Abstract
This study aimed to assess the effect of long noncoding RNAs (lncRNAs) taurine-upregulated gene 1 (TUG1) on cells proliferation and apoptosis as well as its targeting genes in epithelial ovarian cancer (EOC) cells.
Blank mimic, lncRNA TUG1 mimic, blank inhibitor, and lncRNA TUG1 inhibitor plasmids were transfected into SK-OV-3 (SKOV3) cells. Rescue experiment was performed by the transfection of lncRNA TUG1 inhibitor and Aurora kinase A (AURKA) mimic plasmids into SKOV3 cells. Cell counting kit-8 (CKK-8), annexin V-FITC (AV)-propidium iodide (PI) (AV-PI), quantitative polymerase chain reaction (qPCR), and western blot assays were performed to detect cells proliferation, apoptosis, RNA expression, and protein expression respectively.
Cells proliferation was increased in lncRNA TUG1 mimic group and decreased in lncRNA TUG1 inhibitor group than normal control (NC) groups. Cells apoptosis rate was repressed after treatment with lncRNA TUG1 mimic and promoted after treatment with lncRNA TUG1 inhibitor. AURKA expression but not CLDN3, SERPINE1, or ETS1 expression was adversely regulated by lncRNA TUG1 mimic and inhibitor. After transferring lncRNA TUG1 (−) and AURKA (+) plasmids, cells proliferation was increased, while cells apoptosis rate was decreased in AURKA mimic (+)/lncRNA TUG1 inhibitor (−) group than NC (+)/lncRNA TUG1 (−) group, which suggested lncRNA TUG1 regulated cells proliferation and cells apoptosis through targeting AURKA.
LncRNA TUG1 promotes cells proliferation and inhibits cells apoptosis through regulating AURKA in EOC cells.
Keywords: AURKA, cells apoptosis, cells proliferation, epithelial ovarian cancer (EOC), LncRNA TUG1
1. Introduction
Epithelial ovarian cancer (EOC), one of the most deadly gynecological malignancies, affects approximately 239,000 populations and causes estimated 151,900 deaths during 2012 according to 2015 global cancer statistics, accounting for about 5% of all cancer and 4.2% of all cancer deaths among female worldwide.[1,2] Also, estimated 52,100 EOC new cases and 22,500 deaths occur in China during 2015.[3] Although there are great improvement of early diagnosis and optimal treatment, an increasing trend in mortality caused by EOC still has been reported in the last 2 decades due to several factors, such as chemoresistance or complexed pathological processes.[4] Therefore, exploring molecular mechanism underlying EOC progression may help improve the diagnosis and develop novel therapeutic targets.
Long noncoding RNAs (lncRNAs) are a new class of noncoding RNAs (> 200 nucleotides) with limited or no protein-coding capacity, which involve in several biological processes, including cells proliferation, apoptosis, invasion, and differentiation by targeting gene expression.[5,6] The dysregulated expressions of lncRNAs have been reported to contribute to the processes of tumorigenesis and cancer progression.[7] LncRNA taurine-upregulated gene 1 (TUG1), located on chromosome 22q12, is a common lncRNA expressed in human tissue and cancer cells.[8] Accumulating evidences have proven that lncRNA TUG1 plays an oncogenic role in various carcinomas, including breast cancer, gastric cancer, and colorectal cancer.[9–11] However, few studies have been performed to explore the function of lncRNA TUG1 and its potential mechanism in EOC cells. In the early study, we have evaluated the correlation of lncRNAs TUG1 with clinicopathological characteristics as well as overall survival (OS) in EOC patients, and investigate its function in EOC cells proliferation and apoptosis in vitro.[12] In order to further explore the clear molecular mechanism of lncRNA TUG1 underlying EOC progression, following study is necessary to be conducted. Therefore, the purpose of this study was to assess the effect of lncRNA TUG1 on cells proliferation and apoptosis as well as its targeting genes in EOC cells.
2. Methods
2.1. Cells culture
Human EOC cells (SKOV3) were obtained from Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 (Sigma-Aldrich, Whitehouse Station, New Jersey) medium supplemented with 10% fetal bovine serum (FBS) (Gibco, Carlsbad, California), 100 U/mL penicillin, and 100 mg/mL streptomycin at 37°C and 5% CO2 in a humidified incubator after resuscitation. Institutional Review Board of People's Hospital of Lishui City, the Sixth Affiliated Hospital of Wenzhou Medical University approved this study.
