Abstract
Developmental pluripotency associated factor 4 (Dppa4) is a highly specific marker of pluripotent cells, and is also overexpressed in certain cancers, but its function in either of these contexts is poorly understood. In this study, we use ChIP-Seq to identify Dppa4 binding genome-wide in three distinct cell types: mouse embryonic stem cells (mESC), embryonal carcinoma cells, and 3T3 fibroblasts ectopically expressing Dppa4. We find a core set of Dppa4 binding sites shared across cell types, and also a substantial number of sites unique to each cell type. Across cell types Dppa4 shows a preference for binding to regions with active chromatin signatures, and can influence chromatin modifications at target genes. In 3T3 fibroblasts with enforced Dppa4 expression, Dppa4 represses the cell cycle inhibitor Cdkn2c and activates Ets family transcription factor Etv4, leading to alterations in the cell cycle that likely contribute to the oncogenic phenotype. Dppa4 also directly regulates Etv4 in mESC but represses it in this context, and binds with Oct4 to a set of shared targets that are largely independent of Sox2 and Nanog, indicating that Dppa4 functions independently of the core pluripotency network in stem cells. Together these data provide novel insights into Dppa4 function in both pluripotent and oncogenic contexts.
Keywords: Pluripotent stem cells, Chromatin, Histone deacetylase, Cell cycle, Oncogene, Dppa4, Oct4
1. Introduction
Maintenance of a pluripotent state requires complex and precise regulation at many levels, including transcriptionally through the interaction of pluripotency-specific transcription factors within the embryonic stem cell (ESC) chromatin landscape. While a few core pluripotency factors have been well characterized, many open questions remain and there are some pluripotency-specific factors whose functions are largely unknown. One of the best examples is Developmental pluripotency associated factor 4 (Dppa4), which is a putative pluripotency factor that is selectively expressed in ESC compared to differentiated cells. Both it and its homologue Dppa2 are also two of the best pluripotency markers used to validate induced pluripotent stem cells (IPSC) (Kang et al., 2015). However, Dppa4 and Dppa2 protein functions remain largely unknown.
Surprisingly, given its pluripotency-specific expression pattern and tight associations with other pluripotency factors like Oct4 (Chakravarthy et al., 2008; Sperger et al., 2003), Dppa4 has nonetheless been shown to be dispensable for ESC maintenance and for early murine embryonic development (Madan et al., 2009). Targeted disruption of Dppa4, Dppa2, or both in mice, did not produce the predicted early embryonic lethal phenotypes, but rather phenotypes manifested only much later in development in lung and other tissues where these factors are not expressed normally (Madan et al., 2009; Nakamura et al., 2011). Knockdown of Dppa4 in ES cells has also produced results that did not consistently define its role in pluripotency (Ivanova et al., 2006).
The mechanisms of potential Dppa4-mediated transcriptional regulation are also poorly understood. Dppa4 mainly associates with active, euchromatic domains as assessed by cytostaining (Masaki et al., 2007), but it is also a member of a non-canonical Polycomb repressive complex (Oliviero et al., 2015), and it represses transcription in GAL4 assays in vitro (Tung et al., 2013). Dppa4 contains an N-terminal SAP (SAF-A/B, Acinus and PIAS) domain, which is thought to mediate Dppa4 DNA binding, but it also associates with histone H3 through its C-terminal domain (Masaki et al., 2010). Recent work has shown that DPPA4 interacts with ERBB3 binding protein (ERB1) in human pluripotent stem cells, and that this interaction can attenuate DPPA4 mediated gene repression (Somanath et al., 2018), however, the extent to which Dppa4 acts as a transcriptional activator, repressor, or both at endogenous targets in pluripotent cells, and how it associates with chromatin to impact cell biology are major questions that have not been fully elucidated.
In addition to predicted roles in pluripotent stem cells, Dppa4 and Dppa2 are also oncogenic when overexpressed in somatic cells, and they are overexpressed in certain human cancers where they correlate with poor prognosis (Tung et al., 2013; John et al., 2008; Monk and Holding, 2001). Dppa4/2 increase proliferation through upregulation of cyclins and other G1/S transition genes, and induce foci formation and anchorage independent growth (Tung et al., 2013). While several direct transcriptional targets of Dppa4 have been identified using a candidate approach, global, unbiased characterization of Dppa4 direct targets genome-wide in stem cells and cancer cells has not been reported. Such studies would provide a better understanding of the mechanisms of Dppa4 transcriptional regulation and its biological impact.
Here we defined the genomic functions of Dppa4 in both ESC and an oncogenic context. We profiled Dppa4 binding genome-wide by ChIP-Seq in three cell types: E14 ESCs, 3T3 fibroblasts with enforced Dppa4 expression, and P19 embryonal carcinoma cells (ECCs). Comparing Dppa4 binding across cell types, there was substantial overlap of Dppa4-bound targets between the three cell types, particularly strong overlap in P19 and E14 cells, and a shared preference for active chromatin signatures. We in addition identified Dppa4-dependent changes in specific chromatin modifications at a subset of the genes it activates and represses. We also found that some Dppa4-bound target genes can be regulated by Dppa4 in opposing directions in different cell types, suggesting that cell type-specific differences influence the actions of Dppa4 in regulation of its targets. For example, we found that expression of the novel Dppa4 target gene Etv4 was increased both with ec-topic Dppa4 expression in fibroblasts and, conversely, by Dppa4 knockout in mESCs. Our studies also implicate repression of Cdkn2c and the activation of Etv4 as an important downstream effector of Dppa4 biological functions including proliferation in an oncogenic context. Our data also support a specific co-regulatory role for Oct4 and Dppa4 in ESC outside of the conventional Oct4-Sox2-Nanog regulatory context. Overall, our data define roles for direct Dppa4-mediated gene regulation in pluripotent stem cells and in an oncogenic context, and suggest specific epigenomic mechanisms of function.
2. Materials and methods
2.1. ChIP
ChIP was performed largely as described previously (O’Geen et al., 2011). Briefly, cells were crosslinked with 1% formaldehyde, lysed, and sonicated to an average fragment length of 500 bp before being immunoprecipitated with selected antibodies. The resulting chromatin was used for qPCR or library preparation for ChIP-Seq. For each ChIP, 20–50 μg of sonicated chromatin was used, with magnetic Dynabeads (Invitrogen) for immunoprecipitation. For ChIP-qPCR experiments, enrichment was calculated relative to the IgG negative control and then further normalized to an intergenic negative control region. The following antibodies were used: Rabbit IgG (Santa Cruz sc-2027), Goat IgG (Santa Cruz sc2028), H3K27ac (Abcam ab4729), H3K4me3 (Millipore 04–745), Dppa4 (R&D Systems AF3730), OCT4 (Abcam ab19857). HDAC1 (Abcam ab31263), HDAC2 (Abcam ab12169). Primers are listed in Supplemental Table 1.
2.2. ChIP-Seq
Two replicates of Dppa4 ChIP were performed in each of the following cell lines: E14, 3T3, and P19 cells. An input control was also sequenced for each cell line for normalization. Libraries were prepared with the Nextera library prep kit and sequenced on the Illumina Hi-Seq 2500 with fifty base pair single-end sequencing. Bases were called with Casava 1.8 (bcl2fastq 1.8). Raw sequencing data and processed peaks can be accessed with GEO accession number: GSE95055. Gene expression microarray data on Dppa4 overexpression fibroblasts can be accessed with GEO number: GSE58709.
2.3. Bioinformatics
Dppa4 ChIP-Seq reads were aligned to the genome using the Burrows-Wheeler Aligner (BWA), version 0.7.13-r1126 (Li and Durbin, 2010). MACS (version 1.4.2) (Zhang et al., 2008) was used to call peaks, with input samples used as the background control and an FDR of 0.05. Only peaks that overlapped between replicates were used for further analysis. For histone modification and Dppa2 ChIP-Seq, raw data was obtained from ENCODE and GEO, and analyzed using BWA and MACS to be more comparable with our Dppa4 data. DAVID was used for gene ontology analysis (Huang Da et al., 2009; Sherman et al., 2007). Galaxy (Giardine et al., 2005; Goecks et al., 2010) and Cistrome (Liu et al., 2011) were used for all other downstream analysis.
