Supplemental Digital Content is available in the text
Keywords: long noncoding RNA, meta-analysis, ovarian cancer, survival outcome
Abstract
Background:
Previous studies investigating the association between altered long noncoding RNAs (lncRNAs) and survival outcomes in ovarian cancer have obtained controversial results. To comprehensively evaluate the association, we conducted a systematic review and meta-analysis of the studies published on the subject.
Methods:
We performed a systematic search using the databases of the Cochrane Central Register of Controlled Trials, PubMed, and Embase to find all relevant articles from inception to May 7, 2017. Studies that evaluated the association between 1 specific lncRNA and survival outcomes in ovarian cancer were included. Pooled hazard ratios (HRs) and 95% confidence intervals (95% CIs) for overall survival, progression-free survival, and disease-free survival were calculated with a fixed-effects or random-effects model.
Results:
A total of 15 studies involving 1333 patients with ovarian cancer were included in this meta-analysis. Altered lncRNAs were associated with decreased overall survival (HR: 2.29, 95% CI: 1.92–2.75) without heterogeneity (I2 = 0.0%) in ovarian cancer. Altered lncRNAs were also associated with decreased progression-free survival (HR: 2.77, 95% CI: 1.00–7.62, I2 = 76.6%) and disease-free survival (HR: 2.59, 95% CI: 0.89–7.57, I2 = 62.9%) in ovarian cancer.
Conclusion:
Our results supported the strong prognostic value of altered lncRNAs in ovarian cancer. Further large-scale studies should be carried out to verify the clinical applications of altered lncRNAs in the prognosis assessment of ovarian cancer.
1. Introduction
Ovarian cancer is the fifth leading cause of cancer-related death in women worldwide, with 22,280 estimated new cases and 14,240 estimated deaths in the United States alone in 2016.[1] As early-stage ovarian cancer is generally asymptomatic and sensitive screening method is still not available, almost 65% of patients with ovarian cancer are diagnosed at advanced stages.[2] Despite recent advances in surgical techniques and pharmaceutical treatment, the 5-year survival rate of ovarian cancer has not improved accordingly, remaining less than 30%.[3] In order to improve the survival of patients with ovarian cancer, it is imperative to identify effective prognostic factors to predict the survival outcomes. It has been revealed that clinicopathological parameters, such as tumor histology, tumor stage, and residual tumor diameter, are independent prognostic factors for ovarian cancer.[4] However, even for patients with similar status and treatment, the survival outcomes can be different from each other. The underlying mechanism of ovarian cancer is a complicated biological process involving genetic and epigenetic alterations.[5,6] Thus, identifying molecular prognostic factors may enable more accurate prediction of patients’ outcomes and bring novel therapeutic targets.
After decades of studying on RNA biology, long noncoding RNAs (lncRNAs) have been identified as significant regulators involved in various biological processes.[7] LncRNAs are RNA molecules longer than 200 nucleotides and possess no or little protein coding abilities due to their lacking of open reading frames.[8] They can modulate gene expression at the transcriptional, post-transcriptional, and epigenetic levels.[9] It has been found that lncRNAs not only play crucial roles in multiple physiological processes but also participate in various pathological conditions, including cancer.[10] Interestingly, preclinical studies have found that lncRNAs, such as PVT1 and H19, can exert carcinogenic effects via inducing tumor cell proliferation, invasion and migration, and inhibiting tumor cell apoptosis.[11,12] These effects have also been shown in ovarian cancer derived cell lines.[13,14] Accordingly, a body of epidemiologic studies has revealed that altered lncRNAs were associated with poor survival outcomes in several malignancies, including breast, gastric, and colorectal cancer.[15–17]
A number of studies have investigated the relationship between altered lncRNAs and survival outcomes in ovarian cancer; however, the results are inconsistent. Thus, we performed a systematic review based on available evidence to determine whether altered lncRNAs were associated with poor survival outcomes in ovarian cancer.
2. Methods
This meta-analysis was prepared according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses statement.[18] As this study was a review of previous published studies, ethical approval or patient consent was not a requirement.
2.1. Search strategy
We performed a systematic search using the databases of the Cochrane Central Register of Controlled Trials, PubMed, and Embase to find all relevant articles from inception to May 7, 2017. Both subject headings and free text words were used in the search. The detailed search strategies are presented in Appendix A. In addition, the reference lists from all retrieved articles were screened for additional eligible studies. There was no language restriction in our search strategy.
