Abstract
Lysosomal accumulation of undegraded materials is a common feature of lysosomal storage diseases, neurodegenerative disorders, and the aging process. To better understand the role of lysosomal storage in the onset of cell damage, we used human fibroblasts loaded with sucrose as a model of lysosomal accumulation. Sucrose-loaded fibroblasts displayed increased lysosomal biogenesis followed by arrested cell proliferation. Notably, we found that reduced lysosomal catabolism and autophagy impairment led to an increase in sphingolipids (i.e., sphingomyelin, glucosylceramide, ceramide, and the gangliosides GM3 and GD3), at both intracellular and plasma membrane (PM) levels. In addition, we observed an increase in the lysosomal membrane protein Lamp-1 on the PM of sucrose-loaded fibroblasts and a greater release of the soluble lysosomal protein cathepsin D in their extracellular medium compared with controls. These results indicate increased fusion between lysosomes and the PM, as also suggested by the increased activity of lysosomal glycosphingolipid hydrolases on the PM of sucrose-loaded fibroblasts. The inhibition of β-glucocerebrosidase and nonlysosomal glucosylceramidase, both involved in ceramide production resulting from glycosphingolipid catabolism on the PM, partially restored cell proliferation. Our findings indicate the existence of a new molecular mechanism underlying cell damage triggered by lysosomal impairment.—Samarani, M., Loberto, N., Soldà, G., Straniero, L., Asselta, R., Duga, S., Lunghi, G., Zucca, F. A., Mauri, L., Ciampa, M. G., Schiumarini, D., Bassi, R., Giussani, P., Chiricozzi, E., Prinetti, A., Aureli, M., Sonnino, S. A lysosome–plasma membrane–sphingolipid axis linking lysosomal storage to cell growth arrest.
Keywords: glycosphingolipids, glycohydrolases, cell proliferation, cell surface, catabolism
Lysosomes, acidic intracellular organelles containing hydrolytic enzymes, are involved in the degradation and recycling of several macromolecules recruited via endocytosis, phagocytosis, and autophagy degradation pathways (1–3). Although lysosomes have been mainly considered inert catabolic organelles, it is now clear that they are also crucial regulators of cell homeostasis (4). Lysosomal accumulation of uncatabolized material is a hallmark of a large number of diseases, including lysosomal storage disorders, Alzheimer’s disease, Parkinson’s disease, and brain aging (5–7). Moreover, it has been recently demonstrated that diverse lysosomotropic drugs accumulate within the lysosomes, affecting their function (8, 9). However, the molecular mechanism responsible for the onset of cell damage upon lysosomal engulfment remains unknown to date; this represents the main challenge in the field.
Lysosomal stress caused by the aberrant storage of uncatabolized material induces the activation of the coordinated lysosomal enhancement and regulation network responsible for the nuclear translocation of the transcription factor EB (TFEB). TFEB promotes the transcription of multiple genes involved in lysosomal biogenesis and lysosome-related functions, including autophagy and lysosomal exocytosis (10, 11).
A common feature observed upon lysosomal impairment is the secondary accumulation of sphingolipids (SLs) (12). SLs are amphiphilic molecules principally associated with the external leaflet of the plasma membrane (PM) of all eukaryotic cells. Within the PM, SLs are not only structural components but also participate, through their interaction with PM-associated proteins, in controlling several signal transduction pathways that are fundamental to maintaining cell homeostasis (13). Interestingly, the in situ modification of the PM SL composition represents a stimulus able to affect several signaling pathways, including those that control cell death and growth arrest (14–18). Based on these observations, in this work we investigate the involvement of PM glycosphingolipid catabolic pathways in the mechanisms linking lysosomal impairment to the onset of cell damage.
MATERIALS AND METHODS
Cell cultures and treatments
Fibroblast cell lines from healthy subjects (L40, L37, and F1) were cultured as previously described (19–23). Briefly, fibroblasts were cultured in Rosewell Park Memorial Institute (RPMI)-1640 medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin (growth medium) (EuroClone, Pero, Italy). Cells were cultured for 14 d in growth medium with or without 88 mM sucrose (MilliporeSigma, Burlington, MA, USA). The following compounds were added to the cell growth medium for the different treatments: 0.1 µM bafilomycin A1 (MilliporeSigma) for 6 h; 0.5 mM conduritol B epoxide (CBE; MilliporeSigma) for 48 h; and 20 nM adamantane–pentyl-dNM–N-(5-adamantane-1-yl-methoxy-pentyl)-deoxynojirimycin (AMP-DNM) for 48 h.
Evaluation of cell proliferation and viability
Fibroblasts were plated in T25 flasks at a density of 3000 cells/cm2 and sucrose loading was started the day after plating. After 1, 2, 3, 7, 10 and 14 d cell counting by Trypan blue exclusion assay was performed. Data are expressed as number of live cells per square centimeter of growth area.
RNA analysis
Total RNA was extracted using the EuroGoldTriFast reagent (EuroClone) according to the manufacturer’s instructions. RNA concentration was determined using a NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and RNA integrity was assessed on a LabChip GX Touch (PerkinElmer, Waltham, MA, USA). Subsequently, 500 ng of RNA for each sample was used to generate paired-end sequencing libraries with an Illumina TruSeq Stranded mRNA Sample LS Preparation Kit (Illumina, San Diego, CA, USA), according to the manufacturer’s protocol. Sequencing was performed on a NextSeq500 platform (Illumina) at the Humanitas Genomic Facility (Milan, Italy). After quality filtering according to the Illumina pipeline, 76 bp paired-end reads were mapped to the hg19 reference genome and to the Homo sapiens transcriptome [Illumina’s iGenomes reference annotation, downloaded from University of California, Santa Cruz (UCSC); http://support.illumina.com/sequencing/sequencing_software/igenome.html] using Star v.2.5 (24). Differential expression analysis between treated and untreated samples was evaluated with the Wald χ2 test of significance for the negative binomially distributed counts using the DeSeq2 Bioconductor package (25). Differentially expressed (DE) genes were selected using a false discovery rate (FDR) of ≤0.01 and fold change ≥2. Gene ontology (GO) enrichment analyses were performed using Metascape (http://www.metascape.org) with default parameters. Expression profiling data were submitted to the Gene Expression Omnibus repository.
For real-time quantitative RT-PCR (qRT-PCR), random hexamers and the ImProm-II Reverse Transcription System (Promega, Madison, WI, USA) were used to perform first-strand cDNA synthesis starting from 500 ng of RNA. Out of 20 µl obtained from the RT reaction, 1 µl was used as a template for amplifications using the SensiFast SYBR No-Rox Kit (Bioline, London, United Kingdom) on a LightCycler 480 (Roche, Basel, Switzerland), following a touchdown thermal protocol. Expression levels were normalized using HMBS (hydroxymethylbilane synthase) and B2M (β2-microglobulin) as housekeeping genes. In all cases, real-time qRT-PCR assays were performed ≥3 times, and expression levels were analyzed using the GeNorm software (26).
Electron microscopy of cell monolayers
Cell monolayers were fixed in a mixture of 4% paraformaldehyde and 2% glutaraldehyde in cacodylate buffer (0.12 M, pH 7.4) for 4 h at 4°C. Cells were then extensively washed with cacodylate buffer and postfixed for 1 h on ice in a mixture of 1% osmium tetroxide and 1.5% potassium ferrocyanide in cacodylate buffer. After several washes with ultrapure water, samples were stained en bloc with 0.5% uranyl acetate in water overnight at 4°C. Finally, samples were dehydrated in a graded ethanol series, then infiltrated for 2 h in a mixture of ethanol and epon (1:1, v:v), and then in 100% epon, twice for 1 h. Then polymerization was performed for 24 h in an oven at 60°C. Ultra-thin sections (80 nm) were prepared using an ultramicrotome (Leica Ultracut; Leica Microsystems, Wetzlar, Germany) and collected on nickel grids. They were stained with saturated uranyl acetate for 5 min, washed, and then stained again with 3 mM lead citrate for 5 min. Finally, the sections were photographed using a transmission electron microscope Leo 912AB (Advanced Light and Electron Microscopy BioImaging Center, San Raffaele Scientific Institute, Milan, Italy).
LysoTracker staining
LysoTracker Red DND-99 (Molecular Probes, Eugene, OR, USA) was added to the cell medium at 50 nM for 30 min. After 1 wash in PBS, images were acquired with an Olympus IX50 inverted fluorescence microscope equipped with a VarioCam camera (InfraTec, Los Angeles, CA, USA). An LCAch ×20/0.40 PhC objective was used directly on living cells. The fluorescence intensity associated with each cell (n = 70 in 10 different fields) was evaluated by ImageJ software (National Institutes of Health, Bethesda, MD, USA). Data are expressed as a ratio of the fluorescence associated with cells to the total number of cells analyzed.
Autophagosome detection assay
Autophagosomes were detected in controls and cells loaded with sucrose for 14 d using the Cyto-ID Autophagy Detection Kit 2.0 (Enzo Life Sciences, Farmingdale, NY, USA) following the manufacturer’s instructions. Briefly, cells were grown on glass coverslips and then incubated with Cyto-ID Green Detection Reagent for 30 min at 37°C. The fluorescence intensity associated with each cell (n = 70 in 10 different fields) was evaluated by ImageJ software. Data are expressed as a ratio of the fluorescence associated with cells to the total number of cells analyzed.
Nuclear extraction
Nuclear extraction was performed as previously described (27). Briefly, cells were lysed with 0.5 ml of lysis buffer [50 mM Tris-HCl (pH 7.5), 0.5% Triton X-100, 137.5 mM NaCl, 10% glycerol, and 5 mM EDTA] supplemented with Protease Inhibitor Cocktail (MilliporeSigma) and 1 mM Na3VO4 (MilliporeSigma) for 15 min in ice under gentle shaking. Lysates were centrifuged at 15,700 g for 15 min at 4°C. The nuclear pellet was rinsed 3 times with 0.5 ml of lysis buffer, resuspended in 0.1 ml of lysis buffer supplemented with 0.5% SDS (MilliporeSigma), and sonicated in ice. Nuclear extract was collected after centrifugation at 15,700 g for 15 min at 4°C.
