Abstract
Ectomesenchymal chondromyxoid tumor is a rare and benign neoplasm with a predilection for the anterior dorsal tongue. Despite morphologic heterogeneity, most cases are characterized by a proliferation of bland spindle cells with a distinctive reticular growth pattern and myxoid stroma. The immunophenotype of these neoplasms is likewise variable; most cases express glial fibrillary acid protein and S100 protein, with inconsistent reports of keratin and myoid marker expression. The molecular pathogenesis is poorly understood; however, a subset of cases has been reported to harbor EWSR1 gene rearrangement. Following identification of an RREB1-MKL2 fusion gene by RNA-Seq in an index patient, a retrospective review of additional cases of ectomesenchymal chondromyxoid tumors was performed to better characterize the clinical, immunohistochemical and molecular attributes of this neoplasm. A total of 21 cases were included in this series. A marked predisposition for the dorsal tongue was confirmed. Most cases conformed to prior morphologic descriptions; however, hypercellularity, hyalinized stroma and necrosis were rare attributes not previously emphasized. The neoplastic cells frequently co-expressed glial fibrillary acid protein, S100 protein, keratin, smooth muscle actin and/or desmin; a single case was found to contain significant myogenin expression. An RREB1-MKL2 fusion product was identified in 19 tumors (90%), a single tumor (5%) had an EWSR1-CREM fusion product, and the remaining case lacked any known fusion gene by RNA-Seq. The latter two cases subtly differed morphologically from many in the cohort. This series illustrates that recurrent RREB1-MKL2 fusions occur in most, perhaps all, cases of ectomesenchymal chondromyxoid tumor.
Keywords: Ectomesenchymal chondromyxoid tumor, tongue, gene rearrangement, RREB1, MKL2, EWSR1, CREM
INTRODUCTION
Ectomesenchymal chondromyxoid tumor is a rare mesenchymal neoplasm – with fewer than 100 reported cases in the English literature – of uncertain origin, with a striking predilection for the anterior dorsal tongue.(1) Tumors occur across a broad age range, predominating in early-mid adulthood, and equally affect males and females.(2) Clinically, lesions typically present as a small (1-2 cm) painless mass, often of longstanding duration.(2) Simple excision is generally curative; however, a minority of cases have been reported to locally recur.(2, 3)
Morphologically distinctive, tumors are lobulated with thin fibrous septa separating bland polygonal-spindle cells arranged in reticular and globoid patterns.(2, 4, 5) The cytoplasm is pale and eosinophilic. The nuclei are small and ovoid, with scattered atypia, hyperchromasia, pseudonuclear inclusions and binucleation; mitotic activity, if present, is typically sparse.(2) Interspersed between the cells is prominent chondromyxoid stroma;(2) however, overt chondroid differentiation is not generally encountered. The immunophenotype varies—most tumors are characterized by expression of glial fibrillary acid protein and S100 protein, with conflicting reports of immunoreactivity for keratin, smooth muscle actin and CD56.(2) There is a single report of a case with SOX10 expression.(6) Tumors are typically negative for CD34, p63 and epithelial membrane antigen. Ultrastructural examination fails to identify desmosomes or condensations of thin filaments, features that might suggest myoepithelial differentiation.(2) Rearrangement of EWSR1 has been reported in a subset of cases, although a fusion partner has not, to date, been identified.(7)
Following the identification of an RREB1-MKL2 fusion gene in an index case of ectomesenchymal chondromyxoid tumor, we undertook a retrospective review of additional cases to better characterize the clinical, immunohistochemical and molecular attributes of this enigmatic neoplasm.
MATERIALS AND METHODS
Case Selection
An RREB1-MKL2 fusion gene was identified in the index patient (Patient 1) following routine diagnostic RNA Sequencing (RNA-Seq). Based on this unexpected observation retrospective searches were performed by each of the contributors for cases diagnosed as ectomesenchymal chondromyxoid tumor (2007-2018). Each case was reviewed to confirm the diagnosis prior to the initiation of immunohistochemical and RNA-Seq testing. This study received institutional Research Ethics Board approval.
Immunohistochemistry
Representative formalin-fixed paraffin-embedded tissue blocks were selected for each case, and 4 micron sections cut onto positively charged slides. Using standard techniques, staining was performed for glial fibrillary acid protein, S100 protein, keratin (AE1/AE3), smooth muscle actin, desmin and myogenin (Supplementary Table 1). Tumor immunoreactivity was scored based on the percentage of positive cells (0: no staining; 1+: <5%; 2+: 5% to 25%; 3+: 26% to 50%; 4+: 51% to 75%; and 5+: 76% to 100%).