2.2. Eligibility criteria
After conducting the search, 2 independent reviewers removed duplicates and screened the titles and abstracts. Then, they evaluated potentially relevant references in detail to determine the eligibility. Studies that met the following inclusion criteria were included in this meta-analysis: evaluated the association between 1 specific lncRNA and survival outcomes in ovarian cancer; evaluated at least 1 of the outcomes of interest, including overall survival (OS), progression-free survival (PFS), disease-specific survival (DSS), and disease-free survival (DFS); and reported HR and a 95% CI or provided data for their calculation. Articles were excluded if they were reviews, editorials, letters, and case reports; without appropriate data that could be extracted or calculated; and used microarray data. In cases of duplicate publications involving the same population, only the most comprehensive one was included. Any disagreements in study selection were resolved by discussion between the 2 reviewers and, if needed, in consultation with a third reviewer.
2.3. Data extraction and quality assessment
Two reviewers extracted data independently. The following data were collected from each study: publication data (i.e., first author's name, publication year, and study location), specimen, sample size, detective method, internal reference, follow-up, HR and 95% CI, and covariants. When multiple estimates of effect (HR) were presented, the most adjusted one was extracted; when adjusted estimate was not available, crude estimate was extracted. When the HR and 95% CI were not available, we estimated them indirectly from Kaplan–Meier curves using published methods.[19,20]
Three reviewers evaluated the methodological quality of included studies independently. The quality was assessed using the Newcastle–Ottawa scale (NOS).[21] The NOS uses a star system ranging from 0 to 9 stars. Studies that awarded 7 or more stars were considered high quality.
2.4. Statistical analysis
The pooled HR was obtained by combing the HRs from individual eligible study. Heterogeneity was measured using the Chi-square (χ2, or Chi2) and I2 test. When significant heterogeneity (P < .10 or I2 > 50%) was found, a random-effects model was applied to calculate the pooled effect; otherwise, a fixed-effects model was used. All analyses were performed using Stata version 12.0 software (Stata Corporation, College Station, TX). For all tests, a 2-sided P value less than .05 was considered statistically significant.
3. Results
3.1. Study selection
In our initial search, 345 records were identified from the database search. After removing duplicates and screening the titles and abstracts, 27 potentially relevant records were retrieved for further review. Of these, 12 studies were excluded for following reasons: 6 did not have usable data,[22–27] 1 did not report any outcomes of interest,[28] and 5 used microarray data.[29–33] We retrieved no additional studies from reference lists. Finally, 15 studies that met our eligibility criteria were included in the meta-analysis.[14,34–47] The flow diagram summarizing the process of study selection is given in Fig. 1.
Figure 1.
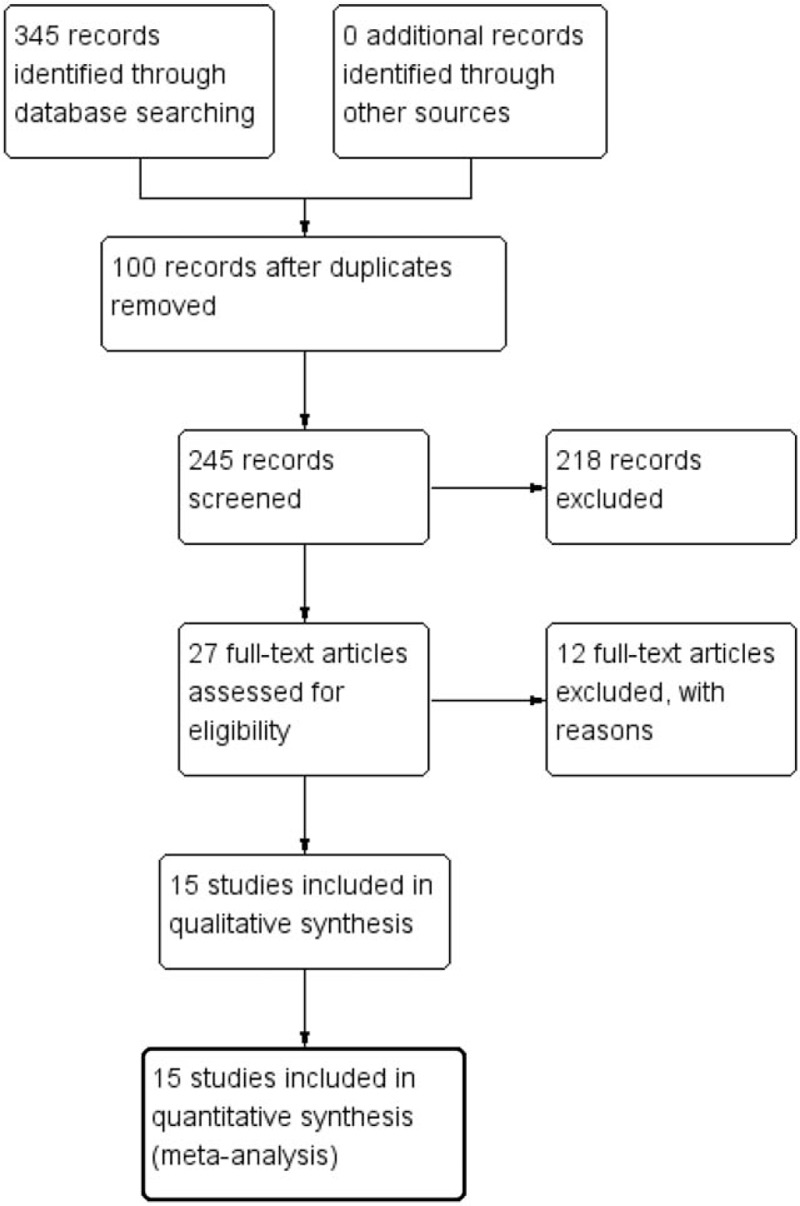
Study flow diagram.