Cell biotinylation
Fibroblasts were fed 38 nM [1-3H]sphingosine (28) for 2 h (pulse), followed by a 48 h chase to allow metabolic labeling of all cellular SLs at the steady state (29). Then cells were loaded or not with sucrose for 14 d. At the end of 14-d loading, cells were washed with PBS and incubated with 1 mg/ml of EZ-Link Sulfo-NHS-Biotin (Thermo Fisher Scientific) in PBS (pH 7.4, 5 ml/T75 flasks) for 30 min at 4°C (30). Under these experimental conditions, the internalization of the biotin derivative does not occur and biotinylation is restricted to the cell surface proteins (31). After biotin labeling, cells were rinsed twice with 100 mM glycine to remove excess unbounded biotin and then washed with ice-cold PBS. Cells were mechanically harvested in PBS and centrifuged at 270 g for 10 min. The cell pellet was lysed in 1 ml of 1% Triton X-100 in 10 mM TNEV buffer [10 mM Tris-HCl, 150 mM NaCl, 5 mM EDTA (pH 7.5)] supplemented with Protease Inhibitor Cocktail (MilliporeSigma) and 1 mM Na3VO4 (MilliporeSigma) for 20 min in ice, and then homogenized with a tight Dounce Homogenizer (DWK Life Sciences, Millville, NJ, USA). Cell lysate was centrifuged at 4°C for 5 min at 800 g to remove nuclei and cellular debris and obtain postnuclear supernatant (PNS). The immunoprecipitation was performed using Dynabeads M-280 Streptavidin (Thermo Fisher Scientific) on the same amount of protein associated with PNS in both control and sucrose-loaded cells. The mixtures were stirred overnight at 4°C, and then the immunoprecipitate (IP) was recovered by both centrifugation and magnets. Under these experimental conditions, we preserved the organization of lipid domains (32, 33). Radioactive lipids associated with the IP were extracted by 2 washes with 200 µl of chloroform:methanol (2:1, v:v) whereas the radioactive lipids associated with PNS and supernatant after immunoprecipitation (SNIP) were extracted following the procedure described for analysis of the endogenous counterpart. The radioactivity associated with IP, PNS, and SNIP was evaluated by liquid scintillation. The radioactive lipids corresponding to the same amount of cellular proteins were separated by monodimensional high performance thin-layer chromatography (HPTLC) (MilliporeSigma) using the solvent systems: chloroform:methanol:water (110:40:6, v:v:v) to separate and quantify neutral glycosphingolipids and sphingomyelin; chloroform:methanol:0.2% aqueous CaCl2 (50:42:11, v:v:v) to separate and quantify gangliosides; and hexane:chloroform:acetone:acetic acid (20:70:20:4, v:v:v:v) to separate and quantify ceramide. Radioactive lipids were visualized by digital autoradiography (TRacer BetaImager; BioSpace Laboratory, Nesles la Vallée, France) and quantified using BioSpace Lab’s M3 Vision software. Identification of lipids after separation was assessed by comigration with radioactive lipid standards. The proteins associated with the IP were recovered once by Laemmli sample buffer at 100°C for 10 min.
Antibodies
Monoclonal mouse anti–Lamp-1 (H4A3; Developmental Studies Hybridoma Bank, Iowa City, IA, USA), monoclonal mouse anti-GD3 (R24; National Cancer Institute, Frederick, MD, USA), polyclonal rabbit anti-TFEB (A303-673A; Bethyl Laboratories, Montgomery, TX, USA), polyclonal rabbit anti-LC3 (L7543; MilliporeSigma), monoclonal rabbit anti-Atg5 (12994; Cell Signaling Technology, Danvers, MA, USA), polyclonal rabbit anti-SQSTM1/p62 (5114; Cell Signaling Technology), polyclonal rabbit anti-cdc2/cyclin-dependent kinase (CDK) 1 (77055; Cell Signaling Technology), monoclonal rabbit anti-GCase (ab128879; Abcam, Cambridge, United Kingdom), polyclonal rabbit anti-GAPDH (G9545; MilliporeSigma), monoclonal rabbit antihistone [3H] (4499; Cell Signaling Technology), monoclonal mouse anti-calnexin (610523; BD Biosciences, Franklin Lakes, NJ, USA), monoclonal rabbit anti-integrin αv (60896; Cell Signaling Technology), monoclonal rabbit anti–protein disulfide isomerase (3501; Cell Signaling Technology), monoclonal mouse anti-TGN38 (sc-166594; Santa Cruz Biotechnology, Dallas, TX, USA), streptavidin horseradish peroxidase (HRP; SA-5004; Vector Laboratories, Burlingame, CA, USA). Polyclonal goat anti-mouse Alexa Fluor 488 (ab150113; Abcam Inc.) was used as the secondary antibody for immunofluorescence. Goat anti-mouse HRP-conjugated (31430; Thermo Fisher Scientific) and goat anti-rabbit HRP-conjugated (7074; Cell Signaling Technology) were used as secondary antibodies for immunoblotting.
Immunofluorescence
Cells were fixed in 4% paraformaldehyde in PBS for 20 min at room temperature. Next, cells were blocked and permeabilized with 5% donkey serum/1% BSA/0.2% Triton X-100/PBS for 1 h at room temperature. After washing with PBS, cells were incubated with the primary antibody overnight at 4°C in a solution containing 1.25% donkey serum/0.25% BSA/0.05% Triton X-100/PBS. After washing with PBS, cells were incubated with Alexa Fluor–conjugated secondary antibodies for 1 h at room temperature. Finally, coverslips were mounted with Dako Fluorescent Mounting Medium (Agilent Technologies, Santa Clara, CA, USA). Images were taken with Olympus BX50 Upright Fluorescence Microscope equipped with a fast-high resolution camera (Colorview 12). A UPlanApo ×100/1.35 oil iris objective was used. For the immunofluorescence experiments in nonpermeabilizing conditions, cells were processed as described, but Triton X-100 was omitted from all solutions.
Immunoblotting
Equivalent amounts of protein determined by a DC Protein Assay (Bio-Rad, Hercules, CA, USA) of total cell lysates, nuclear extracts, PNS, and SNIP, or an equal volume of IP were separated by SDS-PAGE and then transferred to PVDF membranes by electroblotting. Blots were incubated with primary antibodies at 4°C overnight, followed by incubation with HRP-conjugated secondary antibodies and detection with a chemiluminescence kit (Westar ηC; Cyanagen, Bologna, Italy). Digital images were obtained by the chemiluminescence system Alliance Mini HD9 (Uvitec, Cambridge, United Kingdom).
Evaluation of enzymatic activities in cell lysates
The enzymatic activities associated with total cell lysates were determined using a previously described method (16). 4-Methylumbelliferone (MUB)–derived fluorogenic substrates were used, including 4-methylumbelliferyl β-d-glucopyranoside (MUB-β-Glc) for β-glucocerebrosidase (GCase), 4-methylumbelliferyl β-d-galactopyranoside (MUB-β-Gal) for β-galactosidase, and 4-methylumbelliferyl N-acetyl-β-d-glucuronide (MUG) for β-hexosaminidase (all from Glycosynth, Warrington, United Kingdom), as well as 6-hexadecanoylamino 4-MU-phosphoryl-choline for measuring sphingomyelinase (SMase) activity (Moscerdam Substrates, Oegstgeest, The Netherlands). To evaluate GCase activity, aliquots of cell lysates were preincubated for 30 min at room temperature with a reaction mixture comprising 25 µl of McIlvaine buffer (0.4 M citric acid and 0.8 M Na2HPO4, pH 6); the specific inhibitor for nonlysosomal glucosylceramidase (NLGase), AMP-DNM, at a concentration of 5 nM (34); and water, resulting in a final volume of 75 µl. At the end of preincubation, the reaction was started with the addition of 25 µl of MUB-β-Glc at the final concentration of 6 mM. To measure β-galactosidase, β-hexosaminidase, and SMase activity, aliquots of cell lysates were incubated with 25 µl of McIlvaine buffer (pH 5.2) containing the specific fluorogenic substrates at the final concentration of 500 μM, except for 6-hexadecanoylamino 4-MU-phosphoryl-choline 250 µM. Then water was added to reach the final volume of 100 µl. At different time points, the reaction mixtures were blocked by adding 19 volumes of 0.25 M glycine (pH 10.7; MilliporeSigma). For the SMase assay, 0.25 M glycine (pH 10.7) containing 0.3% Triton X-100 was added. The fluorescence was detected by a Victor microplate reader (PerkinElmer). Standard free MUB and hexadecanoylamino-MUB (for SMase) were used to construct calibration curves and quantify substrate hydrolysis. Enzymatic activity is expressed as nanomoles of product per milligram of cell proteins per hour.
Evaluation of enzymatic activity on the surface of living cells
Living cells were plated in 96-well microplates at a density of 20,000 cells/well, and PM-associated activities of GCase, NLGase, β-galactosidase, and β-hexosaminidase were assessed in these cells by a high throughput cell lived-based assay as previously described (14, 16, 35, 36). To distinguish between GCase and NLGase activities, cells were preincubated for 30 min at room temperature in DMEM-F12 without phenol red (Thermo Fisher Scientific) containing 5 nM AMP-DNM or 1 mM CBE, respectively (37). Activities were assayed using the artificial MUB-β-Gal for β-galactosidase, MUG for β-hexosaminidase, and MUB-β-Glc for GCase and NLGase. The fluorigenic substrates were solubilized in DMEM-F12 without phenol red at pH 6, with final concentrations of 250 µM, 1 mM, and 6 mM, respectively. Aliquots of medium (10 µl) were analyzed at different time points by a Victor microplate reader (PerkinElmer), after the addition of 190 µl of 0.25 M glycine (pH 10.7). Standard free MUB was used to construct calibration curves and quantify substrate hydrolysis. Enzymatic activity is expressed as nanomoles of product per 106 cells per hour. The experimental design included the required internal controls. This method is based on the observation that the fluorogenic substrates commonly used for in vitro assays of glycohydrolytic activity are not taken up by living cells. To determine whether substrate hydrolysis occurs only as a result of PM enzyme activity, a series of controls were maintained. In the experimental conditions we set, we did not observe any intracellular fluorescence, which indicated that the substrates were not able to cross the cell membrane. Moreover, we verified that the artificial substrates did not undergo either spontaneous or secreted enzyme-driven hydrolysis. Thus, under these experimental conditions, the fluorescence intensity is exclusively associated with enzymatic hydrolysis on the cell surface.
Lipid analysis by HPTLC
Cell lysates were lyophilized and subjected to lipid extraction with chloroform:methanol:water (2:1:0.1, v:v:v). The total lipid extracts were then subjected to a 2-phase partitioning, resulting in the separation of an aqueous phase (AP) containing gangliosides and an organic phase (OP) containing all other lipids (38). The phospholipid content was determined in the OP as phosphate after perchloric acid mineralization using Bartlett’s method (39). Aliquots of the OP were subjected to alkaline treatment to remove glycerophospholipids. The lipids contained in AP, OP, and alkaline-stable OP were separated by HPTLC (Merck Millipore) using different solvent systems: OP (corresponding to 250 × 103 cells), chloroform:methanol:acetic acid:water (30:20:2:1, v:v:v:v) for phospholipid analysis; alkaline-stable OP (corresponding to 106 cells), chloroform:methanol:water (110:40:6, v:v:v) for neutral glycolipids; alkaline-stable OP (corresponding to 106 cells), hexane:ethyl acetate (3:2, v:v) for cholesterol; alkaline-stable OP (corresponding to 2.5 × 106 cells), hexane:chloroform:acetone:acetic acid (20:70:20:4, v:v:v:v) for ceramide; AP (corresponding to 3 × 106 cells), first run in chloroform:methanol (9:1, v:v) followed by a second run in chloroform:methanol:0.2% aqueous CaCl2 (50:42:11, v:v:v). Lipids were identified after separation by comigration with lipid standards. Phospholipids were detected by spraying the HPTLC with a molybdate reagent (40); neutral glycolipids, cholesterol, and ceramide were visualized by spraying the HPTLC with anisaldehyde; gangliosides were detected by spraying the HPTLC with Ehrlich’s reagent. The relative quantities of lipids were determined by densitometry using ImageJ software.
