ABSTRACT
The importance of circular RNAs (circRNAs) as regulators of muscle development and muscle-associated disorders is becoming increasingly apparent. To explore potential regulators of muscle differentiation, we determined the expression profiles of circRNAs of skeletal muscle C2C12 myoblasts and myotubes using microarray analysis. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed to explore circRNA functions. We also established competing endogenous RNA (ceRNA) networks using bioinformatics methods and predicted the coding potential of differentially expressed circRNAs. We found that 581 circRNAs were differentially regulated between C2C12 myoblasts and myotubes. Bioinformatics analysis suggested that the primary functions of the linear transcripts of the circRNAs were linked with organization of the cytoskeleton, calcium signaling, cell cycle, and metabolic pathways. ceRNA networks showed that the myogenic-specific genes myogenin, myocyte enhancer factor 2a, myosin heavy chain (Myh)-1, Myh7, and Myh7b could combine with 91 miRNAs and the top 30 upregulated circRNAs, forming 239 edges. According to the number of open reading frames and N6-methyladenosine motifs, we identified 224 circRNAs with coding potential, and performed GO and KEGG analyses based on the linear counterparts of 75 circRNAs. We determined that the 75 circRNAs were related to regulation of the actin cytoskeleton and metabolic pathways. We established expression profiles of circRNAs during C2C12 myoblast differentiation and predicted the function of differentially expressed circRNAs, which might be involved in skeletal muscle development. Our study offers new insight into the functions of circRNAs in skeletal muscle growth and development.
Keywords: Circular RNAs (circRNAs), muscle differentiation, microarray analysis, coding potential
1. Introduction
Circular RNAs (circRNAs) are stable, highly conserved covalently closed RNA circles [1–3], and they seem to accomplish noncoding roles [4–6]. Although circRNAs were first discovered in the 1970s [5,7,8], they were initially considered to represent errors of the normal splicing process and were generally considered peculiarities of uncertain biological importance [9]. However, recent studies of circRNAs have resulted in the new understanding that circRNAs play an important role in cellular differentiation, development, and the onset and development of disease. In some cancers [10–12], circRNA expression differed compared with peritumoral tissue, suggesting circRNAs might be involved in cell proliferation, differentiation, and apoptosis. Recent reports have shown that circRNAs play an important role in the differentiation and development of skeletal muscle [13–16], but the mechanism is not well understood [14,17].
It’s reported that circRNAs play their role by acting as competing endogenous RNA (ceRNA) [13,15,16,18]. Coding and noncoding RNAs can regulate one another through their ability to compete for miRNA binding; these molecules have been termed ceRNA [18]. ceRNAs can sequester miRNAs, thereby protecting their target RNAs from repression. Circular RNA sponge for miR-7 (ciRS-7) acting as a miR-7 sponge probably engaged in the functioning of neurons and brain tumour development [6], and circular RNA MTO1 acting as the sponge of miR-9 could suppress hepatocellular carcinoma progression [19]. Recently, there is increasing evidence that circRNAs are important in the muscle regulatory network by ceRNA. circLMO7 was shown to promote primary bovine myoblasts proliferation and protect myoblasts from apoptosis by ceRNA [13]. Both circFUT10 sponging miR-133a [15] and circFGFR4 binding miR-107 [16] were shown to facilitate cell differentiation in bovine primary myoblasts.
While circRNAs were originally considered non-coding RNAs, recent studies have shown that circRNAs can be translated and have an important effect on cellular and muscle differentiation and development [20–22]. circFBXW7 could be translated into the protein FBXW7-185aa, which could induce cell cycle arrest and reduce proliferation in glioma cell [22]. Legnini et al. [14] firstly showed that circZNF609 regulated myoblast proliferation and could be translated, and the flagged protein derived from circZNF609 exhibited mainly nuclear localization, but its function and translation mechanism was unknown. Recently, it was reported that m6A motif was sufficient to drive translation initiation in circRNAs; and m6A motif-driven translation required eukaryotic translation initiation factor 4 gamma 2 (eIF4G2) and m6A reader YTH domain family protein 3 (YTHDF3) in human cells [21]. This was the first report of a circRNA with chemical m6A modification, which undoubtedly represented an important advance in circRNA research. Coding potential of the skeletal muscle circRNAs, and corresponding functions and mechanisms are currently in the development stage; it’s necessary to make further study in the future.
Skeletal muscle is an important part of the motor system that maintains movement and support, and makes up approximately 40% of total body weight. The formation of skeletal muscle fibers is a multistep process that requires the fusion of myoblasts to produce multinucleated muscle fibers with contractile properties [23]. The muscle-specific proteins myogenic differentiation antigen (MyoD), myogenic factor (Myf)-5, myogenin (Myog), myogenic regulatory factor (MRF)-4, and the myocyte enhancer factor 2a-d (Mef2a-d) family, act at numerous points in the muscle lineage to collaboratively establish the skeletal muscle phenotype by regulating proliferation, cell cycle, sarcomeric activation, and muscle-specific genes to promote differentiation and sarcomere assembly [23–25]. Myosin heavy chain (Myh) is a vital component of myosin, which is involved in the formation of the cytoskeleton, providing force for muscular contraction. Myh is divided into different subtypes, and Myh1 is the signature gene of fast-twitch muscle fibers [26], while Myh7 is important for slow-twitch muscle fibers [27]. Myh subtypes can change with external stimuli, and the contraction characteristics of muscle fibers can also change significantly when there is change in Myh subtypes.
While knowledge regarding the importance of circRNAs in the regulation of muscle development and muscle-associated disorders is increasing, little is known about the regulation of circRNAs during C2C12 myoblast differentiation. To explore potential regulators of muscle differentiation, we ascertained the expression profiles of C2C12 myoblast and myotube circRNAs using microarray analysis and predicted the function of differently expressed circRNAs. Our findings provide novel insight into the differentiation and development of skeletal muscle C2C12 myoblasts.
2. Results
2.1. Analysis of differentially expressed circRNAs between myoblasts and myotubes in mouse C2C12 cells
In total, 11,128 expressed circRNAs were detected using an Arraystar mouse circRNA microarray. Based on the circRNA expression profiles, differentially expressed circRNAs were distinguished between mouse C2C12 myoblasts and myotubes. Hierarchical clustering was performed to sort circRNAs according to their expression levels (Figure 1a). In the volcano plots (Figure 1b), differentially expressed circRNAs were grouped by fold change (FC) ≥ 2.0, P < 0.05, and false discovery rate < 0.05, which identified 581 markedly differentially expressed circRNAs in myotubes compared with myoblasts, with 346 upregulated and 235 downregulated circRNAs. The results suggest that the expression of circRNAs in myotubes differs from that in myoblasts.
Figure 1.