3.2. Study characteristics
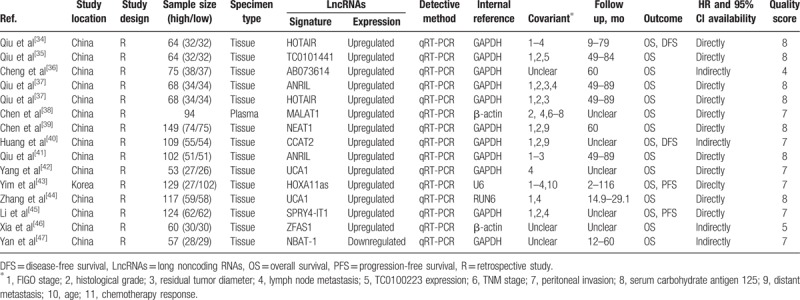
A total of 15 studies involving 1333 patients with ovarian cancer were included in the meta-analysis. The studies were published between 2014 and 2017. Of these, 14 studies were carried out in China[14,34–42,44–47] and 1 in Korea.[43] Fourteen of our included studies were carried out using human tissues[14,34–37,39–47] and 1 using plasma.[38] A total of 12 different lncRNAs were linked with survival outcomes in ovarian cancer. The expression of lncRNAs was upregulated in 14 studies[14,34–46] and downregulated in 1 study.[47] OS, PFS, and DFS were investigated to evaluate survival outcomes in 15 (100.0%), 2 (13.3%), and 2 (13.3%) studies, respectively. The NOS values ranged from 4 to 8 stars: 1 study was awarded 4 stars,[36] 1 study was award 5 stars,[46] and 13 studies were award 7 or more stars.[14,34,35,37–45,47] The characteristics of the included studies are summarized in Table 1.
Table 1.
Characteristics of included studies.
3.3. Meta-analysis of OS
We first investigated the prognostic value of altered lncRNAs for OS in all 15 included studies. The pooled data indicated that altered lncRNAs were associated with decreased OS (15 studies, 1333 patients, HR: 2.29, 95% CI: 1.92–2.75). The Cochran Q test had a P value of .61, and the quantity I2 was 0.0%, both indicating no significant heterogeneity among the 15 included studies (Fig. 2).
Figure 2.
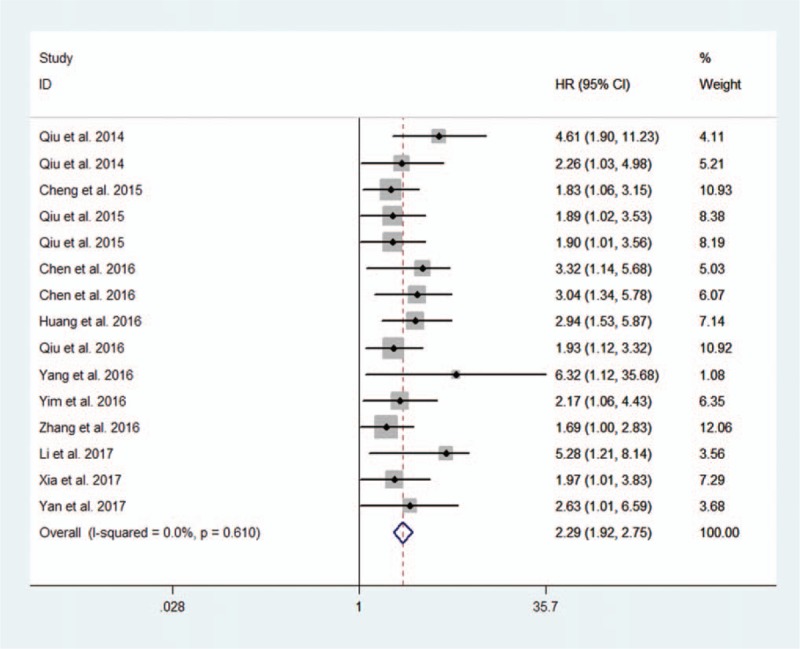
Forest plot of altered lncRNAs and overall survival.