RNA Sequencing
Using formalin-fixed, paraffin-embedded tissue, material was obtained from either scrolls (3-4 cut at 10 microns) or by scrapping unstained sections previously cut onto glass slides (4-5 cut at 4 microns). RNA was extracted using the ExpressArt FFPE Clear RNA Ready kit (Amsbio, Cambridge, MA). RNA-Seq libraries were prepared using 20-100 ng total RNA with the TruSight RNA Fusion Panel (Illumina, San Diego, CA). Each sample was sequenced with 76 base-pair paired-end reads on an Illumina MiSeq at 8 samples per flow cell (~3 million reads per sample). The results were analyzed using both the STAR and BOWTIE2 aligners, and Manta and JAFFA fusion callers, respectively.(8, 9)
Fluorescence In Situ Hybridization
Fluorescence in situ hybridization (FISH) for RREB1 and MKL2 was performed as previously reported in detail.(10) Briefly, custom bacterial artificial chromosome (BAC) clone probes were designed to flank the target genes based on the UCSC genome browser (http://genome.ucsc.edu), and obtained from BACPAC sources of Children’s Hospital of Oakland Research Institute (Oakland, CA; http://bacpac.chori.org) (Supplementary Table 2). DNA from each BAC was isolated and then labeled with fluorochromes by nick translation. Slides were prepared using formalin-fixed, paraffin-embedded tissue cut at 4 microns. The slides were deparaffinized, pretreated, and then hybridized with the denatured probes. Following an overnight incubation, the slides were rinsed, stained with 4′,6-diamidino-2-phenylindole (DAPI), mounted, and examined using a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany).
RESULTS
A total of 21 cases were identified from the contributing institutions; six of the cases (Patients 13, 14, 16, 17, 18, 19) have previously been reported.(7, 11) The mean patient age was 40 years (range: 13-59 years). There were 13 females and 8 males (ratio, 1.6:1). Each of the tumors arose on the dorsal tongue; most were anterior, and a minority posterior. Most cases were treated by simple excision. Despite frequent microscopic extension to inked resection margins, only a single case was associated with local recurrence (Patient 10) (Table 1).
Table 1.
Clinical and demographic information.
| Patient | Age (y) | Sex | Location | Size and clinical findings |
|---|---|---|---|---|
| 1 | 31 | F | Tongue, NOS | 0.9 cm. Excised with negative margin |
| 2 | 48 | M | Tongue, dorsal, tip | 0.6 cm. Shave biopsy. No evidence of recurrence at 1 year |
| 3 | 39 | F | Tongue, left dorsal, posterior | 1.1 cm. < 2 months follow-up |
| 4 | 54 | F | Tongue, dorsal, anterior | 1.4 cm. Re-excision, with positive margin. No report of recurrence |
| 5 | 54 | M | Tongue, dorsal | Not available |
| 6 | 23 | M | Tongue, left anterior | 3.0 cm. No evidence of recurrence at 2 years |
| 7 | 13 | F | Tongue, right midline dorsal | 2.0 cm soft mass |
| 8 | 45 | F | Tongue, left dorsal | Size not specified. 6-7 month asymptomatic mass |
| 9 | 54 | M | Tongue, left anterior | 1.2 cm. Excised with negative margins. No evidence of recurrence at 1.4 years |
| 10 | 35 | M | Tongue, left anterior | 0.7 cm. Excised with positive margins. Recurred at 41 months. No further recurrences after 4 years |
| 11 | 14 | F | Tongue, midline anterior | 1.5 cm. Excised with positive margins. No evidence of recurrence after 8.5 years |
| 12* | 49 | F | Tongue, dorsal lateral | 2.1 cm |
| 13* | 33 | F | Tongue, NOS | Not available |
| 14 | 50 | F | Tongue, dorsal tip | 2.4 cm. Excised with negative margin. LTF |
| 15* | 53 | F | Tongue, NOS | Not available |
| 16* | 59 | M | Tongue, NOS | Not available |
| 17* | 31 | F | Tongue, NOS | Not available |
| 18* | 51 | M | Tongue, midline dorsal, anterior | 2.5 cm. 15-year history prior to excision |
| 19 | 40 | F | Tongue, right dorsal, anterolateral | 1.0 cm. Slow-growing lump x3-5 months. Margin focally positive, but no report of recurrence |
| 20 | 51 | M | Tongue, NOS | 0.7 cm. Margin focally positive, with no report of recurrence |
| 21 | 15 | F | Tongue, right dorsal, posterior | 0.7 cm |
The size of the tumors ranged from 0.6 to 3.0 cm (mean, 1.5 cm); virtually all of the cases were circumscribed, with a pushing border; several tumors contained focal infiltration into the surrounding skeletal muscle. The overlying squamous epithelium was intact in all cases, lacking any well-developed pseudoepitheliomatous hyperplasia. The tumors were frequently multi-lobulated; the lobules were either separated by areas of endogenous stroma, or more frequently by fibrous septa (Figure 1). The cells ranged from polygonal to spindle-stellate. The cytoplasm was pale and eosinophilic to lightly basophilic. The nuclei were round to ovoid and frequently lobulated and hyperchromatic. Scattered cells showed atypia, pseudonuclear inclusions and binucleation. Almost all of the cases lacked conspicuous mitotic activity; however, rare cases contained 1-2 mitoses per 10 high power fields. Similar to prior reports, the architecture included sheets, cords, reticular and globoid patterns; cellular fascicles, microcystic-cystic and papillary patterns were rarely encountered. Occasional interstitial hemorrhage and/or hemosiderin deposition – in the absence of prior sampling – was identified. A single case contained necrosis and dystrophic calcification (Figure 2). The intervening stroma was myxoid and pale and basophilic; occasionally it was hyalinized; very rarely, isolated foci vaguely suggested hyaline cartilage. Sunburst amianthoid fibers were not identified.