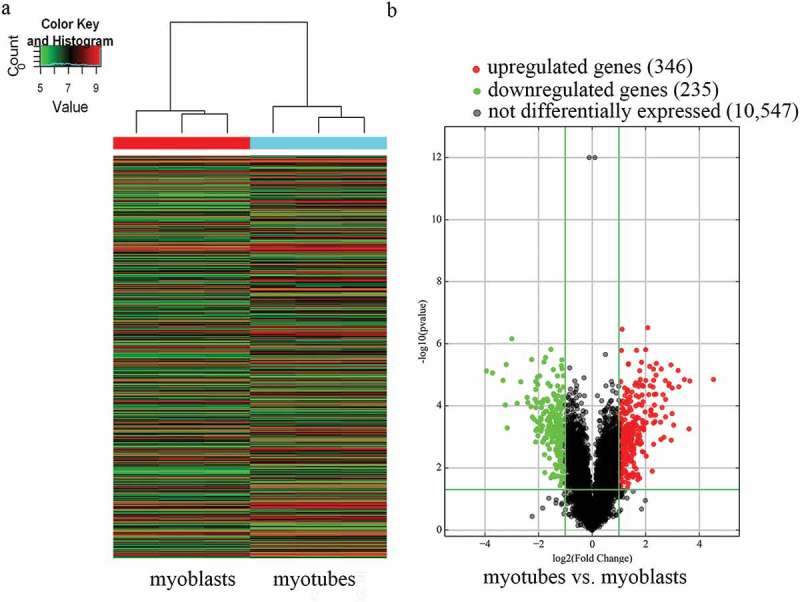
Characterization of circular RNA (circRNA) expression profiles of mouse C2C12 myoblasts and myotubes. a) Hierarchical clustering of the expression profiles of circRNAs. Red and green indicate high and low relative expression, respectively. b) Volcano plots were used to evaluate differences in circRNA expression between myotubes and myoblasts. The horizontal line corresponds to 2.0-fold (log2 scaled) up and down, respectively, and the vertical lines represent a P-value of 0.05 (–log10 scaled). The red and green points in the plot represent the differentially expressed circRNAs with statistical significance.
2.2. Quantitative real-time polymerase chain reaction (qRT-PCR) validation of differentially expressed circrnas between myoblasts and myotubes in mouse C2C12 cells
We selected nine differentially expressed circRNAs, including five upregulated and four downregulated circRNAs, to validate the microarray analysis results in mouse C2C12 cells. The qRT-PCR results were consistent with the microarray assay except for circRNA_007438 (Figures 2a and b), verifying the predictive accuracy of the microarray analysis. These findings provided evidence that these circRNAs might be involved in the differentiation and proliferation of mouse C2C12 myoblasts.
Figure 2.
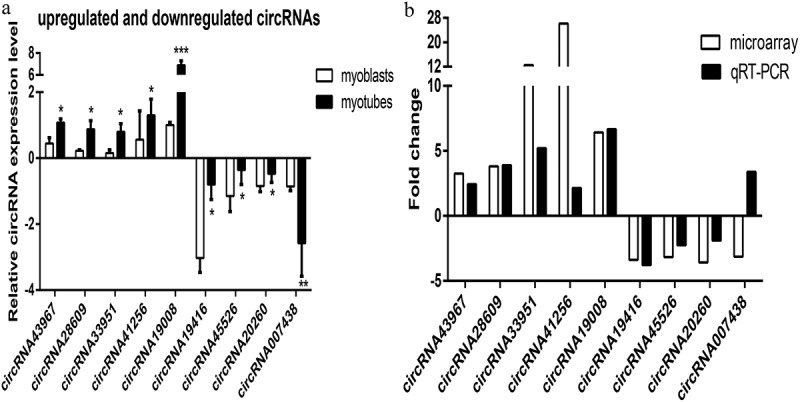
Quantitative real-time polymerase chain reaction (qRT-PCR) validation of nine differentially expressed circRNAs. a) Validation results of five upregulated and four downregulated circRNAs are shown. b) Comparison of qRT-PCR and microarray analysis. The heights of the columns represent fold changes (log2 transformed). Values are the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 myotubes vs. myoblasts.
2.3. Chromosomal distribution and types of differentially expressed circRNAs in mouse C2C12 cells
Based on our statistics, we found the differentially expressed circRNAs were widely distributed on all chromosomes of mouse C2C12 cells (Figure 3a), although they were commonly found on chromosome 1 (chr1), chr2, chr4, chr5, chr7, and chr17. It suggests that each chromosome is related to varying frequency of differentially expressed circRNAs in C2C12 myoblast differentiation.
Figure 3.
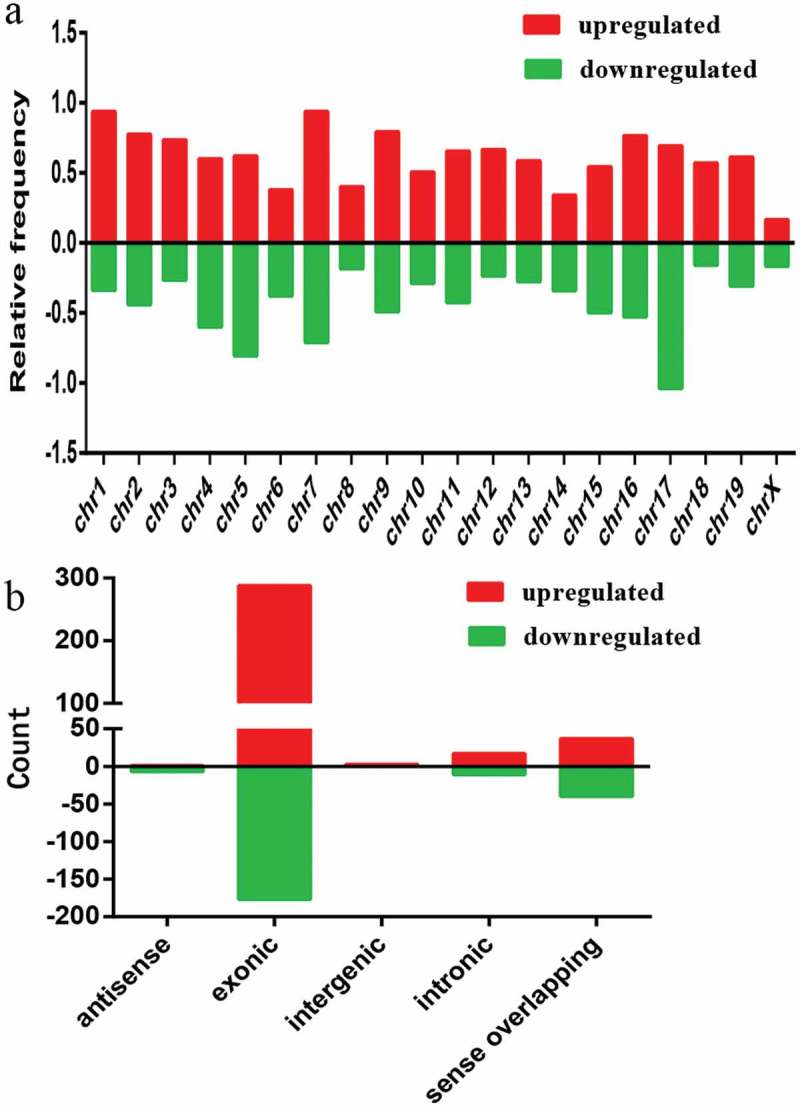
Chromosomal distribution and types of differentially expressed circRNAs. a) Relative frequency of differentially expressed circRNAs on mouse chromosomes. b) Types and counts of differentially expressed circRNAs detected by microarray (fold change ≥ 2.0, P < 0.05). The circRNAs were classified into five or four types according to the relationship and genomic loci with their associated coding genes. Red and green indicate upregulated and downregulated circRNAs, myotubes vs. myoblasts, respectively.