Then, we performed a subanalysis based on the exact expression pattern of lncRNAs in ovarian cancer specimens compared with normal controls. The expression of lncRNAs was upregulated in 14 studies involving 1276 patients with ovarian cancer.[14,34–46] The estimated pooled HR showed that upregulated lncRNA signatures were associated with decreased OS (14 studies, 1276 patients, HR: 2.28, 95% CI: 1.90–2.74). The Cochran Q test had a P value of .539, and the quantity I2 was 0.0%, both of which indicated no significant heterogeneity among the 14 studies (Fig. 3). As only 1 study investigated the association between downregulated lncRNA signature and OS, we were unable to perform an accordingly meta-analysis. In the study by Yan et al,[47] the lncRNA NBAT-1 was downregulated in OC specimens compared with normal controls and was associated with a poor outcome of OC (HR: 2.626, 95% CI: 1.01–6.59).
Figure 3.
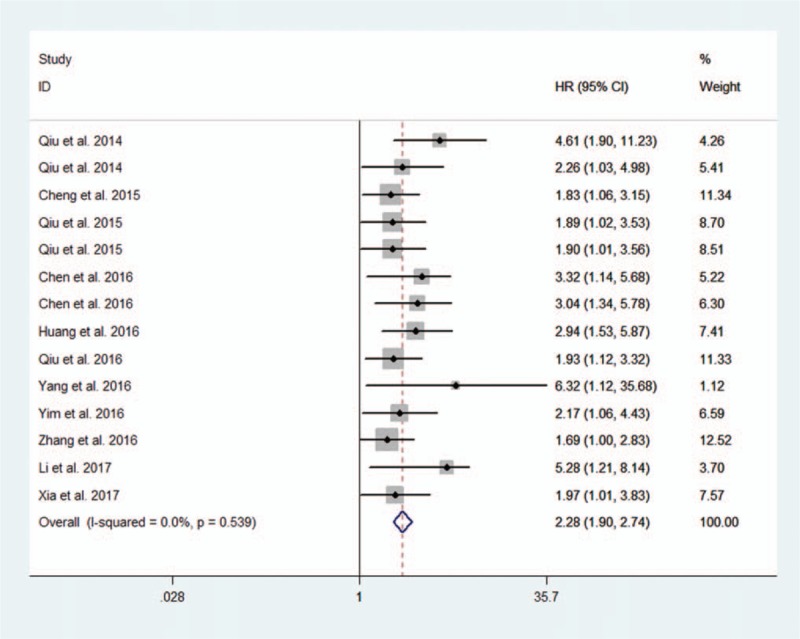
Forest plot of upregulated lncRNA signatures and overall survival.
3.4. Meta-analysis of PFS
The prognostic value of altered lncRNAs in PFS was evaluated in 2 studies with 253 patients.[43,45] The estimated pooled HR indicated that altered lncRNAs were associated with decreased PFS (2 studies, 253 patients, HR: 2.77, 95% CI: 1.00–7.62). The Cochran Q test had a P value of .039, and the quantity I2 was 76.6%, both indicating significant heterogeneity between these 2 studies (Fig. 4).
Figure 4.
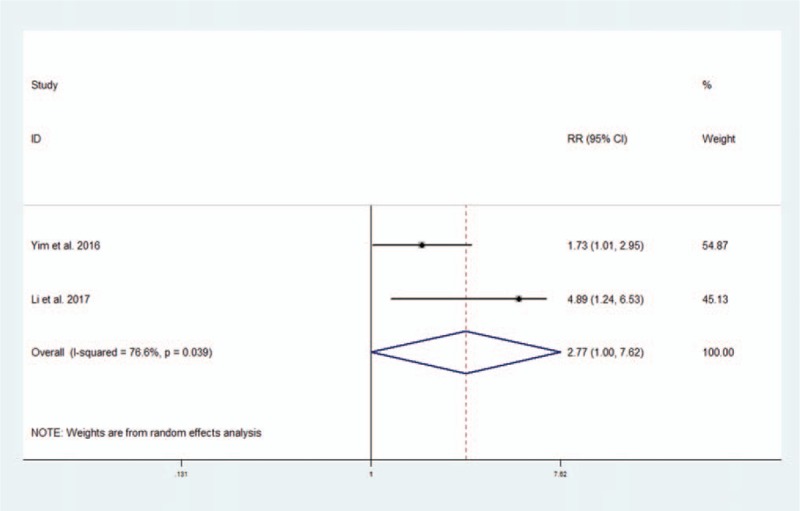
Forest plot of altered lncRNAs and progression-free survival.
