Abstract
The incidence rate of nasopharyngeal cancer (nasopharyngeal carcinoma [NPC]) is much higher in Southeast Asia than in western countries. Interleukin-8 (IL-8), a chemokine produced by macrophages, epithelial cells, airway smooth muscle cells, and endothelial cells, is an important immuno-mediator in the development and progression of many types of cancer. Genetic variations in IL-8 have been associated with the risks of NPC and other cancers. In the current study, we evaluated the role of IL-8 in NPC at the levels of DNA, RNA, and protein in a Taiwanese population. First, in a case-control study, 176 NPC patients and 352 cancer-free controls were genotyped, and the associations of IL-8 T − 251A, C + 781T, C + 1633T, and A + 2767T polymorphisms with NPC risk were evaluated. Second, the NPC tissue samples were assessed for their IL-8 mRNA and protein expression by real-time quantitative reverse transcription polymerase chain reaction (PCR) and Western blotting, respectively. Regarding the IL-8 promoter T − 251A, the TA and AA genotypes were associated with significantly decreased risks of NPC compared with the wild-type TT genotype (adjusted odds ratio = 0.61 and 0.52, 95% confidence interval = 0.47–0.93 and 0.37–0.91, P = .0415 and .0289, respectively). The mRNA and protein expression levels for NPC tissues revealed no significant associations among the 20 NPC samples with different genotypes. These findings suggest that IL-8 may play an important role in the carcinogenesis of NPC in Taiwan.
Keywords: genotype, IL-8, nasopharyngeal cancer, polymorphism, Taiwan
1. Introduction
Nasopharyngeal carcinoma (NPC) is a relatively rare cancer in Western and most countries (age-standardized incidence rate [ASR] of <1/100,000), but its incidence rates are much higher in in southern China (ASR 30–50/100,000), Southeast Asia (ASR 9–12/100,000), and Taiwan (ASR 8.2–8.4/100,000).[1–3] This geographical pattern of NPC incidence suggests an interaction of complicated environmental and genetic factors. The epidemiologic factors that have been associated with increased risks of NPC included Epstein–Barr virus (EBV) infection,[4] tobacco smoking,[5–8] occupational exposure,[9] and unhealthy dietary habits.[10] Previous association studies have indicated that genetic susceptibility also plays an important role in the etiology of NPC.[11–15]
Interleukin-8 (IL-8) is produced by a wide variety of normal cells, including macrophages, epithelial cells, airway smooth muscle cells, endothelial cells, as well as tumor cells. It plays a critical role in the initiation and amplification of acute inflammatory reactions. IL-8 is a major mediator of inflammation, acting as a chemoattractant for neutrophils, basophils, and T cells.[16]IL-8 has been reported to overexpress in various human malignancies,[17–19] and in saliva of patients with oral cancer.[20] Additionally, elevated levels of IL-8 has been reported to correspond to an increased disease severity such as the metastatic potential of melanoma,[21] breast,[22] ovarian,[23] renal,[24] prostate,[25] pancreatic,[26] gastric,[27,28] and colorectal cancers.[29,30] Furthermore, IL-8 overexpression can cause disease progression of bladder cancer[31] and prostate cancer.[32] In the center of solid tumors under hypoxic microenvironments, IL-8 expression may help cancer cells to proliferate, survive, and escape programed cell deaths.[26] To sum up, IL-8 is closed involved in cancer development and progression.
IL-8 gene locates in 4q12-q13 of human genome, consisting of 4 exons.[33] The IL-8 single nucleotide polymorphisms (SNPs) at promoter region A − 251T (rs4073) and C + 781T (rs2227306) have been reported to affect IL-8 expression.[34–36] Previously studies have investigated the associations of IL-8 SNPs with the risks of many cancers including NPC.[37–41] However, the role of IL-8 polymorphisms in NPC ethology in Taiwanese population have not been reported. Thus, in the present study, we performed a case-control study to evaluate the impacts of IL-8 SNPs on the susceptibility of NPC in Taiwan.
2. Materials and methods
2.1. Study population
One hundred and seventy-six patients diagnosed with NPC were recruited at the general surgery outpatient clinics of the study hospital in Taichung, Taiwan, between 2003 and 2009. All patients participated voluntarily, completed a self-administered questionnaire, and provided peripheral blood samples. The questionnaire included questions on history and frequency of alcohol consumption, betel quid chewing, and smoking habits, and “ever” was defined as more than twice a week for at least 1 year. Self-reported alcohol consumption, betel quid chewing, and smoking habits were evaluated and classified as categorical variables.
For each case patient, 2 age- and gender-matched healthy controls, who had no NPC or other types of cancer, were selected from those attending the hospital for a health examination (age matching was done within less than 5 years of the case patient's first diagnosis). These volunteers attended the hospital for regular health assessments by multidisciplinary team approach with registered health practitioners during the years 2002 to 2012; most of the volunteers underwent health examinations every 5 to 6 months. Finally, 352 participants were included for analysis in the present study. The overall agreement rate in this study was more than 85% in collection. The study was approved by the institutional review board of the medical university hospital (DMR101-IRB1-306).