Lipid analyses by HPLC-MS/MS
Quantitative analyses of ceramide, hexosylceramides, and sphingomyelin were performed at the Lipidomics Shared Resource Analytical Unit (Medical University of South Carolina, Charleston, SC, USA). Briefly, SL levels in cells loaded with sucrose for 14 d and control were measured by HPLC-MS (LC-MS/MS) methodology as previously described (41). The equipment consisted of a Vanquish uHPLC system coupled to a Quantum Access Max triple quadrupole mass spectrometer (Thermo Fisher Scientific) equipped with an electrospray ionization probe operating in the multiple reaction monitoring positive ion mode. Chromatographic separations were obtained under a gradient elution with water and methanol as previously described. Cell lysates were fortified with internal standards, extracted into a 1-phase neutral organic solvent system, and analyzed by an LC-MS/MS system. Qualitative analysis (identification) of SLs was performed by a parent ion scan of a common fragment ion characteristic of a particular class of SLs. Quantitative analysis was based on calibration curves generated by spiking an artificial matrix with known amounts of target analyte and synthetic standards and an equal amount of IS. The calibration curves were constructed using a linear regression model by plotting the peak area ratios of analyte to the respective IS against concentration.
Ganglioside MS analyses were carried out using a LCQDeca ion trap mass spectrometer (Thermo Fisher Scientific) equipped with an electrospray ionization source, an Xcalibur data system, and a Jasco PU980 Pump HPLC. Separations of extracted gangliosides were obtained on a 10 μm, 250 × 10 mm Hibar Si100 column (MilliporeSigma). Elution of GM3, GM2, GD3, and GD1a species was carried out at a flow rate of 0.150 ml/min, with a gradient formed by solvent system A, consisting of 2-propanol, and solvent system B, consisting of H2O, both containing 5 mM ammonium acetate. The injection volume for each sample was 20 μl. A gradient with the following time program was used: t = 0 min, 100% solvent A; t = 45 min, 50% solvent A, 50% solvent B; t = 50 min, 50% solvent A, 50% solvent B; t = 60 min, 100% solvent A; t = 80 min, 100% solvent A. Optimum conditions for MS analyses of ganglioside molecular species included sheath gas flow of 60 arbitrary units, spray voltage of 5 kV, capillary voltage of −15 V, and capillary temperature of 250°C. Mass spectra were acquired over a range of m/z 200–2000. For all experiments, source ion optics were adjusted to accomplish desolvation of ions while minimizing fragmentation. Serial dilutions of GM3, GM2, GD3, and GD1a gangliosides were used for calibration curves. Analytical results are expressed as picomoles of lipid per milligram of total cellular protein. Data are representative of 2 independent experiments performed in triplicate.
Evaluation of cholesterol content
Cholesterol content was assessed in control and cells loaded with sucrose for 14 d using Amplex Red Cholesterol Assay Kit (Molecular Probes) according to the manufacturer’s instructions. Briefly, the equivalent of 0.5 µg of cell protein was diluted in 1-time reaction buffer in a final volume of 50 μl. Then 50 μl of the Amplex Red reagent/HRP/cholesterol oxidase/cholesterol esterase working solution was added. The reaction was incubated for 30 min at 37°C. Fluorescence (λex: 544 nm/λem: 590 nm) was detected by the FLUOstar Omega microplate reader (BMG Labtech, Ortenberg, Germany). Cholesterol content is expressed as micromoles of cholesterol per milligram of cell protein.
Evaluation of released cathepsin D by ELISA
Soluble lysosomal cathepsin D released in culture medium after 14 d of sucrose loading was evaluated using Human Cathepsin D ELISA Kit (Abcam) following the manufacturer’s instructions. Briefly, aliquots of culture medium derived from control cells and cells loaded with sucrose for 14 d were incubated in 96-well plates precoated with a cathepsin D specific mouse mAb for 90 min at 37°C. Biotinylated cathepsin D–specific goat pAb from was added, and cells were then washed with TBS buffer. Avidin-Biotin-Peroxidase Complex was added, and unbounded conjugates were washed away with TBS buffer. 3,3′,5,5′-Tetramethylbenzidine was then used to visualize the HRP enzymatic reaction (O.D. absorbance: 450 nm). Released cathepsin D content is expressed as picograms of cathepsin D per milliliter of culture medium.
Treatment of cell cultures with [3-3H(sphingosine)]GM3
Isotopically labeled [3-3H(sphingosine)]GM3 (specific radioactivity 2 Ci/mM) (42) was administered to both control and sucrose-loaded cells as previously described, with some modifications (14, 17). To follow the catabolism of [3-3H(sphingosine)]GM3 at the PM level, dedicated cells were preincubated with 0.1 mM chloroquine (MilliporeSigma) for 1 h in cell medium without serum. Next, medium containing the radioactive lipids were added to each dish and the cells were incubated at 37°C, some with the addition of 0.1 mM chloroquine, for 4 h. At the end of incubation, cells were harvested with PBS and processed for lipid analysis as previously described. The radioactivity associated with lipid extracts was determined by liquid scintillation counting. Total lipid extracts were then separated by HPTLC (MilliporeSigma) with the solvent system chloroform:methanol:0.2% aqueous CaCl2 (50:42:11, v:v:v). Radioactive lipids were detected by digital autoradiography (TRacer Betaimager; BioSpace Laboratory) and quantified using M3 Vision software. Lipids were identified after separation by comigration with radioactive lipid standards.
Statistics
All the experiments were performed in triplicate and repeated ≥3 times. Data are presented as the means ± sd and were tested for significance with Student’s t test using GraphPad Prism software (GraphPad Software, La Jolla, CA, USA).
RESULTS
Sucrose-loaded fibroblasts: an in vitro model of lysosomal accumulation
To create an in vitro model of lysosomal accumulation of uncatabolized substrate, we loaded human skin fibroblasts with 88 mM sucrose for 14 d in culture (43–48). This concentration of sucrose does not induce osmotic stress in fibroblasts (44, 48).
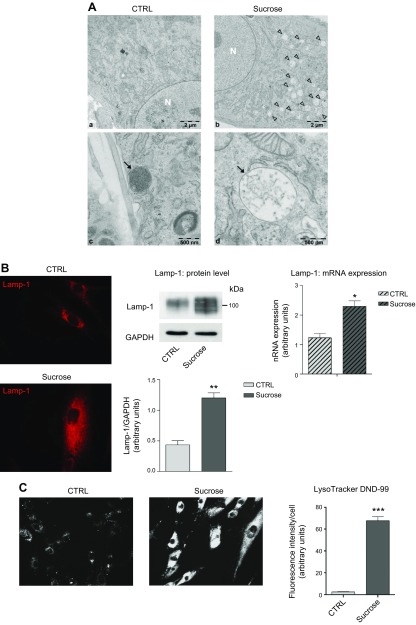
In accordance with the literature (49), we observed via transmission electron microscopy the accumulation of sucrose by lysosomes, thereby forming sucrosomes. As shown in Fig. 1Ab, d, sucrose-loaded cells were characterized by a high number of large, pale, single membrane–bounded organelles (sucrosomes) that were absent in control cells. This morphology, different from that of normal mature lysosomes (Fig. 1Ac), is common in case of saccharides accumulation such as in Pompe disease and other mucopolysaccharidoses (44, 50).
Figure 1.
Effect of sucrose loading on the endolysosomal compartment in human fibroblasts. A) Electron micrographs of human control (a, c) and sucrose-loaded fibroblasts (b, d). Arrowheads (b) and arrow (d) indicate sucrosomes; arrow (c) indicates lysosomes; N (a, b) indicates nucleus. B) Immunofluorescence staining of lysosomal marker Lamp-1 in permeabilized human fibroblasts, or not with sucrose (left). Immunostaining of Lamp-1 and loading control GAPDH accompanied with the semiquantitative graph of normalized Lamp-1/GAPDH (middle). Data are expressed as means ± sd. **P < 0.0021 vs. control (ctrl) (n = 4). Lamp-1 mRNA expression evaluated by quantitative RT-PCR (right). *P < 0.01 vs. ctrl (n = 3) Expression levels were normalized using HMBS and B2M as housekeeping genes. C) LysoTracker Red DND-99 staining performed on living human fibroblasts, loaded or not with sucrose, and a semiquantitative graph of the fluorescence intensity measured in ctrl and sucrose-loaded cells normalized by the number of cells analyzed. ***P < 0.0001 vs. ctrl (n = 70).
As shown in Fig. 1B, the increased number of lysosomes in sucrose-loaded cells was confirmed by immunofluorescent staining, in permeabilized cells, of the lysosome-associated membrane glycoprotein 1 (Lamp-1). The higher expression of Lamp-1 in sucrose-loaded cells with respect to controls was also confirmed by Western blot and qRT-PCR analyses (Fig. 1B).
In addition, we evaluated the relative volume of intracellular acidic organelles using the LysoTracker Red DND-99 fluorescent probe. As shown in Fig. 1C, sucrose-loaded fibroblasts are characterized by more vivid staining with respect to control cells; this result is consistent with an increased endolysosomal network.
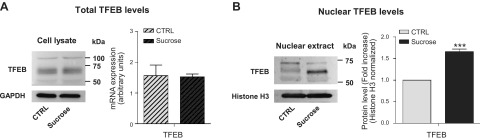
Induced lysosomal biogenesis is an evolutionarily acquired cell response against lysosomal stress controlled by the nuclear translocation of TFEB (51). As shown in Fig. 2B, we found a significant increase of endogenous TFEB protein associated with the nuclear extract of sucrose-loaded fibroblasts compared with controls. In addition, evaluating the total cell content of TFEB and its expression by qRT-PCR, we found that sucrose loading induces a greater TFEB nuclear translocation without affecting its total cellular levels (Fig. 2A).
Figure 2.
TFEB nuclear translocation. A) Immunostaining of TFEB from total cell lysates of control and sucrose-loaded cells (left). TFEB mRNA expression evaluated by quantitative RT-PCR (right). Expression levels were normalized using HMBS and B2M as housekeeping genes. Data are expressed as means ± sd; (n = 3). B) Immunostaining of TFEB in nuclear extracts from ctrl and sucrose-loaded cells (left). Semiquantitative graph of nuclear TFEB normalized as TFEB/histone H3 (right). ***P < 0.0003 vs. ctrl (n = 4).