We summarized the types of circRNAs with different expression characteristics (Figure 3b) according to the relationship and genomic loci with their associated coding genes. The upregulated circRNAs included five distinct types: antisense (1), exonic (288), intergenic (3), intronic (17), and sense overlapping (37) (Figure 3b), while the downregulated circRNAs included four distinct types: antisense (7), exonic (177), intronic (11), and sense overlapping (40) (Figure 3b). Based on our analysis, most of the differentially expressed circRNAs originated from exons.
2.4. Gene ontology (GO) and kyoto encyclopedia of genes and genomes (KEGG) pathway analyses
To determine the involvement of circRNAs in gene regulation, we performed GO analysis of the linear counterparts that resulted in the differentially expressed circRNAs. The results of the GO analysis were classified into the GO terms cellular component (CC), molecular function (MF), and biological process (BP). Our results suggested that the linear counterparts of the differentially expressed circRNAs in myotubes favored the endoplasmic reticulum, nucleic acid binding transcription factor activity, transmembrane transport activity, signal transduction, transcription factor activity, protein binding, and embryonic development compared with myoblasts. Whereas the GO terms of downregulated circRNAs were more commonly linked to chromosomes, DNA/RNA binding, ATPase activity, cell cycle, chromosome organization, cell division, organization of the cytoskeleton, cell proliferation, and translation (Figure 4a). These analyses suggest that the proposed pathways and molecular functions of these circRNAs may promote differentiation and proliferation of mouse myoblasts.
Figure 4.
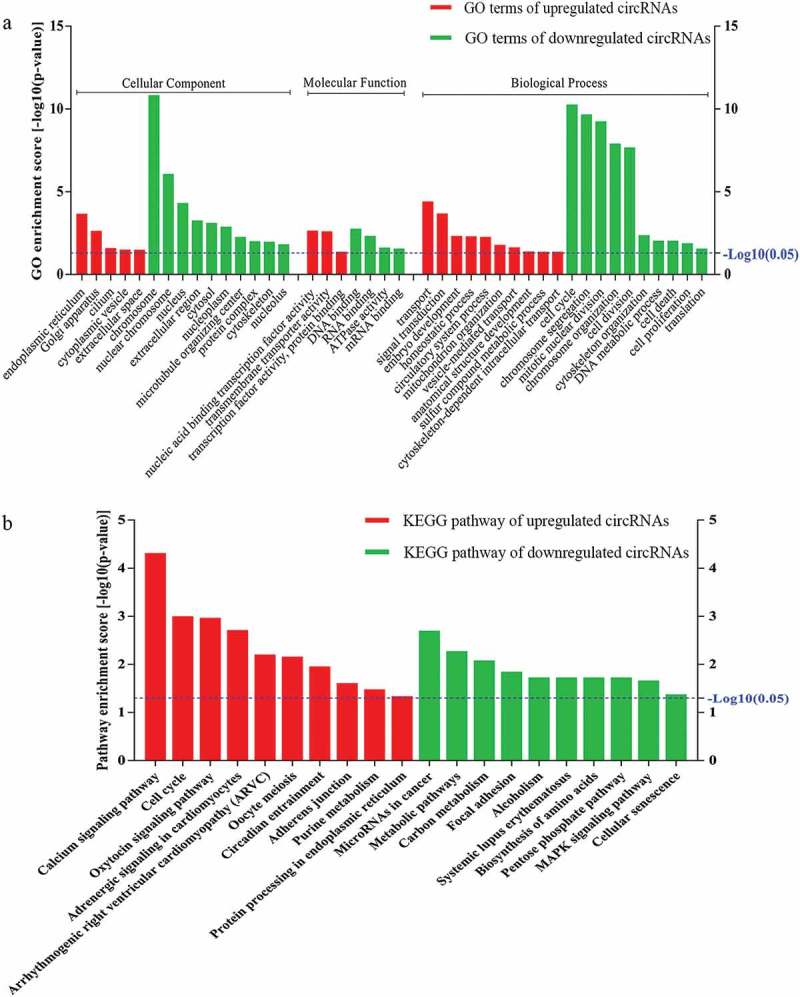
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses. a) GO analysis of genes related to differentially expressed circRNAs. GO annotations of the linear counterparts of upregulated and downregulated circRNAs in the cellular component (CC), molecular function (MF), and biological process (BP) terms. Red and green indicate GO terms of upregulated and downregulated circRNAs, myotubes vs. myoblasts, respectively. b) KEGG pathway enrichment analysis of the genes that produced upregulated and downregulated circRNAs. Red and green indicate KEGG pathways of upregulated and downregulated circRNAs, myotubes vs. myoblasts, respectively. Both upregulated and downregulated circRNAs were significantly altered with fold change ≥ 2.0 and P < 0.05.
We then performed KEGG pathway enrichment analysis for linear transcripts that matched the differentially expressed circRNAs. Our data indicated that the calcium signaling pathway and the cell cycle were the top two related pathways of the upregulated linear transcripts; metabolic and mitogen-activated protein kinase (MAPK) signaling pathways were the related pathways of the downregulated linear transcripts (Figure 4b). In addition, several pathways with significant enrichment scores such as focal adhesion and adherens junction were involved in cell-cell and cell-extracellular matrix interactions, which are known to play crucial roles in modulating cell differentiation, migration, and proliferation [28].