2.4. qPCR
For gene expression analysis, cDNA was prepared from 200 ng of RNA using the iScript cDNA kit, and RT-PCR was performed using Thermo Absolute Blue SYBR Green ROX (Catalog number AB-4162) on the LightCycler 480 (Roche). Mouse PP1A was used as the internal normalization control. RNA was extracted from cells using the Macherey Nagel Nucleospin RNA kit (Catalog number 740955).
For qPCR following ChIP, chromatin was diluted 1:10 and RT-PCR was performed using Thermo Absolute Blue SYBR Green ROX (Catalog number AB-4162) on the LightCycler 480 (Roche). Percent input values were calculated for each sample after subtracting IgG signal, and all values were then normalized to a negative control chromatin region (Crisp3).
Primer sequences in Supplemental Table 1.
2.5. Cell culture, transfections, and transductions
3T3 cells and NT2 clone D1 cells (supplied by Shiro Urayama) were cultured in DMEM supplemented with 10% FBS and 1% Glutamine. E14 cells were cultured under feeder-free conditions in 2i media with 2% FBS on gelatin-coated plates. P19 cells were cultured in DMEM supplemented with 10% FBS and 1% Glutamine. Four siRNA targeting Etv4 (Qiagen GS18612) were pooled and transfected into WT and Dppa4 3T3 cells at a concentration of 25 nM using Lipofectamine RNAi Max (Thermo Fisher Scientific) according to the manufacturer’s directions. Cells were collected 48 h after transfection and assayed for knockdown by qPCR and cell cycle stage by propidium iodide staining and flow cytometry. Full length mouse Cdkn2c and Etv4 were cloned into the pBABE vector and transfected into platE cells to generate virus. WT and Dppa4 overexpressing 3T3 cells were transduced with virus containing media collected from platE cells for each construct. Transduction of cells with virus generated from an empty pBABE vector was used as a control. Cells were selected with hygromycin (100μg/mL) for 7 days, until all 3T3 cells in an untransfected control plate treated with 100μg/mL hygromycin had died.
2.6. TUNEL staining
Cells were seeded onto coverslips and collected after 48 h. The DeadEnd™ Fluorometric TUNEL system (Promega) was used for TUNEL staining. A total of six images (3 images from each of two slides) were collected for each 3T3 cell line (WT empty vector control, WT + Cdkn2c, WT + Etv4, Dppa4 empty vector control, Dppa4 + Cdkn2c, Dppa4 + Etv4). DNase I treatment was used as the positive control.
2.7. 7AAD staining and flow cytometry
One million cells were seeded into 60 mm dishes in triplicate for each 3T3 cell line and collected 48 h after seeding. Cells were washed and stained with 7AAD dye for 30 min at room temperature, before flow cytometry was run on the Attune NxT Flow Cytometer (Thermo Fischer Scientific).
2.8. Flow cytometry/Propidium iodide cell cycle analysis and cell proliferation
Cell cycle analysis and proliferation assays were conducted in parallel using the same samples. One million cells were seeded into 60 mm dishes in triplicate for each 3T3 cell line and collected 48 h after seeding. Cells were washed and fixed in ice cold 70% ethanol, stored overnight at −20 °C, then washed and stained with 50μg/mL PI and 100 μg/mL RNaseA for 30 min at room temperature, before flow cytometry was run on the Attune NxT Flow Cytometer (Thermo Fisher Scientific). Two independent experiments were performed, resulting in N = 6 per sample. FlowJo was used to analyze resulting data. Single cells were also counted concurrently to assess the proliferation rate.
2.9. Co-immunoprecipitation (Co-IP)
NT2 cells were harvested and prepared as nuclear extracts (Abcam nuclear fractionation protocol). Briefly, cells were resuspended in 10 mM HEPES, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT and 0.05% NP40 at pH 7.9. Cells were then pelleted and resuspended in 5 mM HEPES, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 26% glycerol (v/v) and 4.6 M NaCl at pH 7.9. The supernatant was extracted and incubated overnight with 6 μg of primary antibody to OCT4 (Abcam 19,857) or IgG rabbit (Cell Signaling 2729). Protein A/G magnetic beads (Cell Signaling Technology 9006S) were used to collect immunoprecipitated complexes, which were washed four times with buffer (5 mM HEPES, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 26% glycerol (v/v), pH 7.9) and analyzed by Western blot. Experiments were repeated as at least two biological replicates.
2.10. Western blotting
Lysates or immunoprecipitated complexes were electrophoresed on NuPAGE 4–12% Bis-Tris gels (Invitrogen) (Monk and Holding, 2001) and transferred to polyvinylidene fluoride membrane. Membranes were blocked in Blok-FL Fluorescent blocker (Millipore), then incubated with OCT4 (Abcam 19857, 1:1000 dilution) or DPPA4 (Abnova H00055211-B01, 1:300 dilution) antibody overnight. Blots were incubated with goat anti-mouse (1:10,000 dilution IR Dye 680RD, LiCOR 926–68,070) or goat anti-rabbit secondary antibodies (1:10,000 dilution IR Dye 800CW, LiCOR 926–3211) for 1 h at room temperature and imaged using the Odyssey cLX Imaging system (LiCOR).
3. Results
3.1. ChIP-Seq defines Dppa4 genomic binding in murine pluripotent cells and in Dppa4-overexpressing fibroblasts
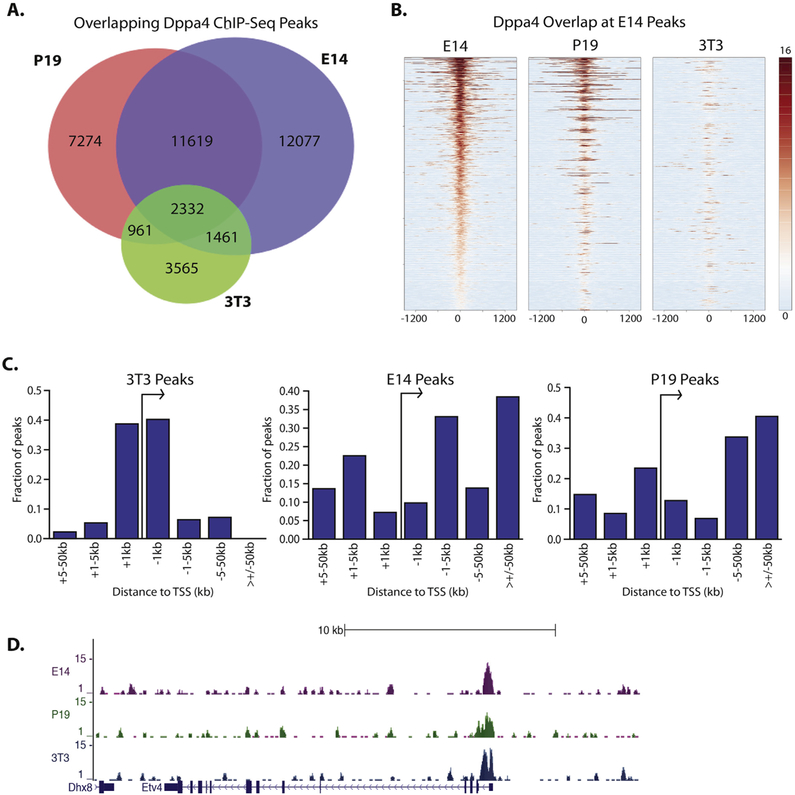
To better understand the role of Dppa4 in gene regulation in ESCs and in oncogenic transformation, we performed ChIP-Seq for Dppa4 in E14 mESCs, P19 ECCs, which represent a combination of an oncogenic and pluripotent cell type, and a 3T3 mouse fibroblast cell line that stably expresses Dppa4 (Supplemental Fig. 1) (Tung et al., 2013), which is normally absent from fibroblasts. With two biological ChIP-Seq replicates for each cell type, we identified 27,489 Dppa4 peaks in E14 mESC, 22,186 peaks in P19, and 8319 peaks in Dppa4-expressing 3T3 cells using MACS (Zhang et al., 2008). A high degree of conservation in binding sites between cell types was observed as Dppa4 bound to 2332 regions in all three cell types (Fig. 1A). There were also > 11,000 additional shared Dppa4 binding sites between E14 and P19 cells (Fig. 1A, B), likely reflective of the similar pluripotent nature of these two cell types. Dppa4 also bound to a substantial number of unique sites in each cell type, which may be indicative of distinct roles of Dppa4 under different cellular and physiological conditions. Within our ChIP-Seq data, we also observed binding of Dppa4 to known targets CDKN1C, SYCE1, and NKX2–5 (Nakamura et al., 2011; Tung et al., 2013) (Supplemental Fig. 2).