2.2. Genotyping protocols
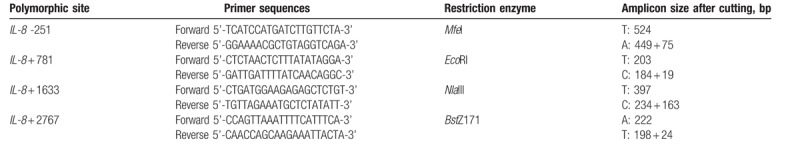
Genomic DNA from the peripheral blood leucocytes of each investigated subject was prepared using the QIAamp Blood Mini Kit (Qiagen, Valencia, CA), further stored in −80°C and subject to polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) methodology as previously described.[42–44] The PCR cycling conditions were: one cycle at 94°C for 5 minutes; 35 cycles of 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds; and a final extension at 72°C for 10 minutes. The sequences of forward and reverse primers and the restriction enzymes for the investigated SNP are summarized in Table 2. The genotype analysis was performed by 2 researchers independently and blindly. About 5% of the samples for each SNP were randomly selected for direct sequencing and the results from PCR-RFLP and direct sequencing were 100% concordant.
Table 2.
Summary of the primers, restriction enzymes and amplicon size after enzyme cutting for interleukin-8 genotyping PCR-RFLP conditions.
2.3. Interleukin-8 mRNA expression pattern
To evaluate the correlation between IL-8 mRNA expression and IL-8 polymorphism, 20 surgically removed NPC tissue samples obtained from sites adjacent to tumors with different genotypes were subjected to extraction of the total RNA using Trizol Reagent (Invitrogen, Carlsbad, CA). Total RNA was measured by real-time quantitative RT-PCR using an FTC-3000 real-time quantitative PCR instrument (Funglyn Biotech Inc., Canada). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal quantitative control. The primers used for amplification of IL-8 mRNA were forward 5’-AAACCACCGGAAGGAACCAT-3’ and reverse 5’-GCCAGCTTGGAAGTCATGT-3’,[45] while for GAPDH the primers were forward 5’-GAAATCCCATCACCATCTTCCAGG-3’ and reverse 5’-GAGCCCCAGCCTTCTCCATG-3’. Fold changes were normalized using the levels of GAPDH expression, and each assay was done at least in triplicate as previously published.[12,46]
2.4. Western blotting analysis
The NPC specimens were homogenized in radio immunoprecipitation assay (RIPA) lysis buffer (Upstate Biotechnology Inc., Lake Placid, NY), the homogenates were centrifuged at 10,000 X g for 30 minutes at 4oC, and the supernatants were used for Western blotting. Samples were denatured by heating at 95oC for 10 minutes, were separated on a 10% sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) gel, and were then transferred to a nitrocellulose membrane (BioRad Laboratories, Hercules, CA). The membrane was blocked with 5% non-fat milk and incubated over-night at 4oC with mouse monoclonal anti-human IL-8 antibody (1:1000; BD Transduction Laboratories; BD Biosciences, Franklin Lakes, NJ), and then with the corresponding horseradish peroxidase-conjugated goat anti-mouse IgG secondary antibody (Chemicon, Temecula, CA) for 1 hour at room temperature. After reaction with enhanced chemiluminescence (ECL) solution (Amersham, Arlington Heights, IL), bound antibody was visualized using a chemiluminescence imaging system (Syngene, Cambridge, UK). Finally, the blots were incubated at 56oC for 18 minutes in stripping buffer (0.0626 M Tris–HCl, pH 6.7, 2% SDS, 0.1 M mer-captoethanol) and re-probed with a mono-clonal mouse anti-β-actin antibody (Sigma, St. Louis, MO) as the loading control. The optical density of each specific band was measured using a computer-assisted imaging analysis system (GeneTools Match software; Syngene).
2.5. Statistical analyses
All 352 controls and 176 NPC cases with genotypic and clinical data were analyzed. To ensure that the controls used were representative of the general population and to exclude the possibility of genotyping error, the deviation of the genotype frequencies of IL-8 SNPs in the control subjects from those expected under the Hardy–Weinberg equilibrium was assessed using the goodness-of-fit test. Pearson's Chi-square test was used to compare the distribution of the IL-8 genotypes between cases and controls. The comparison of the age between control and case group was performed by Student's t test. The associations between the IL-8 polymorphisms and NPC risk were estimated by computing odds ratios (ORs) and their 95% confidence intervals (CIs) from logistic regression analysis with the adjustment for possible confounders. The STATA program was used for haplotype analysis. Any P < .05 was considered statistically significant.
3. Results
3.1. Comparisons of basic characters between the case and control groups
The frequency distributions of age, gender, personal behavioral habits for the 176 NPC patients, and 352 non-cancer controls are summarized in Table 1. The cases and controls were matched on age and gender. There were no significant differences between the cases and controls in the distributions of personal behavioral habits including smoking, alcohol drinking, and betel quid chewing status (Table 1).
Table 1.
Demographic characteristics of investigated 176 nasopharyngeal carcinoma patients and 352 non-cancer healthy controls.