2.2. Plasmids transfection and assays after transfection
Cells were washed with PBS and then transfected with blank mimic, lncRNA TUG1 mimic, blank inhibitor, and lncRNA TUG1 inhibitor plasmids, which were divided into 4 groups, NC1 mimic, lncRNA TUG1 mimic, NC2 inhibitor, and lncRNA TUG1 inhibitor groups. The following assays were carried out: quantitative polymerase chain reaction (qPCR) assay for lncRNA TUG1 expression at 24 hours; cell counting kit-8 (CKK8) assay for assessment of cells proliferation at 0, 24, and 48 hours; annexin V-FITC (AV) -propidium iodide (PI) assay and C-Caspase 3 expression as well as Bcl-2 expression by western blot assay for evaluation of cells apoptosis at 48 hours.
Moreover, qPCR and western blot assays were used for the detection of mRNA and protein expression of candidate target genes, including Aurora kinase A (AURKA), Claudin 3 (CLDN3), plasminogen activator inhibitor 1 (SERPINE1), and E26 transformation-specific 1 (ETS1). Candidate target genes of lncRNA TUG1 in EOC were evaluated: First, RIsearch, RNAplex, and LncTar databases were used for the prediction of target mRNAs of lncRNA TUG1, and target mRNAs with consistent results by these 3 database predicting were selected.[13–15] Subsequently, DisGenet (www.disgenet.org) was used for the evaluation of genes, which were associated with the pathological processes of EOC.[16] After that, potential target genes, including AURKA, CLDN3, SERPINE1, and ETS1, in EOC were evaluated by combining analysis of lncRNA TUG1 predicted target genes and EOC-related genes.
2.3. Rescue experiments
Subsequently, the rescue experiment of AURKA for lncRNA TUG1 in EOC cells was carried out. Cells were transfected with normal control (NC) mimic, AURKA mimic, lncRNA TUG1 inhibitor, and AURKA mimic and lncRNA TUG1 inhibitor plasmids, which were divided into 4 groups, including NC mimic (+)/NC inhibitor (−), AURKA (+)/NC (−), NC (+)/LncRNA TUG1 (−), and AURKA (+)/LncRNA TUG1 (−) groups. The following assays were performed: qPCR assay for lncRNA TUG1 expression at 24 hours; qPCR assay and western blot assay for mRNA and protein expression of AURKA at 24 hours; CKK8 assays at 0, 24, and 48 hours; and AV/PI assay and western blot (C-Caspase 3, Caspase 3, and Bcl-2 proteins) for evaluation of cells apoptosis at 48 hours.
2.4. qPCR
lncRNA and mRNA expressions were evaluated by qPCR. The TRIzol reagent (Invitrogen, Waltham, Massachusetts) was used for the extraction of total RNA. Nonodrop (Thermo Fisher, Wilmington, Massachusetts) was used for quantitation of RNA expression based on the manufacturer's instruction. Subsequently, RNA from each group was used for cDNA synthesis, which was subsequently subjected to qPCR with SYBR Green kit (Takara, Japan). The PCR amplification was subsequently performed: 95°C for 5 minutes, followed by 40 cycles of 95°C for 5 seconds, 61°C for 30 seconds. GAPDH (mRNAs) or U6 (lncRNAs) was served as the internal reference, and 2−ΔΔct method was used for the calculation of qPCR results. The primer sequences were as follows: lncRNA TUG1: Forward 5′ TAGGAGTGGATGTGTTCTGTAGCA 3′ Reverse 5′ TGGTCGTGGAATATGGTCAATGAG 3′; U6: Forward 5′ CTCGCTTCGGCAGCACATA3′ Reverse 5′CTCGCTTCGGCAGCACATA3′; AURKA: Forward 5′ CTGAGGAGGAACTGGCATCAA3′ Reverse 5′ ATTAGGTAGACTCTGGTAGCATCAT3′; CLDN3: Forward 5′ TTCATCGGCAGCAACATCATCA3′ Reverse 5′ AGCAGCGAGTCGTACACCTT3′; SERPINEL: Forward 5′ ACGGCTGGTGCTGGTGAAT3′ Reverse 5′ GCAGTTCCAGGATGTCGTAGTAAT3′; EST1: Forward 5′ TTGCCACCTGAACTTCTTCCT3′ Reverse 5′ GGATGACCAGCCACCATTAGA3′; GAPHD: Forward 5′ GAGTCCACTGGCGTCTTCAC3′ Reverse 5′ ATCTTGAGGCTGTTGTCATACTTCT3′.