Fig. 1.
Patterns of Dppa4 binding in E14 mESC, Dppa4 overexpressing 3T3 fibroblasts, and P19 embryonal carcinoma cells. A) Overlap of Dppa4 peaks between 3T3, P19, and E14 cells. B) Heatmap plot of E14, P19, and 3T3 ChIP-Seq reads centered on E14 Dppa4 peaks. C) Comparison of genomic distribution of Dppa4 peaks in E14, P19, and 3T3 cells. D) Dppa4 ChIP-Seq peaks at the Etv4 promoter.
We also compared our E14 Dppa4 ChIP-Seq data to ChIP-Seq data on Dppa2 generated in CGR8 mouse ESCs (Engelen et al., 2015), and found a substantial overlap of Dppa2 and Dppa4 binding sites, further suggesting the two may co-bind or even heterodimerize (Supplemental Fig. 3, (Tung et al., 2013; Somanath et al., 2018)).
Analyzing the distribution of Dppa4 peaks in each cell type relative to gene promoters, we found a strong preference for Dppa4 binding within 5 kb of transcriptional start sites (TSSs) in 3T3 cells. Approximately 80% of peaks were within 5 kb of a TSS in 3T3, while in contrast in E14 cells Dppa4 bound most frequently to more distal sites, with only about 20% of peaks found within 5 kb of a promoter (Fig. 1C). In P19 cells, Dppa4 bound relatively closer to promoters than in E14s, but the effect was not as pronounced as in the 3T3 cells. Despite the global differences in binding preference relative to TSSs, promoter proximal binding could be observed for all three cell types at specific genes, including transcriptional regulators like the Ets factor Etv4 (Fig. 1D, Supplemental Fig. 4A).
3.2. Dppa4 binding is enriched in domains with active chromatin signatures in all three cell types, and at bivalent domains in mESC
To define the specific types of chromatin that Dppa4 associates with in each cell type, we examined the genome-wide association of Dppa4 with different histone modifications in P19, E14 mESC, and 3T3 cells using published and publicly available ChIP-Seq data (The ENCODE Project Consortium, 2012; Rosenbloom et al., 2012; Zullo et al., 2012; Schick et al., 2015; Zhu et al., 2012; Serandour et al., 2012; Coda et al., 2017), GSE82314. We observed strong overlap between Dppa4 binding and the active histone marks H3K4me3, H3K4me2, and H3K27ac, in all three cell types. We also found a strong overlap of Dppa4 binding with CpG island domains (Supplemental Tables 2–4). Fitting with a preference of Dppa4 for binding to genomic domains enriched in active marks, there was less association of Dppa4 with repressive marks H3K27me3 and H3K9me3 (Supplemental Tables 2–4). In mESCs, approximately half of the Dppa4-H3K27me3 overlapping domains occurred in bivalent domains that also have H3K4me3 (Supplemental Table 3). A smaller overlap of Dppa4 peaks with H3K27me3 and bivalent domains was observed in P19 cells. Consistent with the fact that bivalent domains are much less common in differentiated 3T3 cells, we did not observe a strong overlap of Dppa4 with regions marked by both H3K27me3 and H3K4me3 in those cells (Supplemental Table 2). Overall, these results suggest a strong association of Dppa4 with active chromatin regions, but also a substantial link between Dppa4 and bivalent domains as well as H3K27me3-enriched, potentially repressed domains in a pluripotent context.
3.3. In 3T3 cells, Dppa4 binds both cell cycle factors and other transcriptional regulators, including Ets transcription factor Etv4
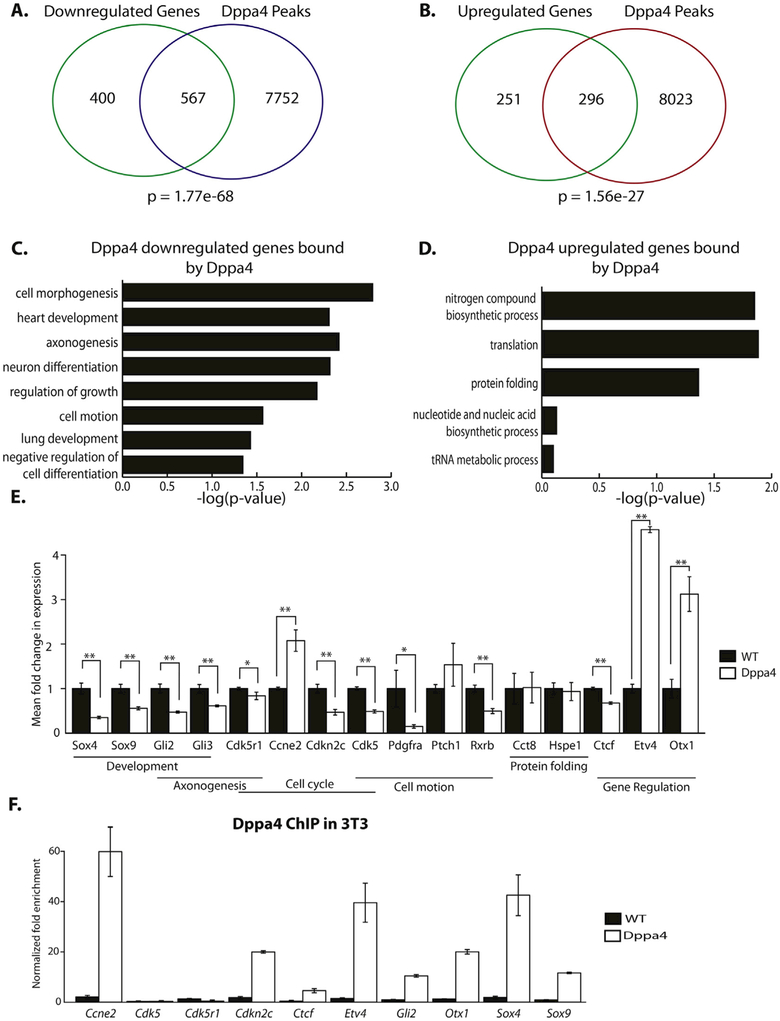
We focused first on the 3T3 data and sought to identify direct targets of Dppa4 by overlapping Dppa4 ChIP-Seq peaks with the promoters of genes either upregulated or downregulated in response to ectopic Dppa4 expression, based on our previously published expression microarray data (Tung et al., 2013). There was a significant overlap of Dppa4 peaks with the promoters of both upregulated and down-regulated genes in Dppa4-overexpressing cells, suggesting Dppa4 may function as both an activator and a repressor depending on the context (Fig. 2A, B). Downregulated genes that were directly bound by Dppa4 were enriched in gene ontology (GO) categories related to development, cell growth, and cell motion (Fig. 2C). Bound, upregulated genes were enriched in GO categories related to protein and RNA metabolism (Fig. 2D). Thus, distinct classes of genes are positively or negatively regulated directly by Dppa4.
Fig. 2.
Dppa4 regulates cell cycle and transcription factor targets in 3T3. A) Overlap of genes downregulated by Dppa4 overexpression in 3T3 and Dppa4 ChIP-Seq peaks in Dppa4 overexpressing 3T3. Hypergeometric distribution was used to calculate probabilities. B) Overlap of genes upregulated by Dppa4 overexpression in 3T3 and Dppa4 ChIP-Seq peaks in Dppa4 overexpressing 3T3. Hypergeometric distribution was used to calculate probabilities. C) Significant gene ontology categories for Dppa4-repressed direct targets. D) Significant gene ontology categories for Dppa4-activated targets. E) qPCR validation of gene expression changes in select Dppa4 targets, n = 4. *p < .05, **p < .01. F) Dppa4 ChIP-qPCR validation of select Dppa4 targets, n = 3. Error bars are the S.E.M.
Among predicted directly-bound and regulated Dppa4 targets were a number of notable genes, including cell cycle regulators Ccne2 and Cdkn2c, signaling receptors Rxrb and Ptch1, and transcription factor coding genes Etv4 and Sox4. We selected sixteen of these predicted Dppa4 targets linked to different cellular functional processes for validation by qRT-PCR. Twelve of these 16 exhibited the expected significant changes associated with ectopic Dppa4 (Fig. 2E). We confirmed binding of Dppa4 to 9 of these target genes by ChIP-qPCR (Fig. 2F, Supplemental Fig. 4B), including Etv4 and cell cycle regulator Ccne2.