Sucrose-induced lysosomal accumulation impairs the autophagic flux
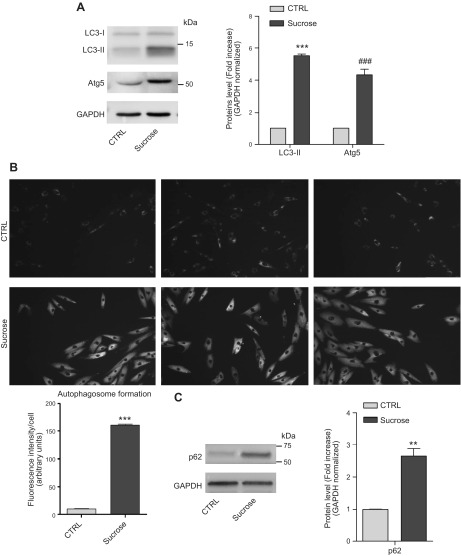
We found an increased level of the autophagic markers, such as microtubule-associated proteins LC3-II (1A/1B light chain 3B) and Atg5 (autophagy protein 5), in sucrose-loaded cells compared with controls (Fig. 3A). In addition, using the autophagosome detection assay, we found that sucrose-loaded cells are characterized by an increased number of autophagosomes with respect to control cells (Fig. 3B). To evaluate the autophagic flux, we treated control and sucrose-loaded cells with bafilomycin A1 (52). As shown in Supplemental Fig. 1, control cells treated with bafilomycin showed a marked increase of LC3-II protein level with respect to untreated control cells. In sucrose-loaded cells we found that bafilomycin induced a greater increase in LC3-II than the untreated sucrose-loaded cells did. The difference in LC3-II levels in the presence or absence of bafilomycin is greater under sucrose loading, indicating that autophagic flux is increased (48, 53). In sucrose-loaded cells, however, we found an increased amount of the autophagy substrate p62 (sequestosome-1) (Fig. 3C), suggesting the blockage of autophagosome degradation (53).
Figure 3.
Macroautophagic markers in sucrose-loaded cells. A) Immunostaining of LC3-I, LC3-II, and Atg5 along with a graph representing the fold change of protein levels over control (ctrl). Data are expressed as means ± sd. ***P < 0.0001 vs. ctrl, ###P < 0.0008 vs. ctrl (n = 3). B) Autophagosomes were evaluated in control and 14-d sucrose-loaded cells using an autophagosome detection assay. The pictures are representative of 3 experiments. The graph represents the fluorescence associated with each cell, data were obtained through the quantification of 70 cells for both sucrose-loaded and ctrl cells. ***P < 0.0001 (n = 3). C) Immunostaining of p62 accompanied by a graph representing the fold change of protein levels over ctrl. Data are expressed as means ± sd. **P < 0.0023 vs. ctrl (n = 3). GAPDH was used as the loading ctrl.
Sucrose loading reduces lysosomal SL catabolism
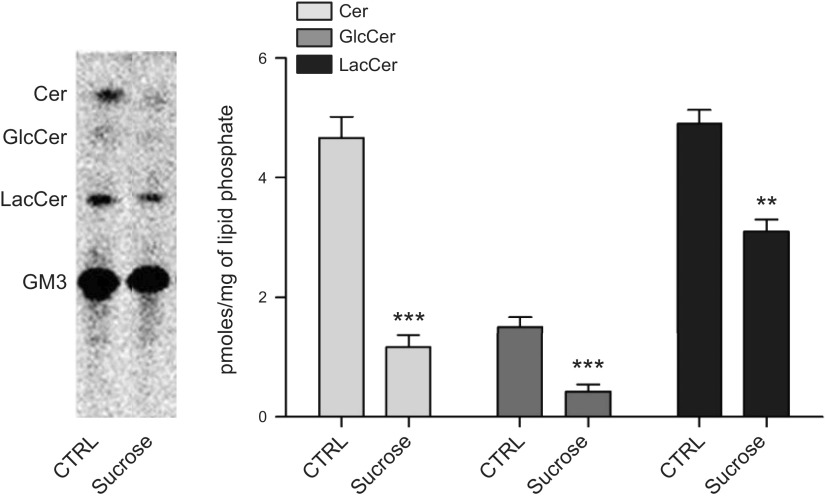
We measured the activity of the lysosomal enzymes GCase, β-galactosidase, β-hexosaminidase, and SMase. As shown in Supplemental Table 1, all lysosomal enzyme activity increased significantly in sucrose-loaded fibroblasts with respect to control cells. GCase activity was 2.6-fold higher, β-galactosidase activity increased 1.9-fold, β-hexosaminidase 1.7-fold, and SMase 3.0-fold. GCase increase was also confirmed by Western blot and qRT-PCR (Supplemental Fig. 2). Because lysosomes are the site of SL catabolism, we assessed lysosome functionality in living cells by feeding radioactive GM3 to control and sucrose-loaded fibroblasts. Cells incorporated the same amount of radioactive GM3 independently from the sucrose loading; however, GM3 catabolism was reduced in sucrose-loaded fibroblasts (Fig. 4). Indeed, quantities of GM3 catabolites lactosylceramide, glucosylceramide, and ceramide were reduced by 1.6-, 3.8-, and 3.8-fold, respectively. These results indicate that, despite the activation of lysosomal biogenesis, catabolism of SLs is strongly impaired in lysosomes of sucrose-loaded cells.
Figure 4.
Sucrose-loaded fibroblasts are characterized by an impairment in SL degradation. Lysosomal catabolism of [3-3H(sphingosine)]GM3 in control and sucrose-loaded fibroblasts. Digital autoradiography of HPTLC (left); graph representing the quantification of GM3 catabolites (right). Data are expressed as picomoles of radioactive lipids per milligram of lipid phosphate. Cer, ceramide; GlcCer, glucosylceramide; LacCer, lactosylceramide. Data are expressed as means ± sd. ***P < 0.0005 vs. control (ctrl), **P < 0.005 vs. ctrl (n = 3).
Impaired lysosomal catabolism induces changes in complex lipid composition
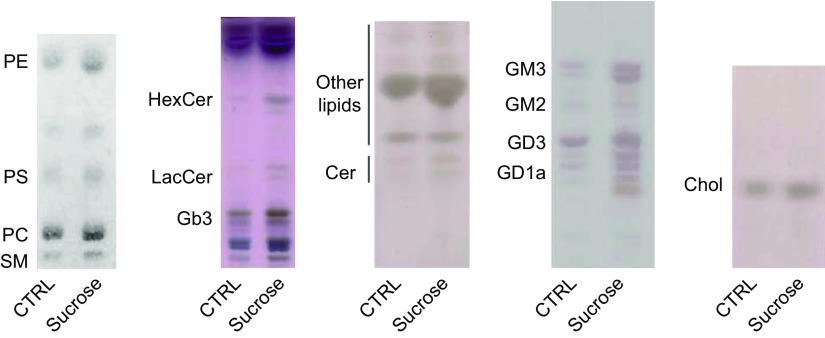
We verified the effect of lysosomal impairment on cell lipid content and pattern. The phospholipid content in sucrose-loaded fibroblasts was characterized by a 3.5-fold increase with respect to control cells (130 ± 12 µg/106 control cells vs. 455 ± 26 µg/106 sucrose-loaded cells). The analyses of the phospholipid pattern by HPTLC revealed an increased content of sphingomyelin, phosphatidylcholine, phosphatidylserine, and phosphatidylethanolamine (Fig. 5, first panel). We performed more detailed analysis by HPLC-MS/MS of the sphingomyelin molecular species. As shown in Supplemental Table 2, we found that all the species revealed were augmented in sucrose-loaded fibroblasts with the highest increase for the C14, C20, C22:1, and C26:1.
Figure 5.
Lipids levels in sucrose-loaded fibroblasts. HPTLC of phospholipids, neutral glycosphingolipids, ceramide, gangliosides, and cholesterol. Cer, ceramide; Chol, cholesterol; Gb3, globotriaosylceramide; HexCer, hexosylceramides; LacCer, lactosylceramide; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PS, phosphatidylserine; SM, sphingomyelin.
Sucrose-loaded fibroblasts were also characterized by higher levels of globotriaosylceramide, lactosylceramide, hexosylceramides (glucosylceramide and galactosylceramide), and ceramide, compared with control cells (Fig. 5, second and third panels). In particular, we used HPLC-MS/MS to analyze the cellular content in terms of the different molecular species of hexosylceramide and ceramide (Supplemental Table 2). MS analyses revealed that sucrose-loaded cells are characterized by a 3.3-fold increase of hexosylceramide content with respect to control cells (Table 1). The molecular species that underwent a major increase are C14, C16, C22, C24:1, C26, and C26:1 (Supplemental Table 2). MS analyses showed a ∼2-fold increase in the ceramide content of sucrose-loaded cells with respect to controls (Table 1). In the case of ceramide, the molecular species that increased significantly are C14, C18, C20, C22, C26, and C26:1 (Supplemental Table 2). Total ganglioside content, evaluated by MS analysis, increased 2.8-fold in sucrose-loaded cells with respect to controls (Fig. 5, fourth panel, and Table 1). In particular, GM3 levels were 3-fold higher, GM2 1.6-fold, GD3 2.8-fold, and GD1a 3-fold in sucrose-loaded cells compared with controls (Supplemental Table 2 and Table 1). In addition, total cholesterol was augmented in sucrose-loaded fibroblasts with respect to controls (Fig. 5, last panel); the quantitation performed using Amplex Red Cholesterol Assay revealed an increase of 2.3-fold.
TABLE 1.
SL content in control and sucrose-loaded fibroblasts
| SLs | Ctrl | Sucrose |
|---|---|---|
| Cer | 797 ± 56 | 1870 ± 90 |
| SM | 6064 ± 682 | 9368 ± 992 |
| HexCer | 345 ± 39 | 1136 ± 93 |
| GM3 | 508 ± 17 | 1508 ± 24 |
| GM2 | 108 ± 4 | 174 ± 8 |
| GD3 | 489 ± 13 | 1383 ± 20 |
| GD1a | 128 ± 3 | 400 ± 5 |
Gangliosides (GM3, GM2, GD3, and GD1a), hexosylceramide (HexCer), sphingomyelin (SM), and ceramide (Cer) content were evaluated by HPLC-MS/MS in control and sucrose-loaded fibroblasts. Data are expressed as mean ± sd pmol/mg cell protein (n = 3). Ctrl, control.
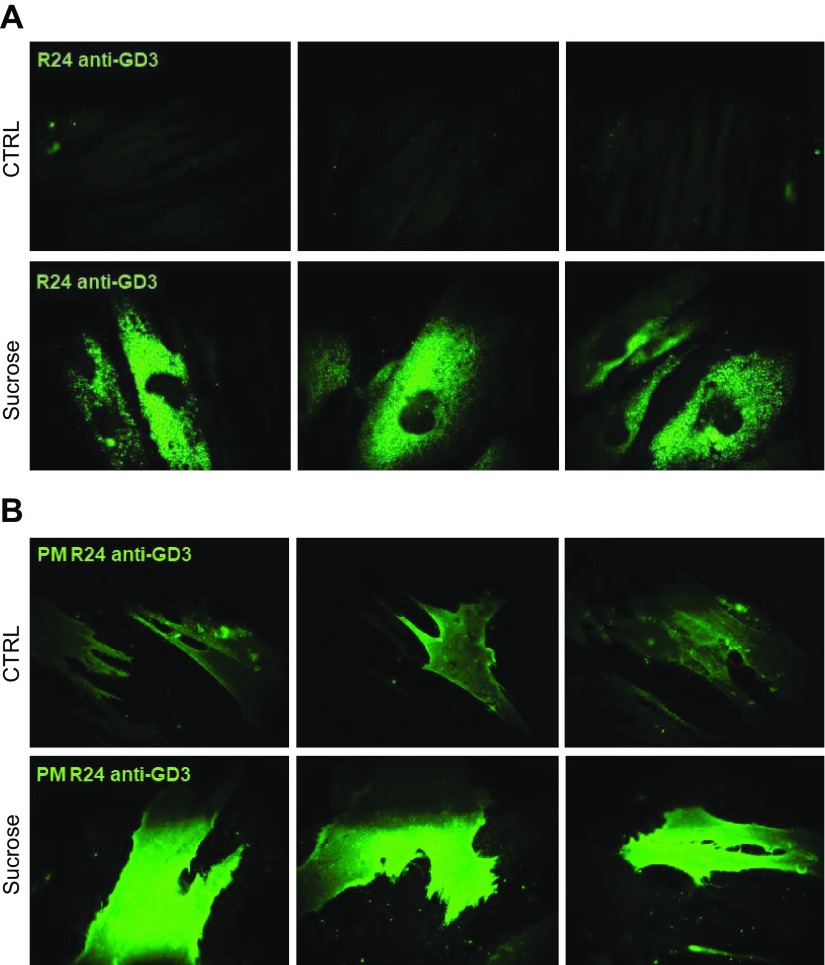
In this fibroblast cell line, GD3 is both particularly abundant and detectable by immunofluorescence, thanks to the specific mAb R24. As shown in Fig. 6A, we found in sucrose-loaded permeabilized cells a marked staining of GD3 mainly associated with intracellular organelles, whereas the fluorescence was scarcely detectable in control permeabilized cells. In nonpermeabilized cells, we observed GD3 staining in both controls and sucrose-loaded cells. However, a stronger signal was clearly associated with the PM of sucrose-loaded cells (Fig. 6B). These data suggest that lysosomal impairment induces changes in SL composition at both intracellular and PM levels.
