetative lifespan. Early flowering is actually often favored when the favorable season is shortened (Franks et al. 2007; Kenney et al. 2014). Ongoing selection for early flowering is clearly widespread at temperate latitudes (Munguía-Rosas et al. 2011) and transitions from perenniality to annuality occur frequently within phylogenies (Kiefer et al. 2017). Testing whether life span reduction associates with an attenuation of immunity gene expression should therefore be possible in many taxa.
The Impact of Life History Evolution on Defense Systems Is Expected across All Kingdoms
In animals, the idea that the optimal investment in immunity depends on the life history of a species was also incorporated in evolutionary models (Jokela et al. 2000). For plants and animals alike, resources available to the organism are limited. Energetic demands on growth may compete with those required for mounting immunity or counteracting the negative effects of parasites and pathogens (van Boven and Weissing 2004; Dowling and Simmons 2009; Lazzaro and Little 2009; Seppälä 2015). Several evolutionary models show that a prolonged lifespan is predicted to favor resource investment into immunity (Jokela et al. 2000; Medzhitov and Janeway 2000; van Boven and Weissing 2004; Miller et al. 2007). As a consequence, changes in life history can mold the evolution of immune systems in animals as well (Van Valen 1973; Sheldon and Verhulst 1996; Schulenburg et al. 2009). This theoretical prediction is supported by analyses of sexual dimorphism in the duration of effective breeding: females with increased reproductive longevity show stronger immune-competence but also by a meta-analysis of selection experiments (Rolff 2007; Nunn et al. 2009; van der Most et al. 2011). In frogs, fast developing species were also shown to be more susceptible to infection by trematodes (Johnson et al. 2012). Yet, such studies cannot exclude that longevity and immunity are constrained in their evolution by common regulatory factors or causal interdependence. To the best of our knowledge, this study is the first to provide evidence that natural variation in the activity of genes that are important for defeating pathogens is assorted with alleles controlling variation in a life history trait of considerable importance for adaptation. Local adaptation for lifespan should therefore be considered as a potentially important contributor to the maintenance of genetic diversity in immune systems.
Materials and Methods
Flowering and Immunity Candidate Genes
Gene Ontology (GO) categories were used to identify functionally related genes whose annotation was inferred from experiments, direct assays, physical interaction, mutant phenotype, genetic interactions or from expression patterns. Based on the keyword “flowering” in the TAIR database, 659 flowering time genes were selected. For immunity genes, we united 17 GO categories yielding 731 genes (supplementary table 2, Supplementary Material online). For flagellin responsive (FlaRe) genes, we took the set of 245 genes that were activated in seedlings described in (Navarro et al. 2004) (supplementary table 2, Supplementary Material online). Subsets of flowering, immunity, and FlaRe genes containing fitness-associated single nucleotide polymorphisms (SNPs) were retrieved from Fournier-Level et al. (2011).
Correlation between Gene Expression and Flowering Time in a Natural Population
We analyzed two published sets of natural ecotypes for which both genome-wide expression profiles and flowering time estimates were available. The first data set comprised 138 lines from Sweden scored for both flowering time (for plants grown at 16-h light–8-h dark at constant 16°C) and gene expression in whole rosette collected at the 9-true-leaf stage (Dubin et al. 2015; Sasaki et al. 2015). For this first data set, gene expression and flowering were determined in the same experiment. The second data set combined data from two sources. RNA extracted from 7-day-old seedlings of 144 genotypes grown on agar plate in long days had been sequenced (Schmitz et al. 2013) and expression levels quantified as quantile normalized fragment numbers per kilobases and million reads (FPKM). For 52 of these genotypes, flowering time, measured in cumulative photothermal units, had been scored in the field (Brachi et al. 2010). Photo-thermal units sum up the combination of temperature and day length and thus provide an estimate of the duration of the favorable season.