2.5. Western blot assay
Total protein was extracted from lysing cells in 1-mL RIPA buffer (Thermo Fisher Scientific, Waltham, Massachusetts). The protein concentration was measured by the bicinchoninic acid (BCA) kit (Thermo Fisher, Wilmington, Massachusetts) (Pierce Biotechnology) and adjudged according to the standard curve. Subsequently, 20 μg protein sample was fractionated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), which was then transferred to polyvinylidene fluoride membranes (Millipore, Bedford, Massachusetts). Protein was blocked by 5% skim milk. After 2 hours, the primary antibody was used for incubation of membranes overnight at 4°C, and then secondary antibody was used for incubation for 1 hour at room temperature. Blots were exposed to x-ray films for chemiluminescence following treatment using an enhanced chemiluminescence (ECL) kit (Millipore). GAPDH was served as the internal reference. The detailed information of antibodies is presented in Table 1.
Table 1.
Antibodies used in this study.
2.6. Cells proliferation assay
Cells were incubated at 37°C with 5% CO2 after adding 10 μL CCK8 and 90 μL RPMI 1640 medium into each plate of cells. Optical density (OD) value was measured by microplate reader (Biotek, Winooski, Vermont). The proliferative-detection was carried out at 0, 24, and 48 hours after transfection.
2.7. AV-PI assay
After digested by pancreatin and washed with PBS, cells were suspended in 100 μL Bling Buffer, and added with 5 μL AV, then incubated at room temperature for 15 minutes in the dark. Subsequently, after adding 5 μL PI, cells were incubated at room temperature for 15 minutes in the dark. The flow cytometry was performed to detect the cells stained with AV and PI.
2.8. Statistics
All operation in vitro experiments were repeated 3 times, Statistics was carried out using SPSS 19.0 (SPSS, New York City, State of New York) and GraphPad prim 6.01 (GraphPad, New York City, State of New York).[17] Data were presented in mean ± standard deviation. Comparison was determined by t test. P < .05 (∗) was considered significant, and P < .01 (†) was considered highly significant.
3. Results
3.1. LncRNA TUG1 expression after lncRNA TUG1 mimic/inhibitor plasmids transfection
After plasmids transfection, we found that transfection rates were all above 90% in NC1 mimic, lncRNA TUG1 mimic, NC2 inhibitor, and lncRNA TUG1 inhibitor groups (Fig. 1A). As presented in Fig. 1B, lncRNA TUG1 expression was upregulated after transfection in lncRNA TUG1 mimic group and downregulated in lncRNA TUG1 inhibitor group.
Figure 1.
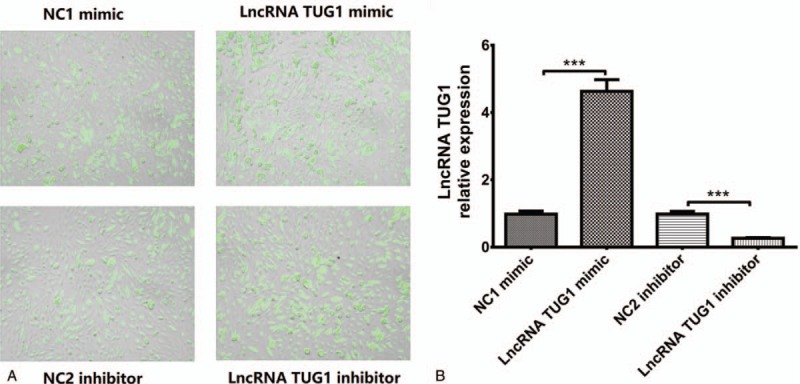
LncRNA TUG1 expression after plasmids transfection. (A) Transfection rate were all above 90% in 4 groups. (B) The expression of lncRNA TUG1 was increased in lncRNA TUG1 mimic group and decreased in lncRNA inhibitor group compared with NCs. ∗P < .05, †P < .01.
3.2. Cells proliferation after lncRNA TUG1 mimic/inhibitor plasmids transfection
CKK8 assay was subsequently performed. At 48 hours post plasmid transfection, cells proliferation was increased in lncRNA TUG1 mimic group compared with NC1 mimic group, and was decreased in lncRNA TUG1 inhibitor group compared with NC2 inhibitor group, which suggested that lncRNA TUG1 promoted EOC cells proliferation (Fig. 2).
Figure 2.