Figure 1.
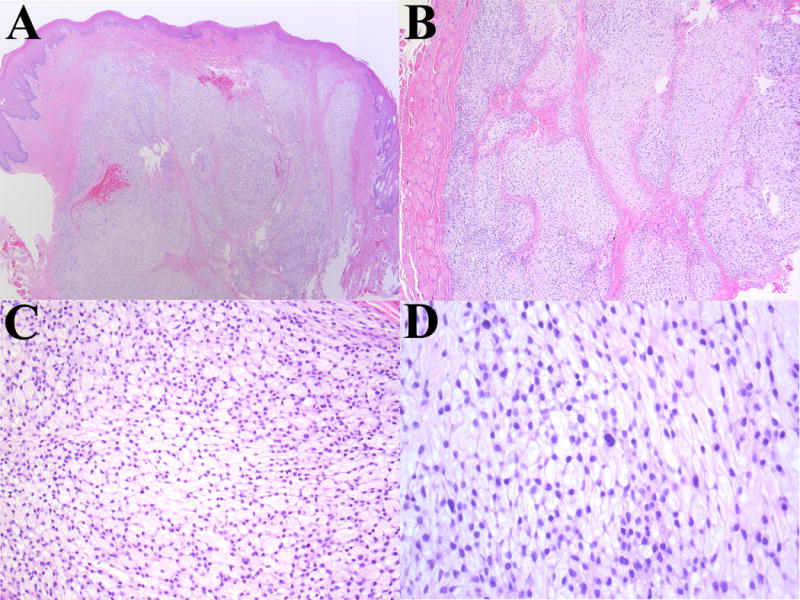
Representative photomicrographs from a conventional case of ectomesenchymal chondromyxoid tumor (Patient 3). (A) Low-power magnification showing circumscribed neoplasm with pushing margin. (B) Intermediate magnification highlighting the presence of fibrous septa separating tumor lobules. (C) Spindle-stellate cells with a reticular, or ‘net-like’ pattern and abundant myxoid stroma. (D) Scattered cytologic atypia and multinucleation; however, mitotic activity is inconspicuous.
Figure 2.
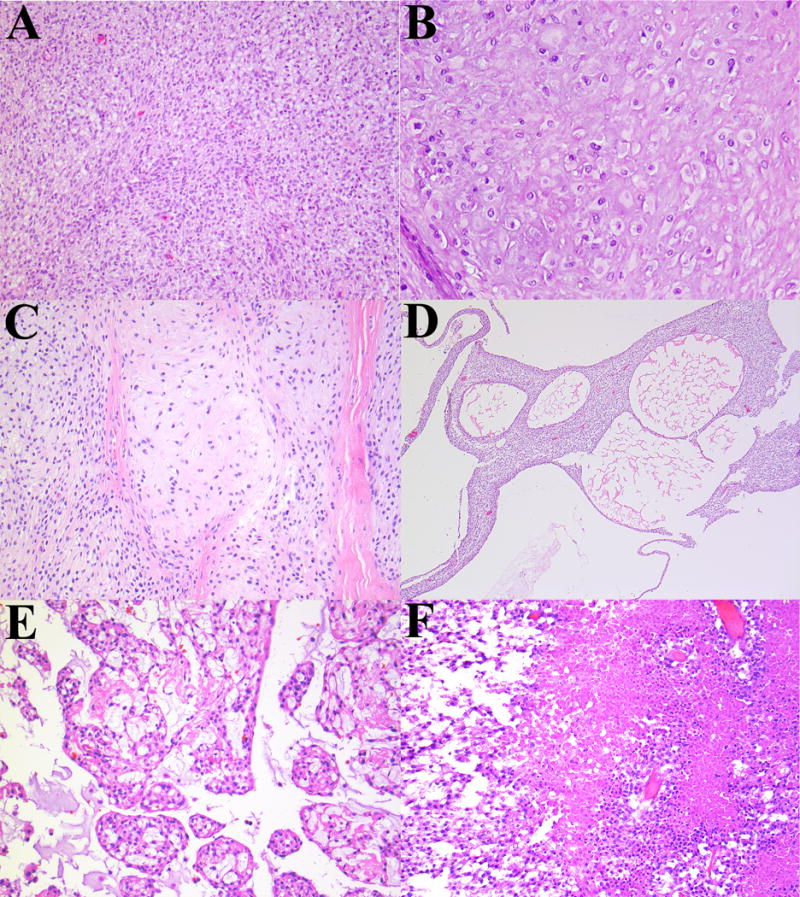
Less common features observed in ectomesenchymal chondromyxoid tumor. (A) Cellular spindle cell areas with a vague fascicular pattern. (B) Eosinophilic hyaline stroma. (C) Stroma vaguely suggestive of hyaline cartilage. (D) Microcystic-cystic pattern. (E) Nodular-cribriform pattern. (F) Necrosis.