2.5. Construction of the circRNA-microRNA(miRNA)-mRNA interaction network
RNA transcripts can compete for the same miRNA reaction elements to regulate one other by using microRNA response elements as letters of a new language [18,29]. According to the ceRNA hypothesis, circRNA could compete for the same miRNA response element and act as a molecular sponge for miRNA; miRNA could target on mRNA 3ʹUTR; hence circRNA could regulate the de-repression of target genes of the respective miRNA family. To investigate the functions of circRNAs during C2C12 myoblast differentiation, we constructed a ceRNA network in C2C12 myotubes based on our microarray data. The differentially expressed circRNAs were predicted based on complementary miRNA matching sequences. We selected the myogenic-specific genes Myog, Mef2a, Myh1, Myh7, and Myh7b to construct a circRNA-miRNA-mRNA network of the differentially expressed circRNAs in mouse C2C12 myotubes. In total, 91 miRNAs could be combined with the selected myogenic-specific genes and the top 30 upregulated circRNAs, forming 239 edges (Figure 5 and Suppl. Table 1). Our data showed Myh1 could target three miRNAs and combine with three circRNAs; Myh7 could target three miRNAs and combine with three circRNAs; Myh7b could target seven miRNAs and combine with seven circRNAs; and Myog could target 42 miRNAs and combine with 22 circRNAs. In particular, Mef2a could target 75 miRNAs and combine with all of the top 30 upregulated circRNAs. Based on our results, circRNA_34451 can interact with Mef2a, Myh7b, and Myog, and circRNA_19008 can interact with Mef2a, Myh1, and Myog. circRNAs might play an important role in the process of C2C12 myoblast differentiation. These RNA interactions will provide novel insight into the differentiation and development of C2C12 myoblasts.
Figure 5.
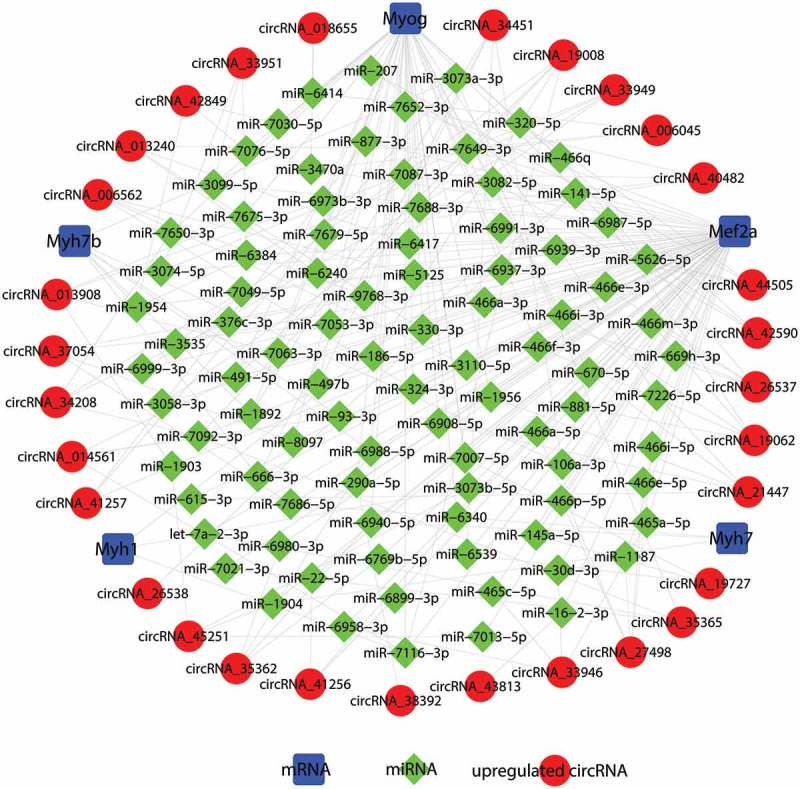
Construction of the circRNA-microRNA(miRNA)-mRNA competing endogenous RNA (ceRNA) regulatory network. In total, 91 miRNAs could be combined with the selected myogenic-specific genes and the top 30 upregulated circRNAs, forming 239 edges. Blue squares indicate myogenic-specific mRNAs, green diamonds indicate miRNAs, and red circles indicate upregulated circRNAs, myotubes vs. myoblasts.
Table 1.
Primers for the detected mRNAs and circRNAs.
| mRNA/circRNA | Forward primer (5ʹ to 3ʹ) | Reverse primer (5ʹ to 3ʹ) |
|---|---|---|
| 18S | GTAACCCGTTGAACCCCATT | CCATCCAATCGGTAGTAGCG |
| Myog | GGCAATGCACTGGAGTTCG | AGCCGCGAGCAAATGATC |
| eIF4G2 | AGTGCGATTGCAGAAGGGG | GTGCTTCGTGCAGGAATCCA |
| YTHDF3 | GATCAGCCTATGCCATATCTGAC | CCCCTGGTTGACTAAAAACACC |
| circRNA_43967 | GAAGGCTCAAAGCAGTGGC | AGGGTCCGAGGTCTGGATG |
| circRNA_28609 | GCTTTAGGAAGGATGATGGGA | TTGGGTAGGTTTTGAGATGGTG |
| circRNA_33951 | AAGAATTTGGAAGACTCATGGTGG | AATCCCATTCCTTTGCTTTGAC |
| circRNA_41256 | TGAAACTCAAGGACATCGTAGGC | TGGTGGTCGTGTTCCCAGTC |
| circRNA_19008 | AATGGGCAACATAGTTAAACCC | GATGCAGCAGGAAGAGCAAA |
| circRNA_19416 | AAGCAGCGGAGGATACAGCC | CCCTTCTCAGATTTCTTCCCAGT |
| circRNA_45526 | GCATGAAATGGACATTCGCTAT | GGTTTGCTGGCTACCCTGAG |
| circRNA_20260 | GCTGTCAGTTTAACGGGTGC | CGGGGTTCTGTCTGGAGGTA |
| circRNA_007438 | GTCAGACACCACAGAACCCTCG | TACCGTCATCATAGACTTTGACTCCT |
2.6. Prediction of coding potential and the presumed encoded polypeptide functions of differentially expressed circRNAs
It was recently reported that circRNAs can be translated into protein [14,20–22]. It is known that one m6A motif is required for circRNAs to encode polypeptides, and this m6A-driven translation requires eIF4G2 and YTHDF3 [21]. In this study, over the course of differentiation during C2C12 myoblast development, the relative mRNA expression level of Myog increased gradually (Figure 6a), verifying that C2C12 cells did undergo differentiation. The relative mRNA expression level of eIF4G2 (Figure 6b) and YTHDF3 (Figure 6c) also increased gradually during C2C12 myoblast differentiation.
Figure 6.