3.2. Association analysis of IL-8 genotypes and NPC risk
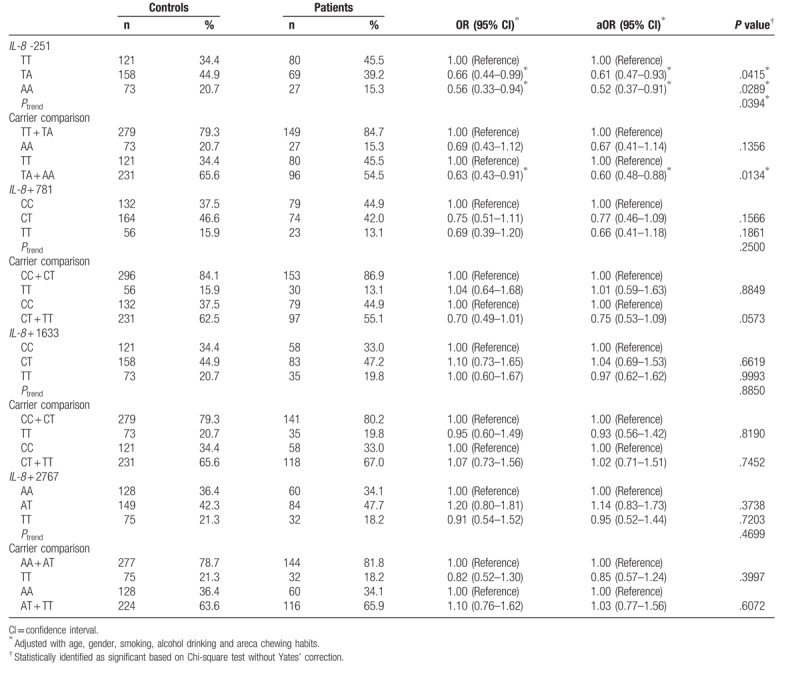
The distributions of the IL-8 promoter T − 251A, C + 781T, C + 1633T, and A + 2767T genotypes among the cases and controls are presented and statistically analyzed in Table 2. The genotypes of IL-8 promoter T − 251A SNP were differently distributed between cases and controls (P for trend = .0394) (Table 3 top panel). In detail, the IL-8 promoter T − 251A heterozygous TA and homozygous AA variant genotypes were associated with decreased NPC risks (OR = 0.66 and 0.56, 95% CI = 0.44–0.99 and 0.33–0.94, P = .0415 and .0289, respectively) (Table 3 top panel). In the dominant model, there was a significant association between the variant genotypes (TA + AA) and NPC risk (OR = 0.63, 95% CI = 0.43–0.91, P = .0134). The significant findings were still observed after adjusting for the potential confounders including age, gender, smoking, alcohol drinking, and areca chewing habits (Table 3 top panel). No significant associations were observed for the other 3 investigated SNPs Table 3.
Table 3.
Distribution of interleukin-8 genotypes among the nasopharyngeal carcinoma patients and non-cancer healthy control subjects.
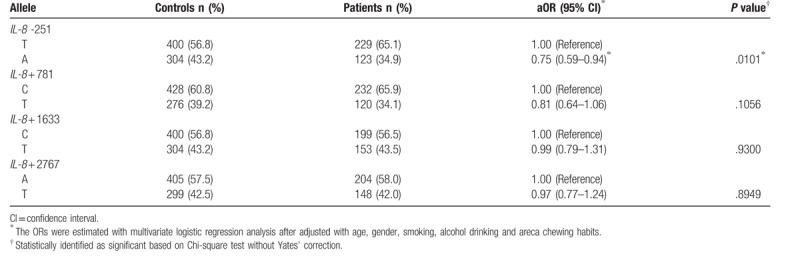
We also performed allelic analysis. Supporting the findings in Table 3, the results showed that the variant allele A was 34.9% in the NPC patient group, significantly much lower than that (43.2%) in the control group (adjusted OR = 0.75, 95%CI = 0.59–0.94, P = .0101). Again, the other 3 SNPs were not significantly associated with NPC risks (Table 4).
Table 4.
Allelic frequency analysis for interleukin-8 (IL-8) polymorphisms and nasopharyngeal carcinoma.
3.3. Stratified analysis of IL-8 genotypes by environmental factors
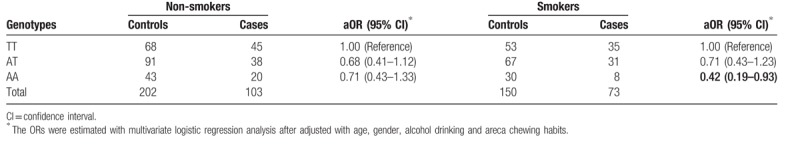
We then performed stratified analyses of IL-8 genotypes with NPC risks by potential environmental risk factors, including cigarette smoking, alcohol drinking, and areca chewing habits. The adjusted ORs for carriers with genotype of TA or AA at IL-8 promoter T − 251A were 0.68 and 0.71 among non-smokers (95% CI = 0.41–1.12 and 0.43–1.33, respectively), and were 0.71 and 0.42 among smokers (95% CI = 0.43–1.23 and 0.19–0.93, respectively) (Table 5). The interaction analysis did not show a significant interaction. Likewise, there was no significant interaction between IL-8 T − 251A genotypes and alcohol drinking or areca chewing habits in modulating NPC risks (data not shown).
Table 5.
Odds ratios for interleukin-8 (IL-8) promoter -251 genotype and nasopharyngeal carcinoma after stratified by smoking status.
3.4. Correlation between IL-8 T–251A genotype and the expression levels of IL-8 mRNA and proteins
Finally, 20 surgically removed NPC tissue samples were collected from sites adjacent to tumors for this analysis. Among these tissues, 10 were of IL-8 T − 251A genotype TT, 8 were of TA, and 2 were of AA. The mRNA expression levels of IL-8 in these patients were examined by real-time quantitative RT-PCR (Fig. 1). The levels of IL-8 mRNA for the TA and AA genotypes were 0.85- and 0.81-fold compared with those of the TT genotype. Combining TA and AA genotypes and compared to TT genotype, there was no significant difference in the mRNA levels of IL-8 (P = .4253). We also examined the IL-8 protein expression levels at the tumor sites of the same NPC patients by Western blotting (Fig. 2A) and did not find significant difference of IL-8 protein expression among different IL-8 T − 251A genotypes either (P = .2197) (Fig. 2B).
Figure 1.