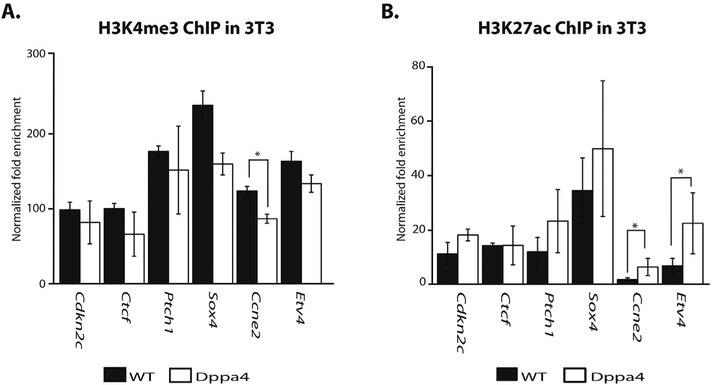
Based on the strong association of Dppa4 with active chromatin signatures, we next looked at the effect of Dppa4 overexpression on H3K4me3 and H3K27ac at validated Dppa4 targets by ChIP-qPCR (Fig. 3). We did not observe any substantial (> 1.5-fold) changes in H3K4me3 at target genes. While there was a statistically significant decrease in H3K4me3 at Ccne2 the magnitude was small (Fig. 3A). H3K27ac was increased at the Dppa4 targets Etv4, and Ccne2 (Fig. 3B).
Fig. 3.
Dppa4 influences levels of histone modifications at target genes. A) ChIP-qPCR of H3K4me3 levels at Dppa4 targets in WT and Dppa4 overexpressing 3T3, n = 3. t-test. B) ChIP-qPCR of H3K27ac levels at Dppa4 targets in WT and Dppa4 overexpressing 3T3, n = 3. t-test. *p < .05, **p < .01. Error bars are the S.E.M.
3.4. Dppa4 acts at least in part through downstream targets Etv4 and Cdkn2c to increase proliferation in fibroblasts
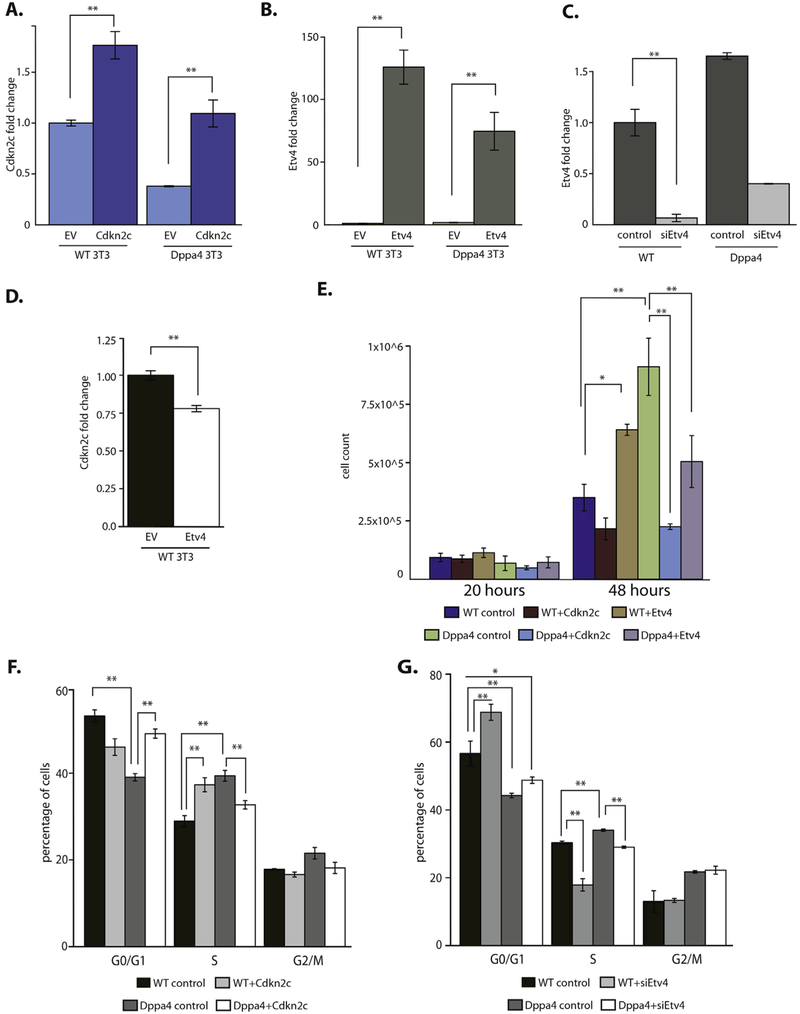
We focused on two newly defined Dppa4 target genes based on their known functions. Since the target Cdkn2c is a cell cycle inhibitor involved in regulation of the G1 phase of the cell cycle (Schafer, 1998), its downregulation by Dppa4 could be a mechanism of Dppa4 impact on proliferation and the cell cycle. We also focused on Etv4, which is overexpressed in many cancers. It promotes invasiveness, proliferation (Pellecchia et al., 2012; Oh et al., 2012), and anchorage independent growth (Hollenhorst et al., 2011), similar to overexpressed Dppa4. To test whether Cdkn2c or Etv4 are important effectors of ectopic Dppa4 in 3T3 cells, we expressed each factor separately in control WT and in Dppa4-overexpressing 3T3 cells (Fig. 4A, B). Because Etv4 is upregulated by Dppa4, we also knocked down Etv4 using siRNA in both WT and Dppa4 expressing cells (Fig. 4C). Etv4 generally did not affect direct Dppa4 target expression, but caused a significant reduction in Cdkn2c expression (Fig. 4D), similar to the effect of Dppa4 overexpression.
Fig. 4.
Characterization of biological impact of downstream targets of Dppa4 in 3T3: Cdkn2c and Etv4. A) Expression of Cdkn2c in WT and Dppa4 expressing 3T3 after transduction and selection for ectopic Cdkn2c expression, compared to WT empty vector (EV) control n = 4. B) Expression of Etv4 in WT and Dppa4 expressing 3T3 after transduction and selection for ectopic Etv4 expression, compared to WT empty vector control n = 4. C) Expression of Etv4 in WT and Dppa4 expressing 3T3 after siRNA knockdown, compared to negative siRNA control in WT 3T3 n = 3. D) Expression of Cdkn2c in WT 3T3 overexpressing Etv4 compared to empty vector control, n = 4. E) Count of number of cells 20 h and 48 h after seeding, n = 3. ANOVA with Post-hoc Tukey HSD test. F) Propidium iodide cell cycle analysis, n = 6. ANOVA with Post-hoc Tukey HSD test. G) Propidium iodide cell cycle analysis, n = 6. ANOVA with Post-hoc Tukey HSD test. *p < .05, **p < .01. Error bars are the S.E.M.
Because of the increased proliferation observed in Dppa4-expressing 3T3 cells (Tung et al., 2013), we examined the effect of Cdkn2c and Etv4 on proliferation in WT 3T3 and Dppa4 overexpressing 3T3 cells. We observed differences in proliferation with Cdkn2c or Etv4 over-expression; in WT 3T3 cells, Cdkn2c overexpression showed a trend toward reduced cell numbers at 48 h, although this did not reach significance (Fig. 4E). In contrast, Etv4 overexpression in WT 3T3 cells resulted in a significant increase in cell proliferation (Fig. 4E), consistent with the link between Etv4 and proliferation in cancer cells. Cdkn2c overexpression in Dppa4-overexpressing 3T3 s significantly decreased cell numbers at 48 h (Fig. 4E), suggesting Dppa4 may increase proliferation through repression of Cdkn2c. Based on 7AAD staining, we did not observe any significant difference in apoptosis between Cdkn2c expressing cells, Etv4 expressing cells and vector controls for WT 3T3 (Supplemental Fig. 5). There was a decrease in apoptosis in Dppa4 expressing 3T3 compared to WT, and small but significant increase in apoptosis levels when either Cdkn2c or Etv4 was expressed in Dppa4 cells (Supplemental Fig. 5B), correlating with the reduced proliferation levels observed in these cells (Fig. 4E). The paradoxical effect of Etv4 on Dppa4-expressing cells may suggest that overexpression of two strong proliferation-promoting factors like Etv4 and Dppa4 causes increased levels of cell stress and apoptosis. However, the relationship between Dppa4 as well as its downstream effectors on cell survival is not entirely clear at this time.