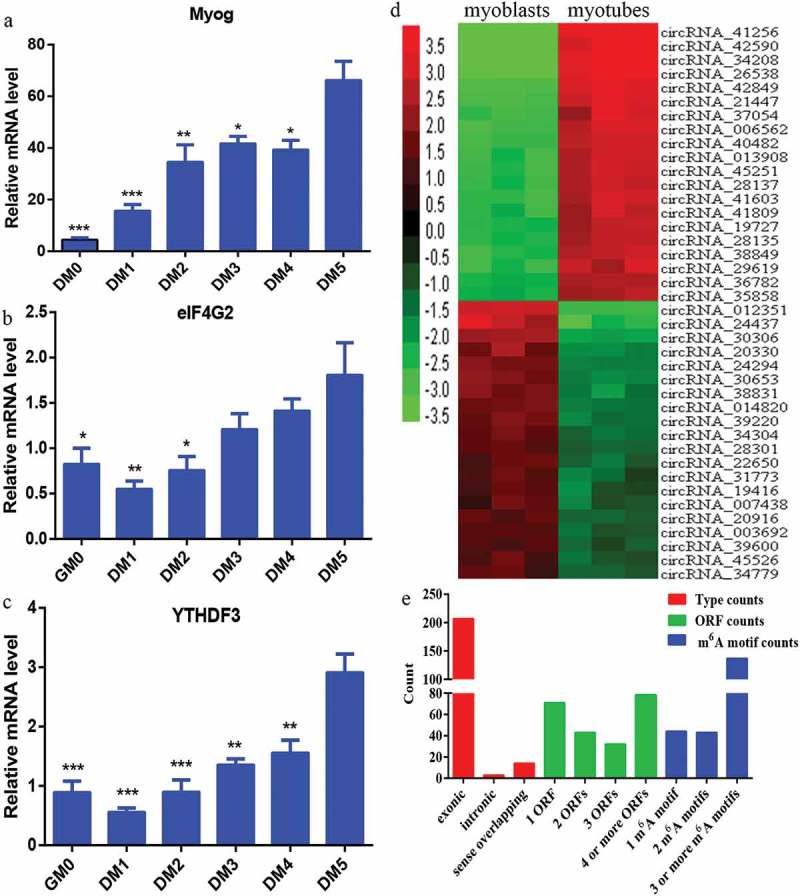
Prediction of the coding potential of differentially expressed circRNAs. a) Expression of the myogenic differentiation marker Myog was examined by qRT-PCR. b, c) eIF4G2 and YTHDF3 were examined by qRT-PCR. d) Hierarchical clustering heat map of the top 20 upregulated and top 20 downregulated circRNAs among the 224 differentially expressed circRNAs with coding potential. Red and green indicate high and low relative expression, respectively. e) The number of types, ORFs, and m6A motifs of the 224 circRNAs with coding potential. *P < 0.05, **P < 0.01, and ***P < 0.001 vs. after day 5 of differentiation.
We attempted to identify conceivable novel polypeptides encoded by putative circRNAs based on our microarray analysis. We screened the identified circRNAs for those with open reading frame (ORF) and at least one m6A motif in the sequence. From this screening, we identified 224 circRNAs with coding potential. A hierarchical clustering heat map was generated to sort the top 20 upregulated and top 20 downregulated circRNAs among the 224 differentially expressed circRNAs with coding potential, according to their expression levels (Figure 6d). We then summarized the classification of the 224 circRNAs with different expressions. The circRNAs with coding potential included three distinct types: exonic (207), intronic (3), and sense overlapping (14) (Figure 6e). Next, we counted the number of ORFs of the 224 circRNAs with coding potential. In total, 71 circRNAs had one ORF, 43 circRNAs had two ORFs, 32 circRNAs had three ORFs, and 78 circRNAs had four or more ORFs (Figure 6e). The number of m6A motifs in the 224 circRNAs with coding potential was also determined, which are required for circRNAs to encode polypeptides [21]. Briefly, 44 circRNAs had one m6A motif, 43 circRNAs had two m6A motifs, and 137 circRNAs had three or more m6A motifs (Figure 6e).
According to the screening of circRNAs with coding potential with known functional domains and more than 30 amino acids, we identified 75 circRNAs to perform GO and KEGG analyses based on the linear counterparts of the circRNAs. The predicted GO functions were ranked according to the number of related genes and classified into the subcategories CC, MF, and BP (Figure 7a). Our results revealed a high proportion of genes of the 75 differentially expressed circRNAs were clustered in binding, catalytic activity, cellular process, and metabolic process categories (Figure 7a). Meanwhile, KEGG analysis of the identified 75 circRNAs revealed a number of pathways primarily related to regulation of the actin cytoskeleton and metabolic pathways (Figure 7b). These annotations could offer new insight into the investigation of specific processes, pathways, and functions of circRNAs in skeletal muscle growth and development.
Figure 7.
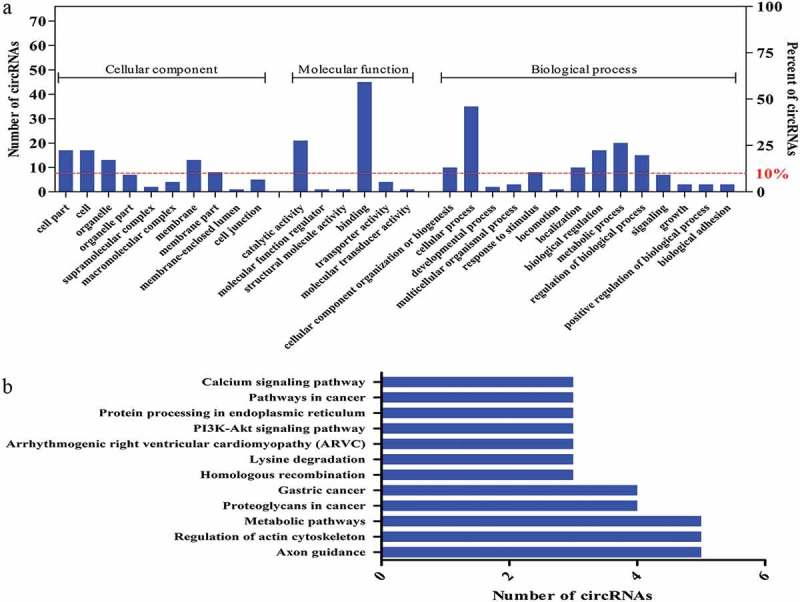
The presumed encoded polypeptide functions of differentially expressed circRNAs. a) GO analysis of the genes related to the 75 selected circRNAs with coding potential. GO annotations of the linear counterparts of the selected 75 circRNAs in the subcategories CC, MF, and BP. The left and right y-axes indicate the number of genes in a category and the percentage of a specific category of genes in the main category, respectively. b) KEGG pathway enrichment analysis of the genes that produced the selected 75 circRNAs. The y- and x-axes indicate the pathways and the number of circRNAs.
3. Discussion
The study of gene regulation concentrated on protein-coding genes until the discovery of numerous non-coding RNAs, including circRNAs [29]. Recently, studies of circRNAs have resulted in the understanding that these non-coding RNAs play an important role in cellular differentiation, development, and the onset and development of disease. The development of skeletal muscle is a comprehensive process that requires the fusion of myoblasts to produce multinucleated muscle fibers with contractile properties [30]. However, analysis of the expression profiles of circRNAs in proliferative C2C12 myoblasts and differentiated C2C12 myotubes had not yet been reported. To explore the functions of circRNAs in C2C12 myoblast differentiation, we determined the expression profiles of circRNAs in proliferative C2C12 myoblasts and differentiated C2C12 myotubes using microarray analysis, and predicted the function of differently expressed circRNAs by GO and KEGG analyses, ceRNA networks, and coding potential.