3.5. Meta-analysis of DFS
The prognostic value of altered lncRNAs in DFS was evaluated in 2 studies with 173 patients.[34,40] The pooled HR indicated that altered lncRNAs were associated with decreased DFS, though the data supporting this association were not as robust (2 studies, 173 patients, HR: 2.59, 95% CI: 0.89–7.57). The Cochran Q test had a P value of .101, and the quantity I2 was 62.9%, both indicating significant heterogeneity between these 2 studies (Fig. 5).
Figure 5.
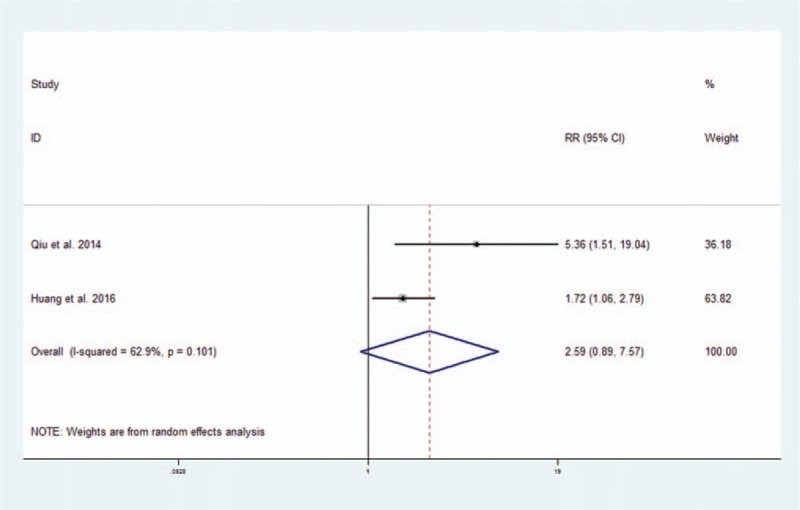
Forest plot of altered lncRNAs and disease-free survival.
4. Discussion
This present meta-analysis supported for a strong association between altered lncRNAs and poor survival outcomes in ovarian cancer. In this meta-analysis of 15 studies, we found that patients with ovarian cancer who had altered lncRNAs showed decreased OS, PFS, and DFS, which is in accordance with the promising findings derived from in vitro studies.[13,14] In addition, our results were in accordance with several other meta-analyses with regard to the association between altered lncRNAs and survival outcomes in cancers of other specific sites. Likewise, they found that altered lncRNAs were associated with poor survival outcomes in renal cell carcinoma, osteosarcoma, and hepatocellular carcinoma.[48–50] Luo et al[51] also performed a meta-analysis to investigate the prognostic value of abnormally expressed lncRNAs in ovarian cancer. In their study, 2 studies using microarray data were included[29,31]; however, we did not include any studies using microarray data. Moreover, while they only performed the meta-analysis of OS, we also performed the meta-analysis of PFS and DFS.
Among the altered lncRNA profiles, 11 lncRNAs, including HOTAIR, TC010441, AB073614, ANRIL, MALAT1, NEAT1, CCAT2, UCA1, HOXA11as, SPRY4-IT1, and ZFAS1, were significantly increased in patients with ovarian cancer, suggesting that these lncRNAs may play oncogenic roles in ovarian cancer development.[14,34–46] However, the expression of some of the above lncRNAs, such as SPRY4-IT1, was decreased in gastric cancer and was associated with a poor prognosis in such disease.[52] On the contrary, the expression of lncRNA NBAT-1 was significantly decreased in patients with ovarian cancer and was associated with a poor prognosis.[47] These findings suggest that the expression of lncRNAs might be tissue-specific, and they play different and distinct roles in cancer development.