To complement the cell growth analysis, we analyzed the cell cycle profiles for each of these cell lines by flow cytometry. At 48 h after seeding, we observed a significant reduction in cells in G0/G1 phases in Dppa4-overexpressing 3T3 compared to WT, and a significant increase in S phase in Dppa4-expressing cells compared to WTs (Fig. 4F), as we have shown previously (Tung et al., 2013). Cdkn2c expression in WT cells caused a trend toward reduced numbers of cells in G0/G1 phases, and a significant increase in cells in S phase (Fig. 4F). In contrast, Cdkn2c expression in Dppa4-overexpressing 3T3 caused an increase in cells in G0/G1, and a reduction in cells in S-phase (Fig. 4F). We did not observe any significant differences between numbers of cells in G2/M phases among the different cell lines (Fig. 4F). Both for G0/G1 and S phase, overexpression of Cdkn2c causes Dppa4-expressing cells to exhibit cell cycle dynamics more similar to WT 3T3, suggesting that Dppa4-mediated downregulation of Cdkn2c levels contributes to the alterations in cell cycle and cell proliferation observed in Dppa4 expressing 3T3 cells. We did not observe any clear patterns of significant changes in cell cycle for either WT or Dppa4 3T3 that overexpress Etv4 (Supplemental Fig. 6). In contrast, Etv4 knockdown in WT 3T3 resulted in a significant increase in cells in G0/G1 and a decrease in cells in S phase (Fig. 4G). Etv4 knockdown in Dppa4-overexpressing 3T3 also reduced the number of cells in S phase and showed a small trend toward increased numbers of cells in G0/G1, causing the cell cycle profile of siEtv4 Dppa4 3T3 cells to more closely resemble the profile of WT 3T3 (Fig. 4G).
3.5. Dppa4 also regulates Etv4 and targets the cell cycle in E14 mESC, with enrichment for active histone marks
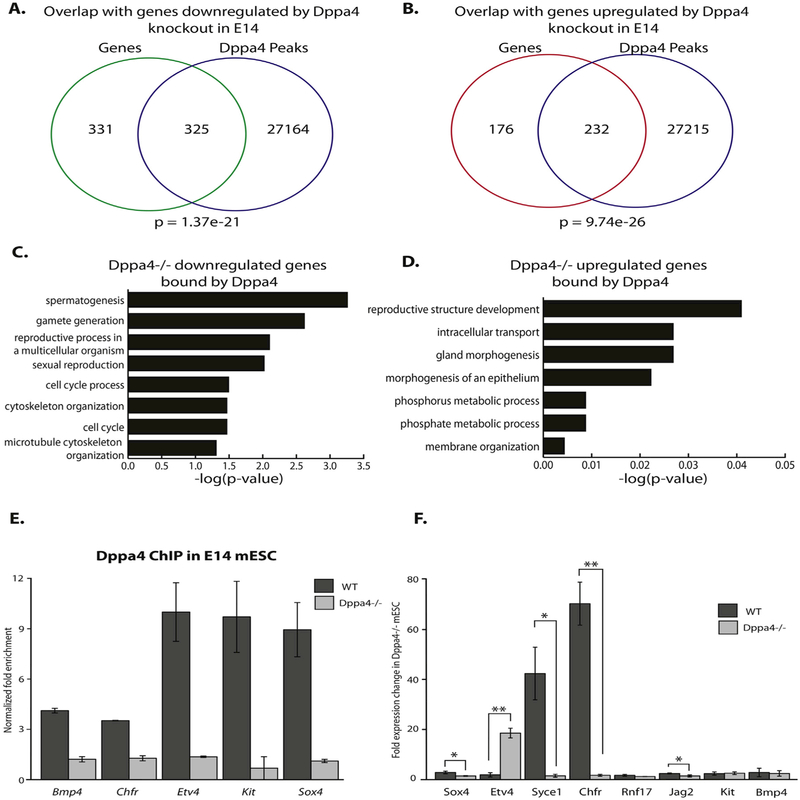
To further define the gene regulatory role of Dppa4 in ESCs and to understand better how the pluripotent functions of Dppa4 relate to oncogenesis, we compared our Dppa4 ChIP-Seq binding data in ESCs to genes affected by disruption of Dppa4 in a published microarray gene expression study on Dppa4 knockout E14 mESC (Madan et al., 2009). Both downregulated and upregulated genes in Dppa4 knockout mESC exhibited substantial overlap with bound Dppa4 peaks in E14 mESC from our ChIP-Seq study (Fig. 5A, B). Direct targets that were down-regulated upon Dppa4 knockout were enriched in GO categories related to cell cycle and gamete production (Fig. 5C). Direct Dppa4 targets that were upregulated in nulls exhibited relatively weak enrichment in developmental GO categories (Fig. 5D). ChIP-qPCR documented significant Dppa4 binding at all tested target gene promoters in mESCs (Fig. 5E). We validated gene expression changes by RT-qPCR in 5 out of 8 predicted target genes in Dppa4−/− versus WT mESC (Fig. 5F) including the transcriptional regulators Etv4 and Sox4, both targets of Dppa4 in 3T3 cells as well, and Chfr, a cell cycle regulator. Surprisingly, the two Dppa4 targets shared between 3T3 and E14 cells, Etv4 and Sox4, are regulated by Dppa4 in opposing directions in each case. Both overexpression of Dppa4 in 3T3 s and Dppa4 knockout in ESCs caused the same pattern of Etv4 upregulation and Sox4 downregulation.
Fig. 5.
Dppa4 also regulates cell cycle and transcription factor targets in E14 mESCs. A) Overlap of genes downregulated by Dppa4 disruption in E14 mESC and Dppa4 ChIP-Seq peaks in WT E14 cells. Hypergeometric distribution was used to calculate probabilities. B) Overlap of genes upregulated by Dppa4 knockout in E14 mESC and Dppa4 ChIP-Seq peaks in WT E14 cells. Hypergeometric distribution was used to calculate probabilities. C) Significant gene ontology categories for Dppa4-activated direct targets. D) Significant gene ontology categories for Dppa4-repressed direct targets. E) Dppa4 ChIP-qPCR validation of select Dppa4 targets, n = 2 (independently-generated knockout lines). F) qPCR validation of gene expression changes in select Dppa4 targets, n = 2 (independently-generated knockout lines). t-test. *p < .05, **p < .01. Error bars are the S.E.M.
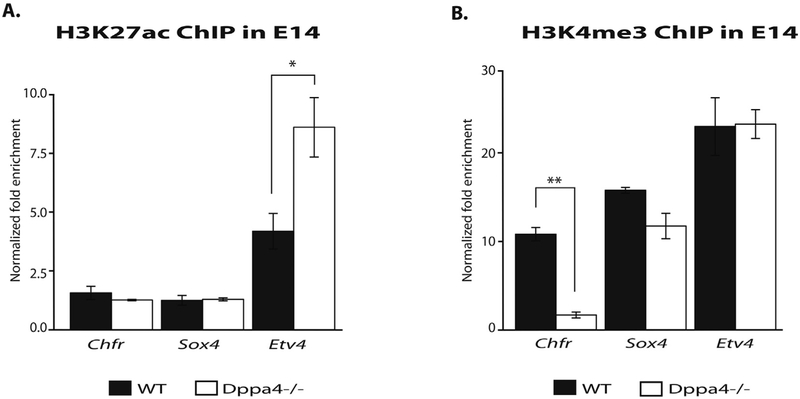
To test Dppa4 regulation of chromatin modifications at targets, we performed ChIP-qPCR in WT, floxed (no Cre), and Dppa4−/− mESC (Madan et al., 2009) for the histone modifications H3K27ac and H3K4me3. While fewer genes exhibited marks affected by Dppa4 knockout in mESCs than were affected by Dppa4 overexpression in 3T3, we did observe a significant increase in H3K27ac at the Etv4 promoter in Dppa4−/− cells (Fig. 6A), fitting with the upregulation of Etv4 seen upon Dppa4 deletion in mESCs. We also saw a significant decrease in H3K4me3 enrichment at Chfr, a gene downregulated by Dppa4 deletion (Fig. 6B). These results suggest that Dppa4 in some cases may function to suppress targets such as Etv4 in E14 mESC by regulating the levels of H3K27ac and potentially other marks at target gene promoters.