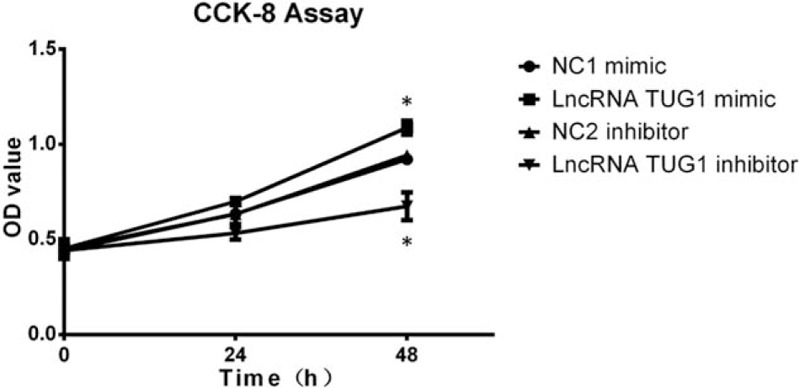
Cells proliferation after transfection. Cells proliferation was induced after treatment with lncRNA TUG1 mimic and was suppressed after treatment with lncRNA TUG1 inhibitor. ∗P < .05.
3.3. Cells apoptosis after lncRNA TUG1 mimic/inhibitor plasmids transfection
Cells apoptosis was repressed in lncRNA TUG1 mimic group compared with NC1 mimic group and was promoted in lncRNA TUG1 inhibitor group compared with NC2 inhibitor group (Fig. 3A, B). In addition, we explored cells apoptosis-related protein expressions and found that lncRNA TUG1 mimic decreased the expression of C-Caspase 3, while lncRNA TUG1 inhibitor increased the expression of C-Caspase 3. As for Bcl-2 expression, lncRNA TUG1 mimic increased Bcl-2 expression and lncRNA TUG1 inhibitor decreased Bcl-2 expression. These indicated that lncRNA TUG1 could repress apoptosis in EOC cells.
Figure 3.
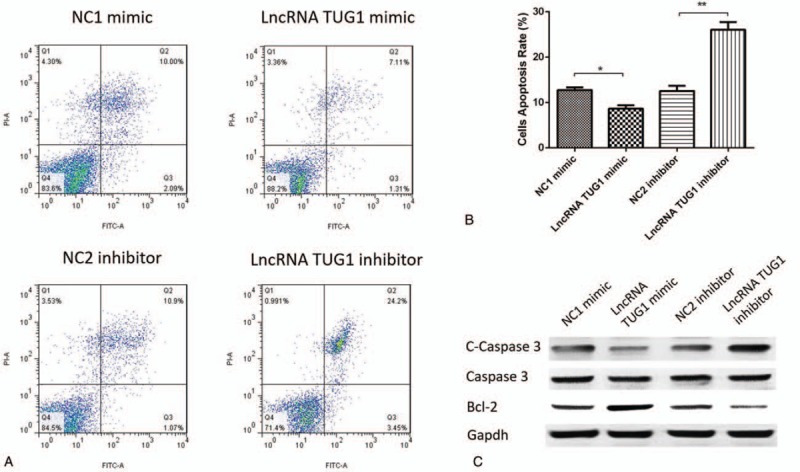
Cells apoptosis after transfection. (A, B) Cells apoptosis rate was decreased in lncRNA TUG1 mimic group compared with NC1 mimic group, and increased in lncRNA TUG1 inhibitor group compared with NC2 inhibitor group. (C) lncRNA TUG1 decreased C-Caspase 3 expression and increased Bcl-2 expression in EOC cells. ∗P < .05, †P < .01.
3.4. Expression of candidate target genes after lncRNA TUG1 mimic/inhibitor plasmids transfection
Both mRNA and protein expressions of AURKA were increased in lncRNA TUG1 mimic group compared with NC1 mimic group and were decreased in lncRNA TUG1 inhibitor group compared with NC2 inhibitor group, indicating the positive regulation of lncRNA TUG1 on AURKA in EOC cells (Fig. 4A, E). No difference of other mRNA and protein expressions, including CLDN3, SERPINE1, and ETS1, between groups was found (Fig. 4B–E).
Figure 4.