In the present study, we identified 581 circRNAs that were differentially regulated during C2C12 myoblast differentiation by microarray analysis. According to the relationship and genomic loci with their associated coding genes, the identified upregulated and downregulated circRNAs can be classified into five distinct types: antisense, exonic, intergenic, intronic, and sense overlapping. The most common type of differentially expressed circRNA was exonic, which is consistent with previous studies [31,32]. The second most common type was sense overlapping, which differs from results obtained during bovine myoblast differentiation [13].
GO analysis of the linear counterparts of differentially expressed circRNAs demonstrated that transmembrane transport activity, signal transduction, protein binding, and embryonic development related to coding genes contributed to C2C12 myoblast differentiation. KEGG pathway analysis uncovered 10 pathways that could have a significant effect on C2C12 myoblast differentiation, including calcium signaling, cell cycle, metabolic pathways, and MAPK signaling, indicating that circRNAs could play an important role by modulating the pathways involved in C2C12 myoblast differentiation. Calcium signaling has been reported to be significant for differentiation-dependent gene expression, and inhibition of the calcium-regulated phosphatase calcineurin could block myogenic gene expression and myogenic differentiation [33]. Lowering intracellular calcium levels could inhibit the differentiation of skeletal myoblasts into mature myotubes [34]. The MAPK signaling pathway could control fundamental cellular processes such as cell proliferation, cell migration, cell differentiation, embryogenesis, and cell death by transmitting largely unknown signals into the cell [35,36]. Liu et al. [37] reported that p38α MAPK signaling was necessary for efficient expression of Myog in C2C12 myoblasts. p38 MAPK signaling impacted the activities of the Mef2 family and MyoD, and was involved in chromatin remodeling at specific muscle-regulatory regions [38].
Several recent studies have reported that some circRNAs targeting miRNAs via the ceRNA network were involved in myoblast differentiation. circLMO7 was shown to inhibit bovine myoblast differentiation, promote cell proliferation, and protect myoblasts from apoptosis by binding miR-378a-3p [13]. circFUT10 was shown to accelerate differentiation and decreased the proliferation of bovine myoblasts by inhibiting miR-133a activity [15]. circFGFR4 by binding miR-107 could promote cell differentiation via targeting Wnt3a in bovine primary myoblasts [16]. To date, the functions of most circRNAs are not well known. In this study, the myogenic-specific genes Myog, Mef2a, Myh1, Myh7, and Myh7b were selected to construct a circRNA-miRNA-mRNA network of differentially expressed circRNAs during mouse C2C12 myoblast differentiation. A total of 91 miRNAs could be combined with the selected myogenic-specific genes and the top 30 upregulated circRNAs, forming 239 edges. In particular, Mef2a could target 75 miRNAs and combine with all of the top 30 upregulated circRNAs. Our analysis also revealed that Myog and Mef2a could combine with circRNA_21447 by targeting miR-670-5p, which has been shown to induce cell proliferation in hepatocellular carcinoma by targeting prospero-related homeobox 1 [39]. Furthermore, circRNA_19008 binding miR-186-5p may promote myoblast differentiation via targeting Myog and Mef2a in our analysis, and it was reported that the reduction in the levels of endogenous miR-186 resulted in increased differentiation of C2C12 and primary muscle cells of mouse through upregulation of Myog [40]. These data indicate that circRNAs could play key regulatory roles in skeletal muscle C2C12 myoblast differentiation. These precursory findings broaden our understanding of circRNA function during myoblast differentiation, although further research will be necessary to confirm our predictions.
Recently, it was reported that circRNAs can be translated into proteins [14,20–22]. m6A motifs were found to be enriched in circRNAs and one m6A motif was shown to be sufficient to drive translation initiation; this m6A-driven translation required the initiation factor eIF4G2 and the m6A reader YTHDF3 [21]. The depletion of eIF4G2 and YTHDF3 significantly reduced protein translation from circRNAs and overexpression of eIF4G2 and YTHDF3 increased protein translation from circRNAs [21]. In this study, the relative mRNA expression level of eIF4G2 and YTHDF3 increased gradually over the course of differentiation during C2C12 myoblast development. Therefore, to explore the coding potential of the differently expressed circRNAs in our microarray analysis, we identified 224 circRNAs with coding potential based on the number of ORFs and m6A motifs, and they could be classified into three types: exonic, intronic, and sense overlapping.
We further identified 75 circRNAs with coding potential, which have known functional domains and more than 30 amino acids [41], to perform GO and KEGG analyses. GO analysis of the 75 differentially expressed circRNAs demonstrated that binding, catalytic activity, cellular process, and metabolic process-related coding genes contributed to C2C12 myoblast differentiation. KEGG analysis of the identified 75 circRNAs revealed a number of pathways mainly related to regulation of the actin cytoskeleton and metabolic pathways. The functions of the actin cytoskeleton were most often aligned, allowing for consolidation and amplification of myofibroblast differentiation signals [42]. Modulation of the actin cytoskeleton by disordering drugs could differentially modify mechanically induced signal transduction and mesenchymal stem cell differentiation [43]. These annotations could offer new insight into the investigation of specific pathways and coding potential of circRNAs in skeletal muscle development.
Studies of the functions of circRNAs are currently in the development stage. The present study predicted the functions and coding potential of differentially expressed circRNAs during skeletal muscle C2C12 myoblast differentiation; however, further work will be necessary to confirm our predictions in the future.
In summary, we established expression profiles of circRNAs during C2C12 myoblast differentiation and predicted the function of differentially expressed circRNAs by GO and KEGG analyses, ceRNA networks, and coding potential based on ORFs and m6A motifs. Our results suggest that circRNAs might be important in skeletal muscle development. Our study also offers new insight into the investigation of circRNA functions in skeletal muscle growth and development.
4. Methods
4.1. Cell culture and establishment of the C2C12 differentiation model
C2C12 cell line is a subclone of the mouse myoblast cell line established by D. Yaffe and O. Saxel in 1977 [44]. It serves as a manageable experimental model system for researching the molecular basis of skeletal muscle cell specification and development [45]. High glucose Dulbecco’s modified Eagle’s medium was used to culture the mouse myoblast cell line C2C12 (Stem Cell Bank, cat no. SCSP-505) in 5% CO2 at 37°C. The markers of C2C12 differentiation model were myogenin and myosin heavy chain [46,47], and the differentiation model was also established in the early work of our laboratory [23,48,49].
4.2. RNA extraction and quantity control
Total RNA was extracted from mouse C2C12 myoblasts and myotubes using TRIzol reagent (Life Technologies, cat no.15596018) according to the manufacturer’s instructions. A NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) was used to assess the quantity and quality of purified RNA. RNA integrity was measured by conventional denaturing agarose gel electrophoresis.