Fig. 6.
Dppa4 influences levels of histone modifications at target genes in E14s. A) ChIP-qPCR of H3K27ac levels at Dppa4 targets in WT and Dppa4−/− E14, n = 2. B) ChIP-qPCR of H3K4me3 levels at Dppa4 targets in WT and Dppa4−/− E14, n = 2. t-test. *p < .05, **p < .01. Error bars are the S.E.M.
3.6. Dppa4 and Oct4 genomic binding substantially overlaps in mESCs
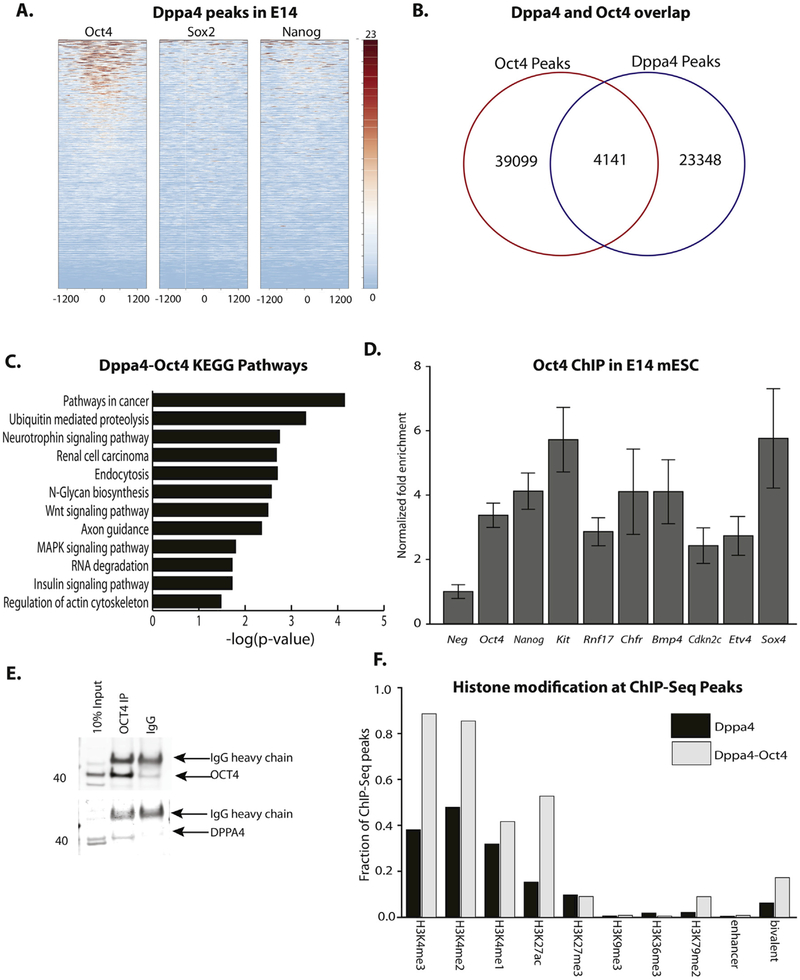
Because of Dppa4’s pluripotency-specific expression, we examined whether Dppa4 binding overlapped with genomic binding of the key pluripotency factors Oct4, Sox2, or Nanog in mESC. While overlap between Sox2 or Nanog and Dppa4 was minimal, there was substantial overlap between Dppa4 and Oct4 peaks (Fig. 7A, B). The genes in closest proximity to these co-bound sites are strongly enriched in GO pathways related to cancer, as well as in Wnt and MAPK signaling pathways, suggesting a co-regulatory role for Dppa4 and Oct4 at genes involved in stem cell function as well as tumorigenesis. ChIP-qPCR validated Oct4 binding to a subset of Dppa4 validated targets, further supporting the potential functional interaction of the two factors (Fig. 7D), but we did not detect a difference in Oct4 binding at these targets in Dppa4−/− mESC (Supplemental Fig. 7), suggesting that Dppa4 is not absolutely required for Oct4 occupancy of these sites. Because of the strong enrichment for cancer pathways among the predicted Oct4-Dppa4 shared targets based on ChIP-Seq, we assessed their potential protein-protein interaction via co-IP studies in human NT2 embryonic carcinoma cells. We determined that DPPA4 interacts with OCT4 (Fig. 7E, Supplemental Fig. 8) in this pluripotent/cancer context, suggesting a potential molecular mechanism for their strongly overlapping genomic binding.
Fig. 7.
Dppa4 binding overlaps with OCT4 in E14 mESC. A) Plot of OCT4, SOX2, and NANOG ChIP-Seq reads at E14 Dppa4 peaks. B) Overlap of OCT4 and Dppa4 peaks. C) Significantly enriched KEGG Pathways at genes bound by both OCT4 and Dppa4. D) ChIP-qPCR validation of OCT4 binding at OCT4 targets (Oct4, Nanog) and at OCT4-Dppa4 shared targets. E) Endogenous OCT4-DPPA4 Co-immunoprecipitation in NT2 cells. F) Overlap of histone modifications with OCT4-Dppa4 co-bound sites, relative to sites only bound by Dppa4. Error bars are the S.E.M.
When we overlapped putative Oct4-Dppa4 co-regulated regions with histone modification ChIP-Seq data, we observed a strong association with active histone marks. For instance, Oct4-Dppa4 co-bound regions were very highly enriched in H3K4 methylation; almost 90% of Oct4-Dppa4 regions contained H3K4me3 and H3K4me2 peaks (Fig. 7F) suggesting regions bound by both factors are sites of high levels of transcriptional activity. Additionally, the association with bivalent domains was even stronger for Oct4-Dppa4 regions than for Dppa4-bound regions alone (Fig. 7F).
4. Discussion
To our knowledge, this work is the first to map Dppa4 binding genome wide and identify direct downstream targets, adding substantially to the knowledge of Dppa4 targets and mechanisms of regulation. By generating and analyzing the data on Dppa4 binding in three unique cell types we are able to provide insights into Dppa4’s role in stem cells, and how this role is carried over or altered in some ways to promote oncogenesis when Dppa4 is overexpressed in a differentiated cell type like fibroblasts.
The distinct genomic binding profiles of Dppa4 in differentiated cells compared to ESCs relative to TSSs was an unexpected finding and could suggest that Dppa4 selectively binds to regions of open chromatin. In ESCs, much of the chromatin is open (Gaspar-Maia et al., 2011; Ahmed et al., 2010), which would allow Dppa4 to bind to more distal sites, as observed in our ChIP-Seq. In differentiated cells, however, the majority of open chromatin is located at the promoters of active genes, which could limit Dppa4 to binding at these sites. Differential access to binding sites based on chromatin accessibility may explain the cohorts of unique genes regulated by Dppa4 only in E14 mESC or only in 3T3. Alternatively, since ectopic expression of Dppa4 in 3T3 results in dramatically higher levels of Dppa4 expression relative to endogenous Dppa4 in E14 and P19 cells (Supplemental Fig. 1), this may in part explain the observed differences in Dppa4 binding patterns. However, there are many similarities between Dppa4 binding patterns in cells with endogenous levels and those with highly overexpressed Dppa4, including a strong preference for active chromatin, and shared targets, suggesting that Dppa4 binding in 3T3 cells is not purely due to the high level of Dppa4 in the cell.
The opposing regulatory dynamics of Dppa4 at a subset of its targets that are shared between differentiated cells and ESCs were also somewhat surprising. Our ChIP-Seq and gene expression data provide evidence that Dppa4 can both activate and repress different gene targets. However, the fact that Dppa4 can bind to the same site and activate expression of a given gene such as Etv4 under one context while repressing its expression in a different cell type suggests that co-factors of Dppa4 or other aspects of the transcriptional regulatory machinery are differentially expressed in differentiated and ESCs, which in turn in-fluences the directionality of Dppa4 regulation at the same gene target. Differences in chromatin organization, including levels and types of histone modification, or structural differences that allow specific sets of enhancers to loop and contact the promoter in specific cell types may also contribute to the directionality of Dppa4 regulation. In addition, it has been hypothesized that part of the role of Dppa4 in pluripotent cells is to organize the chromatin for later gene expression during differentiation, when Dppa4 is no longer expressed. It may be that in differentiated cells, including 3T3, Dppa4 no longer has this chromatin organizing ability due to the absence of co-factors or other chromatin mediators, or changes in the chromatin accessibility, and while it can bind to the same targets, it cannot have the same effect as in the pluripotent context.