Figure 6.
Sucrose-loaded fibroblasts are characterized by an increased content of GD3 at both intracellular and PM levels. A) Indirect immunofluorescence of GD3 ganglioside obtained by staining with R24 antibody in human fibroblasts, loaded or not with sucrose permeabilized with Triton X-100. B) PM levels of GD3 ganglioside evaluated by indirect immunofluorescence in fibroblasts, loaded or not with sucrose with sucrose, in nonpermeabilizing conditions. Images are representative of 10 fields for each sample. Results represent 3 independent experiments performed in triplicate.
To further investigate the possible increase of SL content on the PM after lysosomal impairment, we carried out a procedure that allows the isolation of specific portions of the PM without contamination from subcellular membranes. Cell SLs were metabolically labeled at the steady state using the radioactive precursor [1-3H]sphingosine and then subjected or not to sucrose loading for 14 d. The proteins associated with the external leaflet of the PM were subsequently biotinylated and cells were lysed with 1% Triton X-100 to preserve the lipid–protein interactions. The same quantity of cell lysates (1 mg) was subjected to immunoprecipitation using streptavidin-conjugated magnetic beads. As shown in Supplemental Fig. 3A, we immunoprecipitated >90% of the biotinylated proteins in both control and sucrose-loaded cells. Sucrose-loaded cells were characterized by a stronger staining of biotinylated proteins with respect to controls, indicating an increase in PM-associated proteins. We can exclude differences in the biotinylation process because we used a saturated concentration of Sulfo-NHS-Biotin. We confirmed the immunoprecipitation of PM proteins as indicated by the presence of integrin α5 in IPs (Supplemental Fig. 3B). In addition, we were able to avoid contaminating IP with intracellular proteins, as demonstrated by the immunoblotting of calnexin and protein disulfide isomerase, endoplasmic reticulum markers, and the trans-Golgi network integral membrane protein TGN38, a marker of the Golgi apparatus (Supplemental Fig. 3B).
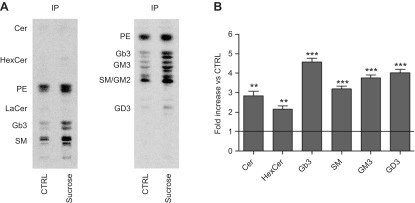
We performed lipid extraction from the PNS, SNIP, and IP. As shown in Supplemental Fig. 4, we found that sucrose-loaded cells were characterized by an increased level of radioactive SLs in accordance with the data obtained from our analyses of the endogenous counterpart. The analyses of sucrose-loaded cells indicated an increase of the SLs associated with both SNIP and IP with respect to controls. Interestingly, the SL changes found in PNS and SNIP were very similar, indicating that the majority of uncatabolized SLs remain inside the cell (Supplemental Fig. 4). From the analysis of SLs associated with the IP, we found that at the PM ceramide increased 3-fold, glucosylceramide 2-fold, Gb3 4-fold, sphingomyelin 3-fold, and the gangliosides 3.5-fold vs. control cells (Fig. 7).
Figure 7.
SL composition and content of detergent-resistant PM regions. A) Representative HPTLC of the radioactive lipids associated with the IP of PM biotinylated proteins performed with 2 solvent systems: chloroform:methanol:water [110:40:6 (v:v:v)] (a) , and chloroform:methanol:0.2% aqueous CaCl2 [50:42:11 (v:v:v)] (b). B) Graph representing the fold increase of each lipid in sucrose-loaded cells with respect to control (ctrl). ***P < 0.0002 vs. ctrl, **P < 0.004 vs. ctrl. The line represents the value fixed at 1 for the ctrl.
Impact of lysosomal impairment on the mRNA expression of lysosome- and cell cycle–related genes
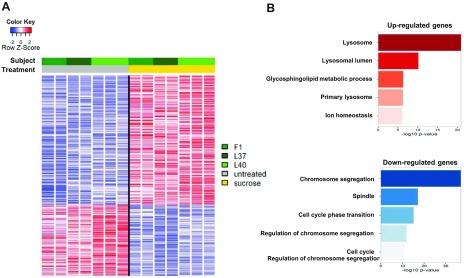
We performed RNA sequencing (RNAseq) experiments on 3 fibroblast cell lines derived from 3 different individuals and subjected or not to sucrose loading (3 biological replicates each performed in duplicate or triplicate, n = 14). In spite of the modest group size and the expected heterogeneity between individuals, sucrose loading led to altered transcription of 532 genes (352 up-regulated and 180 down-regulated, FDR ≤0.01, fold change ≥2 or ≤−2; Fig. 8A, Supplemental Data 1, and Supplemental Fig. 5). GO analysis highlighted the lysosome as the most significantly enriched pathway (P < 1 × 10−20) among up-regulated transcripts and functional terms related to cell cycle and mitosis as the most significantly enriched among down-regulated transcripts (Fig. 8B and Supplemental Data 2).
Figure 8.
Lysosomal accumulation impacts on the expression of lysosome- and cell cycle–related genes. A) Heatmap showing selective gene expression alterations in fibroblasts loaded with sucrose for 14 d (3 biological replicates—L37, L40, F1—each performed in duplicate or triplicate; n = 14). Each row was normalized by z score. B) GO analysis showing enrichment of functional terms among sucrose-induced (red) and sucrose-repressed (blue) genes.
Lysosomal impairment affects lysosome–plasma membrane fusion and promotes SL catabolism at the PM
We evaluated the quantity of Lamp-1 associated with the PM as a marker of lysosomal fusion with the cell surface by immunofluorescence under nonpermeabilizing conditions (54–56). As shown in Fig. 9A, we detected a weak signal in control cells, whereas a more marked staining of PM-associated Lamp-1 was detected in sucrose-loaded cells. In addition, by evaluating Lamp-1 levels in the IP of the biotinylated PM proteins, we detected a more intense signal in sucrose-loaded cells with respect to controls (Fig. 9B). The presence of Lamp-1 on the PM of control cells results from the physiologic fusion between lysosomes and the PM as well as the previously described process for cell-surface damage repair (54, 57).
Figure 9.
Aberrant catabolism of complex glycosphingolipids on the PM. A) Immunofluorescence of PM-associated Lamp-1 in human fibroblasts loaded or not with sucrose. B) Immunostaining of Lamp-1 associated with immunoprecipitated PM biotinylated proteins (biotin-IP) from sucrose-loaded and control cells. C) Released cathepsin D in culture medium from control (ctrl) and sucrose-loaded cells after 14 d, detected by ELISA. ***P < 0.0002 vs. ctrl. D) PM-associated activities of GCase, NLGase, β-galactosidase, and β-hexosaminidase. Activities were normalized to nmol/106 cells/h. Data are expressed as means ± sd. ***P < 0.0007 vs. ctrl (n = 4). E) HPTLC of [3-3H(sphingosine)]GM3 catabolites produced at the PM level in ctrl and sucrose-loaded fibroblasts. The graph represents the quantification of GM3 catabolites. Data are expressed as mean ± sd percentages of radioactivity with respect to the total radioactivity associated with the lipid extracts. ***P < 0.0005 vs. ctrl, **P < 0.005 vs. ctrl. Cer, ceramide; GlcCer, glucosylceramide; LacCer, lactosylceramide.
To further support the increased fusion of lysosomes with the PM, we evaluated by ELISA assay the quantity of the soluble lysosomal enzyme cathepsin D in the culture medium of control cells and cells loaded with sucrose for 14 d. As shown in Fig. 9C, the medium derived from sucrose-loaded cells was characterized by a 1.7-fold increase in cathepsin D content with respect to that found in control cells.
It is currently accepted that several lysosomal glycohydrolytic enzymes are also associated with the cell surface because of the fusion between lysosomes and the PM (35, 36, 58). For this reason, we measured the activity of GCase, β-galactosidase, and β-hexosaminidase on the PM of living cells. In addition, we evaluated the activity of NLGase, which mainly operates on the PM. We found that GCase was increased 6.4-fold, NLGase 2-fold, β-galactosidase 12.6-fold, and β-hexosaminidase 18.8-fold in sucrose-loaded cells compared with controls (Fig. 9D and Supplemental Table 3).
Western blot revealed an increase in GCase associated with the IP of sucrose-loaded cells with respect to controls (Supplemental Fig. 6). This result is in line with the GCase activity measured on the PM of control and sucrose-loaded fibroblasts (Fig. 9D). The presence of GCase in the IP of control cells is in accordance with previous findings regarding its association with the PM of cells of different origin (16, 35, 36).
We further investigated the ability of glycohydrolytic enzymes to exert their activity directly on natural substrates associated with the PM. To this end, control and sucrose-loaded fibroblasts were treated with chloroquine and were then fed [3-3H(sphingosine)]GM3. In this condition, [3-3H(sphingosine)]GM3 can be catabolized on the cell surface only through the action of PM-associated glycohydrolases (17). Cells incorporated in the PM absorbed the same amount of radioactive GM3 independent of sucrose loading; nevertheless, as shown in Fig. 9E, we found that [3-3H(sphingosine)]GM3 catabolism on the PM was increased in sucrose-loaded fibroblasts with respect to controls. Indeed, the percentage of radioactivity associated with GM3 catabolites was significantly augmented in sucrose-loaded cells: lactosylceramide 1.6-fold, glucosylceramide 5.3-fold, and ceramide 11.5-fold.
Taken together, these results indicate that sucrose loading induces increased fusion between lysosomes and the PM and determines the aberrant association of SLs and SL hydrolases with the cell surface.
