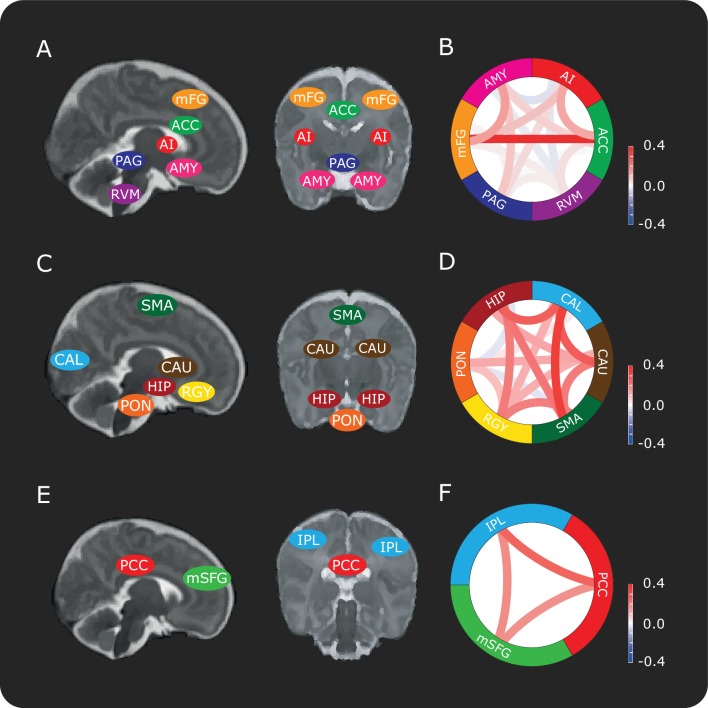
Figure 1. Connectivity between brain regions in the DPMS and control networks.
Schematic representation showing approximate locations of brain regions in sagittal and coronal slices in the (A) DPMS Network, (C) Control Network and (E) Default Mode Network. Each anatomical region of interest is identified in Figure 1—figure supplement 1 and the source data is provided in Figure 1—source data 1. Figure 1—figure supplement 2 shows the registration of two example masks from template to functional space and example time series. Network schematics of the mean pre-stimulus functional connectivity between pairs of regions in the (B) DPMS Network, (D) Control Network and (F) Default Mode Network. For abbreviations see main text.
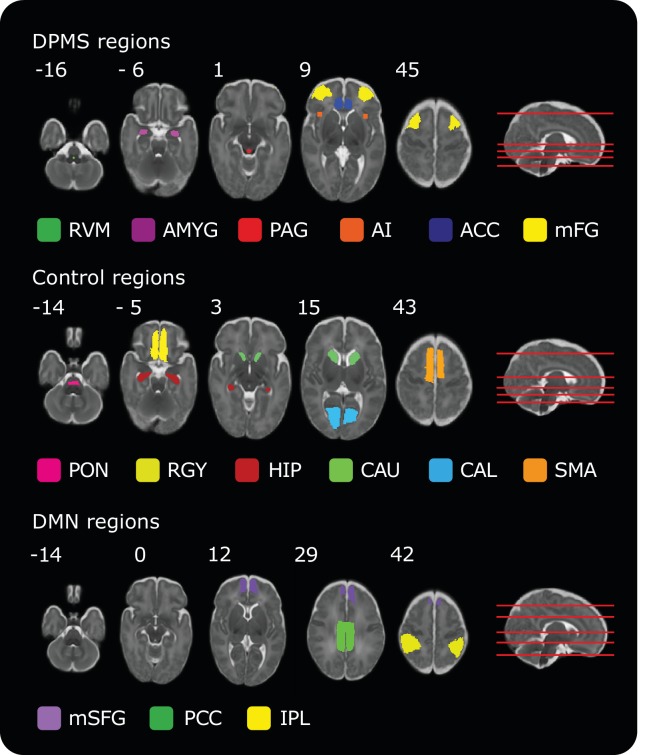
Figure 1—figure supplement 1. Masks of regions included in the DPMS, Control Network and Default Mode Network.
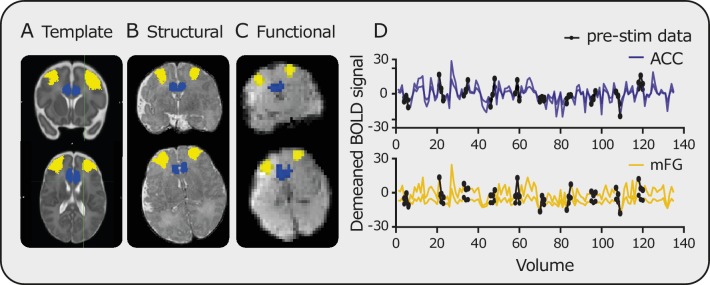
Figure 1—figure supplement 2. Registration and time series data.
Figure 1—figure supplement 3. Pre-stimulus connectivity is stable.