Expression counts were loge +1-transformed to include null values of expression and a Spearman correlation coefficient between flowering time and expression level was computed for each gene. P values were adjusted for false discovery rate using the p.adjust function in R (Benjamini and Hochberg 1995; Yekutieli and Benjamini 1999). A Kolmogorov–Smirnov test was used to compare the distribution of Spearman correlation coefficients ρ of flowering time and immunity genes with the distribution of ρ for 22,686 genes for which gene expression was quantified. Gene enrichments were tested using hypergeometric tests in R. The GO enrichment analysis was performed with the Gene Set Enrichment Analysis (GSEA) test akin to nonparametric Kolmogorov–Smirnov tests, first described by Subramanian et al. (2005), and implemented in the “topGO” R package (Alexa and Rahnenfuhrer 2010). We further applied the elim procedure, available in this package, which calculates enrichment significance of parent nodes after eliminating genes of significant children nodes. This controls for the dependency among nested parent–child GO categories so that the significance of each enrichment can be interpreted without overconservative P value corrections for multiple-testing (Alexa et al. 2006). To test the impact of population structure on the correlation, we ran a mixed model with the help of the R package lmekin. For each gene, we used gene expression level as a dependent variable. Flowering time was used as independent variable and a kinship matrix, generated with a matrix of SNPs segregating among Swedish genotypes (Duin et al. 2015), was included as random effect. The estimate of the flowering time effect was extracted. This allowed compared the distribution of estimates observed for the whole genome, the subset of flowering time genes, or the subsets of defense genes.
Analysis of Gene Expression in Segregant Pools Bulked by Flowering Time
Seeds of Bur-0, Col-0, and 278 Bur-0xCol-0 Recombinant Inbred Lines (RIL) obtained after 8 generations of selfing were provided by the Arabidopsis Stock Center at INRA Versailles (France, Simon et al. 2008). Each line was grown individually in six replicates, each in 6 cm diameter pots randomly allocated to 24 trays, each containing 35 pots. Seeds were stratified at 5°C for 3 days and grown in growth chambers (Elbanton BV, Holland, equipped with Sylvania Gro-Lux F36W/Gro [T8] fluorescent tubes and Osram 25 W 220 Lumen light bulbs) under long-day conditions (21°C, 16 h light, 18°C, 8 h dark). Trays were rotated within the chamber every other day. Flowering time was scored as the day to the first open flower. Genotypes of individuals lines were retrieved from Simon et al. (2008) and mapping of flowering time recovered the same QTL (not shown).
We selected the 40 RIL in the 15% and 85% quantiles of flowering time for RNA sequencing. Each RIL and the two parental lines were planted in 20 replicates in the conditions described earlier. At days 14 and 28, the oldest true leaf was flash-frozen in liquid nitrogen. Three pools, each combining 13 RIL, were produced at each time point for early and late lines, for a total of 3 biological replicates, 2 pool types (early and late RIL) and 2 time points (14 and 28 days). For each of the two parental lines, leaves of 12 replicates were pooled for each time point.
RNA was isolated using the TRIzol extraction protocol (ThermoFisher Scientific). DNA traces were removed with the Ambion DNA-free kit (ThermoFisher Scientific) and purified RNA was stored in TE buffer at −80°C. RNA quality and integrity was confirmed with the 2100 Expert Software on a Bioanalyzer (Agilent Technologies, Inc. Waldbronn, Germany). All samples had RNA integrity index (RIN) >8. Single-read libraries were prepared with 1 μg of total RNA per sample using the Illumina TruSeq RNA Sample Preparation Kit v2 (Illumina Inc. San Diego) based on poly-A RNA purification. Sequencing of 75-bp single reads was performed on the Illumina HighScan SQ system of the Core Facility of the Department of Genetic Epidemiology, Institute of Human Genetics, University of Münster, Germany. Raw data have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al. 2002) and are accessible through GEO Series accession number GSE97664.