The results of immunohistochemical staining are summarized in Table 2 and Figure 3. Briefly, similar to prior reports, there was relatively consistent expression of glial fibrillary acid protein. Immunoreactivity for S100 protein, pancytokeratin, smooth muscle actin and desmin expression was common, but tended to be weak and/or focal. A single case contained significant myogenin expression. Immunohistochemistry was performed for SOX10 (N=7) and p63 (N=2), which were negative (data not shown).
Table 2.
Summary of immunohistochemical and molecular attributes of ectomesenchymal chondromyxoid tumor.
| Patient | Immunohistochemistry | Molecular | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| GFAP | S100 | Keratin | Desmin | Myogenin | SMA | Method | Result | |
| 1 | – | 3+ | – | 1+ | N/P | 1+ | RNAseq | RREB1-MKL2 |
| 2 | – | – | – | – | – | – | RNAseq | EWSR1-CREM |
| 3 | 5+ | 5+ | 1+ | 1+ | – | 3+ | RNAseq | RREB1-MKL2 |
| 4 | 4+ | 3+ | 1+ | – | – | 1+ | RNAseq | Negative |
| 5 | N/P | N/P | N/P | N/P | N/P | N/P | RNAseq | RREB1-MKL2 |
| 6 | 5+ | 5+ | 2+ | 1+ | 2+ | 2+ | RNAseq | RREB1-MKL2 |
| 7 | 5+ | 2+ | – | N/P | N/P | - | RNAseq | RREB1-MKL2 |
| 8 | 5+ | 3+ | – | N/P | N/P | N/P | RNAseq | RREB1-MKL2 |
| 9 | 1+ | 3+ | – | – | – | 3+ | RNAseq | RREB1-MKL2 |
| 10 | 2+ | 3+ | – | – | – | 3+ | RNAseq & FISH | RREB1-MKL2 |
| 11 | 5+ | 3+ | 2+ | 4+ | – | 2+ | RNAseq | RREB1-MKL2 |
| 12 | 4+ | 2+ | 1+ | 1+ | – | N/P | FISH | RREB1-MKL2 |
| 13 | 5+ | 2+ | 1+ | 1+ | – | N/P | FISH | RREB1-MKL2 |
| 14 | 4+ | 1+ | – | 1+ | – | N/P | FISH | RREB1-MKL2 |
| 15 | 4+ | N/P | – | N/P | – | N/P | FISH | RREB1-MKL2 |
| 16 | 4+ | 1+ | – | – | – | N/P | FISH | RREB1-MKL2 |
| 17 | 2+ | N/P | – | N/P | – | N/P | FISH | RREB1-MKL2 |
| 18 | 5+ | 5+ | 2+ | 1+ | – | 3+ | FISH | RREB1-MKL2 |
| 19 | 5+ | 2+ | 2+ | 1+ | – | N/P | FISH | RREB1-MKL2 |
| 20 | 4+ | 1+ | 1+ | 3+ | 1+ | 3+ | FISH | RREB1-MKL2 |
| 21 | 5+ | 2+ | 2+ | – | – | – | FISH | RREB1-MKL2 |
Abbreviations: - (negative)’ FISH (fluorescence in situ hybridization); GFAP (glial fibrillary acid protein); N/P (not performed); SMA (smooth muscle actin).
Figure 3.
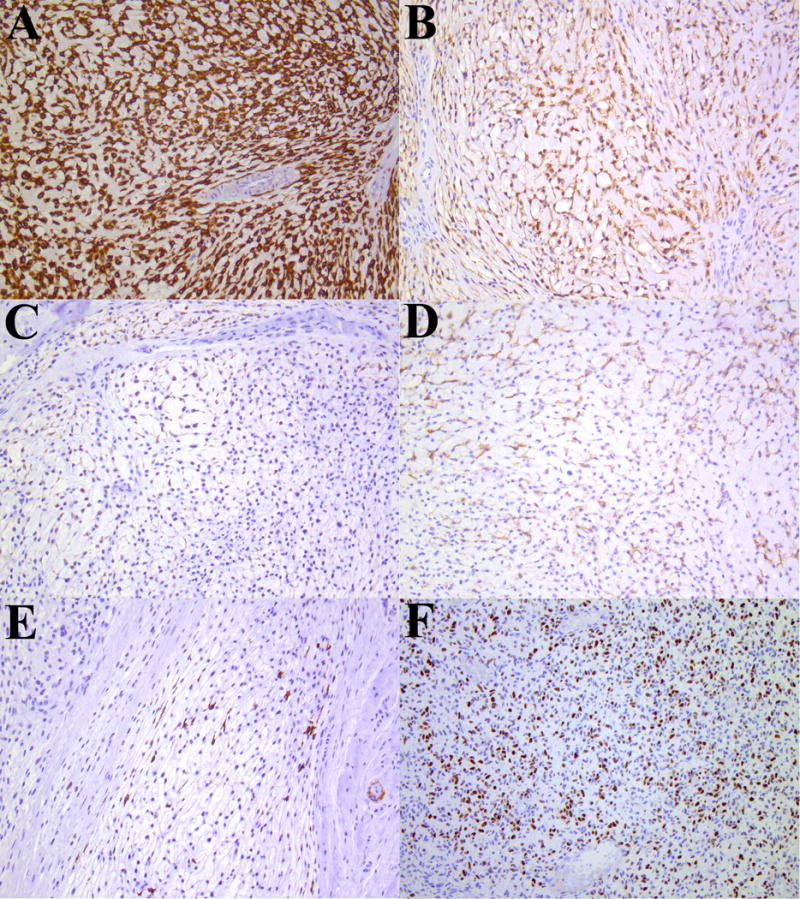
Representative photomicrographs of immunophenotype of ectomesenchymal chondromyxoid tumor (Patient 3). (A) Glial fibrillary acid protein, (B) S100 protein, (C) keratin (AE1/AE3), (D) desmin, (E) smooth muscle actin and (F) myogenin; all photomicrographs x200.