4.3. Microarray analysis
To determine differentially expressed circRNAs, three pairs of mouse C2C12 myoblasts and differentiated myotubes were used for the microarray analysis. Sample labeling and mouse circRNA array hybridization were performed based on the standard protocols of the Arraystar Mouse circRNA Array V2 (Arraystar, Inc., Rockville, MD, USA). In brief, RNase R was used to digest total RNAs to remove linear RNAs and enrich circRNAs. Then, a random priming method was used to amplify the enriched circRNAs, which were transcribed into fluorescent cRNA using the Super RNA Labeling kit (Arraystar, Inc.). The labeled cRNAs were hybridized onto the Arraystar Mouse circRNA Array V2. The slides were washed and the arrays were scanned using the Agilent Scanner G2505C (Agilent Technologies, Santa Clara, CA, USA). Agilent Feature Extraction software (version 11.0.1.1) was used to analyze the acquired array images. The Gene Expression Omnibus accession number is GSE112014.
4.4. QRT-PCR
The myogenic-specific gene Myog was examined to verify the degree of muscle differentiation. eIF4G2 and YTHDF3 have been shown to be required for the translation of circRNAs. Five upregulated (mmu_circRNA_43967, mmu_circRNA_28609, mmu_circRNA_33951, mmu_circRNA_41256, and mmu_circRNA_19008) and four downregulated (mmu_circRNA_19416, mmu_circRNA_45526, mmu_circRNA_20260, and mmu_circRNA_007438) circRNAs were randomly selected for validation by qRT-PCR. Briefly, total RNA was extracted from myoblasts and myotubes and 2.5 μg of RNA was reverse-transcribed to cDNA using the PrimeScript RT Master Mix (TaKaRa, cat no. RR036A). According to the manufacturer’s instructions, the abundance of circRNAs was measured with Takara SYBR Green Mix (TaKaRa, cat no. RR820A). The qRT-PCR protocol was initiated at 95°C for 30 s, followed by 40 cycles of denaturation at 95°C for 5 s, annealing at 60°C for 30 s, and extension at 60°C for 1 min. 18S ribosomal RNA was used as a reference. The 2–ΔΔCt method was used to calculate the relative expression level of selected circRNAs. The examined mRNA and circRNA cross-junction site primers are shown in Table 1.
4.5. GO and KEGG pathway analyses
GO analysis based on the linear counterparts of the circRNAs was performed to investigate attributes of the differentially expressed circRNAs, including the GO terms CC, MF, and BP. The – log10 (P-value) indicated the enrichment score, representing the significance of GO term enrichment among genes producing differentially expressed circRNAs.
Pathway enrichment analysis helped identify significantly enriched metabolic or signal transduction pathways in differentially expressed genes compared with the whole genome background [50]. KEGG pathway analysis based on linear counterparts was used to harvest pathway clusters covering our knowledge of the molecular interaction and reaction networks in genes producing differentially expressed circRNAs [51].
4.6. CeRNA analysis
miRNA could target on mRNA 3ʹUTR; circRNA could act as a molecular sponge to regulate the de-repression of target genes of the respective miRNA family. In the present study, circRNA-miRNA-mRNA interactions were predicted with Arraystar’s in-house miRNA target prediction software based on TargetScan (http://www.targetscan.org/vert_71/) and miRanda (http://34.236.212.39/microrna/home.do). We selected the myogenic-specific genes Myog, Mef2a, Myh1, Myh7, and Myh7b to construct a ceRNA network of the differentially expressed circRNAs in mouse C2C12 myoblasts and myotubes. An entire circRNA-miRNA-mRNA interaction network was defined using Cytoscape.
4.7. Prediction of coding potential of differentially expressed circRNAs based on the number of ORFs and m6A motifs
In a recent study, one m6A motif was determined to be sufficient to drive efficient initiation of circRNA protein translation in cells and m6A was required for circRNAs to encode polypeptides [21]. We attempted to identify possible novel polypeptides encoded by putative circRNAs based on our microarray analysis. The circRNAs were screened to identify sequences with at least one ORF and at least one m6A motif. This screening revealed 224 circRNAs with coding potential for the translation of polypeptides. To further predict the functions of the polypeptides presumably encoded by the circRNAs, we performed GO and KEGG analyses with the linear counterparts of the circRNAs with known functional domains and more than 30 amino acids [41].
4.8. Statistical analysis
GraphPad Prism 5.0 software (La Jolla, CA, USA) was used to perform statistical analysis. Student’s t-test was applied for the comparison of two groups and P < 0.05 was considered to indicate statistical significance.
Supplementary Material
Funding Statement
This work was supported by the Natural Science Foundation of Guangdong Province [2018A030313591]; the Administration of Traditional Chinese Medicine of Guangdong Province [20172004, 20181014]; the Science Foundation of Guangdong Second Provincial General Hospital [YN-2017-002, YN-2017-003].
Acknowledgments
This work was supported by the Natural Science Foundation of Guangdong Province under Grant no. 2018A030313591; the Administration of Traditional Chinese Medicine of Guangdong Province under Grant no. 20172004 and 20181014; and the Science Foundation of Guangdong Second Provincial General Hospital under Grant no. YN-2017-002 and YN-2017-003.
Disclosure statement
The authors declare that they have no competing interests.