Data Analysis of RNA-Seq from Bulk Segregant Pools
In total, 24 RNA libraries were sequenced. Raw sequences were demultiplexed and read quality validated with FastQC. Bad quality base calls were trimmed using the fastx-toolkit (Version 0.013). Trimmed reads (FastQ, quality score 33, quality threshold 20 and minimum length 30 base pair) were mapped to the A. thaliana TAIR10 annotated transcriptome using Bowtie 2 (version 2-2.0.0-beta6; Langmead and Salzberg 2012). Tophat (version-2.0.5.Linux_x86_64) was used to discover splice sites and Cufflinks for assembling the transcriptome (Trapnell et al. 2010). In total 411, 5-M sequence reads were obtained, with a mean read count per sample of 17, 1-M reads. After trimming, 96.5% of the reads were mapped uniquely with a final average coverage of 66 reads per base pair.
We used a custom R script to verify that coverage was uniform across transcripts and confirmed that the RNA sequenced was not degraded. Read counts were calculated by counting the number of reads that mapped uniquely to the corresponding gene (isoforms were not considered). Lowly expressed genes with <20 reads over all samples were excluded from the analysis. The samples clustered by time point of sampling (fig. 1), with the exception of RNA samples from the Col-0 at 28 days, which resembled more expression levels measured at 14 days, probably because of its early shift to flowering. Differentially expressed (DE) genes were identified by running a nested analysis of sampling time effects within parental genotype (and/or early- and late-flowering leaf pools) with DESeq2 version 1.2.5 (Anders et al. 2013; Love et al. 2014). P values were corrected for false discovery rate (Benjamini–Hochberg correction; Benjamini and Hochberg 1995). DE genes were defined as having an adjusted P value < 0.05. This analysis allowed the identification of genes showing differential expression between the parents (supplementary table 3, Supplementary Material online) and genes showing flowering time dependent expression (differential expression between early and late flowering RIL pools, i.e., FT-regulated genes supplementary table 4, Supplementary Material online) both at day 14 and at day 28. We performed further analyses to disentangle significant sources of gene expression variation. To test whether gene expression was significantly modified at each time point, separate tests were performed for each parental genotype and RIL pool type. Genes differentially regulated at 14 and 28 days in Bur-0 (adjusted P value < 0.05) were defined as age-regulated genes (supplementary table 5, Supplementary Material online). To determine whether one or both sampling time points drove significant differential expression, separate tests were performed for each time point (not shown).
Confirmation with qRT-PCR
We confirmed gene expression levels for 11 selected immunity genes with differential expression between Bur-0 and Col-0 or early versus late flowering pools (log2-fold change > 1.5) using RT-PCR. We followed standard protocols and used RNA Helicase (AT1G58060), Protein Phosphatase 2A Subunit A3 (PP2AA3) and transcript AT5G12240 as control genes. Gene expression based on RNA sequencing and RT-PCR were strongly correlated (Pearson correlation, 0.58<R < 0.96, max P < 0.01).
Supplementary Material
Acknowledgments
This research was supported by the Deutsche Forschung Gesellschaft (DFG) in the realm of SPP1530 grant ME 2742/2-1, and by the European Research Council with grant 648617 “AdaptoSCOPE.”