Following RNA-Seq the index patient was found to have an RREB1-MKL2 fusion gene. RNA-Seq was subsequently performed on Patients 2-10. As a mean of independent verification of the RNA-Seq fusion product, fluorescence in situ hybridization testing was applied to the recurrent lesion of Patient 10. The remaining cases (Patients 11-22) were examined by FISH alone (Figure 4). All but 2 cases (90.5%) were found to harbor RREB1-MKL2 fusion genes. One patient was found to have an EWSR1-CREM fusion gene (Patient 2), while the remaining case lacked any fusion candidates, despite suitable RNA (Patient 4) (Figure 5). RNA-Seq of the cases with an RREB1-MKL2 fusion product demonstrated identical breakpoints involving exon 8 of RREB1 (NCBI Reference Sequence: NM_001003699.3) and exon 11 of MKL2 (NM_001308142.1). The single case with an EWSR1-CREM fusion had breakpoints involving exon 8 of EWSR1 (NM_013986.3), and either exon 7 or 3 of CREM (NM_181571.2, or NM_001352467.1) depending on whether CREM transcript variant 1 or 32.
Figure 4.
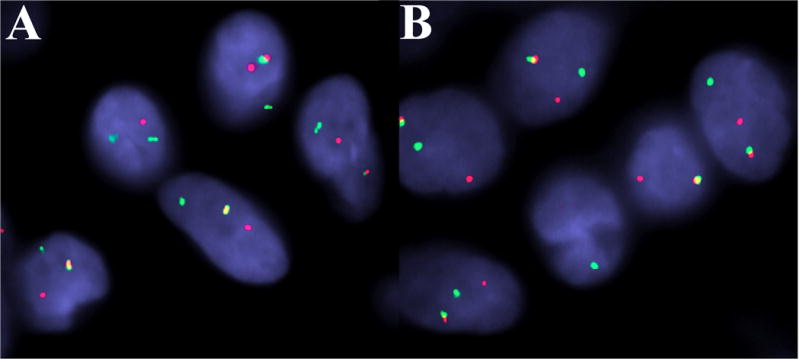
Representative fluorescence in situ hybridization in ectomesenchymal chondromyxoid tumor (Patient 20). Rearrangement of: (A) MKL2, and (B) RREB1. (Both images: red, centromeric; green, telomeric.)
Figure 5.
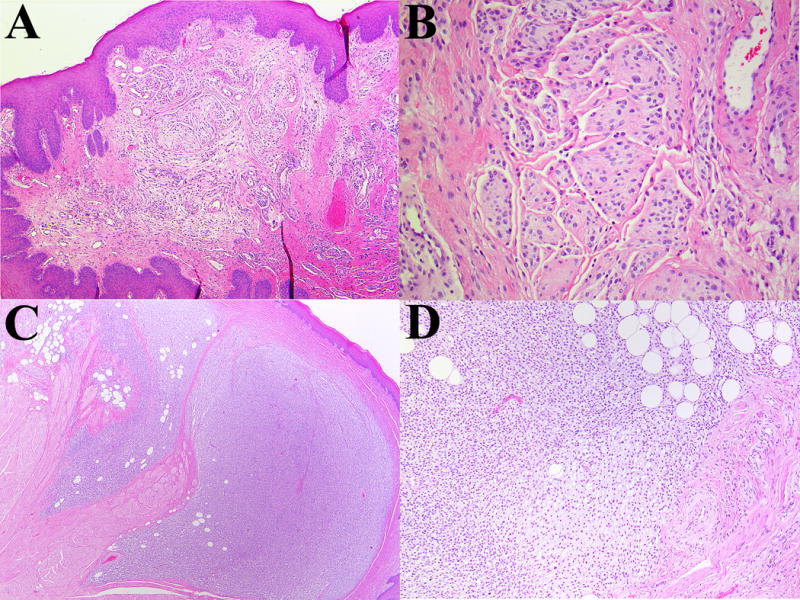
Representative photomicrographs of ectomesenchymal chondromyxoid tumor lacking RREB1-MKL2 fusion partners. (A/B) A neoplasm with an EWSR1-CREM fusion product contains lobules of bland cells. (C/D) A neoplasm lacking any gene fusions by RNA-Seq. The tumor is infiltrative and contains an adipocytic component. The lesion is cellular with less conspicuous myxoid component.