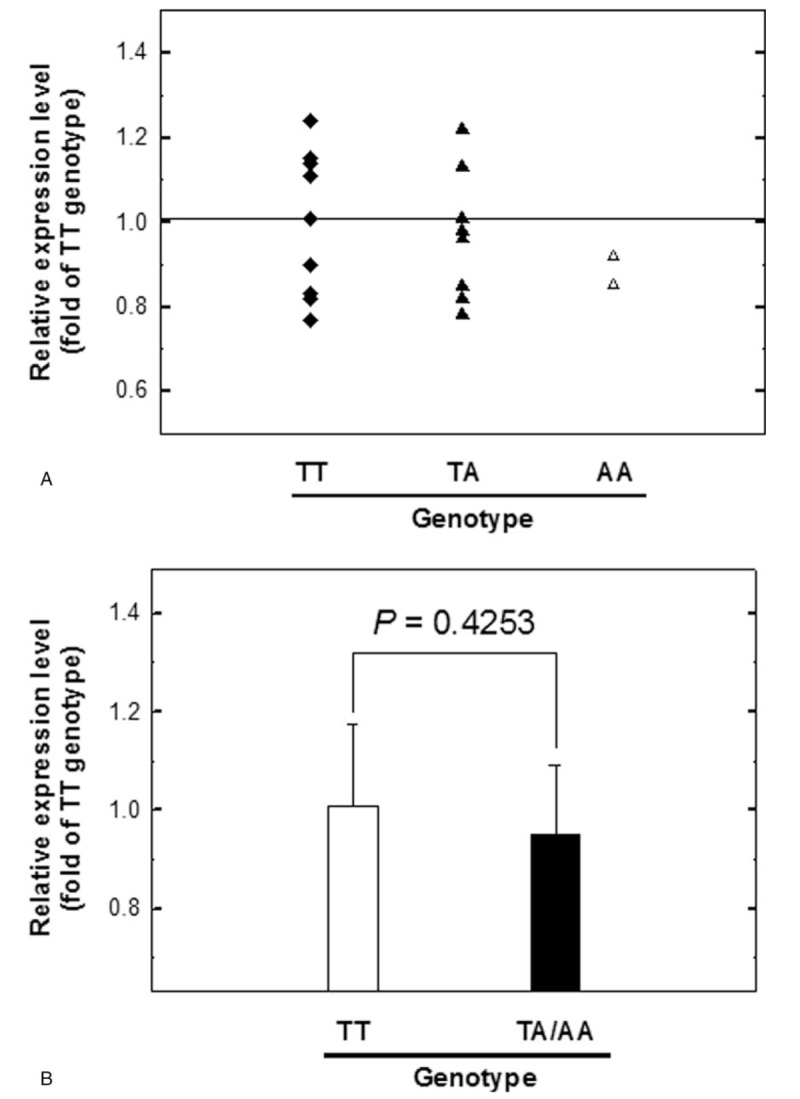
Analysis of IL-8 mRNA expression levels among nasopharyngeal carcinoma (NPC) patients. (A) Quantitative RT-PCR of NPC tissue samples for the three genotypes of IL-8 promoter T–251A was performed. GAPDH was used as an internal quantitative control. Fold changes were normalized using the levels of GAPDH expression, and each assay was performed at least in triplicate. (B) The TA and AA groups were combined and compared with the TT group. GAPDH = glyceraldehyde.
Figure 2.
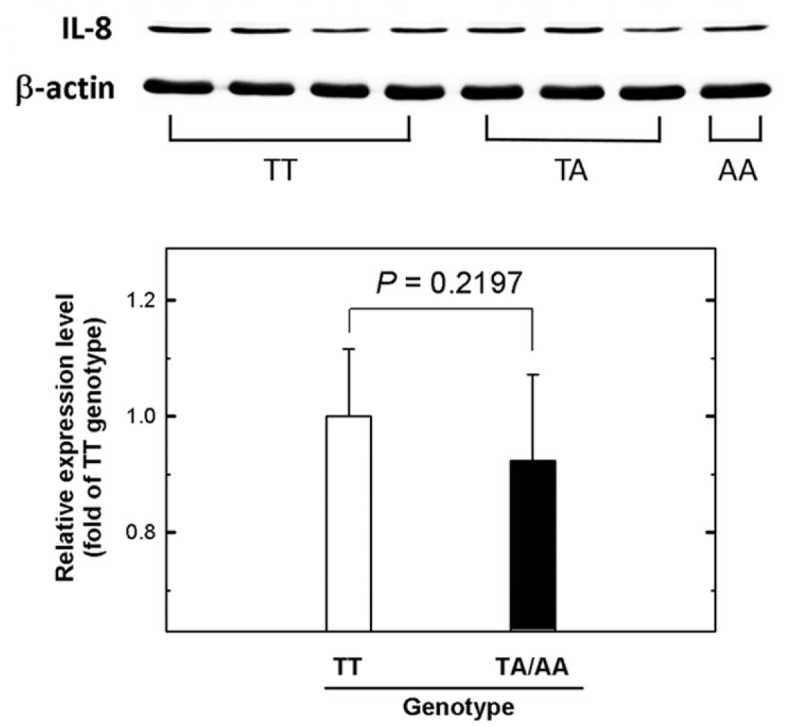
The expression levels of IL-8 in nasopharyngeal carcinoma (NPC) tissues from patients with different IL-8 promoter T–251A genotypes. (A) Western blotting analysis of IL-8 expression in tumor tissues from cases with TT, TA, and AA IL-8 promoter T–251A genotypes. (B) Quantification of the Western blotting data from (A). β-actin was used as the loading control. Data were averaged from at least 3 repeat analyses of the tissues of each group, with 15 μg total sample protein for each lane.