Literature Cited
- Alcázar R, Reymond M, Schmitz G, de Meaux J.. 2011. Genetic and evolutionary perspectives on the interplay between plant immunity and development. Curr Opin Plant Biol. 14(4):378–384. [DOI] [PubMed] [Google Scholar]
- Alexa A, Rahnenfuhrer J. 2010. topGO: enrichment analysis for Gene Ontology. R package version 2.20.0
- Alexa A, Rahnenfuhrer J, Lengauer T. 2006. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22:1600–1607. [DOI] [PubMed] [Google Scholar]
- Anders S, et al. 2013. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc. 8(9):1765–1786. [DOI] [PubMed] [Google Scholar]
- Andrés F, Coupland G.. 2012. The genetic basis of flowering responses to seasonal cues. Nat Rev Genet. 13(9):627–639. [DOI] [PubMed] [Google Scholar]
- Ashworth MB, Walsh MJ, Flower KC, Vila Aiub MM, Powles SB.. 2016. Directional selection for flowering time leads to adaptive evolution in Raphanus raphanistrum (wild radish). Evol Appl. 9(4):619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, et al. 2006. The PHYTOCHROME C photoreceptor gene mediates natural variation in flowering and growth responses of Arabidopsis thaliana. Nat Genet. 38(6):711–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton KE, Boege K.. 2017. Future directions in the ontogeny of plant defence: understanding the evolutionary causes and consequences. Ecol Lett. 20(4):403–411. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y.. 1995. Controlling the false discovery rate – a practical and powerful approach to multiple testing. J R Stat Soc B 57:289–300. [Google Scholar]
- Bergelson J, Dwyer G, Emerson JJ.. 2001. Models and data on plant-enemy coevolution. Annu Rev Genet. 35:469–499. [DOI] [PubMed] [Google Scholar]
- Bergelson J, Roux F.. 2010. Towards identifying genes underlying ecologically relevant traits in Arabidopsis thaliana. Nat Rev Genet. 11(12):867–879. [DOI] [PubMed] [Google Scholar]
- Boccara M, et al. 2014. The Arabidopsis miR472-RDR6 silencing pathway modulates PAMP- and effector-triggered immunity through the post-transcriptional control of disease resistance genes. PLoS Pathog. 10(1):e1003883–e1003816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachi B, et al. 2010. Linkage and association mapping of Arabidopsis thaliana flowering time in nature. PLoS Genet. 6(5):e1000940.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachi B, et al. 2013. Investigation of the geographical scale of adaptive phenological variation and its underlying genetics in Arabidopsis thaliana. Mol Ecol. 22(16):4222–4240. [DOI] [PubMed] [Google Scholar]
- Burghardt LT, Metcalf CJE, Wilczek AM, Schmitt J, Donohue K.. 2015. Modeling the influence of genetic and environmental variation on the expression of plant life cycles across landscapes. Am Nat. 185(2):212–227. [DOI] [PubMed] [Google Scholar]
- Carella P, Wilson DC, Cameron RK.. 2015. Some things get better with age: differences in salicylic acid accumulation and immunity signaling in young and mature Arabidopsis. Front Plant Sci. 5:1001.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona D, Lajeunesse MJ, Johnson MTJ.. 2011. Plant traits that predict resistance to herbivores. Funct Ecol. 25(2):358–367. [Google Scholar]
- Chiang GCK, et al. 2013. Pleiotropy in the wild: the dormancy gene DOG1 exerts cascading control on life cycles. Evolution 67(3):883–893. [DOI] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ.. 2006. Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124(4):803–814. [DOI] [PubMed] [Google Scholar]
- Davila Olivas NH, et al. 2017. Natural variation in life history strategy of Arabidopsis thaliana determines stress responses to drought and insects of different feeding guilds. Mol Ecol. 26(11):2959–2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Meaux J, Mitchell-Olds T.. 2003. Evolution of plant resistance at the molecular level: ecological context of species interactions. Heredity 91(4):345–352. [DOI] [PubMed] [Google Scholar]
- Debieu M, et al. 2013. Co-variation between seed dormancy, growth rate and flowering time changes with latitude in Arabidopsis thaliana. PLoS One 8(5):e61075.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Develey-Riviere MP, Galiana E.. 2007. Resistance to pathogens and host developmental stage: a multifaceted relationship within the plant kingdom. New Phytol. 175(3):405–416. [DOI] [PubMed] [Google Scholar]