LncRNAs have been shown to participate extensively in tumor initiation, progression, and metastasis during the development of ovarian cancer. For example, the lncRNA NEAT1 can regulate carcinogenesis and progression of ovarian cancer via interactions with HuR and miR-124–3p and may therefore serve as a potential antineoplastic therapeutic target.[53] In addition, the lncRNA TUG1 can promote ovarian cancer metastasis by affecting the epithelial-mesenchymal transition.[54] Abnormal expression of HOTAIR is associated with chemoresistance in ovarian cancer.[55] HOTAIR can interact with polycomb repressive complex 2 (PRC2) and is essential for histone H3 lysine-27 trimethylation of the HOD locus32.[56]
In our analysis, a strength deserved to be mentioned is that no significant heterogeneity was present in meta-analysis of OS. This reinforced the reliability of our pooled results. Still, there are several limitations in our analysis that should be noted. First, several lncRNAs were included to evaluate their prognostic value in ovarian cancer; however, a specific ovarian cancer related lncRNA was absent for clinical assessment. Second, the probable action mechanism of lncRNAs has not been clarified yet. Third, on some occasions, we calculated HRs ourselves on the basis of the data provided in the article, which may not be the most accurate estimate of HR. However, significant difference has not been shown yet comparing our calculation with direct estimation of HR. Fourth, as most patients enrolled in this meta-analysis were Chinese and the total sample size was relatively small, the overall result of this study may not be able to be extended to all populations. As a result, in order to confirm these results, multicenter and larger-size studies investigating the prognostic value of lncRNAs in ovarian cancer should be conducted in the future.
5. Conclusion
Our results supported the strong prognostic value of altered lncRNAs in ovarian cancer. Further large-scale studies should be carried out to verify the clinical applications of altered lncRNAs in the prognosis assessment of ovarian cancer.
Author contributions
NL: study design, literature search, systematic review and data collection, statistical analysis, interpretation of results, and preparation of the manuscript; H-YC and WS: a contribution to critical review of the manuscript; L-JH: checked and corrected the typographical and grammatical errors, and confirmed all contributing authors gave permission to be named.
Conceptualization: Li Ning.
Software: Yingchao Hu.
Validation: Shu Wang.
Writing – original draft: Li Ning.
Writing – review & editing: Jinghe Lang.
Author name: ORCID number.
Supplementary Material
Footnotes
Abbreviations: CI = confidence interval, DFS = disease-free survival, DSS = disease-specific survival, HR = hazard ratio, LncRNAs = long noncoding RNAs, NOS = Newcastle–Ottawa scale, OS = overall survival, PFS = progression-free survival.
Funding/support: This study had no funding.
The authors have no conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
References
- [1].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7–30. [DOI] [PubMed] [Google Scholar]
- [2].Narod S. Can advanced-stage ovarian cancer be cured? Nat Rev Clin Oncol 2016;13:255–61. [DOI] [PubMed] [Google Scholar]
- [3].Liu CF, Liu SY, Min XY, et al. The prognostic value of CXCR4 in ovarian cancer: a meta-analysis. PLoS One 2014;9:e92629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Landrum LM, Java J, Mathews CA, et al. Prognostic factors for stage III epithelial ovarian cancer treated with intraperitoneal chemotherapy: a Gynecologic Oncology Group study. Gynecol Oncol 2013;130:12–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ramalingam P. Morphologic, immunophenotypic, and molecular features of epithelial ovarian cancer. Oncology (Williston Park) 2016;30:166–76. [PubMed] [Google Scholar]
- [6].Balch C, Fang F, Matei DE, et al. Minireview: epigenetic changes in ovarian cancer. Endocrinology 2009;150:4003–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Li X, Wu Z, Fu X, et al. lncRNAs: insights into their function and mechanics in underlying disorders. Mutat Res Rev Mutat Res 2014;762:1–21. [DOI] [PubMed] [Google Scholar]
- [8].Kazemzadeh M, Safaralizadeh R, Orang AV. LncRNAs: emerging players in gene regulation and disease pathogenesis. J Genet 2015;94:771–84. [DOI] [PubMed] [Google Scholar]
- [9].Zhu J, Fu H, Wu Y, et al. Function of lncRNAs and approaches to lncRNA-protein interactions. Sci China Life Sci 2013;56:876–85. [DOI] [PubMed] [Google Scholar]
- [10].Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci 2016;73:2491–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yang YR, Zang SZ, Zhong CL, et al. Increased expression of the lncRNA PVT1 promotes tumorigenesis in non-small cell lung cancer. Int J Clin Exp Pathol 2014;7:6929–35. [PMC free article] [PubMed] [Google Scholar]