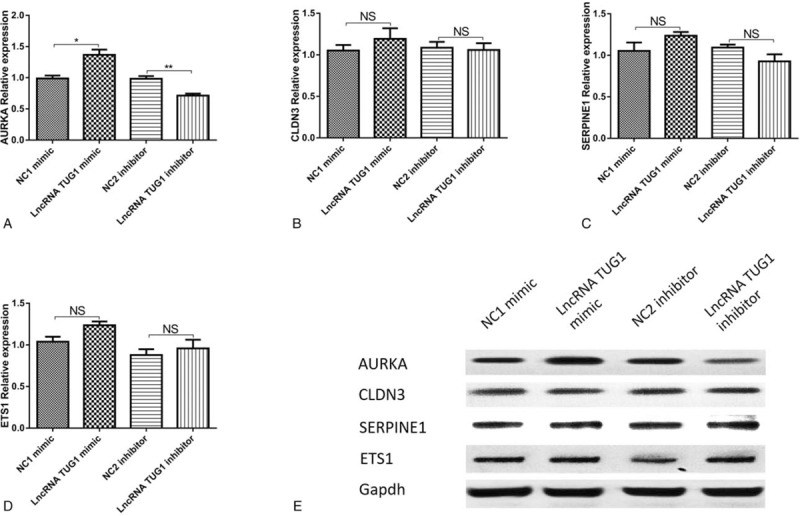
Expression of candidate target genes after lncRNA TUG1 (mimic/inhibitor) plasmids transfection. Both mRNA (A) and protein (E) expressions of AURKA were increased in lncRNA TUG1 mimic group than NC1 mimic group, and were decreased in lncRNA TUG1 inhibitor group than NC2 inhibitor. No difference was found in CLDN3 (B), SERPINE1 (C), and ETS1 (D) expression. ∗P < .05, †P < .01. No significance (NS) was considered no statistical significance.
3.5. Expressions of lncRNA TUG1 and AURKA in rescue experiment
Rescue experiment was performed to identify whether lncRNA TUG1 regulated EOC cells functions via targeting AURKA. LncRNA TUG1 expression was decreased in NC(+)/lncRNA TUG1(−) group compared with NC(+)/NC (−) group, while no difference was found between NC(+)/NC (−) and AURKA(+)/NC(−) groups as well as NC(+)/lncRNA TUG1(−) group and AURKA(+)/lncRNA TUG1(−) group, suggesting that AURKA could not regulate the expression of lncRNA TUG1 (Fig. 5A).
Figure 5.
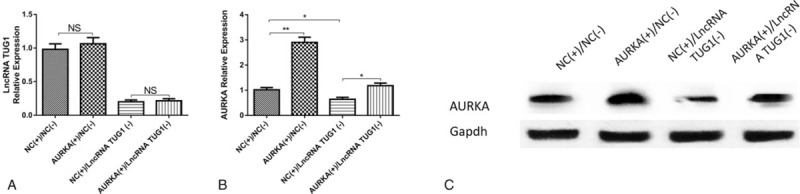
Expression of lncRNA TUG1 and AURKA after lncRNA TUG1 (−) and AURKA (+) plasmids transfection. LncRNA TUG1 expression was not influenced by AURKA (+) (A); LncRNA TUG1 enhanced the expressions of mRNA (B) and protein (C) of AURKA. ∗P < .05, †P < .01. No significance (NS) was considered no statistical significance.
As presented in Fig. 5B and C, both mRNA and protein expressions of AURKA were increased in AURKA(+)/NC(−) group compared with NC(+)/NC (−) group, and in AURKA(+)/lncRNA TUG1(−) group compared with NC(+)/lncRNA TUG1(−) group, while their expressions were lower in NC(+)/lncRNA TUG1(−) group than in NC(+)/NC(−) group. These indicated that lncRNA TUG1(−) inhibited the expression of AURKA, while AURKA(+) decreased the influence.
3.6. Cells proliferation in rescue experiment
CCK8 assay revealed that cells proliferation was increased in AURKA(+)/NC(−) group compared with NC(+)/NC(−) group, and in AURKA(+)/lncRNA TUG1(−) group compared with NC(+)/lncRNA TUG1(−) group, suggesting that lncRNA TUG1 regulated cells proliferation through targeting AURKA (Fig. 6).
Figure 6.
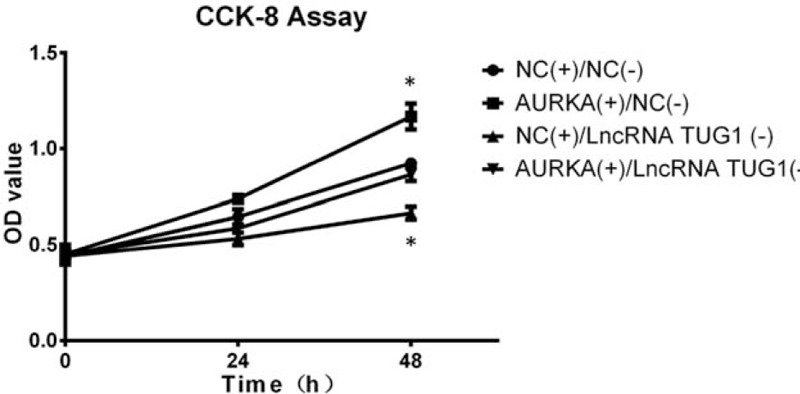
Cells proliferation after lncRNA TUG1 (−) and AURKA (+) plasmids transfection. Cells proliferation was increased in AURKA (+)/NC (−) group than NC (+)/NC (−) group, as well as in AURKA (+)/lncRNA TUG1 (−) group than NC (+)/lncRNA TUG1 (−) group. P < .05.