Ectopic glycosphingolipid catabolism is associated with the onset of cell cycle arrest
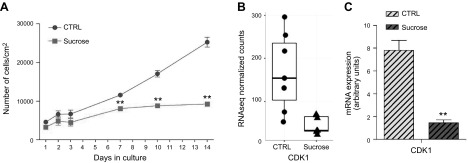
At different time points, we evaluated the effect of progressive lysosomal impairment on cell viability. As shown in Fig. 10A, no significant effects of sucrose loading were observed until d 7, indicating the absence of acute toxic effects triggered directly by sucrose. Comparing doubling times, on d 14 of sucrose loading, cells reached a 75–78% reduction in cell growth with respect to controls. At the same time point, we found by Hoechst assay that sucrose-loaded cells were characterized by a 15% increase in apoptotic cells compared with unloaded ones (unpublished results). The long-term effect of sucrose loading on cell growth is in line with the results obtained from RNAseq, which indicated significant enrichment of genes involved in controlling the cell cycle among down-regulated genes (Fig. 8B). Indeed, after 14 d of sucrose loading, ∼28% (51/180) of down-regulated genes encode for proteins involved in cell cycle regulation (Supplemental Data 1). Among these, it emerged that CDK1 was down-regulated (Fig. 10B); this data was also confirmed by Western blot (Fig. 11E).
Figure 10.
Cell growth arrest in sucrose-loaded cells. A) Cell growth curve of control and sucrose-loaded fibroblasts on d 1–3, 7, 10, and 14. Data are expressed as number of living cells per square centimeter of growth area. **P < 0.0094. B) CDK1 expression evaluated by RNAseq on fibroblasts from 3 individuals (n = 7), FDR-corrected P = 0.86 × 10−5 (left). CDK1 mRNA expression evaluated by qPCR. Expression levels were normalized using HMBS and B2M as housekeeping genes (right). **P < 0.0022 (n = 3).
Figure 11.
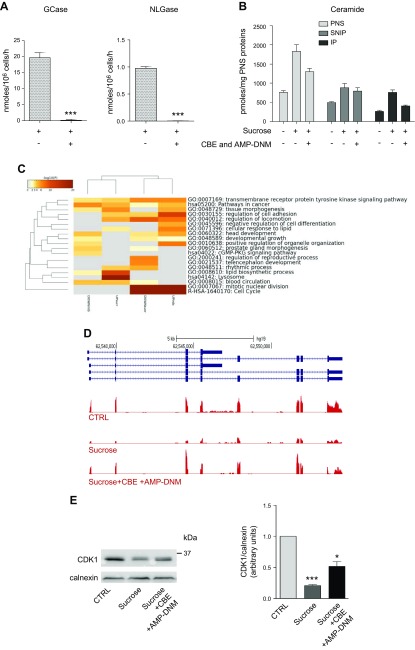
Cell growth arrest is rescued by the inhibition of glycosphingolipid catabolism on the PM. A) PM-associated activities of GCase and NLGase in sucrose-loaded cells treated or untreated with GCase and NLGase inhibitors (CBE and AMP-DNM, respectively). Activities were normalized to nmol/106 cells/h ± sd. ***P < 0.0005 vs. sucrose (n = 3). B) Graph representing the picomoles of ceramide associated with the fraction obtained by streptavidin separation after cell-surface biotinylation from PNS preparation in conditions that preserved the lipid–protein interactions. Bars refer to control, sucrose-loaded cells, and sucrose-loaded cells treated with inhibitors. *P < 0.04 vs. ctrl, **P < 0.006 vs. ctrl, #P < 0.03 vs. sucrose, ##P < 0.009 vs. sucrose (n = 2). Data are expressed as pmol/mg PNS protein. C) Comparison of GO enrichment analysis among DE genes in sucrose-loaded cells untreated or treated with GCase and NLGase inhibitors after 14 d. The heatmap reports P values for significance of enrichment; gray cells indicate a lack of enrichment. D) Snapshot of RNAseq expression data for the CDK1 gene, showing down-regulated and enhanced mRNA levels, respectively, in fibroblasts treated with sucrose alone or with sucrose and GCase and NLGase inhibitors (CBE + AMP-DNM). E) Immunostaining of CDK1 evaluated in control (ctrl), 14-d sucrose-loaded cells (sucrose), and 14-d sucrose-loaded cells treated with GCase and NLGase inhibitors (sucrose + CBE + AMP-DNM); calnexin was used as the loading ctrl (left). A semiquantitative graph of normalized CDK1/calnexin (right). ***P < 0.0001 vs. ctrl, *P < 0.02 vs. sucrose (n = 3).
To verify the possible involvement of PM-associated glycohydrolases with aberrant ectopic catabolism of glycosphingolipids in cell cycle arrest, after 12 d of sucrose loading, fibroblasts were treated for 48 h with CBE and AMP-DNM, which specifically inhibit GCase and NLGase, respectively. As shown in Fig. 11A, after treatment with the inhibitors, GCase and NLGase activity associated with the PM was almost undetectable.
The analyses of the radioactive ceramide associated with the lipid environment of the immunoprecipitated biotinylated proteins reveal a decrease in its content in sucrose-loaded cells treated with β-glucocerebrosidase inhibitors with respect to that found in not inhibited cells (Fig. 11B).
Using RNAseq, we analyzed the global effect of CBE and AMP-DNM treatment on transcriptional output. Compared with cells that were only loaded with sucrose for the same period (14 d), fibroblasts that received GCase and NLGase inhibitors for the last 2 d of culture showed dramatic changes in gene expression. Indeed, >3000 genes were significantly DE between the 2 conditions (3143 DE genes, 1222 up-regulated and 1921 down-regulated; FDR ≤0.01, fold change ≥2 or ≤−2; unpublished results). We found DE genes upon sucrose loading that were also deregulated between sucrose-loaded and CBE+AMP-DNM–treated sucrose-loaded cells (FDR ≤0.01, fold change ≥2 or ≤−2). In particular, 427 DE genes were shared between the 2 conditions, 307 (71.8%) of which changed the direction of expression. Specifically, 165 sucrose-induced genes were repressed by β-glucocerebrosidase inhibitors, whereas 142 genes were repressed by sucrose and induced by inhibitor treatment (Fig. 11C and Supplemental Fig. 7). GO analysis indicated that functional terms related to cell cycle were the most significantly enriched (P < 1 × 10−37) among transcripts that switched from down-regulated (by sucrose) to up-regulated (by CBE+AMP-DNM), suggesting a generalized and significant reversion in the inhibition of cell proliferation (Fig. 11D) that was confirmed by the increase in CDK1 mRNA expression and protein content (Fig. 11E).
These results indicate that ectopic glycosphingolipid catabolism on the cell surface could be responsible for the activation of downstream signaling pathways involved in the control of cell proliferation.
DISCUSSION
In this study we propose a molecular mechanism responsible for the onset of cell damage upon lysosomal accumulation of undigested materials. Increasing evidence reported in the literature supports the etiopathological role of lysosomal storage and dysfunction, independent of the nature of the stored material, in several diseases ranging from neurodegenerative disorders to aneuploidy in cancer (5–7, 59, 60). One of the main challenges in conducting these studies is developing cell models characterized by the accumulation of high levels of uncatabolized molecules within the lysosomes as occurs in the cells of affected organs. Indeed, our data and those obtained from other groups highlight the need to overcome a threshold of accumulation to induce the onset of cell damage (59).
To this purpose, we used a simple cellular model of lysosomal accumulation represented by human fibroblasts subjected to sucrose loading for 14 d (43–48). Sucrose is not degraded in fibroblasts because of the lack of invertase and does not induce osmotic intracellular stress at 88 mM, the concentration we used (44, 48).
As already reported by others, in sucrose-loaded cells we observed the activation of lysosomal biogenesis controlled by TFEB nuclear translocation (10). After a whole-transcriptome analysis using RNAseq technology, however, we found that sucrose loading also leads to altered transcription of genes not under the direct control of TFEB. These data suggest the involvement of a more complex genetic program coordinated by other transcription factors. GO analyses identified lysosome-related genes as the most significantly enriched among the up-regulated transcripts. In fact, sucrose-loaded fibroblasts were characterized by the production of new lysosomes, suggesting an adaptive cell response against sucrose loading aimed at removing accumulated material. Nevertheless, the newly formed lysosomes did not stop the accumulation of the undigested molecule.
Lysosomes are commonly considered as “waste recycling” organelles because they catabolize complex macromolecules to produce catabolites mainly reutilized in biosynthetic pathways. However, lysosomes that accumulate uncatabolized substrates are no longer able to refill cell with nutrients, mimicking an “apparent starvation” (57). In this condition, as seen in sucrose-loaded fibroblasts, cells activate rescue pathways such as macroautophagy, but such an effort is ultimately futile due to a lack of lysosomal functionality, as demonstrated by the accumulation of the autophagic substrate p62.
The reduced catabolic activity of engulfed lysosomes was also suggested by their inability to degrade complex SLs, thus resulting in SL accumulation. We excluded the contribution of the biosynthetic pathway because we did not observe any significant change in the transcription of the key enzymes involved in SL biosynthesis, such as serine palmitoyltransferase 1 and 2, sphingomyelin synthase 1 and 2, glucosylceramide synthase, and GM3 synthase (unpublished results).
Alteration in SL pattern and composition is a common feature of several lysosome-dependent diseases, and SLs are considered as secondary accumulated materials in several lysosomal storage disorders independent of the primary ones (61, 62). Nevertheless, the localization of SL secondary storage is still poorly understood. Interestingly, in our model we found that SL accumulation occurred not only in the intracellular compartments but also on the PM.
Several processes could interact to determine a different SL composition on the cell surface. In sucrose-loaded cells, we observed the induction of lysosomal fusion with the PM, as indicated by the increased content of Lamp-1 associated with the cell surface, by the augmented activity of PM-associated glycosphingolipid hydrolases of lysosomal origin, as well as by an increased amount of the soluble lysosomal protein cathepsin D in the extracellular media. Based on this evidence, it is reasonable to speculate that the fusion between lysosomes and the cell surface could directly modify the PM glycosphingolipid composition by the inclusion of uncatabolized SLs and their catabolic enzymes.
Interestingly, aside from the increased activity of PM-associated GCase, in sucrose-loaded fibroblasts we also found increased activity of NLGase. GCase and NLGase are the enzymes responsible for the last step of glycosphingolipid catabolism that leads to ceramide production. Ceramide formation via aberrant glycosphingolipid catabolism on the PM has been proposed to have a role in the onset of cell damage (14, 17, 63–67). In line with this evidence, we found that, starting on d 7 of sucrose loading, lysosomal impairment induced a significant decrease in cell proliferation that resulted in cell growth arrest by d 14. In sucrose-loaded cells we found that several genes coding for proteins regulating the cell cycle, including CDK1, were down-regulated. Notably, a similar phenotype was also observed upon aberrant protein accumulations in aneuploid cells (59, 60, 68). To investigate the possible effect of ceramide production due to the ectopic catabolism of PM glycosphingolipids in cell cycle arrest, we treated sucrose-loaded fibroblasts with the 2 specific inhibitors for GCase and NLGase. In the presence of these 2 inhibitors, GCase and NLGase activity was almost undetectable; therefore, they must not contribute to the PM ceramide production from glycosphingolipid hydrolysis. We also found that the inhibition of GCase and NLGase induced a partial rescue of cell cycle arrest, as indicated by the up-regulation of both CDK1 protein levels and genes responsible for cell proliferation.