Our work also identified a role for Dppa4 in modulating levels of histone modifications at the genes it regulates. This is particularly interesting in light of the studies that have demonstrated an interaction between the C-terminal end of Dppa4 and histone H3 (Masaki et al., 2010); such an interaction may allow for Dppa4 to bring chromatin modifying enzymes in proximity to the histone tail for subsequent modification of specific residues therein. Indeed, multiple mass-spec-trometry studies have demonstrated interactions between DPPA4 and a wide variety of different chromatin factors; these include HP1γ (Zaidan et al., 2018), and polycomb repressive complex protein PCGF1, as well as EBP1 (Somanath et al., 2018), which is known to recruit histone modifying complexes like SIN3 (Zhang et al., 2005).
As observed in previous studies (Masaki et al., 2007), we find that Dppa4 associates most often with DNA regions that are marked by active chromatin signatures both in differentiated cells and in ESCs. We also find that Dppa4 associates with a substantial number of bivalent sites in E14 mESC, but not in differentiated 3T3 cells. Bivalent domains often occur at genes that are turned on early in the differentiation process of pluripotent stem cells. Given that Dppa4 appears dispensable for normal ESC function, but its loss causes defects in later differentiation processes in lung and skeleton, we speculate that Dppa4 may act to regulate certain aspects of the stem cell differentiation process at bivalent sites upon commitment to differentiation. Interestingly, our findings on differential Dppa4 regulation of the same individual target genes in pluripotent versus somatic cells also suggest that during early differentiation of pluripotent cells prior to full repression of Dppa4 expression, Dppa4 transcriptional function may strongly change even as it continues to bind some of the same target genes.
Our characterization of Dppa4 downstream targets suggests that some of the oncogenic effects of Dppa4 in somatic cells are likely mediated by regulation of cell cycle inhibitor Ckdn2c and Ets family member Etv4. We find that Cdkn2c is a direct target of Dppa4 mediated repression in 3T3 cells when Dppa4 is overexpressed in this context, and that the effects of Dppa4 on the cell growth rate and cell cycle dynamics can be reverted back to levels more comparable to WT cells when Cdkn2c expression is reintroduced into Dppa4 overexpressing 3T3 cells. In contrast, we find a proliferation promoting role for Dppa4 activated target Etv4; siRNA knockdown of Etv4 in Dppa4 expressing 3T3 is also able to shift proliferation and cell cycle dynamics back toward WT. Additionally, the link between Etv4 and cancer is strong. Etv4 is over-expressed in a wide range of cancer types, including breast, prostate, colon, and lung (Oh et al., 2012; Hakuma et al., 2005; Yuan et al., 2014; Moss et al., 2006), and has links to metastasis and cell proliferation (Pellecchia et al., 2012; Hollenhorst et al., 2011). In the somatic context, Dppa4 upregulation of Etv4 expression through alterations in histone acetylation levels likely contributes to the increase in proliferation observed in Dppa4 overexpressing 3T3 (Supplemental Fig. 9). In a pluripotent context, we find that Dppa4 has the opposite effect on Etv4; Dppa4 acts to reduce levels of Etv4, potentially through the observed reductions in histone acetylation. In mESC, loss of Etv4 and Etv5 has been shown to decrease levels of proliferation and affect expression of early ectodermal genes upon induction of differentiation (Akagi et al., 2015). In this context, it is possible that Dppa4 may act through suppression of Etv4 to modulate levels of proliferation to maintain the pluripotent state (Supplemental Fig. 9).
In addition, we observed an overlap between Dppa4 and Oct4 binding, but not between Dppa4 and either of the two additional core pluripotency factors (Nanog and Sox2), suggesting that Dppa4 does not interact with the core pluripotency transcription factor complex, but rather binds with Oct4 in a unique multiprotein complex to regulate shared gene targets (Supplemental Fig. 9). These shared targets are highly enriched for cancer, cell signaling and stem cell related gene ontology categories, fitting with data linking Oct4 to numerous cancer types, and to the regulation of stem cell-like properties of tumors (Murakami et al., 2015; Kaufhold et al., 2016; Villodre et al., 2016). These findings are also relevant in the converse context of the tumorigenic properties of pluripotent stem cells, which form teratomas when injected into mice, and suggest that Dppa4 gene targets regulate a number of the shared properties at the intersection of stem cells and cancer. Given the role of Dppa4 in both stem cells and tumorigenesis, it is possible that Dppa4 and Oct4 proteins also interact at these shared targets in an oncogenic context to promote stem-cell like properties of cancer cells.
5. Conclusion
In conclusion, our study provides extensive new insights into the function and regulatory dynamics of Dppa4. We have characterized the genomic binding profiles of Dppa4 in three different cell types. Many targets of Dppa4 are shared across cell types, but a number of differences suggest that Dppa4 binding is context dependent. Dppa4 binds predominantly to promoters in 3T3 cells, but more distally in pluripotent cells, possibly related to chromatin accessibility. We identify direct targets of Dppa4 in 3T3 and in E14, including two gene, Cdkn2c and Etv4, which we find act downstream of Dppa4 to regulate proliferation in 3T3 cells. We also identify a role for Dppa4 in regulating levels of histone modifications H3K4me3 and H3K27ac at a subset of its targets. We further show that DPPA4 and OCT4 interact in human pluripotent cells, in a multiprotein complex distinct from the OCT4-SOX2-NANOG complex. In short, our work has elucidated important previously uncharacterized aspects of Dppa4-mediated gene regulation in both pluripotent stem cells and in an oncogenic context.
Supplementary Material
Acknowledgments
We thank the UC Davis Genome Center for their assistance with ChIP-Seq. We thank Michael Chen and Kelly Bush for reading the manuscript and providing feedback. This work was supported by funding from NIH Grants 1R01GM100782 and 1R01GM116919 (PK) and CIRM Grant RN2–00922-1 (to PK). HJF has been funded through SFB497-A7.
Footnotes
Disclosure of potential conflicts of interest
The authors have no conflicts of interest to declare.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2018.07.009.