3.5. Interaction of IL-8 T–251A genotype and the EBV infection status on NPC risk
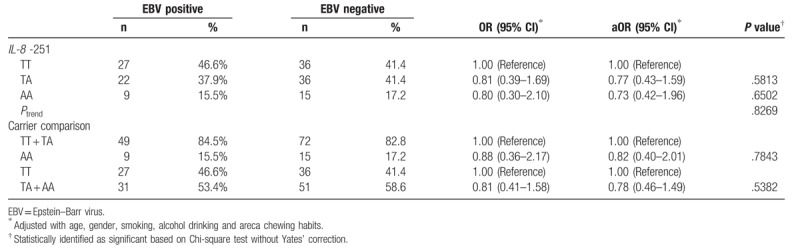
EBV infection was reported to be associated with NPC development and clinical outcomes in Taiwan.[47–50] However, the early predictive rates were of very wide range in accuracy according to the detection methodology in the targets including LMP-1, EBNA-1, EBNA-2, which are still in development.[47,48,51] In our investigated population, 145 of 176 NPC patients have the complete records in their detectable plasma EBV DNA, and the distributions of their TT, AT, AA genotypes at IL-8 − 251 were 27 (46.6%), 22 (37.9%), 9 (15.5%) in EBV positive NPC patients and 36 (41.4%), 36 (41.4%), 15 (17.2%) in EBV negative NPC patients. The results showed that there was not a positive interaction between IL-8 − 251 genotypes and EBV infection (P = .8269). The control subjects were lacking of their EBV infection status that the effects of EBV infection on IL-8 − 251 genotypes as for early prediction of NPC could not be evaluated in this study (Table 6).
Table 6.
Distribution of interleukin-8 genotypes among the EBV positive and negative nasopharyngeal carcinoma patients.
3.6. Haplotype of IL-8 genotypes and stratified analysis by environmental factors
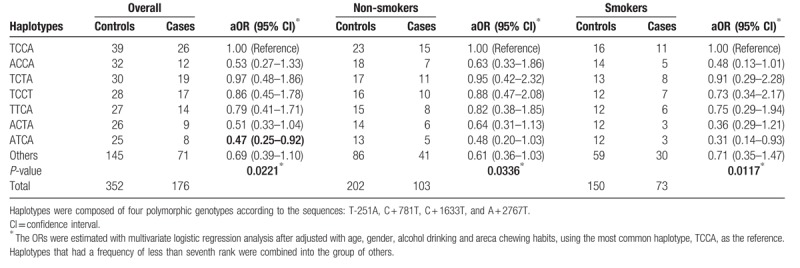
We have performed the IL-8 T − 251A-C + 781T-C + 1633T-A + 2767T haplotype analysis, finding that the haplotypes of IL-8 T − 251A-C + 781T-C + 1633T-A + 2767T were differentially distributed between case and control groups (P = .0221). Among the haplotypes for IL-8 T − 251A-C + 781T-C + 1633T-A + 2767T, the distributions of ATCA haplotype were significantly of lower percentage for the cases than the controls (adjusted OR = 0.47, 95% CI = 0.25–0.92). Furthermore, the haplotypes of IL-8 T − 251A-C + 781T-C + 1633T-A + 2767T were differentially distributed among the subgroups of non-smokers (P = .0336) and smokers (P = .0117). Most interesting, the smokers of ATCA haplotype for IL-8 T − 251A-C + 781T-C + 1633T-A + 2767T were of significantly lower risk of NPC (adjusted OR = 0.31, 95% CI = 0.14–0.93) (Table 7). There were no any significant differences in the cases while analyzing the interaction between status of alcohol drinking, betel quid chewing, and IL-8 T − 251A-C + 781T-C + 1633T-A + 2767T haplotypes (data not shown). The protective effects of IL-8 − 251A and + 781T seemed to be additive for those carrying ATCA haplotype, especially among those smokers.
Table 7.
Distribution of interleukin-8 (IL-8) haplotypes among nasopharyngeal carcinoma patients and control subjects after stratified by smoking status.
4. Discussion
In the current study, the role of IL-8 in NPC was evaluated from the levels of DNA, RNA, and protein. The contributions of IL-8 promoter T − 251A, C + 781T, C + 1633T, and A + 2767T SNPs to NPC risk were evaluated and the results showed that the TA and AA genotypes of T − 251A were significantly associated with the risks of NPC. This study is the first to analyze the association between IL-8 T − 251A genotype and NPC susceptibility.
It has been reported that chronic smoking and EBV infection contributed to the etiology of NPC development and decreased the survival rates of the patients,[52–54] but the detailed mechanisms are not clear. In this study, we found that IL-8 promoter T − 251A genotypes were associated to NPC risk, suggesting that IL-8 may mediate the effect of infection and inflammation on NPC development. The IL-8 promoter T − 251A SNP has been studied extensively previously in relation to cancer risk and the results were heterogeneous.[37] Four studies have been published with regard to this SNP and risks of NPC, 1 in African population,[40] 2 in Chinese population,[38,39] and 1 in Europeans population.[41] The meta-analysis of these 4 studies showed that the variant genotypes were associated with increased risks of NPC. The discrepancy of our results to the previous publications may be attributed to different populations, different exposures, or the relative small sample sizes of all these publications. Future large validation studies are needed to clarify the association of IL-8 − 251A/T SNP with NPC risk in different populations.
One limitation of this study is that we defined ever smokers as those who smoked more than twice a week for at least 1 year. This is not a traditional definition, which may have obscured the true association of smoking and NPC risk and resulted in the lack of interaction between IL-8 genotypes and smoking in elevating NPC risk. Future larger studies with detailed smoking information are warranted to clarify the interaction between IL-8 genotypes and smoking status in modulating NPC risk.