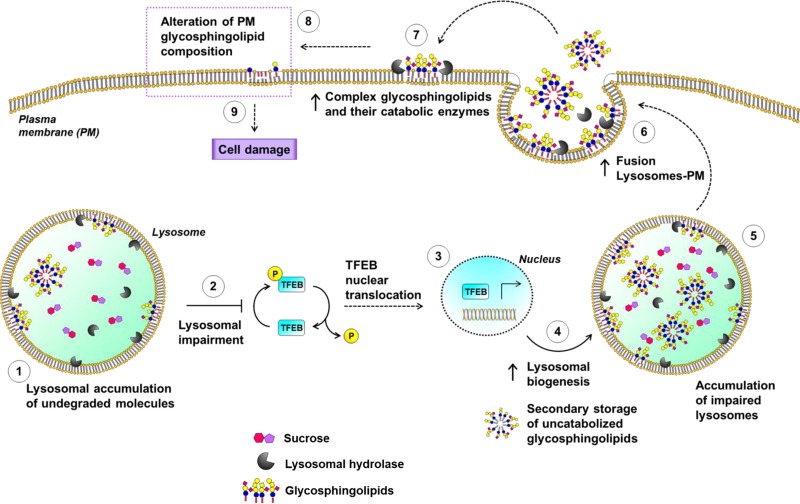
In summary, our results suggest that the alteration in SL composition on the PM triggered by lysosomal impairment is involved in the onset of cell damage in an in vitro model of lysosomal accumulation. The data obtained led us to speculate on the model schematized in Fig. 12, where the primary accumulation of a substrate within the lysosomes leads to a more general lysosomal impairment that results in the storage of other undigested materials, such as SLs. Lysosomal impairment causes the nuclear translocation of TFEB, which in turn is responsible for increased biogenesis of lysosomes and the activation of futile macroautophagy. In addition, the cell promotes the fusion between lysosomes and the PM, consistent with an increase in both PM SLs and PM-associated glycosphingolipid hydrolases. These events lead to an abnormal catabolism of the PM glycosphingolipids with the production of ceramide, events that are strictly related to the onset of cell cycle arrest.
Figure 12.
A suggested molecular mechanism linking lysosomal engulfment to the onset of cell damage: crosstalk between lysosomes and PM.
Supplementary Material
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
ACKNOWLEDGMENTS
The authors thank Dr. Maria Carla Panzeri (Advanced Light and Electron Microscopy BioImaging Center, San Raffaele Scientific Institute, Milan, Italy) for her expert assistance in electron microscopy imaging, and Prof. J. M. F. G. Aerts (Leiden University, Leiden, The Netherlands) for providing AMP-DNM. This work was supported by Fondazione Cariplo Grant 2015–1017 to M.A., and was partially supported by the Italian National Research Council’s flagship “InterOmics” Project (PB.P05), part of the 2012–2014 program on aging sponsored by the National Research Program and the National Research Council (Italian Ministry of Education, Universities and Research). Lipidomic analyses were partially supported by the Medical University of South Carolina’s Lipidomics Shared Resource [Hollings Cancer Center, Medical University of South Carolina (MUSC); Grant P30 CA138313]; the Lipidomics Shared Resource of the South Carolina Lipidomics and Pathobiology Centers of Biomedical Research Excellence (COBRE); MUSC Department of Biochemistry (Grant P20 RR017677); and the U.S. National Institutes of Health, National Center for Research Resources and Office of the Director (Grant C06 RR018823) for the occupancy of space for the Lipidomics Shared Resource, 505 Children’s Research Institute (CRI) Analytical Unit. The authors declare no conflicts of interest.
Glossary
- AMP-DNM
adamantane–pentyl-dNM–N-(5-adamantane-1-yl-methoxy-pentyl)-deoxynojirimycin
- AP
aqueous phase
- B2M
β2-microglobulin
- CBE
conduritol B epoxide
- CDK
cyclin-dependent kinase
- DE
differentially expressed
- FDR
false discovery rate
- GCase
β-glucocerebrosidase
- GO
gene ontology
- HMBS
hydroxymethylbilane synthase
- HPTLC
high performance thin-layer chromatography
- HRP
horseradish peroxidase
- IP
immunoprecipitate
- MUB
4-methylumbelliferone
- NLGase
nonlysosomal glucosylceramidase
- OP
organic phase
- PM
plasma membrane
- PNS
postnuclear supernatant
- qRT-PCR
quantitative RT-PCR
- RNAseq
RNA sequencing
- SL
sphingolipid
- SMase
sphingomyelinase
- SNIP
supernatant after immunoprecipitation
- TFEB
transcription factor EB
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
M. Samarani performed the experiments, analyzed the data, and participated in writing the manuscript; N. Loberto performed endogenous lipid analyses; G. Soldà, L. Straniero, R. Asselta, and S. Duga performed RNAseq analyses, analyzed the data, and wrote the related paragraph of the Results section; G. Lunghi conducted the total enzymatic activity; F. A. Zucca performed transmission electron microscope analyses; L. Mauri and M. G. Ciampa performed and analyzed the mass spectrometry experiments; D. Schiumarini conducted the PM enzymatic activity; P. Giussani helped in the setup of immunofluorescence experiments and in the analyses of the data; R. Bassi, E. Chiricozzi, and A. Prinetti equally contributed in editing the content of this manuscript; and M. Aureli and S. Sonnino designed and coordinated the study, analyzed the data, and wrote the manuscript.
REFERENCES
- 1.Appelmans F., Wattiaux R., De Duve C. (1955) Tissue fractionation studies. 5. The association of acid phosphatase with a special class of cytoplasmic granules in rat liver. Biochem. J. 59, 438–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Duve C., Pressman B. C., Gianetto R., Wattiaux R., Appelmans F. (1955) Tissue fractionation studies. 6. Intracellular distribution patterns of enzymes in rat-liver tissue. Biochem. J. 60, 604–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saftig P., Klumperman J. (2009) Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat. Rev. Mol. Cell Biol. 10, 623–635 [DOI] [PubMed] [Google Scholar]
- 4.Perera R. M., Zoncu R. (2016) The lysosome as a regulatory hub. Annu. Rev. Cell Dev. Biol. 32, 223–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Platt F. M., Boland B., van der Spoel A. C. (2012) The cell biology of disease: lysosomal storage disorders: the cellular impact of lysosomal dysfunction. J. Cell Biol. 199, 723–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L., Sheng R., Qin Z. (2009) The lysosome and neurodegenerative diseases. Acta Biochim. Biophys. Sin. (Shanghai) 41, 437–445 [DOI] [PubMed] [Google Scholar]
- 7.Carmona-Gutierrez D., Hughes A. L., Madeo F., Ruckenstuhl C. (2016) The crucial impact of lysosomes in aging and longevity. Ageing Res. Rev. 32, 2–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Duve C., de Barsy T., Poole B., Trouet A., Tulkens P., Van Hoof F. (1974) Commentary. Lysosomotropic agents. Biochem. Pharmacol. 23, 2495–2531 [DOI] [PubMed] [Google Scholar]
- 9.Lu S., Sung T., Lin N., Abraham R. T., Jessen B. A. (2017) Lysosomal adaptation: how cells respond to lysosomotropic compounds. PLoS One 12, e0173771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sardiello M., Palmieri M., di Ronza A., Medina D. L., Valenza M., Gennarino V. A., Di Malta C., Donaudy F., Embrione V., Polishchuk R. S., Banfi S., Parenti G., Cattaneo E., Ballabio A. (2009) A gene network regulating lysosomal biogenesis and function. Science 325, 473–477 [DOI] [PubMed] [Google Scholar]
- 11.Palmieri M., Impey S., Kang H., di Ronza A., Pelz C., Sardiello M., Ballabio A. (2011) Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum. Mol. Genet. 20, 3852–3866 [DOI] [PubMed] [Google Scholar]
- 12.Prinetti A., Prioni S., Chiricozzi E., Schuchman E. H., Chigorno V., Sonnino S. (2011) Secondary alterations of sphingolipid metabolism in lysosomal storage diseases. Neurochem. Res. 36, 1654–1668 [DOI] [PubMed] [Google Scholar]
- 13.Sonnino S., Prinetti A., Mauri L., Chigorno V., Tettamanti G. (2006) Dynamic and structural properties of sphingolipids as driving forces for the formation of membrane domains. Chem. Rev. 106, 2111–2125 [DOI] [PubMed] [Google Scholar]
- 14.Aureli M., Bassi R., Prinetti A., Chiricozzi E., Pappalardi B., Chigorno V., Di Muzio N., Loberto N., Sonnino S. (2012) Ionizing radiations increase the activity of the cell surface glycohydrolases and the plasma membrane ceramide content. Glycoconj. J. 29, 585–597 [DOI] [PubMed] [Google Scholar]
- 15.Aureli M., Gritti A., Bassi R., Loberto N., Ricca A., Chigorno V., Prinetti A., Sonnino S. (2012) Plasma membrane-associated glycohydrolases along differentiation of murine neural stem cells. Neurochem. Res. 37, 1344–1354 [DOI] [PubMed] [Google Scholar]
- 16.Aureli M., Loberto N., Lanteri P., Chigorno V., Prinetti A., Sonnino S. (2011) Cell surface sphingolipid glycohydrolases in neuronal differentiation and aging in culture. J. Neurochem. 116, 891–899 [DOI] [PubMed] [Google Scholar]
- 17.Valaperta R., Chigorno V., Basso L., Prinetti A., Bresciani R., Preti A., Miyagi T., Sonnino S. (2006) Plasma membrane production of ceramide from ganglioside GM3 in human fibroblasts. FASEB J. 20, 1227–1229 [DOI] [PubMed] [Google Scholar]
- 18.Valaperta R., Valsecchi M., Rocchetta F., Aureli M., Prioni S., Prinetti A., Chigorno V., Sonnino S. (2007) Induction of axonal differentiation by silencing plasma membrane-associated sialidase Neu3 in neuroblastoma cells. J. Neurochem. 100, 708–719 [DOI] [PubMed] [Google Scholar]
- 19.Sciannamblo M., Chigorno V., Passi A., Valaperta R., Zucchi I., Sonnino S. (2002) Changes of the ganglioside pattern and content in human fibroblasts by high density cell population subculture progression. Glycoconj. J. 19, 181–186 [DOI] [PubMed] [Google Scholar]
- 20.Chigorno V., Pitto M., Cardace G., Acquotti D., Kirschner G., Sonnino S., Ghidoni R., Tettamanti G. (1985) Association of gangliosides to fibroblasts in culture: a study performed with GM1 [14C]-labelled at the sialic acid acetyl group. Glycoconj. J. 2, 279–291 [Google Scholar]
- 21.Chigorno V., Riva C., Valsecchi M., Nicolini M., Brocca P., Sonnino S. (1997) Metabolic processing of gangliosides by human fibroblasts in culture--formation and recycling of separate pools of sphingosine. Eur. J. Biochem. 250, 661–669 [DOI] [PubMed] [Google Scholar]
- 22.Chigorno V., Tettamanti G., Sonnino S. (1996) Metabolic processing of gangliosides by normal and Salla human fibroblasts in culture. A study performed by administering radioactive GM3 ganglioside. J. Biol. Chem. 271, 21738–21744 [DOI] [PubMed] [Google Scholar]