- [12].Li H, Yu B, Li J, et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget 2014;5:2318–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yiwei T, Hua H, Hui G, et al. HOTAIR interacting with MAPK1 regulates ovarian cancer skov3 cell proliferation, migration, and invasion. Med Sci Monit 2015;21:1856–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Qiu JJ, Lin YY, Ding JX, et al. Long non-coding RNA ANRIL predicts poor prognosis and promotes invasion/metastasis in serous ovarian cancer. Int J Oncol 2015;46:2497–505. [DOI] [PubMed] [Google Scholar]
- [15].Zhang XF, Liu T, Li Y, et al. Overexpression of long non-coding RNA CCAT1 is a novel biomarker of poor prognosis in patients with breast cancer. Int J Clin Exp Pathol 2015;8:9440–5. [PMC free article] [PubMed] [Google Scholar]
- [16].Xu C, Shao Y, Xia T, et al. lncRNA-AC130710 targeting by miR-129-5p is upregulated in gastric cancer and associates with poor prognosis. Tumour Biol 2014;35:9701–6. [DOI] [PubMed] [Google Scholar]
- [17].Shi D, Zheng H, Zhuo C, et al. Low expression of novel lncRNA RP11-462C24.1 suggests a biomarker of poor prognosis in colorectal cancer. Med Oncol 2014;31:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int J Surg 2010;8:336–41. [DOI] [PubMed] [Google Scholar]
- [19].Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med 1998;17:2815–34. [DOI] [PubMed] [Google Scholar]
- [20].Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 2007;8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wells GA, Shea B, O’Connell D, et al. The Newcastle-Ottawa Scale (NOS) for Assessing the Quality of Nonrandomized Studies in Meta-analyses. Available at: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp. Accessed March 1, 2017. [Google Scholar]
- [22].Qiu JJ, Hua KQ. Long non-coding RNA aHIF predicts poor prognosis in epithelial ovarian cancer and affects cell proliferation through the regulation of cell cycle, apoptosis and senescence. J Minim Invasive Gynecol 2016;23(suppl 1):S173. [Google Scholar]
- [23].Li J, Huang H, Li Y, et al. Decreased expression of long non-coding RNA GAS5 promotes cell proliferation, migration and invasion, and indicates a poor prognosis in ovarian cancer. Oncol Rep 2016;36:3241–50. [DOI] [PubMed] [Google Scholar]
- [24].Rupaimole R, Previs R, Lee J, et al. SiRNA therapy against novel lncRNA NRCP: shutting down the fuel for cancer cells. Cancer Res 2015;75(suppl 1):142. [Google Scholar]
- [25].Zhang X, Qiu J, Hua K. Long non-coding RNA RAD51-AS1 predicts negative prognosis and promotes proliferation in ovarian cancer. J Minim Invasive Gynecol 2016;23(suppl 1):S164. [Google Scholar]
- [26].Lawrenson K, Spindler T, Gayther SA. A novel long non-coding RNA associated with poor prognosis in epithelial ovarian cancer. Cancer Res 2014;74(suppl 1):1497. [Google Scholar]
- [27].Qiu JJ, Hua KQ. Long non-coding RNA TC0101441 predicts poor prognosis and promotes cell metastasis by upregulating KISS-1 to induce EMT in epithelial ovarian cancer. J Minim Invasive Gynecol 2016;23(suppl 1):S30–1. [Google Scholar]
- [28].Zhou M, Wang X, Li J, et al. Prioritizing candidate disease-related long non-coding RNAs by walking on the heterogeneous lncRNA and disease network. Mol Biosyst 2015;11:760–9. [DOI] [PubMed] [Google Scholar]
- [29].Guo Q, Cheng Y, Liang T, et al. Comprehensive analysis of lncRNA-mRNA co-expression patterns identifies immune-associated lncRNA biomarkers in ovarian cancer malignant progression. Sci Rep 2015;5:17683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Szafron LM, Balcerak A, Grzybowska EA, et al. The putative oncogene, CRNDE, is a negative prognostic factor in ovarian cancer patients. Oncotarget 2015;6:43897–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Teschendorff AE, Lee SH, Jones A, et al. HOTAIR and its surrogate DNA methylation signature indicate carboplatin resistance in ovarian cancer. Genome Med 2015;7:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Martini P, Paracchini L, Caratti G, et al. lncRNAs as novel indicators of patients’ prognosis in stage i epithelial ovarian cancer: a retrospective and multicentric study. Clin Cancer Res 2017;23:2356–66. [DOI] [PubMed] [Google Scholar]
- [33].Zhao L, Ji G, Le X, et al. Long noncoding RNA LINC00092 acts in cancer-associated fibroblasts to drive glycolysis and progression of ovarian cancer. Cancer Res 2017;77:1369–82. [DOI] [PubMed] [Google Scholar]
- [34].Qiu JJ, Lin YY, Ye LC, et al. Overexpression of long non-coding RNA HOTAIR predicts poor patient prognosis and promotes tumor metastasis in epithelial ovarian cancer. Gynecol Oncol 2014;134:121–8. [DOI] [PubMed] [Google Scholar]