DISCUSSION
Ectomesenchymal chondromyxoid tumor is a rare mesenchymal neoplasm of unknown histogenesis. Our series, amongst the largest to date, confirms prior reports of the relatively narrow clinical distribution of this enigmatic tumor; in addition, we expand the morphologic and immunohistochemical spectrum of this neoplasm. Finally, we demonstrate that the majority of ectomesenchymal chondromyxoid tumors are characterized by RREB1-MKL2 fusions.
In this series tumors were found in patients from across a relatively broad age range (13-59 years), with a slight female predilection. All of our cases originated in the tongue. Indeed, virtually all prior reports of ectomesenchymal chondromyxoid tumor have occurred in this location. While a hard palate origin has been suggested in two reports,(12, 13) the first has been previously questioned,(3, 14–16) and the second is also noted to lack a prototypic morphology and immunophenotype. Most of the tumors in our series arose in the anterior tongue, but a minority involved the base of tongue.(4, 17, 18) Where clinically specified, all cases were dorsally situated. Despite several cases in this series with positive margins following excision, only a single case locally recurred and none metastasized. This would appear to confirm a clinically benign behavior.
Ectomesenchymal chondromyxoid tumor has a distinct morphology and immunophenotype. Since its initial report by Smith, et al. in 1995,(2) there have been few revisions to the original comprehensive histopathologic description; indeed, our findings were largely similar. The cells ranged from polygonal to spindle and stellate. Most cases contained, at least focally, the prototypic ‘net-like’ (reticular) arrangement of cells with abundant myxoid stroma. Clefting, papillary, cystic and more solid growth patterns were occasionally observed. Hyalinized eosinophilic stroma, calcification, hypercellularity, and necrosis were rare events. Similar to the earlier reports, immunoreactivity for glial fibrillary acid protein, S100 protein, keratin and actin were frequently observed;(2) glial fibrillary protein notwithstanding, immunoreactivity tended to be patchy and/or weak. While there are conflicting reports on desmin expression,(18–20) our results suggest this is relatively common, although focal. We observed significant nuclear myogenin expression in a single, otherwise unremarkable, case.
Until now the molecular pathogenesis of ectomesenchymal chondromyxoid tumor has remained elusive. In a series of 11 cases Argyris, et al. identified EWSR1 rearrangement in 3 cases (27%); 2 subsequent cases tested by Laco, et al. failed to show EWSR1 rearrangement.(6) Following the discovery of a RREB1-MKL2 fusion gene in our index patient, we confirmed, using a combination of RNA-Seq and fluorescence in situ hybridization, this fusion product in 19 of 21 cases (90%). The two cases lacking an RREB1-MKL2 fusion did not possess an archetypical morphology. Nevertheless, as they currently fall within the accepted clinical and pathologic spectrum of ectomesenchymal chondromyxoid tumor, there is currently insufficient evidence to warrant rejection of the original diagnosis. One case (Patient 4) was cellular with infiltrative tongues resembling a somewhat atypical case presented by Portnof, et al..(3) The second case (Patient 2) was found to have an EWSR1-CREM fusion product by RNA-Seq; this case contained small nodules of polygonal cells with indistinct cell borders and prominent slit-like spaces. This was morphologically similar to at least two of the prior cases of ectomesenchymal chondromyxoid tumor with EWSR1 rearrangement.(7) While further study is required, it is possible that cases with EWSR1 rearrangement in this context may ultimately be more appropriately classified under the recently characterized family of myxoid mesenchymal tumors with EWSR1 – CREB family gene fusions.(10) In contrast to that seminal report, our patient was considerably older than the patients in that series, and our tumor was extracranial. The limited literature on myxoid mesenchymal neoplasms with EWSR1 – CREM makes it difficult to draw meaningful inferences to our case; moreover, one of us (BCD) has recently encountered a malignant small round cell neoplasm with an EWSR1 – CREM fusion gene in an extra cranial location.
Interestingly, a recent case of so-called ‘biphenotypic “oropharyngeal” sarcoma’ was reported to possess an RREB1-MKL2 gene fusion.(21) In their article, Siegfried et al. describe a 3.5 cm parapharyngeal mass comprised of fascicles of bland spindle-ovoid cells with uniform nuclei and minimal mitotic activity, and occasional pseudoangiectatic and cystic spaces; immunohistochemistry demonstrated patchy staining for S100 protein, desmin, smooth muscle actin and myogenin. Anatomic proximity notwithstanding, this case does not appear to involve the base of tongue. And, while some of the cases in our series contained areas of increased cellularity and a fascicular-herringbone pattern, this was not a dominant finding amongst our cases.(21) Whether biphenotypic “oropharyngeal” sarcoma represents an atypical example of ectomesenchymal chondromyxoid tumor, or a distinct entity with a pleiotropic fusion gene – analogous to the relationship of EWSR1-ATF1 in clear cell sarcoma, angiomatoid fibrous histiocytoma and hyalinizing clear cell carcinoma of salivary gland – is presently unclear.