- 23.Giglioni A., Pitto M., Chigorno V., Zorzino L., Ghidoni R. (1990) Subcellular metabolism of exogenous GM1 ganglioside in normal human fibroblasts. Biochem. Int. 22, 85–94 [PubMed] [Google Scholar]
- 24.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Love M. I., Huber W., Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Settembre C., Medina D. L. (2015) TFEB and the CLEAR network. Methods Cell Biol. 126, 45–62 [DOI] [PubMed] [Google Scholar]
- 28.Toyokuni T., Nisar M., Dean B., Hakomori S.-I. (1991) A facile and regiospecific tritiation of sphingosine: synthesis of (2S,3R,4E)-2-amino-4-octadecene-1,3-diol-1-3H. J. Labelled Comp. Radiopharm. 29, 567–574 [Google Scholar]
- 29.Schiumarini D., Loberto N., Mancini G., Bassi R., Giussani P., Chiricozzi E., Samarani M., Munari S., Tamanini A., Cabrini G., Lippi G., Dechecchi M. C., Sonnino S., Aureli M. (2017) Evidence for the involvement of lipid rafts and plasma membrane sphingolipid hydrolases in Pseudomonas aeruginosa infection of cystic fibrosis bronchial epithelial cells. Mediators Inflamm. 2017, 1730245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altin J. G., Pagler E. B. (1995) A one-step procedure for biotinylation and chemical cross-linking of lymphocyte surface and intracellular membrane-associated molecules. Anal. Biochem. 224, 382–389 [DOI] [PubMed] [Google Scholar]
- 31.Cole S. R., Ashman L. K., Ey P. L. (1987) Biotinylation: an alternative to radioiodination for the identification of cell surface antigens in immunoprecipitates. Mol. Immunol. 24, 699–705 [DOI] [PubMed] [Google Scholar]
- 32.Prinetti A., Prioni S., Chigorno V., Karagogeos D., Tettamanti G., Sonnino S. (2001) Immunoseparation of sphingolipid-enriched membrane domains enriched in Src family protein tyrosine kinases and in the neuronal adhesion molecule TAG-1 by anti-GD3 ganglioside monoclonal antibody. J. Neurochem. 78, 1162–1167 [DOI] [PubMed] [Google Scholar]
- 33.Brown D. A., Rose J. K. (1992) Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell 68, 533–544 [DOI] [PubMed] [Google Scholar]
- 34.Boot R. G., Verhoek M., Donker-Koopman W., Strijland A., van Marle J., Overkleeft H. S., Wennekes T., Aerts J. M. (2007) Identification of the non-lysosomal glucosylceramidase as β-glucosidase 2. J. Biol. Chem. 282, 1305–1312 [DOI] [PubMed] [Google Scholar]
- 35.Aureli M., Bassi R., Loberto N., Regis S., Prinetti A., Chigorno V., Aerts J. M., Boot R. G., Filocamo M., Sonnino S. (2012) Cell surface associated glycohydrolases in normal and Gaucher disease fibroblasts. J. Inherit. Metab. Dis. 35, 1081–1091 [DOI] [PubMed] [Google Scholar]
- 36.Aureli M., Masilamani A. P., Illuzzi G., Loberto N., Scandroglio F., Prinetti A., Chigorno V., Sonnino S. (2009) Activity of plasma membrane β-galactosidase and β-glucosidase. FEBS Lett. 583, 2469–2473 [DOI] [PubMed] [Google Scholar]
- 37.Overkleeft H. S., Renkema G. H., Neele J., Vianello P., Hung I. O., Strijland A., van der Burg A. M., Koomen G. J., Pandit U. K., Aerts J. M. (1998) Generation of specific deoxynojirimycin-type inhibitors of the non-lysosomal glucosylceramidase. J. Biol. Chem. 273, 26522–26527 [DOI] [PubMed] [Google Scholar]
- 38.Folch J., Lees M., Sloane Stanley G. H. (1957) A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 39.Bartlett G. R. (1959) Phosphorus assay in column chromatography. J. Biol. Chem. 234, 466–468 [PubMed] [Google Scholar]
- 40.Vaskovsky V. E., Kostetsky E. Y. (1968) Modified spray for the detection of phospholipids on thin-layer chromatograms. J. Lipid Res. 9, 396. [PubMed] [Google Scholar]
- 41.Bielawski J., Pierce J. S., Snider J., Rembiesa B., Szulc Z. M., Bielawska A. (2009) Comprehensive quantitative analysis of bioactive sphingolipids by high-performance liquid chromatography–tandem mass spectrometry. Methods Mol. Biol. 579, 443–467 [DOI] [PubMed] [Google Scholar]
- 42.Sonnino S., Chigorno V., Tettamanti G. (2000) Preparation of radioactive gangliosides, 3H or 14C isotopically labeled at oligosaccharide or ceramide moieties. Methods Enzymol. 311, 639–656 [DOI] [PubMed] [Google Scholar]
- 43.Kato T., Okada S., Yutaka T., Yabuuchi H. (1984) The effects of sucrose loading on lysosomal hydrolases. Mol. Cell. Biochem. 60, 83–98 [DOI] [PubMed] [Google Scholar]
- 44.Karageorgos L. E., Isaac E. L., Brooks D. A., Ravenscroft E. M., Davey R., Hopwood J. J., Meikle P. J. (1997) Lysosomal biogenesis in lysosomal storage disorders. Exp. Cell Res. 234, 85–97 [DOI] [PubMed] [Google Scholar]
- 45.Helip-Wooley A., Thoene J. G. (2004) Sucrose-induced vacuolation results in increased expression of cholesterol biosynthesis and lysosomal genes. Exp. Cell Res. 292, 89–100 [DOI] [PubMed] [Google Scholar]
- 46.Isaac E. L., Karageorgos L. E., Brooks D. A., Hopwood J. J., Meikle P. J. (2000) Regulation of the lysosome-associated membrane protein in a sucrose model of lysosomal storage. Exp. Cell Res. 254, 204–209 [DOI] [PubMed] [Google Scholar]
- 47.Hamer I., Jadot M. (2005) Endolysosomal transport of newly-synthesized cathepsin D in a sucrose model of lysosomal storage. Exp. Cell Res. 309, 284–295 [DOI] [PubMed] [Google Scholar]
- 48.Higuchi T., Nishikawa J., Inoue H. (2015) Sucrose induces vesicle accumulation and autophagy. J. Cell. Biochem. 116, 609–617 [DOI] [PubMed] [Google Scholar]
- 49.Jahraus A., Storrie B., Griffiths G., Desjardins M. (1994) Evidence for retrograde traffic between terminal lysosomes and the prelysosomal/late endosome compartment. J. Cell Sci. 107, 145–157 [DOI] [PubMed] [Google Scholar]
- 50.Hug G., Soukup S., Ryan M., Chuck G. (1984) Rapid prenatal diagnosis of glycogen-storage disease type II by electron microscopy of uncultured amniotic-fluid cells. N. Engl. J. Med. 310, 1018–1022 [DOI] [PubMed] [Google Scholar]
- 51.Settembre C., Di Malta C., Polito V. A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S. U., Huynh T., Medina D., Colella P., Sardiello M., Rubinsztein D. C., Ballabio A. (2011) TFEB links autophagy to lysosomal biogenesis. Science 332, 1429–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klionsky D. J., Elazar Z., Seglen P. O., Rubinsztein D. C. (2008) Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy 4, 849–850 [DOI] [PubMed] [Google Scholar]
- 53.Mizushima N., Yoshimori T., Levine B. (2010) Methods in mammalian autophagy research. Cell 140, 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reddy A., Caler E. V., Andrews N. W. (2001) Plasma membrane repair is mediated by Ca2+-regulated exocytosis of lysosomes. Cell 106, 157–169 [DOI] [PubMed] [Google Scholar]
- 55.Medina D. L., Fraldi A., Bouche V., Annunziata F., Mansueto G., Spampanato C., Puri C., Pignata A., Martina J. A., Sardiello M., Palmieri M., Polishchuk R., Puertollano R., Ballabio A. (2011) Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev. Cell 21, 421–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mencarelli S., Cavalieri C., Magini A., Tancini B., Basso L., Lemansky P., Hasilik A., Li Y. T., Chigorno V., Orlacchio A., Emiliani C., Sonnino S. (2005) Identification of plasma membrane associated mature β-hexosaminidase A, active towards GM2 ganglioside, in human fibroblasts. FEBS Lett. 579, 5501–5506 [DOI] [PubMed] [Google Scholar]
- 57.Settembre C., Fraldi A., Medina D. L., Ballabio A. (2013) Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell Biol. 14, 283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aureli M., Samarani M., Loberto N., Bassi R., Murdica V., Prioni S., Prinetti A., Sonnino S. (2014) The glycosphingolipid hydrolases in the central nervous system. Mol. Neurobiol. 50, 76–87 [DOI] [PubMed] [Google Scholar]
- 59.Santaguida S., Amon A. (2015) Aneuploidy triggers a TFEB-mediated lysosomal stress response. Autophagy 11, 2383–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Santaguida S., Vasile E., White E., Amon A. (2015) Aneuploidy-induced cellular stresses limit autophagic degradation. Genes Dev. 29, 2010–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walkley S. U. (2004) Secondary accumulation of gangliosides in lysosomal storage disorders. Semin. Cell Dev. Biol. 15, 433–444 [DOI] [PubMed] [Google Scholar]
- 62.Walkley S. U., Vanier M. T. (2009) Secondary lipid accumulation in lysosomal disease. Biochim. Biophys. Acta 1793, 726–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kolesnick R. (2002) The therapeutic potential of modulating the ceramide/sphingomyelin pathway. J. Clin. Invest. 110, 3–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morales A., Lee H., Goñi F. M., Kolesnick R., Fernandez-Checa J. C. (2007) Sphingolipids and cell death. Apoptosis 12, 923–939 [DOI] [PubMed] [Google Scholar]
- 65.Westwick J. K., Bielawska A. E., Dbaibo G., Hannun Y. A., Brenner D. A. (1995) Ceramide activates the stress-activated protein kinases. J. Biol. Chem. 270, 22689–22692 [DOI] [PubMed] [Google Scholar]
- 66.Wolff R. A., Dobrowsky R. T., Bielawska A., Obeid L. M., Hannun Y. A. (1994) Role of ceramide-activated protein phosphatase in ceramide-mediated signal transduction. J. Biol. Chem. 269, 19605–19609 [PubMed] [Google Scholar]
- 67.Schütze S., Tchikov V., Schneider-Brachert W. (2008) Regulation of TNFR1 and CD95 signalling by receptor compartmentalization. Nat. Rev. Mol. Cell Biol. 9, 655–662 [DOI] [PubMed] [Google Scholar]
- 68.Williams B. R., Prabhu V. R., Hunter K. E., Glazier C. M., Whittaker C. A., Housman D. E., Amon A. (2008) Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science 322, 703–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.