- Dowling DK, Simmons LW.. 2009. Reactive oxygen species as universal constraints in life-history evolution. Proc Biol Sci. 276(1663):1737–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin MJ, et al. 2015. DNA methylation in Arabidopsis has a genetic basis and shows evidence of local adaptation. eLife 4:e05255.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dybdahl MF, Jenkins CE, Nuismer SL.. 2014. Identifying the molecular basis of host-parasite coevolution: merging models and mechanisms. Am Nat. 184(1):1–13. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE.. 2002. Gene Expression Omnibus: nCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30(1):207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endara M-J, Coley PD.. 2011. The resource availability hypothesis revisited: a meta-analysis. Funct Ecol. 25(2):389–398. [Google Scholar]
- Eulgem T. 2005. Regulation of the Arabidopsis immunity transcriptome. Trends Plant Sci. 10(2):71–78. [DOI] [PubMed] [Google Scholar]
- Fan M, et al. 2014. The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern-triggered immunity in Arabidopsis. Plant Cell 26(2):828–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier-Level A, et al. 2011. A map of local adaptation in Arabidopsis thaliana. Science 334(6052):86–89. [DOI] [PubMed] [Google Scholar]
- Fournier-Level A, et al. 2013. Paths to selection on life history loci in different natural environments across the native range of Arabidopsis thaliana. Mol Ecol. 22(13):3552–3566. [DOI] [PubMed] [Google Scholar]
- Fournier-Level A, et al. 2016. Predicting the evolutionary dynamics of seasonal adaptation to novel climates in Arabidopsis thaliana. Proc Natl Acad Sci USA. 113(20):E2812–E2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks SJ, Sim S, Weis AE.. 2007. Rapid evolution of flowering time by an annual plant in response to a climate fluctuation. Proc Natl Acad Sci USA. 104(4):1278–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier E. 1992. Growth analysis of congeneric annual and perennial grass species. J Ecol. 80(4):665. [Google Scholar]
- Hancock AM, et al. 2011. Adaptation to climate across the Arabidopsis thaliana genome. Science 334(6052):83–86. [DOI] [PubMed] [Google Scholar]
- Herms DA, Mattson WJ.. 1992. The dilemma of plants – to grow or defend. Q Rev Biol. 67(3):283–335. [Google Scholar]
- Hu J, Lei L, de Meaux J.. 2017. Temporal fitness fluctuations in experimental Arabidopsis thaliana populations. PLoS One 12(6):e0178990.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Góngora T, Kim S-K, Lozano-Durán R, Zipfel C.. 2015. Flg22-triggered immunity negatively regulates key BR biosynthetic genes. Front Plant Sci. 6:303.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PTJ, et al. 2012. Living fast and dying of infection: host life history drives interspecific variation in infection and disease risk. Ecol Lett. 15(3):235–242. [DOI] [PubMed] [Google Scholar]
- Jokela J, Schmid-Hempel P, Rigby MC.. 2000. Dr. Pangloss restrained by the Red Queen – steps towards a unified defence theory. Oikos 89(2):267–274. [Google Scholar]
- Jones JDG, Dangl JL.. 2006. The plant immune system. Nature 444(7117):323–329. [DOI] [PubMed] [Google Scholar]
- Karasov TL, et al. 2014. The long-term maintenance of a resistance polymorphism through diffuse interactions. Nature 512(7515):436–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney AM, Mckay JK, Richards JH, JUENGER TE.. 2014. Direct and indirect selection on flowering time, water‐use efficiency (WUE, δ13C), and WUE plasticity to drought in Arabidopsis thaliana. Ecol Evol. 4(23):4505–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerwin R, et al. 2015. Natural genetic variation in Arabidopsis thaliana immunity metabolism genes modulates field fitness. eLife 4:e05604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer C, et al. 2017. Divergence of annual and perennial species in the Brassicaceae and the contribution of cis-acting variation at FLC orthologues. Mol Ecol. 26(13):3437–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooyers NJ, Blackman BK, Holeski LM.. 2017. Optimal immunity theory explains deviations from latitudinal herbivory immunity hypothesis. Ecology 98(4):1036–1048. [DOI] [PubMed] [Google Scholar]