The histogenesis of ectomesenchymal chondromyxoid tumor has been a subject of debate. It has been suggested to originate from an uncommitted ectomesenchymal cell derived from the neural crest.(1,2) Alternatively, it has been proposed to represent a myoepithelial-derived tumor of either minor salivary glands or soft tissue.(2, 22) Our molecular findings, combined with the unique clinical and histopathologic attributes of this neoplasm, suggest ectomesenchymal chondromyxoid tumor represents a distinct entity. MKL2 encodes Myocardin Like 2 – a transcription coactivator of serum response factor – that is involved in smooth and skeletal muscle differentiation,(23, 24) and neuronal development.(25) Siegfried, et al. eloquently describe how the RREB1-MKL2 fusion gene – which is associated with increased MKL2 expression – accounts for a biphenotypic immunophenotype.(21) There is a precedent for MKL2 fusions in another mesenchymal neoplasm; C11orf95-MKL2 fusions occur in chondroid lipoma(26, 27) which, similar to ectomesenchymal chondromyxoid tumor, is variably characterized by myxoid-chondroid matrix. Similar to many other translocation-associated neoplasms that undergo fundamental molecular re-reprogramming(28) – with a concomitant absence of an overt line of differentiation (e.g., Ewing sarcoma and synovial sarcoma) – it is conceivable that a cell of origin may remain elusive for some time; that being said, Smith et al.’s original hypothesis of an uncommitted ectomesenchymal cell progenitor remains a distinct possibility.(2)
In summary, this study represents the largest series of molecularly interrogated ectomesenchymal chondromyxoid tumors to date. We confirm the strong predilection of this neoplasm for the dorsal aspect of the tongue, and expand the morphologic and immunohistochemical spectrum of findings for this tumor. Finally, we demonstrate that the overwhelming majority – if not all, if two morphologic outliers can be excluded – of cases of ectomesenchymal chondromyxoid tumor are characterized by an RREB1-MKL2 fusion gene.
Supplementary Material
Acknowledgments
The authors are grateful to Mr. Anthony Wing and Ms. Jasmine Wong for their expertise in immunohistochemistry; Ms. Grace Murray (Illumina, San Diego, CA) for generously providing test kits; and, Ms. Evangeline Agro and Ms. Sharon Crafter for facilitating RNA-Seq testing.
Supported in part by: P50 CA140146-01 (CRA); P30-CA008748 (CRA); Kristen Ann Carr Foundation (CRA); Cycle for Survival (CRA)
Footnotes
Conflicts of Interest: None
References
- 1.Bishop JA, Gnepp DR, Ro JY. Ectomesenchymal chondromyxoid tumour. In: El-Naggar AK, Chan JKC, Grandis JR, et al., editors. WHO Classification of Head and Neck Tumours. Lyon: IARC; 2017. pp. 119–120. [Google Scholar]
- 2.Smith BC, Ellis GL, Meis-Kindblom JM, et al. Ectomesenchymal chondromyxoid tumor of the anterior tongue. Nineteen cases of a new clinicopathologic entity. Am J Surg Pathol. 1995;19:519–530. doi: 10.1097/00000478-199505000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Portnof JE, Friedman JM, Reich R, et al. Oral ectomesenchymal chondromyxoid tumor: case report and literature review. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2009;108:e20–24. doi: 10.1016/j.tripleo.2009.05.040. [DOI] [PubMed] [Google Scholar]
- 4.Seo SH, Shin DH, Kang HJ, et al. Reticulated myxoid tumor of the tongue: 2 cases supporting an expanded clinical and immunophenotypic spectrum of ectomesenchymal chondromyxoid tumor of the tongue. Am J Dermatopathol. 2010;32:660–664. doi: 10.1097/DAD.0b013e3181d7d3bf. [DOI] [PubMed] [Google Scholar]
- 5.Ide F, Mishima K, Saito I. Ectomesenchymal chondromyxoid tumor of the anterior tongue with myxoglobulosislike change. Virchows Arch. 2003;442:302–303. doi: 10.1007/s00428-003-0781-7. [DOI] [PubMed] [Google Scholar]
- 6.Laco J, Mottl R, Hobling W, et al. Cyclin D1 Expression in Ectomesenchymal Chondromyxoid Tumor of the Anterior Tongue. Int J Surg Pathol. 2016;24:586–594. doi: 10.1177/1066896916652221. [DOI] [PubMed] [Google Scholar]
- 7.Argyris PP, Bilodeau EA, Yancoskie AE, et al. A subset of ectomesenchymal chondromyxoid tumours of the tongue show EWSR1 rearrangements and are genetically linked to soft tissue myoepithelial neoplasms: a study of 11 cases. Histopathology. 2016;69:607–613. doi: 10.1111/his.12973. [DOI] [PubMed] [Google Scholar]
- 8.Liu S, Tsai WH, Ding Y, et al. Comprehensive evaluation of fusion transcript detection algorithms and a meta-caller to combine top performing methods in paired-end RNA-seq data. Nucleic Acids Res. 2016;44:e47. doi: 10.1093/nar/gkv1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Schulz-Trieglaff O, Shaw R, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–1222. doi: 10.1093/bioinformatics/btv710. [DOI] [PubMed] [Google Scholar]