The transcriptional and translational impacts of different genotypes at IL-8 promoter T − 251A were investigated in this current study but no correlation between genotypes and gene expression was found. Also, the serum level of IL-8 was detected in 20 NPC patients, and the levels were not differentially distributed among patients of IL-8 − 251 TT, TA, and AA genotypes, similar to those at mRNA and protein levels. Further, we have stratified them according to the EBV infection, finding that there was neither correlation between IL-8 − 251 genotypes and IL-8 serum level nor no interaction between EBV infection status with IL-8 − 251 genotypes on IL-8 serum level. Only 20 samples from NPC patients may have limited our power to find significant correlations between genotypes and gene expression. In addition, the tissue samples from normal subjects were not available for analysis. Further investigations of IL-8 mRNA and/or protein expression in relation to genotypes are warranted. In addition, the enlargement of the sample size is encouraged in the future and may alter the current conclusion.
In conclusion, our study provided evidence that the TA and AA genotypes at IL-8 promoter T − 251A SNP are associated with decreased risks of NPC in Taiwan, supporting a role of inflammation in the etiology of NPC. Importantly, the novel genomic biomarkers can add to the traditional methodology depending on the EBV infection in NPC risk and prognosis outcome prediction. It would be valuable to investigate additional SNPs in other inflammatory mediators followed by mechanistic study to understand the roles of inflammation in NPC pathogenesis.
Author contributions
Conceptualization: Chung-Yu Huang, Te-Chun Shen, Da-Tian Bau, Hao-Ai Shui.
Funding acquisition: Te-Chun Hsia, Da-Tian Bau.
Investigation: Chung-Yu Huang, Wen-Shin Chang, Chia-Wen Tsai, Te-Chun Shen, Da-Tian Bau, Hao-Ai Shui.
Methodology: Chung-Yu Huang, Wen-Shin Chang, Chia-Wen Tsai, Te-Chun Shen, Da-Tian Bau, Hao-Ai Shui.
Resources: Te-Chun Hsia, Da-Tian Bau.
Writing – original draft: Chung-Yu Huang, Te-Chun Shen, Da-Tian Bau, Hao-Ai Shui.
Te-Chun Hsia orcid: 0000-0002-9427-1068
Footnotes
Abbreviations: ASR = age-standardized incidence rate, CI = confidence interval, EBV = Epstein–Barr virus, ECL = enhanced chemiluminescence, GAPDH = glyceraldehyde 3-phosphate dehydrogenase, IL-8 = interleukin-8, NPC = nasopharyngeal carcinoma, OR = odds ratios, PCR-RFLP = polymerase chain reaction-restriction fragment length polymorphism, RIPA = radio immunoprecipitation assay, SDS-PAGE = sodium dodecyl sulphate polyacrylamide gel electrophoresis, SNP = single nucleotide polymorphism.
T-CS, D-TB, and H-AS contributed equally to this work.
This study was supported mainly by grants from the Taiwan Ministry of Science and Technology to Professor Bau (MOST-106-2320-B-039-035), from Taoyuan Armed Forces General Hospital to Dr Huang (ATTYGH-10628 and ATTYGH-10738) and partially from Taiwan Ministry of Health and Welfare Clinical Trial and Research Center of Excellence (MOHW107-TDU-B-212-123004).
The authors report no conflicts of interest.
References
- [1].Cao SM, Simons MJ, Qian CN. The prevalence and prevention of nasopharyngeal carcinoma in China. Chin J Cancer 2011;30:114–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Chan AT, Teo PM, Johnson PJ. Nasopharyngeal carcinoma. Ann Oncol 2002;13:1007–15. [DOI] [PubMed] [Google Scholar]
- [3].Yu MC, Yuan JM. Epidemiology of nasopharyngeal carcinoma. Semin Cancer Biol 2002;12:421–9. [DOI] [PubMed] [Google Scholar]
- [4].Zong YS, Sham JS, Ng MH, et al. Immunoglobulin A against viral capsid antigen of Epstein-Barr virus and indirect mirror examination of the nasopharynx in the detection of asymptomatic nasopharyngeal carcinoma. Cancer 1992;69:3–7. [DOI] [PubMed] [Google Scholar]
- [5].Xue WQ, Qin HD, Ruan HL, et al. Quantitative association of tobacco smoking with the risk of nasopharyngeal carcinoma: a comprehensive meta-analysis of studies conducted between 1979 and 2011. Am J Epidemiol 2013;178:325–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hsu WL, Chen JY, Chien YC, et al. Independent effect of EBV and cigarette smoking on nasopharyngeal carcinoma: a 20-year follow-up study on 9,622 males without family history in Taiwan. Cancer Epidemiol Biomarkers Prev 2009;18:1218–26. [DOI] [PubMed] [Google Scholar]
- [7].Chen CJ, Liang KY, Chang YS, et al. Multiple risk factors of nasopharyngeal carcinoma: Epstein-Barr virus, malarial infection, cigarette smoking and familial tendency. Anticancer Res 1990;10:547–53. [PubMed] [Google Scholar]
- [8].Cheng YJ, Hildesheim A, Hsu MM, et al. Cigarette smoking, alcohol consumption and risk of nasopharyngeal carcinoma in Taiwan. Cancer Causes Control 1999;10:201–7. [DOI] [PubMed] [Google Scholar]
- [9].Mirabelli MC, Hoppin JA, Tolbert PE, et al. Occupational exposure to chlorophenol and the risk of nasal and nasopharyngeal cancers among U.S. men aged 30 to 60. Am J Ind Med 2000;37:532–41. [DOI] [PubMed] [Google Scholar]