- Korves TM, Bergelson J.. 2003. A developmental response to pathogen infection in Arabidopsis. Plant Physiol. 133(1):339–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korves TM, et al. 2007. Fitness effects associated with the major flowering time gene FRIGIDA in Arabidopsis thaliana in the field. Am Nat. 169(5):E141–E157. [DOI] [PubMed] [Google Scholar]
- Laine A-L, Burdon JJ, Dodds PN, Thrall PH.. 2011. Spatial variation in disease resistance: from molecules to metapopulations. J Ecol. 99(1):96–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasky JR, et al. 2012. Characterizing genomic variation of Arabidopsis thaliana: the roles of geography and climate. Mol Ecol. 21(22):5512–5529. [DOI] [PubMed] [Google Scholar]
- Lazzaro BP, Little TJ.. 2009. Immunity in a variable world. Philos Trans R Soc B 364(1513):15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Corre V. 2005. Variation at two flowering time genes within and among populations of Arabidopsis thaliana: comparison with markers and traits. Mol Ecol. 14(13):4181–4192. [DOI] [PubMed] [Google Scholar]
- Lee YK, Mazmanian SK.. 2010. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science 330(6012):1768–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempe J, et al. 2005. Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genet. 1(1):e6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, et al. 2014. Multiple FLC haplotypes defined by independent cis-regulatory variation underpin life history diversity in Arabidopsis thaliana. Genes Dev. 28(15):1635–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liow LH, Van Valen L, Stenseth NC.. 2011. Red Queen: from populations to taxa and communities. Trends Ecol Evol. 26(7):349–358. [DOI] [PubMed] [Google Scholar]
- Lochmiller RL, Deerenberg C.. 2000. Trade‐offs in evolutionary immunology: just what is the cost of immunity? Oikos 88(1):87–98. [Google Scholar]
- Love MI, Huber W, Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15(12):550.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Durán R, Zipfel C.. 2015. Trade-off between growth and immunity: role of brassinosteroids. Trends Plant Sci. 20(1):12–19. [DOI] [PubMed] [Google Scholar]
- Lyons R, et al. 2015. Investigating the association between flowering time and defense in the Arabidopsis thaliana-Fusarium oxysporum interaction. PLoS One 10(6):e0127699.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A, Sun X, Bergelson J.. 2016. Genetic architecture and pleiotropy shape costs of Rps2-mediated resistance in Arabidopsis thaliana. Nat Plants 2:16110–16118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T, Kufer TA, Schulze-Lefert P.. 2011. NLR functions in plant and animal immune systems: so far and yet so close. Nat Immunol. 12(9):817–826. [DOI] [PubMed] [Google Scholar]
- Martinez C, Pons E, Prats G, Leon J.. 2004. Salicylic acid regulates flowering time and links defence responses and reproductive development. Plant J. 37(2):209–217. [DOI] [PubMed] [Google Scholar]
- Matthew Ogburn R, Edwards EJ.. 2015. Life history lability underlies rapid climate niche evolution in the angiosperm clade Montiaceae. Mol Phylogenet Evol. 92:181–192. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Janeway C.. 2000. The Toll receptor family and microbial recognition. Trends Microbiol. 8(10):452–456. [DOI] [PubMed] [Google Scholar]
- Metcalf CJE, Mitchell-Olds T.. 2009. Life history in a model system: opening the black box with Arabidopsis thaliana. Ecol Lett. 12(7):593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez-Vigo B, Picó FX, Ramiro M, Martínez-Zapater JM, Alonso-Blanco C.. 2011. Altitudinal and climatic adaptation is mediated by flowering traits and FRI, FLC, and PHYC genes in Arabidopsis. Plant Physiol. 157(4):1942–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger CMJA, Luijckx P, Bento G, Mariadassou M, Ebert D.. 2016. The Red Queen lives: epistasis between linked resistance loci. Evolution 70(2):480–487. [DOI] [PubMed] [Google Scholar]