- 10.Kao YC, Sung YS, Zhang L, et al. EWSR1 Fusions With CREB Family Transcription Factors Define a Novel Myxoid Mesenchymal Tumor With Predilection for Intracranial Location. Am J Surg Pathol. 2017;41:482–490. doi: 10.1097/PAS.0000000000000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schep LA, Bullock MJ, Taylor SM. Ectomesenchymal Chondromyxoid Tumour of the Dorsal Tongue Presenting with Impaired Speech. Case Rep Otolaryngol. 2016;2016:7342910. doi: 10.1155/2016/7342910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nigam S, Dhingra KK, Gulati A. Ectomesenchymal chondromyxoid tumor of the hard palate–a case report. J Oral Pathol Med. 2006;35:126–128. doi: 10.1111/j.1600-0714.2006.00368.x. [DOI] [PubMed] [Google Scholar]
- 13.Gouvea AF, Diaz KP, Leon JE, et al. Nodular lesion in the anterior hard palate. Oral Surg Oral Med Oral Pathol Oral Radiol. 2012;114:154–159. doi: 10.1016/j.oooo.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 14.Kato MG, Erkul E, Brewer KS, et al. Clinical features of ectomesenchymal chondromyxoid tumors: A systematic review of the literature. Oral Oncol. 2017;67:192–197. doi: 10.1016/j.oraloncology.2017.02.021. [DOI] [PubMed] [Google Scholar]
- 15.Ide F. Chondromyxoid tumor of palate. J Oral Pathol Med. 2006;35:523–524. doi: 10.1111/j.1600-0714.2006.00452_1.x. author reply 524. [DOI] [PubMed] [Google Scholar]
- 16.Allen CM. The ectomesenchymal chondromyxoid tumor: a review. Oral Dis. 2008;14:390–395. doi: 10.1111/j.1601-0825.2008.01447.x. [DOI] [PubMed] [Google Scholar]
- 17.Cardin MJ, Fiset PO, Zeitouni AG, et al. Ectomesenchymal chondromyxoid tumour of the posterior tongue. Head Neck Pathol. 2014;8:329–333. doi: 10.1007/s12105-013-0500-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aldojain A, Jaradat J, Summersgill K, et al. Ectomesenchymal Chondromyxoid Tumor: A Series of Seven Cases and Review of the Literature. Head Neck Pathol. 2015;9:315–322. doi: 10.1007/s12105-014-0578-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pires FR, Abrahao AC, Cabral MG, et al. Clinical, histological and immunohistochemical features of ectomesenchymal chondromyxoid tumor. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2009;108:914–919. doi: 10.1016/j.tripleo.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 20.de Visscher JG, Kibbelaar RE, van der Waal I. Ectomesenchymal chondromyxoid tumor of the anterior tongue. Report of two cases. Oral Oncol. 2003;39:83–86. doi: 10.1016/s1368-8375(01)00117-8. [DOI] [PubMed] [Google Scholar]
- 21.Siegfried A, Romary C, Escudie F, et al. RREB1-MKL2 fusion in biphenotypic “oropharyngeal” sarcoma: New entity or part of the spectrum of biphenotypic sinonasal sarcomas? Genes Chromosomes Cancer. 2017 doi: 10.1002/gcc.22521. [DOI] [PubMed] [Google Scholar]
- 22.Nikitakis NG, Argyris P, Sklavounou A, et al. Oral myoepithelioma of soft tissue origin: report of a new case and literature review. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2010;110:e48–51. doi: 10.1016/j.tripleo.2010.05.074. [DOI] [PubMed] [Google Scholar]
- 23.Cen B, Selvaraj A, Prywes R. Myocardin/MKL family of SRF coactivators: key regulators of immediate early and muscle specific gene expression. J Cell Biochem. 2004;93:74–82. doi: 10.1002/jcb.20199. [DOI] [PubMed] [Google Scholar]
- 24.Selvaraj A, Prywes R. Megakaryoblastic leukemia-1/2, a transcriptional co-activator of serum response factor, is required for skeletal myogenic differentiation. J Biol Chem. 2003;278:41977–41987. doi: 10.1074/jbc.M305679200. [DOI] [PubMed] [Google Scholar]
- 25.Mokalled MH, Johnson A, Kim Y, et al. Myocardin-related transcription factors regulate the Cdk5/Pctaire1 kinase cascade to control neurite outgrowth, neuronal migration and brain development. Development. 2010;137:2365–2374. doi: 10.1242/dev.047605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang D, Sumegi J, Dal Cin P, et al. C11orf95-MKL2 is the resulting fusion oncogene of t(11;16)(q13;p13) in chondroid lipoma. Genes Chromosomes Cancer. 2010;49:810–818. doi: 10.1002/gcc.20788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flucke U, Tops BB, de Saint Aubain Somerhausen N, et al. Presence of C11orf95-MKL2 fusion is a consistent finding in chondroid lipomas: a study of eight cases. Histopathology. 2013;62:925–930. doi: 10.1111/his.12100. [DOI] [PubMed] [Google Scholar]
- 28.Garcia CB, Shaffer CM, Alfaro MP, et al. Reprogramming of mesenchymal stem cells by the synovial sarcoma-associated oncogene SYT-SSX2. Oncogene. 2012;31:2323–2334. doi: 10.1038/onc.2011.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