References
- Ahmed K, Dehghani H, Rugg-Gunn P, Fussner E, Rossant J, Bazett-Jones DP, 2010. Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLoS One 5, e10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi T, Kuure S, Uranishi K, Koide H, Costantini F, Yokota T, 2015. ETS-related transcription factors ETV4 and ETV5 are involved in proliferation and induction of differentiation-associated genes in embryonic stem (ES) cells. J. Biol. Chem 290, 22460–22473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy H, Boer B, Desler M, Mallanna SK, McKeithan TW, Rizzino A, 2008. Identification of DPPA4 and other genes as putative Sox2:Oct-3/4 target genes using a combination of in silico analysis and transcription-based assays. J. Cell. Physiol 216, 651–662. [DOI] [PubMed] [Google Scholar]
- Coda DM, Gaarenstroom T, East P, Patel H, Miller DS, Lobley A, Matthews N, Stewart A, Hill CS, 2017. Distinct modes of SMAD2 chromatin binding and remodeling shape the transcriptional response to NODAL/Activin signaling. eLife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelen E, Brandsma JH, Moen MJ, Signorile L, Dekkers DHW, Demmers J, Kockx CEM, Ozgür Z, van Ijcken WFJ, van den Berg DLC, Poot RA, 2015. Proteins that bind regulatory regions identified by histone modification chromatin immunoprecipitations and mass spectrometry. Nat. Commun 6, 7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A, Alajem A, Meshorer E, Ramalho-Santos M, 2011. Open chromatin in pluripotency and reprogramming. Nat. Rev. Mol. Cell Biol. 12, 36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, Miller W, Kent WJ, Nekrutenko A, 2005. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 15, 1451–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J, 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11, R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakuma N, Kinoshita I, Shimizu Y, Yamazaki K, Yoshida K, Nishimura M, Dosaka-Akita H, 2005. E1AF/PEA3 activates the Rho/Rho-associated kinase pathway to increase the malignancy potential of non-small-cell lung cancer cells. Cancer Res. 65, 10776–10782. [DOI] [PubMed] [Google Scholar]
- Hollenhorst PC, Paul L, Ferris MW, Graves BJ, 2011. The ETS gene ETV4 is required for anchorage-independent growth and a cell proliferation gene expression program in PC3 prostate cells. Genes Cancer 1, 1044–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Da W, Sherman BT, Lempicki RA, 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, Decoste C, Schafer X, Lun Y, Lemischka IR, 2006. Dissecting self-renewal in stem cells with RNA interference. Nature 442, 533–538. [DOI] [PubMed] [Google Scholar]
- John T, Caballero OL, Svobodova SJ, Kong A, Chua R, Browning J, Fortunato S, Deb S, Hsu M, Gedye CA, Davis ID, Altorki N, Simpson AJ, Chen YT, Monk M, Cebon JS, 2008. ECSA/DPPA2 is an embryo-cancer antigen that is co-expressed with cancer-testis antigens in non-small cell lung cancer. Clin. Cancer Res. 14, 3291–3298. [DOI] [PubMed] [Google Scholar]
- Kang R, Zhou Y, Tan S, Zhou G, Aagaard L, Xie L, Bunger C, Bolund L, Luo Y, 2015. Mesenchymal stem cells derived from human induced pluripotent stem cells retain adequate osteogenicity and chondrogenicity but less adipogenicity. Stem Cell Res Ther 6, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufhold S, Garban H, Bonavida B, 2016. Yin Yang 1 is associated with cancer stem cell transcription factors (SOX2, OCT4, BMI1) and clinical implication. J. Exp. Clin. Cancer Res. 35, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R, 2010. Fast and accurate long-read alignment with burrows-wheeler transform. Bioinformatics 26, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Ortiz JA, Taing L, Meyer CA, Lee B, Zhang Y, Shin H, Wong SS, Ma J, Lei Y, Pape UJ, Poidinger M, Chen Y, Yeung K, Brown M, Turpaz Y, Liu XS, 2011. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 12, R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan B, Madan V, Weber O, Tropel P, Blum C, Kieffer E, Viville S, Fehling HJ, 2009. The pluripotency-associated gene Dppa4 is dispensable for embryonic stem cell identity and germ cell development but essential for embryogenesis. Mol. Cell. Biol 29, 3186–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaki H, Nishida T, Kitajima S, Asahina K, Teraoka H, 2007. Developmental pluripotency-associated 4 (DPPA4) localized in active chromatin inhibits mouse embryonic stem cell differentiation into a primitive ectoderm lineage. J. Biol. Chem. 282, 33034–33042. [DOI] [PubMed] [Google Scholar]
- Masaki H, Nishida T, Sakasai R, Teraoka H, 2010. DPPA4 Modulates Chromatin Structure Via Association with DNA and Core Histone H3 in Mouse Embryonic Stem Cells, Genes to Cells : Devoted to Molecular & Cellular Mechanisms. vol. 15. pp. 327–337. [DOI] [PubMed] [Google Scholar]
- Monk M, Holding C, 2001. Human embryonic genes re-expressed in cancer cells. Oncogene 20, 8085–8091. [DOI] [PubMed] [Google Scholar]
- Moss AC, Lawlor G, Murray D, Tighe D, Madden SF, Mulligan AM, Keane CO, Brady HR, Doran PP, MacMathuna P, 2006. ETV4 and Myeov knockdown impairs colon cancer cell line proliferation and invasion. Biochem. Biophys. Res. Commun 345, 216–221. [DOI] [PubMed] [Google Scholar]
- Murakami S, Ninomiya W, Sakamoto E, Shibata T, Akiyama H, Tashiro F, 2015. SRY and OCT4 are required for the acquisition of cancer stem cell-like properties and are potential differentiation therapy targets. Stem Cells 33, 2652–2663. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Nakagawa M, Ichisaka T, Shiota A, Yamanaka S, 2011. Essential roles of ECAT15–2/Dppa2 in functional lung development. Mol. Cell. Biol. 31, 4366–4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Geen H, Echipare L, Farnham PJ, 2011. Using ChIP-seq technology to generate high-resolution profiles of histone modifications. Methods Mol. Biol. 791, 265–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Shin S, Janknecht R, 2012. ETV1, 4 and 5: an oncogenic subfamily of ETS transcription factors. Biochim. Biophys. Acta 1826, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliviero G, Munawar N, Watson A, Streubel G, Manning G, Bardwell V, Bracken AP, Cagney G, 2015. The variant Polycomb repressor complex 1 component PCGF1 interacts with a pluripotency sub-network that includes DPPA4, a regulator of embryogenesis. Sci. Rep 5, 18388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellecchia A, Pescucci C, De Lorenzo E, Luceri C, Passaro N, Sica M, Notaro R, De Angioletti M, 2012. Overexpression of ETV4 is oncogenic in prostate cells through promotion of both cell proliferation and epithelial to mesenchymal transition. Oncogene 1, e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, Wong MC, Maddren M, Fang R, Heitner SG, Lee BT, Barber GP, Harte RA, Diekhans M, Long JC, Wilder SP, Zweig AS, Karolchik D, Kuhn RM, Haussler D, Kent WJ, 2012. ENCODE data in the UCSC genome browser: year 5 update. Nucleic Acids Res. 41, D56–D63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer KA, 1998. The cell cycle: a review. Vet. Pathol 35, 461–478. [DOI] [PubMed] [Google Scholar]
- Schick S, Fournier D, Thakurela S, Sahu SK, Garding A, Tiwari VK, 2015. Dynamics of chromatin accessibility and epigenetic state in response to UV damage. J. Cell Sci. 128, 4380–4394. [DOI] [PubMed] [Google Scholar]
- Serandour AA, Avner S, Oger F, Bizot M, Percevault F, Lucchetti-Miganeh C, Palierne G, Gheeraert C, Barloy-Hubler F, Peron CL, Madigou T, Durand E, Froguel P, Staels B, Lefebvre P, Metivier R, Eeckhoute J, Salbert G, 2012. Dynamic hydroxymethylation of deoxyribonucleic acid marks differentiation-associated enhancers. Nucleic Acids Res. 40, 8255–8265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman BT, Huang Da W, Tan Q, Guo Y, Bour S, Liu D, Stephens R, Baseler MW, Lane HC, Lempicki RA, 2007. DAVID knowledgebase: a gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinformatics 8, 426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somanath P, Bush KM, Knoepfler PS, 2018. ERBB3-binding protein 1 (EBP1) is a novel developmental pluripotency-associated-4 (DPPA4) cofactor in human pluripotent cells. Stem Cells 36 (5), 671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperger JM, Chen X, Draper JS, Antosiewicz JE, Chon CH, Jones SB, Brooks JD, Andrews PW, Brown PO, Thomson JA, 2003. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc. Natl. Acad. Sci. U. S. A 100, 13350–13355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung PY, Varlakhanova NV, Knoepfler PS, 2013. Identification of DPPA4 and DPPA2 as a novel family of pluripotency-related oncogenes. Stem Cells 31, 2330–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villodre ES, Kipper FC, Pereira MB, Lenz G, 2016. Roles of OCT4 in tumorigenesis, cancer therapy resistance and prognosis. Cancer Treat. Rev 51, 1–9. [DOI] [PubMed] [Google Scholar]
- Yuan ZY, Dai T, Wang SS, Peng RJ, Li XH, Qin T, Song LB, Wang X, 2014. Overexpression of ETV4 protein in triple-negative breast cancer is associated with a higher risk of distant metastasis. OncoTargets Ther. 7, 1733–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidan NZ, Walker KJ, Brown JE, Schaffer LV, Scalf M, Shortreed MR, Iyer G, Smith LM, Sridharan R, 2018. Compartmentalization of HP1 proteins in pluripotency acquisition and maintenance. Stem Cell Rep. 10, 627–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Akinmade D, Hamburger AW, 2005. The ErbB3 binding protein Ebp1 interacts with Sin3A to repress E2F1 and AR-mediated transcription. Nucleic Acids Res. 33, 6024–6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS, 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, van Essen D, Saccani S, 2012. Cell-type-specific control of enhancer activity by H3K9 trimethylation. Mol. Cell 46, 408–423. [DOI] [PubMed] [Google Scholar]
- Zullo JM, Demarco IA, Pique-Regi R, Gaffney DJ, Epstein CB, Spooner CJ, Luperchio TR, Bernstein BE, Pritchard JK, Reddy KL, Singh H, 2012. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell 149, 1474–1487. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.