- [10].Armstrong RW, Imrey PB, Lye MS, et al. Nasopharyngeal carcinoma in Malaysian Chinese: salted fish and other dietary exposures. Int J Cancer 1998;77:228–35. [DOI] [PubMed] [Google Scholar]
- [11].Tsai CW, Chang WS, Gong CL, et al. Contribution of matrix metallopeptidase-1 genotypes, smoking, alcohol drinking and areca chewing to nasopharyngeal carcinoma susceptibility. Anticancer Res 2016;36:3335–40. [PubMed] [Google Scholar]
- [12].Huang CY, Tsai CW, Hsu CM, et al. The role of XRCC6/Ku70 in nasopharyngeal carcinoma. Int J Oral Maxillofac Surg 2015;44:1480–5. [DOI] [PubMed] [Google Scholar]
- [13].Tsai CW, Tsai MH, Shih LC, et al. Association of interleukin-10 (IL10) promoter genotypes with nasopharyngeal carcinoma risk in Taiwan. Anticancer Res 2013;33:3391–6. [PubMed] [Google Scholar]
- [14].Shih LC, Tsai CW, Tsai MH, et al. Association of cyclin D1 genotypes with nasopharyngeal carcinoma risk. Anticancer Res 2012;32:1093–8. [PubMed] [Google Scholar]
- [15].Tsou YA, Tsai CW, Tsai MH, et al. Association of caveolin-1 genotypes with nasopharyngeal carcinoma susceptibility in Taiwan. Anticancer Res 2011;31:3629–32. [PubMed] [Google Scholar]
- [16].Rizzo A, Losacco A, Carratelli CR. Lactobacillus crispatus modulates epithelial cell defense against Candida albicans through Toll-like receptors 2 and 4, interleukin 8 and human beta-defensins 2 and 3. Immunol Lett 2013;156:102–9. [DOI] [PubMed] [Google Scholar]
- [17].Gunter MJ, Canzian F, Landi S, et al. Inflammation-related gene polymorphisms and colorectal adenoma. Cancer Epidemiol Biomarkers Prev 2006;15:1126–31. [DOI] [PubMed] [Google Scholar]
- [18].Lerebours F, Vacher S, Andrieu C, et al. NF-kappa B genes have a major role in inflammatory breast cancer. BMC Cancer 2008;8:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zabaleta J, Su LJ, Lin HY, et al. Cytokine genetic polymorphisms and prostate cancer aggressiveness. Carcinogenesis 2009;30:1358–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Nagler RM. Saliva as a tool for oral cancer diagnosis and prognosis. Oral Oncol 2009;45:1006–10. [DOI] [PubMed] [Google Scholar]
- [21].Huang S, Mills L, Mian B, et al. Fully humanized neutralizing antibodies to interleukin-8 (ABX-IL8) inhibit angiogenesis, tumor growth, and metastasis of human melanoma. Am J Pathol 2002;161:125–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Miller LJ, Kurtzman SH, Wang Y, et al. Expression of interleukin-8 receptors on tumor cells and vascular endothelial cells in human breast cancer tissue. Anticancer Res 1998;18:77–81. [PubMed] [Google Scholar]
- [23].Venkatakrishnan G, Salgia R, Groopman JE. Chemokine receptors CXCR-1/2 activate mitogen-activated protein kinase via the epidermal growth factor receptor in ovarian cancer cells. J Biol Chem 2000;275:6868–75. [DOI] [PubMed] [Google Scholar]
- [24].Slaton JW, Inoue K, Perrotte P, et al. Expression levels of genes that regulate metastasis and angiogenesis correlate with advanced pathological stage of renal cell carcinoma. Am J Pathol 2001;158:735–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Uehara H, Troncoso P, Johnston D, et al. Expression of interleukin-8 gene in radical prostatectomy specimens is associated with advanced pathologic stage. Prostate 2005;64:40–9. [DOI] [PubMed] [Google Scholar]
- [26].Shi Q, Abbruzzese JL, Huang S, et al. Constitutive and inducible interleukin 8 expression by hypoxia and acidosis renders human pancreatic cancer cells more tumorigenic and metastatic. Clin Cancer Res 1999;5:3711–21. [PubMed] [Google Scholar]
- [27].Zhang XY, Chan WY, Whitney BM, et al. Changes of interleukin expression correlate with Helicobacter pylori infection and lymph node metastases in gastric carcinoma. Diagn Mol Pathol 2002;11:135–9. [DOI] [PubMed] [Google Scholar]
- [28].Lee KH, Bae SH, Lee JL, et al. Relationship between urokinase-type plasminogen receptor, interleukin-8 gene expression and clinicopathological features in gastric cancer. Oncology 2004;66:210–7. [DOI] [PubMed] [Google Scholar]
- [29].Li A, Varney ML, Singh RK. Expression of interleukin 8 and its receptors in human colon carcinoma cells with different metastatic potentials. Clin Cancer Res 2001;7:3298–304. [PubMed] [Google Scholar]
- [30].Chung YC, Chang YF. Significance of inflammatory cytokines in the progression of colorectal cancer. Hepatogastroenterology 2003;50:1910–3. [PubMed] [Google Scholar]
- [31].Sheryka E, Wheeler MA, Hausladen DA, et al. Urinary interleukin-8 levels are elevated in subjects with transitional cell carcinoma. Urology 2003;62:162–6. [DOI] [PubMed] [Google Scholar]
- [32].Kim SJ, Uehara H, Karashima T, et al. Expression of interleukin-8 correlates with angiogenesis, tumorigenicity, and metastasis of human prostate cancer cells implanted orthotopically in nude mice. Neoplasia 2001;3:33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Mukaida N, Shiroo M, Matsushima K. Genomic structure of the human monocyte-derived neutrophil chemotactic factor IL-8. J Immunol 1989;143:1366–71. [PubMed] [Google Scholar]