- Michaels SD, He Y, Scortecci KC, Amasino RM.. 2003. Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc Natl Acad Sci USA. 100(17):10102–10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MR, White A, Boots M.. 2007. Host life span and the evolution of resistance characteristics. Evolution 61(1):2–14. [DOI] [PubMed] [Google Scholar]
- Mitchell CE, Blumenthal D, Jarošík V, Puckett EE, Pyšek P.. 2010. Controls on pathogen species richness in plants’ introduced and native ranges: roles of residence time, range size and host traits. Ecol Lett. 13(12):1525–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell CE, Power AG.. 2003. Release of invasive plants from fungal and viral pathogens. Nature 421(6923):625–627. [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds T. 1996. Pleiotropy causes long-term genetic constraints on life-history evolution in Brassica rapa. Evolution 50(5):1849–1858. [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds T, Schmitt J.. 2006. Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature 441(7096):947–952. [DOI] [PubMed] [Google Scholar]
- Moeller DA, Tiffin P.. 2005. Genetic diversity and the evolutionary history of plant immunity genes in two species of Zea. Mol Biol Evol. 22(12):2480–2490. [DOI] [PubMed] [Google Scholar]
- Montesinos-Navarro A, Wig J, Picó FX, Tonsor SJ.. 2011. Arabidopsis thaliana populations show clinal variation in a climatic gradient associated with altitude. New Phytol. 189(1):282–294. [DOI] [PubMed] [Google Scholar]
- Munguía-Rosas MA, Ollerton J, Parra-Tabla V, De-Nova JA.. 2011. Meta-analysis of phenotypic selection on flowering phenology suggests that early flowering plants are favoured. Ecol Lett. 14(5):511–521. [DOI] [PubMed] [Google Scholar]
- Navarro L, et al. 2004. The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent immunity responses and bacterial pathogenesis. Plant Physiol. 135(2):1113–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn CL, Lindenfors P, Pursall ER, Rolff J.. 2009. On sexual dimorphism in immune function. Philos Trans R Soc B Biol Sci. 364(1513):61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagan I, Alonso-Blanco C, Garcia-Arenal F.. 2008. Host responses in life-history traits and tolerance to virus infection in Arabidopsis thaliana. PLoS Pathog. 4(8):e1000124.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajerowska-Mukhtar K, et al. 2009. Single nucleotide polymorphisms in the allene oxide synthase 2 gene are associated with field resistance to late blight in populations of tetraploid potato cultivars. Genetics 181(3):1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker IM, et al. 2015. Phylogenetic structure and host abundance drive disease pressure in communities. Nature 520(7548):542–544. [DOI] [PubMed] [Google Scholar]
- Parker IM, Gilbert GS.. 2004. The evolutionary ecology of novel plant-pathogen interactions. Annu Rev Ecol Evol Syst. 35(1):675–700. [Google Scholar]
- Purrington CB. 2000. Costs of resistance. Curr Opin Plant Biol. 3(4):305–308. [DOI] [PubMed] [Google Scholar]
- Ravensdale M, Nemri A, Thrall PH, Ellis JG, Dodds PN.. 2011. Co-evolutionary interactions between host resistance and pathogen effector genes in flax rust disease. Mol Plant Pathol. 12(1):93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolff J. 2007. Why did the acquired immune system of vertebrates evolve? Dev Comp Immunol. 31(5):476–482. [DOI] [PubMed] [Google Scholar]
- Roux F, Bergelson J.. 2016. The genetics underlying natural variation in the biotic interactions of Arabidopsis thaliana: the challenges of linking evolutionary genetics and community ecology. Curr Top Dev Biol. 119:111–156. [DOI] [PubMed] [Google Scholar]
- Rusterucci C, et al. 2005. Age-related resistance to Pseudomonas syringae pv. tomato is associated with the transition to flowering in Arabidopsis and is effective against Peronospora parasitica. Physiol Mol Plant Pathol. 66(6):222–231. [Google Scholar]