- [35].Qiu JJ, Ye LC, Ding JX, et al. Expression and clinical significance of estrogen-regulated long non-coding RNAs in estrogen receptor (-positive ovarian cancer progression. Oncol Rep 2014;31:1613–22. [DOI] [PubMed] [Google Scholar]
- [36].Cheng Z, Guo J, Chen L, et al. A long noncoding RNA AB073614 promotes tumorigenesis and predicts poor prognosis in ovarian cancer. Oncotarget 2015;6:25381–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Qiu JJ, Wang Y, Ding JX, et al. The long non-coding RNA HOTAIR promotes the proliferation of serous ovarian cancer cells through the regulation of cell cycle arrest and apoptosis. Exp Cell Res 2015;333:238–48. [DOI] [PubMed] [Google Scholar]
- [38].Chen Q, Su Y, He X, et al. Plasma long non-coding RNA MALAT1 is associated with distant metastasis in patients with epithelial ovarian cancer. Oncol Lett 2016;12:1361–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Chen ZJ, Zhang Z, Xie BB, et al. Clinical significance of up-regulated lncRNA NEAT1 in prognosis of ovarian cancer. Eur Rev Med Pharmacol Sci 2016;20:3373–7. [PubMed] [Google Scholar]
- [40].Huang S, Qing C, Huang Z, et al. The long non-coding RNA CCAT2 is up-regulated in ovarian cancer and associated with poor prognosis. Diagn Pathol 2016;11:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Qiu JJ, Wang Y, Liu YL, et al. The long non-coding RNA ANRIL promotes proliferation and cell cycle progression and inhibits apoptosis and senescence in epithelial ovarian cancer. Oncotarget 2016;7:32478–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Yang Y, Jiang Y, Wan Y, et al. UCA1 functions as a competing endogenous RNA to suppress epithelial ovarian cancer metastasis. Tumour Biol 2016;37:10633–41. [DOI] [PubMed] [Google Scholar]
- [43].Yim GW, Kim HJ, Kim LK, et al. Long non-coding RNA HOXA11 antisense promotes cell proliferation and invasion and predicts patient prognosis in serous ovarian cancer. Cancer Res Treat 2017;49:656–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zhang L, Cao X, Zhang L, et al. UCA1 overexpression predicts clinical outcome of patients with ovarian cancer receiving adjuvant chemotherapy. Cancer Chemother Pharmacol 2016;77:629–34. [DOI] [PubMed] [Google Scholar]
- [45].Li H, Liu C, Lu Z, et al. Upregulation of the long non-coding RNA SPRY4-IT1 indicates a poor prognosis and promotes tumorigenesis in ovarian cancer. Biomed Pharmacother 2017;88:529–34. [DOI] [PubMed] [Google Scholar]
- [46].Xia B, Hou Y, Chen H, et al. Long non-coding RNA ZFAS1 interacts with miR-150-5p to regulate Sp1 expression and ovarian cancer cell malignancy. Oncotarget 2017;8:19534–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Yan C, Jiang Y, Wan Y, et al. Long noncoding RNA NBAT-1 suppresses tumorigenesis and predicts favorable prognosis in ovarian cancer. Onco Targets Ther 2017;10:1993–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Chen J, Chen Y, Gu L, et al. LncRNAs act as prognostic and diagnostic biomarkers in renal cell carcinoma: a systematic review and meta-analysis. Oncotarget 2016;7:74325–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Yang Y, Wang S, Li T. Altered long non-coding RNAs predict worse outcome in osteosarcoma patients: evidence from a meta-analysis. Oncotarget 2017;8:35234–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Qu Z, Yuan CH, Yin CQ, et al. Meta-analysis of the prognostic value of abnormally expressed lncRNAs in hepatocellular carcinoma. Onco Targets Ther 2016;9:5143–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Luo P, Liu XF, Wang YC, et al. Prognostic value of abnormally expressed lncRNAs in ovarian carcinoma: a systematic review and meta-analysis. Oncotarget 2017;8:23927–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Xie M, Nie FQ, Sun M, et al. Decreased long noncoding RNA SPRY4-IT1 contributing to gastric cancer cell metastasis partly via affecting epithelial-mesenchymal transition. J Transl Med 2015;13:250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Chai Y, Liu J, Zhang Z, et al. HuR-regulated lncRNA NEAT1 stability in tumorigenesis and progression of ovarian cancer. Cancer Med 2016;5:1588–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Wang J, Chen D, He X, et al. Downregulated lincRNA HOTAIR expression in ovarian cancer stem cells decreases its tumorgeniesis and metastasis by inhibiting epithelial-mesenchymal transition. Cancer Cell Int 2015;15:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Özeş AR, Miller DF, Özeş ON, et al. NF-(B-HOTAIR axis links DNA damage response, chemoresistance and cellular senescence in ovarian cancer. Oncogene 2016;35:5350–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007;129:1311–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