3.7. Cells apoptosis in rescue experiment
AV/PI assay disclosed that cells apoptosis rate was decreased in AURKA (+)/NC(−)group compared with NC(+)/NC(−) group, and also in AURKA(+)/lncRNA TUG1(−) group compared with NC(+)/lncRNA TUG1(−) group, indicating that lncRNA TUG1 regulated cells apoptosis in EOC cells via mediating AURKA (Fig. 7A, B).
Figure 7.
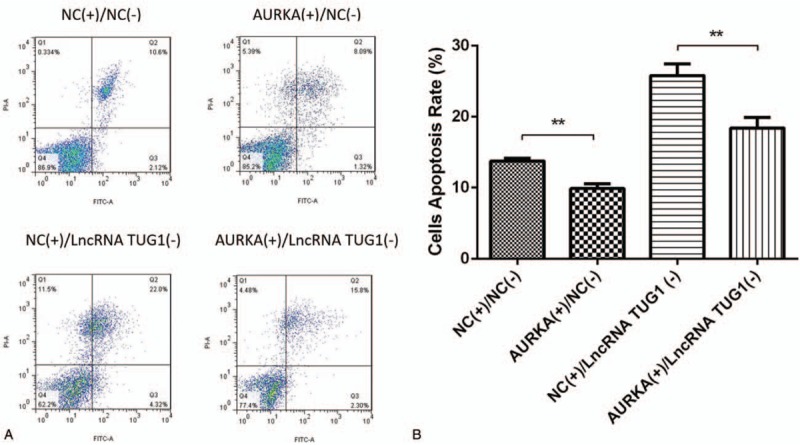
Cells apoptosis after lncRNA TUG1 (−) and AURKA (+) plasmids transfection. Cells apoptosis rate was decreased in AURKA (+)/NC (−) group than NC (+)/NC (−) group, as well as in AURKA (+)/lncRNA TUG1 (−) group than NC (+)/lncRNA TUG1 (−) group (A-B). ∗P < .05, †P < .01.
4. Discussion
In this study, we observed that lncRNA TUG1 promoted cells proliferation and inhibited cells apoptosis in EOC cells. Rescue experiment illustrated that lncRNA TUG1 mediated cells proliferation and cells apoptosis via regulating AURKA in EOC cells.
LncRNAs have been verified to involve in the processes of tumorigenesis and progression through regulating several oncogenic or tumor suppressed pathways via multiple potential mechanisms, including chromatin modification; regulation of transcription factors and coactivators recruitment; alternative splicing of pre-mRNAs; repression of RNA polymerase II activity; microRNAs sequestration; and mediation of mRNA stability.[18–22]
LncRNA TUG1, a common lncRNA with 6.7-kb nucleotides, is considered as an oncogene to various carcinomas, and plays an important role in pathological processes, such as cells proliferation, cells apoptosis, cells invasion, and cells differentiation.[23–25] Recent data have proven that lncRNA TUG1 induces cells proliferation and cells invasion by inhibiting miR-219 or inducing formin-like protein 2 (FMNL2) in oral squamous cell carcinoma cells.[24] As to bladder cancer cells, lncRNA TUG1 promotes cells proliferation and inhibit cells apoptosis through accumulating zinc finger E-box-binding homeobox 2 (ZEB2) regulated by miR-142 via suppressing Wnt/β-catenin pathway.[23] Also, as for papillary thyroid cancer cells, lncRNA TUG1 induces cells proliferation and repress cells invasion by targeting miR-145/ZEB1 signal pathway.[25] Although there are several studies that have explored the functions of lncRNA TUG1 in some kinds of cancer cells, little is known about the role of lncRNA TUG1 in EOC cells. Thus, we investigated the function of lncRNA TUG1 in EOC cells and found that lncRNA TUG1 promoted cells proliferation and inhibited cells apoptosis in EOC cells. These indicated that lncRNA TUG1 affects EOC pathology through regulating cells proliferation and apoptosis.