- Sasaki E, Zhang P, Atwell S, Meng D, Nordborg M.. 2015. G x E variation controls flowering time in Arabidopsis thaliana. PLoS Genet. 11(10):e1005597. ‘Missing’ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, et al. 2013. Patterns of population epigenomic diversity. Nature 495(7440):193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulenburg H, Kurtz J, Moret Y, Siva-Jothy MT.. 2009. Introduction. Ecological immunology. Philos Trans R Soc B Biol Sci. 364(1513):3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppälä O. 2015. Natural selection on quantitative immune defence traits: a comparison between theory and data. J Evol Biol. 28(1):1–9. [DOI] [PubMed] [Google Scholar]
- Sheldon BC, Verhulst S.. 1996. Ecological immunology: costly parasite defences and trade-offs in evolutionary ecology. Trends Ecol Evol. 11(8):317–321. [DOI] [PubMed] [Google Scholar]
- Siddle KJ, Quintana-Murci L.. 2014. The Red Queen’s long race: human adaptation to pathogen pressure. Curr Opin Genet Dev. 29:31–38. [DOI] [PubMed] [Google Scholar]
- Simon M, et al. 2008. Quantitative trait loci mapping in five new large recombinant inbred line populations of Arabidopsis thaliana genotyped with consensus single-nucleotide polymorphism markers. Genetics 178(4):2253–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stich B, et al. 2008. Comparison of mixed-model approaches for association mapping. Genetics 178(3):1745–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tank DC, Olmstead RG.. 2008. From annuals to perennials: phylogeny of subtribe Castillejinae (Orobanchaceae). Am J Bot. 95(5):608–625. [DOI] [PubMed] [Google Scholar]
- Tellier A, Brown JKM.. 2007. Polymorphism in multilocus host parasite coevolutionary interactions. Genetics 177(3):1777–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines BC, Youn Y, Duarte MI, Harmon FG.. 2014. The time of day effects of warm temperature on flowering time involve PIF4 and PIF5. J Exp Bot. 65(4):1141–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Traw MB, Chen JQ, Kreitman M, Bergelson J.. 2003. Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423(6935):74–77. [DOI] [PubMed] [Google Scholar]
- Toomajian C, et al. 2006. A nonparametric test reveals selection for rapid flowering in the Arabidopsis genome. PLoS Biol. 4(5):e137.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, et al. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 28(5):511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Boven M, Weissing FJ.. 2004. The evolutionary economics of immunity. Am Nat. 163(2):277–294. [DOI] [PubMed] [Google Scholar]
- van der Most PJ, de Jong B, Parmentier HK, Verhulst S.. 2011. Trade-off between growth and immune function: a meta-analysis of selection experiments. Funct Ecol. 25(1):74–80. [Google Scholar]
- Van Valen L. 1973. A new evolutionary law. Evol Theory 1:1–30. [Google Scholar]
- Vetter MM, et al. 2012. Flagellin perception varies quantitatively in Arabidopsis thaliana and its relatives. Mol Biol Evol. 29(6):1655–1667. [DOI] [PubMed] [Google Scholar]
- Vidigal DS, et al. 2016. Altitudinal and climatic associations of seed dormancy and flowering traits evidence adaptation of annual life cycle timing in Arabidopsis thaliana. Plant Cell Environ. 39(8):1737–1748. [DOI] [PubMed] [Google Scholar]
- Weinig C, Stinchcombe JR, Schmitt J.. 2003. Evolutionary genetics of resistance and tolerance to natural herbivory in Arabidopsis thaliana. Evolution 57(6):1270–1280. [DOI] [PubMed] [Google Scholar]
- Whalen MC. 2005. Host defence in a developmental context. Mol Plant Pathol. 6(3):347–360. [DOI] [PubMed] [Google Scholar]
- Wilczek AM, et al. 2009. Effects of genetic perturbation on seasonal life history plasticity. Science 323(5916):930–934. [DOI] [PubMed] [Google Scholar]
- Wilson DC, Carella P, Isaacs M, Cameron RK.. 2013. The floral transition is not the developmental switch that confers competence for the Arabidopsis age-related resistance response to Pseudomonas syringae pv. tomato. Plant Mol Biol. 83(3):235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekutieli D, Benjamini Y.. 1999. Resampling-based false discovery rate controlling multiple test procedures for correlated test statistics. J Stat Plan Inference 82(1–2):171–196. [Google Scholar]
- Yu J, et al. 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet. 38(2):203–208. [DOI] [PubMed] [Google Scholar]
- Zou YP, et al. 2017. Adaptation of Arabidopsis thaliana to the Yangtze River basin. Genome Biol. 18:239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.