- [34].Fey MF, Tobler A. An interleukin-8 (IL-8) cDNA clone identifies a frequent HindIII polymorphism. Hum Genet 1993;91:298. [DOI] [PubMed] [Google Scholar]
- [35].Hacking D, Knight JC, Rockett K, et al. Increased in vivo transcription of an IL-8 haplotype associated with respiratory syncytial virus disease-susceptibility. Genes Immun 2004;5:274–82. [DOI] [PubMed] [Google Scholar]
- [36].Ohyauchi M, Imatani A, Yonechi M, et al. The polymorphism interleukin 8-251 A/T influences the susceptibility of Helicobacter pylori related gastric diseases in the Japanese population. Gut 2005;54:330–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wang Z, Liu Y, Yang L, et al. The polymorphism interleukin-8 -251A/T is associated with a significantly increased risk of cancers from a meta-analysis. Tumour Biol 2014;35:7115–23. [DOI] [PubMed] [Google Scholar]
- [38].Tai SH, Wei YS, Wang P, et al. Genetic polymorphisms of interferon-gamma and interleukin-8 in patients with nasopharyngeal carcinoma. Med J Sichuan Univ 2007;38:862–5. [PubMed] [Google Scholar]
- [39].Wei YS, Lan Y, Tang RG, et al. Single nucleotide polymorphism and haplotype association of the interleukin-8 gene with nasopharyngeal carcinoma. Clin Immunol 2007;125:309–17. [DOI] [PubMed] [Google Scholar]
- [40].Ben Nasr H, Chahed K, Mestiri S, et al. Association of IL-8 (-251)T/A polymorphism with susceptibility to and aggressiveness of nasopharyngeal carcinoma. Hum Immunol 2007;68:761–9. [DOI] [PubMed] [Google Scholar]
- [41].Campa D, Hashibe M, Zaridze D, et al. Association of common polymorphisms in inflammatory genes with risk of developing cancers of the upper aerodigestive tract. Cancer Causes Control 2007;18:449–55. [DOI] [PubMed] [Google Scholar]
- [42].Chang WS, Yueh TC, Tsai CW, et al. Contribution of DNA repair xeroderma pigmentosum group D genotypes to colorectal cancer risk in Taiwan. Anticancer Res 2016;36:1657–63. [PubMed] [Google Scholar]
- [43].Lai CY, Chang WS, Hsieh YH, et al. Association of tissue inhibitor of metalloproteinase-1 genotypes with lung cancer risk in Taiwan. Anticancer Res 2016;36:155–60. [PubMed] [Google Scholar]
- [44].Pei JS, Chang WS, Hsu PC, et al. The association of flap endonuclease 1 genotypes with the risk of childhood leukemia. Cancer Genomics Proteomics 2016;13:69–74. [PubMed] [Google Scholar]
- [45].Huang FC. Differential regulation of interleukin-8 and human beta-defensin 2 in Pseudomonas aeruginosa-infected intestinal epithelial cells. BMC Microbiol 2014;14:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Hsu CM, Chang WS, Hwang JJ, et al. The role of apurinic/apyrimidinic endonuclease DNA repair gene in endometriosis. Cancer Genomics Proteomics 2014;11:295–301. [PubMed] [Google Scholar]
- [47].Lin JC, Chen KY, Wang WY, et al. Detection of Epstein-Barr virus DNA in the peripheral-blood cells of patients with nasopharyngeal carcinoma: relationship to distant metastasis and survival. J Clin Oncol 2001;19:2607–15. [DOI] [PubMed] [Google Scholar]
- [48].Wang WY, Lin TY, Twu CW, et al. Long-term clinical outcome in nasopharyngeal carcinoma patients with post-radiation persistently detectable plasma EBV DNA. Oncotarget 2016;7:42608–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Chen CY, Lin YS, Chen CL, et al. Targeting annexin A2 reduces tumorigenesis and therapeutic resistance of nasopharyngeal carcinoma. Oncotarget 2015;6:26946–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Chen CY, Lin YS, Chen CH, et al. Annexin A2-mediated cancer progression and therapeutic resistance in nasopharyngeal carcinoma. J Biomed Sci 2018;25:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Lao TD, Nguyen DH, Nguyen TM, et al. Molecular screening for Epstein-Barr virus (EBV): detection of genomic EBNA-1, EBNA-2, LMP-1, LMP-2 among Vietnamese patients with nasopharyngeal brush samples. Asian Pac J Cancer Prev 2017;18:1675–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lv JW, Chen YP, Zhou GQ, et al. Cigarette smoking complements the prognostic value of baseline plasma Epstein-Barr virus deoxyribonucleic acid in patients with nasopharyngeal carcinoma undergoing intensity-modulated radiation therapy: a large-scale retrospective cohort study. Oncotarget 2016;7:16806–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Ouyang PY, Su Z, Mao YP, et al. Prognostic impact of cigarette smoking on the survival of patients with established nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers Prev 2013;22:2285–94. [DOI] [PubMed] [Google Scholar]
- [54].Xu FH, Xiong D, Xu YF, et al. An epidemiological and molecular study of the relationship between smoking, risk of nasopharyngeal carcinoma, and Epstein-Barr virus activation. J Natl Cancer Inst 2012;104:1396–410. [DOI] [PubMed] [Google Scholar]