AURKA, a serine-threonine kinase, contributes to the regulation of G2/M transition, chromosome segregation, and centrosome functions.[26,27] Accumulating evidence have proven that AURKA plays a critical role in the oncogenic processes of many types of human cancers and cancer cell lines.[27–30] For example, AURKA accelerates cells migration and invasion in head and neck squamous cell carcinoma via upregulating focal adhesion kinase (FAK).[31] Also, AURKA enhances cells proliferation, apoptosis, invasion, and metastasis via the activity of FAK/phosphatidyl inositol-3-hydroxykinase (PI3K)/protein kinase B (PKB) pathway in laryngeal squamous cell carcinoma.[32] Another study in EOC reveals that AURKA promotes cells proliferation, migration, and invasion of HO8910 and SKOV3 cells in vitro.[33] These data suggest that AURKA contributes to the development and progress of several carcinomas, including EOC, through affecting cells activities via regulating multiple genes or pathways. In the current study, we found that lncRNA TUG1 mediates EOC cells functions through targeting AURKA. In order to confirm this hypothesis, we subsequently carried out the rescue experiment and found that lncRNA TUG1 mediated cells proliferation and cells apoptosis via targeting AURKA in EOC cells. However, the detailed mechanism of AURKA related to lncRNA TUG1 in EOC was not explored in this experiment, and further experiment is necessary.
In conclusion, lncRNA TUG1 promotes cells proliferation and inhibits cells apoptosis through regulating AURKA in EOC cells.
Author contributions
Conceptualization: Shaoxiao Liu.
Data curation: Yan Chen.
Formal analysis: Tonghuai Li, Yan Chen.
Investigation: Yan Chen.
Methodology: Formal analysis.
Resource: Jingjing Zhang.
Supervision: Shaoxiao Liu.
Writing – original draft: Formal analysis.
Writing – review and editing: Jingjing Zhang, Shaoxiao Liu.
Footnotes
Abbreviations: AURKA = Aurora kinase A, AV = annexin V-FITC, BCA = bicinchoninic acid, CLDN3 = Claudin 3, ECL = enhanced chemiluminescence, EOC = epithelial ovarian cancer, ETS1 = E26 transformation-specific 1, FAK = focal adhesion kinase, FBS = fetal bovine serum, FMNL2 = formin-like protein 2, lncRNAs = long noncoding RNAs, NC = normal control, OS = overall survival, PI = propidium iodide, PI3K = phosphatidyl inositol-3-hydroxykinase, PKB = protein kinase B, qPCR = quantitative polymerase chain reaction, RPMI = Roswell Park Memorial Institute, SDS-PAGE = sodium dodecyl sulfate-polyacrylamide gel electrophoresis, TUG1 = taurine-upregulated gene 1, ZEB2 = zinc finger E-box-binding homeobox 2.
The authors have no funding and conflicts of interest to disclose.
References
- [1].Jayson GC, Kohn EC, Kitchener HC, et al. Ovarian cancer. Lancet 2014;384:1376–88. [DOI] [PubMed] [Google Scholar]
- [2].Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- [3].Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115–32. [DOI] [PubMed] [Google Scholar]
- [4].Cramer DW. The epidemiology of endometrial and ovarian cancer. Hematol Oncol Clin North Am 2012;26:1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature 2012;489:101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature 2012;482:339–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Qiu MT, Hu JW, Yin R, et al. Long noncoding RNA: an emerging paradigm of cancer research. Tumour Biol 2013;34:613–20. [DOI] [PubMed] [Google Scholar]
- [8].Li Z, Shen J, Chan MT, et al. TUG1: a pivotal oncogenic long non-coding RNA of human cancers. Cell Prolif 2016;49:471–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li T, Liu Y, Xiao H, et al. Long non-coding RNA TUG1 promotes cell proliferation and metastasis in human breast cancer. Breast Cancer 2017;24:535–43. [DOI] [PubMed] [Google Scholar]
- [10].Zhang E, He X, Yin D, et al. Increased expression of long noncoding RNA TUG1 predicts a poor prognosis of gastric cancer and regulates cell proliferation by epigenetically silencing of p57. Cell Death Dis 2016;7:e2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sun J, Ding C, Yang Z, et al. The long non-coding RNA TUG1 indicates a poor prognosis for colorectal cancer and promotes metastasis by affecting epithelial-mesenchymal transition. J Transl Med 2016;14:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Li TH, Zhang JJ, Liu SX, et al. Long non-coding RNA taurine-upregulated gene 1 predicts unfavorable prognosis, promotes cells proliferation, and inhibits cells apoptosis in epithelial ovarian cancer. Medicine (Baltimore) 2018;97:e0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wenzel A, Akbasli E, Gorodkin J. RIsearch: fast RNA-RNA interaction search using a simplified nearest-neighbor energy model. Bioinformatics 2012;28:2738–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Tafer H, Hofacker IL. RNAplex: a fast tool for RNA-RNA interaction search. Bioinformatics 2008;24:2657–63. [DOI] [PubMed] [Google Scholar]