References
- [1].Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Vicens Q, Westhof E.. Biogenesis of circular RNAs. Cell. 2014;159:13–14. [DOI] [PubMed] [Google Scholar]
- [3].Chen LL, Yang L.. Regulation of circRNA biogenesis. RNA BIOL. 2015;12:381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. DEVELOPMENT. 2016;143:1838–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. NAT STRUCT MOL BIOL. 2015;22:256–264. [DOI] [PubMed] [Google Scholar]
- [6].Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. [DOI] [PubMed] [Google Scholar]
- [7].Sanger HL, Klotz G, Riesner D, et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73:3852–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339–340. [DOI] [PubMed] [Google Scholar]
- [9].Kristensen LS, Hansen TB, Veno MT, et al. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bachmayr-Heyda A, Reiner AT, Auer K, et al. Correlation of circular RNA abundance with proliferation – exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis and normal human tissues. SCI REP-UK. 2015;5:8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. CLIN CHIM ACTA. 2015;444:132–136. [DOI] [PubMed] [Google Scholar]
- [12].Li J, Yang J, Zhou P, et al. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. AM J CANCER RES. 2015;5:472–480. [PMC free article] [PubMed] [Google Scholar]
- [13].Wei X, Li H, Yang J, et al. RNA profiling reveals an abundant circLMO7 that regulates myoblasts differentiation and survival by sponging miR-378a-3p. Cell Death Dis. 2017;8:e3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Legnini I, Di Timoteo G, Rossi F, et al. Is a circular rna that can be translated and functions in Myogenesis. MOL CELL. 2017;66:22–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Li H, Yang J, Wei X, et al. CircFUT10 reduces proliferation and facilitates differentiation of myoblasts by sponging miR-133a. J CELL PHYSIOL. 2018;233:4643–4651. [DOI] [PubMed] [Google Scholar]
- [16].Li H, Wei X, Yang J, et al. circFGFR4 promotes differentiation of Myoblasts via binding miR-107 to relieve its inhibition of Wnt3a. Mol Ther Nucleic Acids. 2018;11:272–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Abdelmohsen K, Panda AC, De S, et al. RNAs in monkey muscle: age-dependent changes. Aging (Albany NY). 2015;7:903–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. HEPATOLOGY. 2017;66:1151–1164. [DOI] [PubMed] [Google Scholar]
- [20].Pamudurti NR, Bartok O, Jens M, et al. Translation of circRNAs. MOL CELL. 2017;66:9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. CELL RES. 2017;27:626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yang Y, Gao X, Zhang M, et al. Novel role of FBXW7 circular RNA in Repressing Glioma Tumorigenesis. JNCI: J Natl Cancer Inst. 2018;110:304–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chen R, Jiang T, She Y, et al. Comprehensive analysis of lncRNAs and mRNAs with associated co-expression and ceRNA networks in C2C12 myoblasts and myotubes. Gene. 2018;647:164–173. [DOI] [PubMed] [Google Scholar]
- [24].Hernandez-Hernandez JM, Garcia-Gonzalez EG, Brun CE, et al. The myogenic regulatory factors, determinants of muscle development, cell identity and regeneration. SEMIN CELL DEV BIOL. 2017;72:10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Doynova MD, Markworth JF, Cameron-Smith D, et al. JM. linkages between changes in the 3D organization of the genome and transcription during myotube differentiation in vitro. SKELET MUSCLE. 2017;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Dugdale HF, Hughes DC, Allan R, et al. The role of resveratrol on skeletal muscle cell differentiation and myotube hypertrophy during glucose restriction. MOL CELL BIOCHEM. 2018;444:109–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Meredith C, Herrmann R, Parry C, et al. Mutations in the slow skeletal muscle fiber myosin heavy chain Gene (MYH7) Cause Laing Early-Onset Distal Myopathy (MPD1). Am J Hum Genet. 2004;75:703–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sun J, Yuan X, Li X, et al. Comparative transcriptome analysis of the global circular RNAs expression profiles between SHEE and SHEEC cell lines. AM J TRANSL RES. 2017;9:5169–5179. [PMC free article] [PubMed] [Google Scholar]
- [29].Huang M, Zhong Z, Lv M, et al. Comprehensive analysis of differentially expressed profiles of lncRNAs and circRNAs with associated co-expression and ceRNA networks in bladder carcinoma. Oncotarget. 2016;7:47186–47200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Leikina E, Melikov K, Sanyal S, et al. Extracellular annexins and dynamin are important for sequential steps in myoblast fusion. J Cell Biol. 2013;200:109–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wu H, Zhang C, Zhang S, et al. Microarray expression profile of circular RNAs in heart tissue of mice with Myocardial infarction-induced heart failure. CELL PHYSIOL BIOCHEM. 2016;39:205–216. [DOI] [PubMed] [Google Scholar]
- [32].Liu W, Zhang J, Zou C, et al. Microarray expression profile and functional analysis of circular RNAs in Osteosarcoma. CELL PHYSIOL BIOCHEM. 2017;43:969–985. [DOI] [PubMed] [Google Scholar]
- [33].Nasipak BT, Padilla-Benavides T, Green KM, et al. Opposing calcium-dependent signalling pathways control skeletal muscle differentiation by regulating a chromatin remodelling enzyme. NAT COMMUN. 2015;6:7441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Porter GA, Makuck RF, Rivkees SA. Reduction in intracellular calcium levels inhibits Myoblast differentiation. J BIOL CHEM. 2002;277:28942–28947. [DOI] [PubMed] [Google Scholar]
- [35].Segales J, Perdiguero E, Munoz-Canoves P. Regulation of muscle stem cell functions: A focus on the p38 MAPK signaling pathway. Front Cell Dev Biol. 2016;4:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chakraborty C, Sharma AR, Patra BC, et al. MicroRNAs mediated regulation of MAPK signaling pathways in chronic myeloid leukemia. Oncotarget. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Liu Q, Zha X, Faralli H, et al. Comparative expression profiling identifies differential roles for Myogenin and p38α MAPK signaling in myogenesis. J MOL CELL BIOL. 2012;4:386–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Keren A, Tamir Y, Bengal E. The p38 MAPK signaling pathway: a major regulator of skeletal muscle development. MOL CELL ENDOCRINOL. 2006;252:224–230. [DOI] [PubMed] [Google Scholar]
- [39].Shi C, Xu X. MiR-670-5p induces cell proliferation in hepatocellular carcinoma by targeting PROX1. BIOMED PHARMACOTHER. 2016;77:20–26. [DOI] [PubMed] [Google Scholar]
- [40].Antoniou A, Mastroyiannopoulos NP, Uney JB, et al. miR-186 inhibits muscle cell differentiation through Myogenin regulation. J BIOL CHEM. 2014;289:3923–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Matsumoto A, Pasut A, Matsumoto M, et al. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature. 2017;541:228–232. [DOI] [PubMed] [Google Scholar]
- [42].Sandbo N, Dulin N. Actin cytoskeleton in myofibroblast differentiation: ultrastructure defining form and driving function. TRANSL RES. 2011;158:181–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Muller P, Langenbach A, Kaminski A, et al. Modulating the actin cytoskeleton affects mechanically induced signal transduction and differentiation in mesenchymal stem cells. PLoS One. 2013;8:e71283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Yaffe D, Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977;270:725–727. [DOI] [PubMed] [Google Scholar]
- [45].Kislinger T, Gramolini AO, Pan Y, et al. Proteome dynamics during C2C12 Myoblast differentiation. MOL CELL PROTEOMICS. 2005;4:887–901. [DOI] [PubMed] [Google Scholar]
- [46].Chen J, Mandel EM, Thomson JM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. NAT GENET. 2006;38:228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Naguibneva I, Ameyar-Zazoua M, Polesskaya A, et al. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. NAT CELL BIOL. 2006;8:278–284. [DOI] [PubMed] [Google Scholar]
- [48].Chen R, Jiang T, She Y, et al. Effects of Cobalt Chloride, a Hypoxia-Mimetic Agent, on Autophagy and Atrophy in Skeletal C2C12 Myotubes. BIOMED RES INT. 2017;2017:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Chen R, Xu J, She Y, et al. Necrostatin-1 protects C2C12 myotubes from CoCl2-induced hypoxia. INT J MOL MED. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Xie M, Huang Y, Zhang Y, et al. Transcriptome profiling of fruit development and maturation in Chinese white pear (Pyrus bretschneideri Rehd). BMC Genomics. 2013;14:823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zeng Y, Xu Y, Shu R, et al. Altered expression profiles of circular RNA in colorectal cancer tissues from patients with lung metastasis. INT J MOL MED. 2017;40:1818–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