- [15].Li J, Ma W, Zeng P, et al. LncTar: a tool for predicting the RNA targets of long noncoding RNAs. Brief Bioinform 2015;16:806–12. [DOI] [PubMed] [Google Scholar]
- [16].Pinero J, Bravo A, Queralt-Rosinach N, et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res 2017;45:D833–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gao J, Zhu M, Liu RF, et al. Cardiac hypertrophy is positively regulated by microRNA24 in rats. Chin Med J (Engl) 2018;131:1333–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Fu X, Zhang L, Dan L, et al. LncRNA EWSAT1 promotes ovarian cancer progression through targeting miR-330-5p expression. Am J Transl Res 2017;9:4094–103. [PMC free article] [PubMed] [Google Scholar]
- [19].Gooding AJ, Zhang B, Jahanbani FK, et al. The lncRNA BORG drives breast cancer metastasis and disease recurrence. Sci Rep 2017;7:12698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhai HY, Sui MH, Yu X, et al. Overexpression of long non-coding RNA TUG1 promotes colon cancer progression. Med Sci Monit 2016;22:3281–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Li CH, Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. Int J Biochem Cell Biol 2013;45:1895–910. [DOI] [PubMed] [Google Scholar]
- [22].Yu X, Li Z. Long non-coding RNA growth arrest-specific transcript 5 in tumor biology. Oncol Lett 2015;10:1953–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Liu Q, Liu H, Cheng H, et al. Downregulation of long noncoding RNA TUG1 inhibits proliferation and induces apoptosis through the TUG1/miR-142/ZEB2 axis in bladder cancer cells. Onco Targets Ther 2017;10:2461–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yan G, Wang X, Yang M, et al. Long non-coding RNA TUG1 promotes progression of oral squamous cell carcinoma through upregulating FMNL2 by sponging miR-219. Am J Cancer Res 2017;7:1899–912. [PMC free article] [PubMed] [Google Scholar]
- [25].Lei H, Gao Y, Xu X. LncRNA TUG1 influences papillary thyroid cancer cell proliferation, migration and EMT formation through targeting miR-145. Acta Biochim Biophys Sin (Shanghai) 2017;49:588–97. [DOI] [PubMed] [Google Scholar]
- [26].Carmena M, Earnshaw WC. The cellular geography of aurora kinases. Nat Rev Mol Cell Biol 2003;4:842–54. [DOI] [PubMed] [Google Scholar]
- [27].Eterno V, Zambelli A, Villani L, et al. AurkA controls self-renewal of breast cancer-initiating cells promoting wnt3a stabilization through suppression of miR-128. Sci Rep 2016;6:28436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Treekitkarnmongkol W, Katayama H, Kai K, et al. Aurora kinase-A overexpression in mouse mammary epithelium induces mammary adenocarcinomas harboring genetic alterations shared with human breast cancer. Carcinogenesis 2016;37:1180–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chuang TP, Wang JY, Jao SW, et al. Over-expression of AURKA, SKA3 and DSN1 contributes to colorectal adenoma to carcinoma progression. Oncotarget 2016;7:45803–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Schnepp RW, Khurana P, Attiyeh EF, et al. A LIN28B-RAN-AURKA signaling network promotes neuroblastoma tumorigenesis. Cancer Cell 2015;28:599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wu J, Yang L, Shan Y, et al. AURKA promotes cell migration and invasion of head and neck squamous cell carcinoma through regulation of the AURKA/Akt/FAK signaling pathway. Oncol Lett 2016;11:1889–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yang LY, He CY, Chen XH, et al. Aurora kinase A revives dormant laryngeal squamous cell carcinoma cells via FAK/PI3K/Akt pathway activation. Oncotarget 2016;7:48346–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wang C, Yan Q, Hu M, et al. Effect of AURKA gene expression knockdown on angiogenesis and tumorigenesis of human ovarian cancer cell lines. Target Oncol 2016;11:771–81. [DOI] [PubMed] [Google Scholar]


